Abstract
Human herpesvirus 8 (HHV-8) is the etiologic agent of Kaposi's sarcoma and primary effusion lymphoma. Activation of the cellular transcription factor nuclear factor-kappa B (NF-κB) is essential for latent persistence of HHV-8, survival of HHV-8-infected cells, and disease progression. We used reverse-transfected cell microarrays (RTCM) as an unbiased systems biology approach to systematically analyze the effects of HHV-8 genes on the NF-κB signaling pathway. All HHV-8 genes individually (n = 86) and, additionally, all K and latent genes in pairwise combinations (n = 231) were investigated. Statistical analyses of more than 14,000 transfections identified ORF75 as a novel and confirmed K13 as a known HHV-8 activator of NF-κB. K13 and ORF75 showed cooperative NF-κB activation. Small interfering RNA-mediated knockdown of ORF75 expression demonstrated that this gene contributes significantly to NF-κB activation in HHV-8-infected cells. Furthermore, our approach confirmed K10.5 as an NF-κB inhibitor and newly identified K1 as an inhibitor of both K13- and ORF75-mediated NF-κB activation. All results obtained with RTCM were confirmed with classical transfection experiments. Our work describes the first successful application of RTCM for the systematic analysis of pathofunctions of genes of an infectious agent. With this approach, ORF75 and K1 were identified as novel HHV-8 regulatory molecules on the NF-κB signal transduction pathway. The genes identified may be involved in fine-tuning of the balance between latency and lytic replication, since this depends critically on the state of NF-κB activity.
Human herpesvirus 8 (HHV-8), also called Kaposi's sarcoma-associated herpesvirus, is the causative agent of Kaposi's sarcoma, an endothelial-cell-derived tumor which is characterized by neoangiogenesis and infiltration of inflammatory cells, and of the lymphoproliferative diseases primary effusion lymphoma (PEL) and multicentric Castleman's disease (6-8, 12, 14, 23, 34, 47, 54). The HHV-8 genome is approximately 170 kbp in length and contains more than 80 genes (40). Open reading frames with homology to genes of herpesvirus saimiri (HVS) were numbered according to their position on the HVS genome. HHV-8 genes without homologous counterparts in HVS were numbered separately and given the prefix “K” (“K genes”) (16, 36, 40, 43).
Infection with HHV-8 constitutively activates the transcription factor nuclear factor-kappa B (NF-κB) in endothelial cells and lymphocytes (25, 44). The activation of NF-κB is crucial for the development and progression of HHV-8-associated diseases. It protects HHV-8-infected cells against spontaneous apoptosis (25) and maintains the latent viral life cycle (5, 67). The latter is mandatory for the establishment of viral persistence. In agreement with this, the inhibition of NF-κB signaling delays the growth of HHV-8-associated lymphomas in a mouse model (24) and regulates the production of infectious HHV-8 virions (5, 50).
Only a few HHV-8 genes have been studied for their impact on NF-κB signaling. The genes K13 (9), K15 (4), and ORF74 (48) were described as activators and K10.5 (49) as an inhibitor of NF-κB, whereas conflicting results were obtained on the activity of K1 (28, 39). Presently, it is not known whether these genes are the only HHV-8 genes which act on NF-κB or whether different HHV-8 genes cooperate positively or negatively in the regulation of this important signaling pathway. Systematic analyses of all HHV-8 genes for the effects of single genes and gene combinations on NF-κB activity are not available due to the large genome of HHV-8, containing at least 86 genes (36, 43). Systematic analyses of all single-gene and pairwise-combination effects of HHV-8 genes on NF-κB require almost 4,000 transfection experiments. Therefore, this approach demands high-throughput transfection technology, which has only recently become available.
In 2001, Ziauddin and Sabatini succeeded in scaling down high-throughput gene function analysis to the microarray level (70). Different cDNA expression plasmids are spotted onto slides by using a microarray robot. The dried slides are exposed to a transfection reagent, placed in a culture dish, and covered with adherent mammalian cells in medium. Alternatively, DNA and transfection reagent can be mixed at once and applied to the slide (70). Both methods of application create microarrays of cell clusters simultaneously transfected with different plasmids in distinct and defined areas in a lawn of cells. The procedure creating a microarray of clusters of transfected cells was called transfected cell microarray. The transfection method was named reverse transfection, because, in contrast to conventional transfection protocols, DNA was “seeded” first and the cells were added subsequently. Reverse-transfected cell microarrays (RTCM), also called “cell chip analyses,” allow the carrying out of several hundred to several thousand transfection experiments in parallel using eukaryotic cells on a single glass slide. Cotransfections of appropriate reporter plasmids can be used to establish quantitative measures of gene effects on signaling pathways (for a review, see reference 53).
For the work reported herein, we used the RTCM technology as an unbiased systems biology approach to determine the effects of all HHV-8 genes and of selected combinations of genes (all K and latent genes) on NF-κB activation in HEK 293T cells. Through the performance of more than 14,000 transfections and the subsequent biostatistical analyses, K13 and ORF75 were identified as the major activators of NF-κB. Both genes cooperated in the activation of NF-κB in HHV-8-infected cells. In addition, K1 was demonstrated to be a novel inhibitor of K13- and ORF75-induced NF-κB activation.
MATERIALS AND METHODS
Plasmids and small interfering RNA (siRNA).
The cloning of 85 HHV-8 genes has been previously described (45). Additionally, the following open reading frames and/or splice variants (GenBank nucleotide sequence accession numbers are given in parentheses) were cloned in pcDNA4-Myc/His, with a Myc/His tag at the 3′ end (pcDNA; Invitrogen, Karlsruhe, Germany): K8 (U93872), K11 (U75698), genomic K11 (AF148805, with introns), ORF29b (U93872), ORF40 (U93872), ORF41 (U93872), and ORF57 (U75698). All cloned constructs were confirmed by full-length sequencing. The sequences were aligned with GenBank sequences U75698 (43), U93872 (36), and AF148805 (16, 40). The reporter plasmids pNF-κB-EGFP and NF-κB-Luc were previously described (35, 45). Both reporter plasmids contain four consecutive repeats of consensus NF-κB binding sites (5′-GGG AAT TTC C-3′) linked to a thymidine kinase minimal promoter and enhanced green fluorescent protein (GFP) gene or luciferase gene as the reporter. The reporter plasmid EF-1α-Luc was obtained by inserting the HindIII fragment of the elongation factor 1α (EF-1α) promoter from the plasmid pEF1/Myc-His (Invitrogen) into the plasmid pGL3-Basic (Promega, Mannheim, Germany). Superrepressor IκBα S32/36A, the kinase-dead NF-κB-inducing kinase (KD-NIK) (K429/430A), and KD-TGF-β-activated kinase I (TAK1) (K63W) plasmids were previously described (30, 37, 61). Expression plasmids containing p65 or p50 under the control of a cytomegalovirus promoter were gifts from J. Hiscott (McGill University, Quebec, Canada) (1). A plasmid constitutively expressing GFP (pFRED) and an expression plasmid for latent membrane protein 1 (LMP-1) from Epstein-Barr virus (EBV) under the control of a simian virus 40 promoter were previously described (17). Plasmids encoding red fluorescent protein (RFP) (pDsRed1-N1; Clontech, BD Biosciences, Heidelberg, Germany) and Myc-tagged LacZ (pcDNA4-Myc/His-LacZ, Invitrogen) were obtained commercially and used as controls.
A synthetic siRNA molecule (5′-ACG UAA AGG UCG AAG GCA A dTdT-3′) targeting ORF75 mRNA was selected by using the siRNA design tool siDESIGN Center (Dharmacon, Inc., Chicago, IL). A nonsense siRNA (5′-GAC UAC CGU UGU AUA GUA G dTdT-3′), which showed no homology to any known human or HHV-8 gene, was used as a control. Both siRNA molecules were purchased from IBA (Göttingen, Germany).
Cell culture.
HEK 293T cells were cultivated in Dulbecco's modified Eagle's medium (DMEM; PAA Laboratories, Pasching, Austria) supplemented with 10% fetal calf serum (FCS; Biochrom, Berlin, Germany) at 37°C and 8.5% CO2. For reverse transfection, 100 U/ml penicillin and 100 μg/ml streptomycin (both from PAA Laboratories) were added to the medium.
RTCM. (i) Sample preparation and printing.
The method of RTCM was initially described by Ziauddin and Sabatini (70). Our protocol differed in the following steps: 0.5 μg of pNF-κB-EGFP was mixed with two different HHV-8 gene expression plasmids (0.5 μg each; for combinatory gene approach) or one HHV-8 gene expression plasmid and the empty vector (0.5 μg each; for single-gene approach), resulting in 1.5 μg total DNA. The volume was adjusted to 5 μl with double-distilled water. Three microliters Opti-MEM (Invitrogen) containing 0.4 M sucrose (Merck, Darmstadt, Germany) and 3.5 μl Lipofectamine 2000 (Invitrogen) were added and mixed. After 20 min of incubation at room temperature, 7.25 μl gelatin (0.2% in double-distilled water; Sigma-Aldrich, Seelze, Germany) was added to the transfection mixture. The final concentration of the 18.75-μl transfection mixture used for spotting contained 80 ng/μl DNA, 0.077% gelatin, and 18.7% Lipofectamine 2000. The mixture was transferred into a well of a 384-well plate that served as the source plate for slide printing (gamma aminopropyl silane II-coated slides; Corning, Schiphol-Rijk, The Netherlands). Samples were printed with a VersArray ChipWriter Pro (Bio-Rad, Munich, Germany) and PTS600 pins (600-μm diameter; Point Technologies, Boulder, CO) with a center-to-center distance of 1,120 μm. The source plate was cooled to 12°C, and the relative humidity was set to 65%. Slides were stored after printing in a desiccator with drying pearls. The slides were dried for at least 12 h prior to being overlaid with cells.
(ii) Cell seeding and RTCM processing.
The slides were placed printed side up in a sterile dish and were gently overlaid with HEK 293T cells (1 × 105 cells/cm2) in DMEM containing 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were grown at 37°C and 8.5% CO2 for 48 h. After the medium was removed, the slides were carefully taken out of the dishes and washed gently with phosphate-buffered saline (PBS; Biochrom), incubated for 15 min with 4% paraformaldehyde (Sigma-Aldrich), and washed again with PBS. The slides were dried for 5 min and then mounted with fluorescence mounting medium (Dako, Hamburg, Germany) and a coverslip.
(iii) Scanning and quantification of the signal intensities.
The reverse-transfected slides were scanned with a Fuji FLA-5000 laser scanner (Fujifilm, Düsseldorf, Germany) using a 473-nm excitation laser (blue) and a Y510 filter (long pass blue) at a resolution of 25 μm. The fluorescence intensity of every spot was quantified with the AIDA software package (version 4.15; raytest, Straubenhardt, Germany). Circular measurement areas of identical size were placed over each spot, and the fluorescence intensities were quantified and compared.
Luciferase gene reporter assay.
Twenty-four hours prior to transfection, 3 × 105 HEK 293T cells were seeded in 2 ml DMEM with 10% FCS per 6-well dish. Calcium phosphate [Ca3(PO4)2] transfection was performed with 3 μg DNA (amounts of different plasmids are indicated in the figures or figure legends) for each well. At 6 h posttransfection, the cells were washed once with PBS and fresh medium was added. At 48 h after transfection, the cells were washed again with PBS, harvested, and lysed with 200 μl passive lysis buffer (Promega) according to the manufacturer's instructions. The samples were centrifuged at 18,000 × g and 4°C for 1 min. The firefly luciferase activity of the supernatant was measured with a luciferase assay system (Promega), using a Luminoskan Ascent instrument (Thermo Scientific, Langenselbold, Germany). The values obtained were normalized according to their protein content as determined by a DC protein assay (Bio-Rad). The luciferase activities obtained in the experiments, in which the plain vector was added to the indicator plasmid, were set to 1. Student's t test and SPSS (version 15.0; SPSS, Inc., Chicago, IL) software for Windows were used to determine statistical differences, which were regarded as significant at a P value of ≤0.05.
Transfection of HHV-8-infected HEK 293 cells.
Recombinant Kaposi's sarcoma-associated herpesvirus rKSHV.219 (kindly provided by J. Vieira, Department of Laboratory Medicine, University of Washington, Seattle, WA) (60, 62) was latently propagated in HEK 293 cells in DMEM supplemented with 10% FCS and 1 μg/ml puromycin (Roth, Karlsruhe, Germany). Infected cells were seeded 24 h prior to transfection in 1 ml medium at a density of 1 × 105 cells per 12-well dish. Reporter plasmid NF-κB-Luc (0.2 μg) and siRNA (2.6 μg) were mixed with 50 μl of Opti-MEM and 2 μl of PLUS reagent (Invitrogen) and incubated for 15 min at room temperature. Amounts of 50 μl of Opti-MEM and 2 μl of Lipofectamine (Invitrogen) were added and incubated for another 15 min at room temperature. The addition of 300 μl Opti-MEM completed the transfection mixture. Cells were washed once with Opti-MEM prior to the addition of the transfection mixture. A 400-μl amount of DMEM supplemented with 20% FCS was added 4 h after transfection. Cells were harvested after 48 h with 150 μl passive lysis buffer, and the luciferase assay was performed as described above.
RNA isolation and reverse transcription-PCR.
RNA isolation was carried out with a NucleoSpin RNA isolation kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's protocol. Reverse transcription was carried out in a total reaction mixture volume of 20 μl, with 1.1 μg of total RNA, 100 U Superscript III reverse transcriptase (Invitrogen), and 50 ng of random hexamer primer (Bioline, Luckenwalde, Germany). PCR was carried out in a total reaction mixture volume of 20 μl in 1× PCR buffer with 1 μl cDNA, 0.25 U platinum Taq DNA polymerase (Invitrogen), 1.5 mM MgCl2, 0.25 mM of each deoxynucleoside triphosphate, and 0.5 μM oligonucleotide primers for amplification of a specific fragment of ORF75 (forward, 5′-AGG TCC TTC CCT TCA ACT CGG T-3′, and reverse, 5′-AAC AAG CAT GAA CAG GCT GGC-3′) and β-actin (forward, 5′-CCA TCT ACG AGG GGT ATG C-3′, and reverse, 5′-CGT GGC CAT CTC TTG CTC-3′). The cycle parameters were initial denaturation at 94°C for 2 min and 25 cycles for β-actin and 38 cycles for ORF75 of 94°C for 15 s, 60°C for 30 s, and 72°C for 40 s.
Immunocytochemistry of reverse-transfected cells.
Reverse-transfected cells were treated as described above (see “Cell seeding and RTCM processing”), including paraformaldehyde fixation. Afterwards, the slides were washed with Tris-buffered saline (TBS). Cells were permeabilized with 0.1% saponin in TBS for 20 min. After the permeabilization, unspecific binding sites were blocked with 10% goat normal serum (GNS; Dianova, Hamburg, Germany) in TBS for 10 min, and then the cells were incubated for 3 h at room temperature with a mouse anti-Myc tag monoclonal antibody (9B11; Cell Signaling, Danvers, MA) diluted 1:5,000 in 5% GNS in TBS or a rat anti-ORF73/LANA-1 antibody (Tebu-bio, Offenbach, Germany) diluted 1:500. Subsequently, the slides were washed two times with TBS and incubated for 45 min with the secondary antibody (Alexa Fluor 488-conjugated goat anti-mouse or goat anti-rat immunoglobulin G; Invitrogen) diluted 1:500 in 5% GNS. After the slides were washed two times with TBS, the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml; Invitrogen) for 10 min. Finally, the slides were washed two times with TBS and mounted with fluorescence mounting medium (Dako) and a coverslip. The covered and dried slides were scanned with a Fuji FLA-5000 laser scanner (Fujifilm). A 473-nm excitation laser (blue) with a Y510 filter (long pass blue) was used to measure the GFP and Alexa Fluor 488 signals, and a 532-nm excitation laser (green) with a BPG1 filter (green; 570DF20) was used to measure the RFP signals.
Statistical analyses.
The preprocessing of the GFP signals was done in three steps: (i) the natural logarithm (ln) of the signal was taken; (ii) replications were standardized [the ln(GFP signals) of each replication were centered by the median and divided by the ratio of the interquartile ranges and the mean interquartile range]; and (iii) when two replications were on one slide, the average of the two values of the standardized ln(GFP) signals was computed.
Two comparisons were done. First, the results for all HHV-8 genes and gene combinations were compared to those for the empty vector controls. Second, the results for all gene combinations with LMP-1 were compared to those for empty vector (pcDNA) plus LMP-1 as a control. The differences between the preprocessed signals and the median of the preprocessed signals for the empty vector controls and the empty vector plus the LMP-1 gene as a control were analyzed by using one-sided and two-sided tests, respectively, for the one-sample Wilcoxon rank sum statistic. The results for multiple testing of 339 or 22 single tests for the empty vector controls or the empty vector-plus-LMP-1 gene control, respectively, were adjusted by the Bonferroni method.
Statistically significant test results were defined as adjusted P values smaller than (i) 0.001 and (ii) 0.01. All computations for the screening of HHV-8 genes and gene combinations in NF-κB activation were done by using R version 2.6.1. (R is a language and environment for statistical computing; R Development Core Team, R Foundation for Statistical Computing, Vienna, Austria, 2007, ISBN 3-900051-07-0, http://www.R-project.org).
RESULTS
Detection of NF-κB activation by using RTCM.
The RTCM was optimized with a plasmid (pFRED) constitutively expressing enhanced GFP. Increasing concentrations of pFRED (from 0.05 μg to 5.5 μg in 18.75 μl transfection mixture), gelatin (from 0.02% to 0.31%), and Lipofectamine 2000 (9.3% and 18.7%) were used. The optimal transfection efficiency was determined according to the maximal GFP signal intensity of the different transfection spots. The highest transfection efficiency was observed using 80 ng/μl DNA, 0.077% gelatin, and 18.7% Lipofectamine 2000 (data not shown).
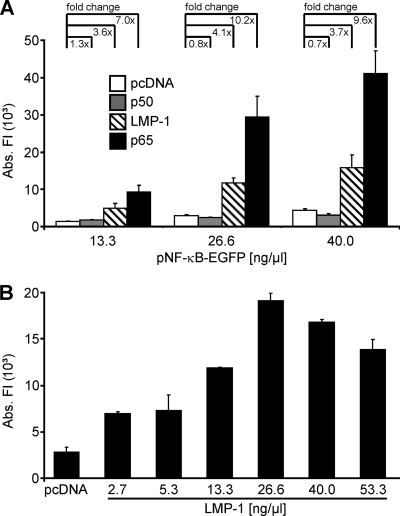
In order to detect activation of NF-κB, the reporter plasmid pNF-κB-EGFP, containing four consecutive NF-κB sites upstream of GFP, was used. It has been previously shown that the GFP fluorescence intensity is directly proportional to the abundance of GFP mRNA (51). As the first step, transfection parameters were established which allowed the most sensitive detection of NF-κB activation. For this purpose, three different concentrations of the reporter plasmid pNF-κB-EGFP (13.3 ng/μl, 26.6 ng/μl, and 40.0 ng/μl) were cotransfected with constant concentrations (13.3 ng/μl) of plasmids encoding LMP-1 of EBV and the p65 subunit of NF-κB, both of which strongly activate NF-κB (1, 17). The empty vector (pcDNA) and the p50 subunit of NF-κB (35) were used as negative controls (Fig. 1A). GFP expression in consequence of NF-κB activation was clearly increased in the presence of the two activators (Fig. 1A) compared to its levels in transfections with the negative controls at all concentrations of the reporter plasmid (Fig. 1A). However, the relative increase of promoter activity in the presence of the activators was highest with 26.6 ng/μl of the reporter plasmid. Therefore, 26.6 ng/μl of the reporter plasmid were used in all further approaches. Next, constant concentrations of the reporter plasmid (pNF-κB-EGFP; 26.6 ng/μl) were combined with increasing concentrations of the LMP-1-expressing activator plasmid (2.7 ng/μl to 53.3 ng/μl). Promoter activity peaked at a concentration of 26.6 ng/μl of the activator plasmid (Fig. 1B). Thus, in the following experiments this concentration was used for all plasmids carrying HHV-8 genes.
FIG. 1.
Detection of NF-κB activation in RTCM. (A) An NF-κB-inducible GFP reporter plasmid (pNF-κB-EGFP) in three different concentrations (as shown) and constant amounts (13.3 ng/μl) of plasmids encoding two different activators of NF-κB (LMP-1 and p65) or control plasmids (pcDNA and p50) were cotransfected into HEK 293T cells by using RTCM. (B) The reporter plasmid pNF-κB-EGFP (26.6 ng/μl) was cotransfected with increasing amounts (as shown) of an LMP-1-encoding plasmid into HEK 293T cells by using RTCM. As a negative control, cells were transfected with empty vector (pcDNA). (A, B) In all transfections, the total amount of plasmid DNA was adjusted to 80.0 ng/μl by the addition of pcDNA. Experiments were carried out in quadruplicate (A) or duplicate (B). GFP fluorescence intensities of the transfection spots were determined with a laser scanner (473 nm) and were quantified with the AIDA software package. Mean values and standard deviations of the absolute fluorescent intensities (Abs. FI) are depicted. The fc values compared to the results for the empty-vector control (pcDNA) are indicated in panel A.
Expression of HHV-8 genes using RTCM.
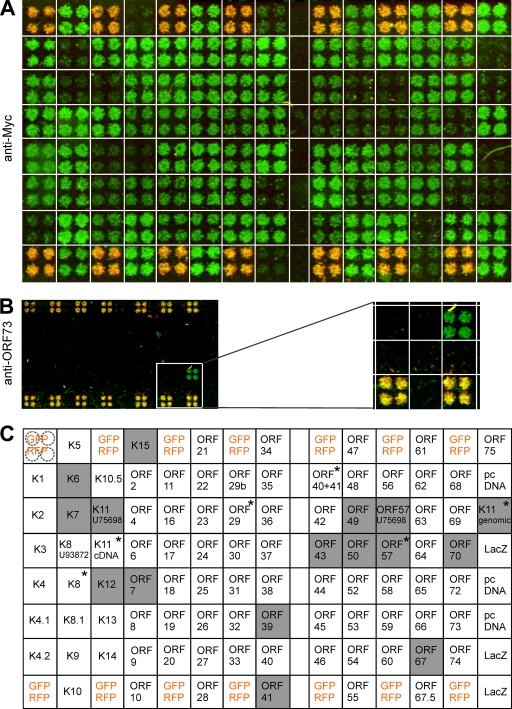
The coding sequences of all HHV-8 genes and known splice variants with the exception of ORF73/LANA-1 were cloned in expression plasmids with a 3′-end Myc tag sequence for immunochemical detection of the respective proteins (n = 91) (see Material and Methods). LANA-1 was expressed without a Myc tag due to difficulties with cloning. These 92 plasmids were transfected into HEK 293T cells by using RTCM. In addition, a plasmid encoding a Myc-tagged LacZ, the empty vector (pcDNA), and a mixture of plasmids for constitutive expression of GFP and RFP (positive controls and positional markers) was applied. The application pattern on the chip is shown in Fig. 2C. RTCM were subjected to immunocytochemical staining with an anti-Myc antibody. Fluorescence signals were detected with a laser scanner and quantified with the AIDA software (Fig. 2A). Another RTCM chip was stained in parallel with an anti-ORF73/LANA-1 antibody (Fig. 2B). Signal evaluation of different reverse-transfected slides showed that 83% (76 of 92) of the transfected HHV-8 gene-carrying plasmids were robustly expressed in the RTCM (Fig. 2). Only 17% (16 of 92) of the transfected HHV-8 gene-carrying plasmids were not expressed at detectable levels using this technique (signal intensity equal to or lower than that of pcDNA) (Fig. 2A and C). However, the expression of all of these 16 plasmids could be confirmed by immunocytochemical staining of classical transfected HeLa cells (45) or by Western blot analyses of total cell lysates obtained from classical transfected HEK 293T cells (data not shown).
FIG. 2.
Expression of HHV-8 genes in RTCM. (A) All HHV-8 gene-encoded proteins (except the ORF73-encoded LANA-1) were expressed with a Myc tag in HEK 293T cells by using RTCM and were detected by immunocytochemistry with an antibody against the Myc tag. Myc immunofluorescence is green. The pattern of application of plasmids on the chip is shown in panel C. (B) Expression of ORF73-encoded LANA-1 was detected with a specific antibody against ORF73-encoded LANA-1 (green). The relevant part of the slide is enlarged on the right side of panel B and positioned underneath the corresponding area in panel A. (C) Application pattern of transfected plasmids in RTCM shown in panels A and B. Each plasmid was spotted in quadruplicate (see upper left, dashed circles). As a negative control, cells were transfected with empty vector (pcDNA). A mix of constitutively GFP- and RFP-expressing plasmids was spotted at several different positions in order to evaluate transfection efficiency on the chip and to provide positional markers (yellow in panels A, B, and C). A plasmid expressing a Myc-tagged LacZ protein was used as a positive control. The secondary antibody used in immunofluorescence analyses was conjugated with Alexa Fluor 488. Fluorescence signals were determined with a laser scanner (473 nm for Alexa Fluor 488 and GFP signals and 532 nm for RFP signals; overlays of the two scans are depicted in panels A and B) and quantitatively analyzed with the AIDA software package. Proteins with fluorescence intensities equal to or lower than that of the empty vector control (pcDNA) were defined as not detectable on RTCM (shaded gray; white squares show proteins that were robustly expressed). Transfection mixtures contained 80.0 ng/μl of each plasmid (except for GFP and RFP, used at 40 ng/μl each). Recently reported splice variants of HHV-8 genes (indicated by asterisks) were included in the immunocytochemical analysis but not in the RTCM screen of NF-κB activation (see Fig. 3).
Single-gene and combination effects of HHV-8 genes on NF-κB activation.
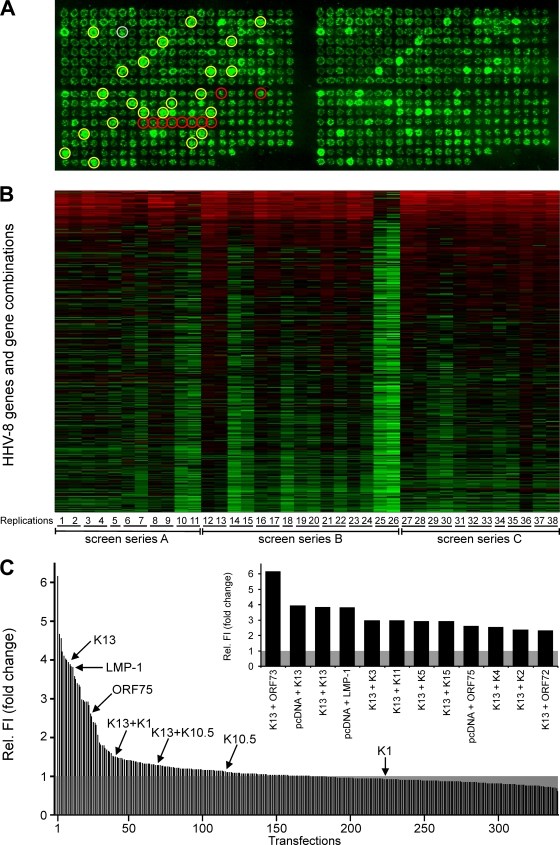
The effects of 86 individual HHV-8 genes on NF-κB activation were investigated with RTCM. Recently identified splice variants (Fig. 2C) were not included in this screen but were investigated in a supplementary separate screen (data not shown). In addition, the effects of selected HHV-8 genes in combination were examined. This selection included all K genes (n = 19), which have been originally regarded to be specifically present in the HHV-8 genome and not in other herpesviruses, and all latently expressed genes. Besides various K genes (K2, K10, K10.5, K11, K12, K13, and K15), only ORF72 and ORF73 are classified as latent genes of HHV-8 (13, 23, 38, 47). Therefore, these two “open reading frame” genes were included in our combinatory approach. Accordingly, 21 genes (19 K genes, ORF72, and ORF73) were expressed in all possible pairwise combinations in the RTCM (n = 231) to screen for NF-κB modulators (Fig. 3A). In order to detect potential inhibitors of NF-κB, the LMP-1 gene of EBV was used as an activator of NF-κB (Fig. 3A) and was combined with the K genes, ORF72, and ORF73 (n = 21). Transfections with pcDNA were used as negative controls (n = 10) (Fig. 3A). In addition, pFRED was randomly spotted at different positions on the slide as a positional marker and in order to determine the variability of the transfection procedure (n = 22) (Fig. 3A). Altogether, every RTCM experiment consisted of 371 transfection areas (single HHV-8 genes [n = 86]; combinations of K genes, ORF72, and ORF73 [n = 231]; the LMP-1 gene combined with K genes, ORF72, and ORF73 [n = 21]; GFP positional controls [n = 22]; negative control pcDNA [n = 10]; and the positive-control LMP-1 gene [n = 1]). All of these samples were spotted in duplicate, resulting in 742 transfections overall on one slide (Fig. 3A, compare left and right).
FIG. 3.
Single-gene and combination effects of HHV-8 genes on NF-κB activation. (A) Image of a representative RTCM chip with 371 transfections in duplicate (left and right). Expression plasmids for all HHV-8 genes individually (n = 86) and, additionally, all K and latent genes (19 K genes, ORF72, and ORF73) in all possible pairwise combinations (n = 231) were spotted onto the slide. A plasmid encoding LMP-1 was used as a positive control (gray circle; n = 1) and empty-vector pcDNA (red circles; n = 10) as a negative control. Moreover, the LMP-1 gene was combined with the K genes, ORF72, and ORF73 (n = 21). The reporter plasmid pNF-κB-EGFP was added to all of the described settings. A constitutively GFP-expressing plasmid (pFRED) was spotted at several different positions in order to control transfection efficiency on the chip and to provide a positional marker (yellow circles; n = 22). HEK 293T cells were seeded on RTCM chips. GFP fluorescence was determined with a laser scanner (473 nm) after 48 h. (B) Heat map of 38 replications of RTCM analyses. The natural logarithms of the fluorescence intensity values (corresponding to NF-κB activation) are depicted. The median value of GFP signals obtained with the empty-vector control (pcDNA) for each RTCM replication was set to 1 (black). Signals were sorted according to decreasing average fc. The color scheme from red to black to green corresponds to the fluorescence intensity values from high [ln(fc) = 3] to low [ln(fc) = −1.18]. The numbers in the bottom line (1 to 38) indicate the different replications. Replications with the same underlining were carried out on one slide. Three different preparations of spotted slides (screen series A to C) were used. (C) GFP signals corresponding to NF-κB activity were determined for all 38 replications by using a laser scanner (473 nm) and analyzed with the AIDA software package. Median values of relative fluorescence intensities (Rel. FI) compared to that of the empty-vector control pcDNA (set to 1; shaded gray) were calculated and are arranged in decreasing order (except for constitutive GFP signals and combinations of HHV-8 genes with the LMP-1 gene). Values obtained with certain genes which are of relevance in this report are indicated. Genes and gene combinations which resulted in more-than-twofold (fc ≥ 2.0) and highly significant (P ≤ 0.001) activation of NF-κB are given in the inset.
Three independent series of slides (Fig. 3B, series A, B, and C) were spotted, and reverse transfection was performed at least six times with each series of slides. Overall, 21 RTCM slides (resulting in 42 replications) were used (Fig. 3B). Four replications were excluded because of partial detachment of the cell layer. Altogether, 38 replications with over 14,000 transfections were carried out (Fig. 3B). GFP fluorescence as a readout of NF-κB activity was determined with a laser scanner, and the signals were quantified with the AIDA software. The signals of constitutive GFP expression due to transfection with pFRED were omitted. All other GFP signals, indicating NF-κB activation of all RTCM, are shown in a heat map (Fig. 3B). The median value of GFP signals obtained in transfections with the empty vector control (pcDNA) was calculated and set to 1. High GFP signal intensities are shown in red, and low signal intensities in green (Fig. 3B). The relative levels of activation observed in the different replications were highly reproducible, except for replications 25 and 26 (both on the same slide), which showed overall low NF-κB activation, possibly due to lower transfection efficiencies in these two experiments (Fig. 3B).
In order to reveal HHV-8 genes that are NF-κB activators, the median intensities of all GFP signals shown in Fig. 3B (except for combinations of HHV-8 genes with the LMP-1 gene) were calculated and compared to those of transfections with the empty vector controls (Fig. 3C), and the results are given as the factor of change (fc), sorted decreasingly (Fig. 3C). All genes and gene combinations with greater-than-twofold (fc ≥ 2.0) and highly significant (P ≤ 0.001) average induction of NF-κB are shown in the inset in Fig. 3C and in Table 1. K13 was the gene which best fulfilled this criterion. It was present individually (pcDNA plus K13) and in eight pairwise combinations with different HHV-8 genes (Fig. 3C, inset, and Table 1). Only the combination of K13 with ORF73 had a level of activation higher than that observed for K13 alone (pcDNA plus K13) in most of the RTCM. However, this difference (pcDNA plus K13 compared to K13 plus ORF73) was not statistically significant (Fig. 3C, inset, and Table 1). Therefore, the combination of K13 and ORF73 was not further investigated.
TABLE 1.
NF-κB activation by HHV-8 genes/gene combinations
| Genes | NF-κB activation
|
Adjusted P value | |
|---|---|---|---|
| Fca | Rangeb | ||
| K13 + ORF73 | 6.2 | 2.8-14.8 | 0.0002 |
| pcDNA + K13 | 3.9 | 0.6-13.2 | 0.0003 |
| K13 + K13 | 3.8 | 1.9-10.1 | 0.0002 |
| pcDNA + LMP-1c | 3.8 | 1.2-6.2 | 0.0002 |
| K13 + K3 | 3.0 | 0.9-3.8 | 0.0003 |
| K13 + K11 | 3.0 | 1.6-5.8 | 0.0002 |
| K13 + K5 | 2.9 | 1.5-5.2 | 0.0002 |
| K13 + K15 | 2.9 | 1.5-4.7 | 0.0002 |
| pcDNA + ORF75 | 2.6 | 1.5-4.7 | 0.0002 |
| K13 + K4 | 2.5 | 0.8-4.3 | 0.0005 |
| K13 + K2 | 2.4 | 0.9-6.3 | 0.0003 |
| K13 + ORF72 | 2.3 | 1.4-7.5 | 0.0002 |
The fc values were determined in comparison to the results for transfections with pcDNA alone.
RTCM results were obtained for 38 replications.
The LMP-1 gene was used as a positive control for NF-κB activation.
K13 has been previously described as an NF-κB activator (9, 11, 19, 57, 60). The data shown here confirmed that K13 is the strongest NF-κB activator of all HHV-8 genes, with an average fc of 3.9 (Fig. 3C, inset, and Table 1). The only HHV-8 gene in addition to K13 which showed strong and highly significant (P ≤ 0.001) activation of NF-κB when applied alone was ORF75 (Fig. 3C, inset). The activation of NF-κB by ORF75 was consistently greater than twofold in this experimental setup (average fc, 2.6) (Fig. 3C, inset, and Table 1). LMP-1 was used as a positive control for NF-κB activation (Fig. 3C, inset, and Table 1). Evaluation of a separate screen revealed that none of the recently described splice variants (Fig. 2C) activated NF-κB more highly than K13 or ORF75 (data not shown).
Identification of HHV-8 genes that are NF-κB inhibitors with RTCM.
In order to identify inhibitors of NF-κB activation, the GFP signals corresponding to NF-κB activities of the transfections with LMP-1 alone (positive control) and of all combinations of this activator with the K genes, ORF72, and ORF73 were determined. The median fc values from these experiments were calculated in comparison to the values obtained with the positive control (set to 1). Transfections with highly significant (P ≤ 0.01) and >50%-decreased (fc ≤ 0.5) NF-κB activation are shown in Table 2. Four genes (K1, K9, K10.5, and K14) inhibited LMP-1-induced NF-κB activation (Table 2). Of these genes, only K10.5 has been previously described as an inhibitor of NF-κB activation (49).
TABLE 2.
NF-κB inhibition by HHV-8 genes
| Analytical method | Gene
|
NF-κB inhibition
|
P value | ||
|---|---|---|---|---|---|
| Activating | Inhibitory | Fca | Rangeb | ||
| RTCM | LMP-1 | K14 | 0.50 | 0.22-1.62 | 0.01 |
| K10.5 | 0.48 | 0.28-1.39 | 0.01 | ||
| K9 | 0.41 | 0.18-2.09 | 0.01 | ||
| K1 | 0.38 | 0.26-1.25 | 0.01 | ||
| Classical transfection | LMP-1 | K14 | 0.41 | 0.35-0.49 | 0.001 |
| K1 | 0.35 | 0.32-0.39 | 0.001 | ||
| K9 | 0.27 | 0.21-0.31 | 0.001 | ||
| K10.5 | 0.17 | 0.14-0.20 | 0.001 | ||
| K13 | K9 | 0.65 | 0.61-0.69 | 0.001 | |
| K14 | 0.62 | 0.58-0.65 | 0.001 | ||
| K10.5 | 0.48 | 0.44-0.50 | 0.001 | ||
| K1 | 0.40 | 0.37-0.43 | 0.001 | ||
The fc values were determined in comparison to those for transfections with the activator (the LMP-1 gene or K13) alone. The threshold for significant inhibition of LMP-1 gene- or K13-induced NF-κB activation was set to an fc of ≤0.50 and a P value of ≤0.05.
RTCM results were obtained for 38 replications.
ORF75 and K13 are the major activators of NF-κB encoded by HHV-8.
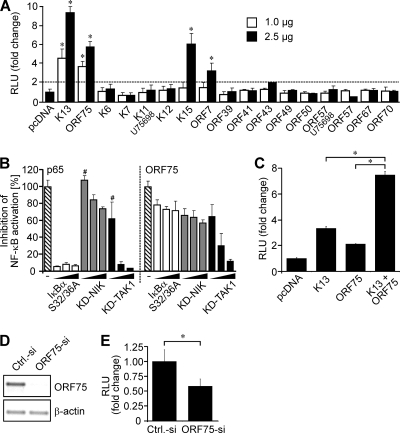
NF-κB activation by K13 and ORF75 was confirmed in classical transfection experiments (Fig. 4A). Small (1.0 μg) and large amounts (2.5 μg) of K13 or ORF75 were cotransfected with a reporter plasmid encoding luciferase under the control of an NF-κB-responsive promoter (NF-κB-Luc). K13 and ORF75 plasmids in both concentrations significantly (P ≤ 0.05) activated NF-κB in HEK 293T cells compared to the activity in the cells transfected with empty vector (pcDNA; negative control, set to 1) (Fig. 4A). NF-κB activation by K13 was quantitatively higher (fc = 4.6 and fc = 9.4 for 1.0 μg and 2.5 μg plasmid, respectively) than NF-κB activation by ORF75 (fc = 3.7 and fc = 5.8 for 1.0 μg and 2.5 μg plasmid, respectively) (Fig. 4A), which was in clear agreement with the results obtained using RTCM (Fig. 3C).
FIG. 4.
ORF75 and K13 are the major activators of NF-κB in HHV-8 infected cells. (A) Luciferase activity in HEK 293T cells cotransfected with reporter plasmid NF-κB-Luc (0.5 μg) and two different amounts (1.0 μg and 2.5 μg) of the indicated plasmids. Transfection with NF-κB-Luc (0.5 μg) and the empty vector (pcDNA; 2.5 μg) served as a negative control. The dashed line shows the threshold level. (B) HEK 293T cells were cotransfected with reporter plasmid NF-κB-Luc (0.5 μg) and p65 (0.1 μg) or ORF75 (1.0 μg) either alone (hatched bars) or in combination with increasing amounts (0.1 μg, 1.0 μg, and 1.5 μg) of the indicated inhibitors (IκBα S32/36A, KD-NIK, and KD-TAK1). All inhibitions were significant (P ≤ 0.05) except those indicated with the symbol #. (C) HEK 293 cells were cotransfected with reporter plasmid NF-κB-Luc (0.5 μg) and K13 (1 μg), ORF75 (1 μg), or both combined (1 μg each). Transfection of the empty vector (pcDNA) served as a negative control. (A to C) The total amount of plasmid was adjusted to 3.0 μg by the addition of pcDNA. (D, E) HHV-8-infected HEK 293 cells were cotransfected with reporter plasmid NF-κB-Luc (0.2 μg) and siRNA against ORF75 (ORF75-si) or control siRNA (Ctrl.-si) (2.6 μg each). Cells were divided in two parts. One part was used for reverse transcription-PCR to control the downregulation of ORF75 (D), and the other was subjected to the luciferase assay (E). (D) Reverse transcription-PCR analysis of ORF75 and β-actin. Equal amounts of cDNA were subjected to PCR after reverse transcription of mRNA isolated from the samples described above. Densiometric analysis (values obtained were normalized according to the respective β-actin signal) revealed a 95% downregulation of ORF75 in the ORF75 siRNA-transfected cells. (E) Luciferase activity in siRNA-treated HHV-8-infected cells. (A to C, E) In all luciferase studies, NF-κB promoter activation was analyzed by measurement of luciferase 48 h after transfection. Values obtained were normalized according to protein content, and the results are expressed as either fc (A, C, and E) or percent (B) in comparison to the results for the control (pcDNA, set to 1, in panels A and C; p65 and ORF75 [hatched bars], set to 100%, in panel B; and Ctrl.-siRNA, set to 1, in panel E). Luciferase assays were performed at least three times; mean values and standard deviations of relative light units (RLU) are shown. *, P ≤ 0.05.
As described above, 16 plasmids consistently showed only a low level of expression in the RTCM (Fig. 2A). In order to investigate whether the low level of expression in RTCM may have averted the detection of additional NF-κB activators, 15 of these plasmids were subjected to the luciferase assay with classical transfection in two different concentrations (1.0 μg and 2.5 μg) (Fig. 4A). The plasmid “genomic K11” was omitted because “K11 cDNA” encodes the identical protein but was expressed at higher levels (Fig. 2A). In this screen, K15 and ORF7 also activated NF-κB significantly (for K15, fc = 6.1 and for ORF7, fc = 3.2; P ≤ 0.05). In agreement with these results, it has been shown previously that K15 can activate NF-κB (4). However, both K15 and ORF7 only activated NF-κB when larger amounts of plasmid DNA (2.5 μg) were used (Fig. 4A). None of the 15 plasmids induced significant activation above the threshold level (twofold above background) (Fig. 4A) when used in smaller (1.0 μg) amounts, which clearly differentiated them from K13 and ORF75 (Fig. 4A). Therefore, we concentrated on ORF75 in the following experiments.
ORF75 and K13 cooperate in the activation of NF-κB in HHV-8-infected cells.
In order to characterize the effect of ORF75 on NF-κB activation in more detail, we investigated whether it may act through the classical/canonical or the alternative/noncanonical pathway. To this end, three different inhibitors of the NF-κB pathway were used in increasing amounts (0.1, 1.0, and 1.5 μg): (i) a KD mutant of TAK1 which inhibits the classical pathway (10, 37, 63); (ii) a KD mutant of NIK which inhibits the alternative pathway (10, 30, 66); and (iii) inhibitor of NF-κBα S32/36A (IκBα S32/36A), a constitutively active IκBα mutant, which acts downstream of TAK1 (61). Both p65-induced (a control of the classical pathway) and ORF75-induced NF-κB activation were potently inhibited by KD-TAK1 (Fig. 4B) and only slightly inhibited by KD-NIK (Fig. 4B). We found that, as a control of KD-NIK activity, LMP-1-induced NF-κB activation was potently inhibited (∼90%) (data not shown), as has been described previously by others (58). These results suggested that ORF75 activates NF-κB through the classical pathway.
Interestingly, ORF75-mediated activation was only slightly inhibited (∼30%) even with large amounts (1.5 μg) of the IκBα S32/36A plasmid (Fig. 4B, right panel). In contrast, p65-induced (Fig. 4B, left panel) and K13-induced (data not shown; also described by others [56]) NF-κB activation was efficiently inhibited already at very small amounts (0.1 μg) of transfected IκBα S32/36A plasmid (90% and 70% inhibition, respectively). These results indicated that ORF75 and K13 may use partly different routes of the classical pathway downstream of TAK-1.
Interestingly, the combination of K13 and ORF75 resulted in a level of activation of NF-κB that was significantly higher than the levels of activation by the single genes (Fig. 4C). This indicated that both genes could cooperate in the activation of NF-κB. Accordingly, we investigated the impact of ORF75-mediated NF-κB activation in HHV-8-infected cells using an siRNA approach. To this end, HHV-8-infected HEK 293 cells (62) were transfected with the NF-κB luciferase reporter plasmid and either a control siRNA or a specific siRNA against ORF75 mRNA. Due to the lack of a specific antibody, the expression of ORF75 was investigated with reverse transcription-PCR. ORF75 mRNA was found to be expressed in HHV-8-infected HEK 293 cells in the absence of lytic stimulation (control siRNA) (Fig. 4D), and its expression was found to be efficiently (95%) inhibited by the ORF75 siRNA (Fig. 4D). Of note, NF-κB activity was significantly lower (∼40%; P ≤ 0.05) in cells transfected with the ORF75 siRNA than in cells transfected with the control siRNA (Fig. 4E). These results show that ORF75 significantly contributes to the NF-κB activation in HHV-8-infected cells. The residual NF-κB activity was most likely due to K13, which has been shown to be expressed and to contribute to increased NF-κB activation in HHV-8-infected cells (2, 9, 19, 60).
K1 is an inhibitor of NF-κB activation.
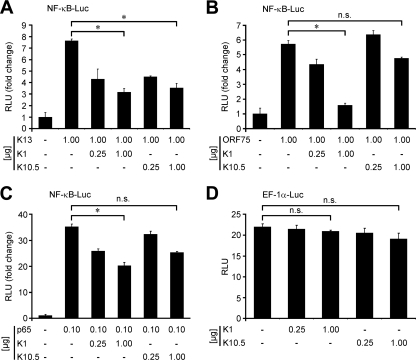
As described for the activators, the putative inhibitors of NF-κB activation identified by RTCM were also subjected to confirmation experiments using classical transfection methodology. The effects of K1, K9, K10.5, and K14 on LMP-1-induced NF-κB activation were examined. In this study, all four HHV-8 genes that were inhibitors (K1, K9, K10.5, and K14) repressed LMP-1-induced NF-κB activation (fc ≤ 0.5; P ≤ 0.05), confirming the results of the RTCM analysis (Table 2). The switch to the major HHV-8 NF-κB activator revealed that all four of these genes also significantly inhibited K13-induced NF-κB activation, but the results for K9 and K14 did not reach the threshold level (fc ≤ 0.5) (Table 2). Therefore, K9 and K14 were not evaluated in further studies. Next, the effects of K1 and K10.5 on NF-κB activation by K13, ORF75, and p65 were investigated. NF-κB activation induced by all of the three activators was significantly (P ≤ 0.05) inhibited by K1 in a dose-dependent manner (Fig. 5A, B, and C). K10.5, which has been previously shown to inhibit NF-κB activation (49), repressed NF-κB activation by K13 but had only slight, nonsignificant effects on NF-κB activation by ORF75 and p65 (Fig. 5A, B, and C). Under these conditions, neither of the two inhibitors (K1 and K10.5) had an impact on the activity of the EF-1α promoter (Fig. 5D).
FIG. 5.
K1 is an inhibitor of NF-κB activation. Luciferase assays of NF-κB promoter activation in HEK 293T cells. (A to C) NF-κB-Luc (0.5 μg) was cotransfected with different activators of NF-κB (K13, ORF75, and p65) and K1 or K10.5 in the indicated amounts. (D) In an additional control experiment, a reporter plasmid (EF-1α-Luc; 0.5 μg) expressing luciferase under the control of the constitutively active promoter of EF-1α was used in the absence or presence of K1 and K10.5 under identical conditions. (A to D) In all studies, the total amount of plasmid in each transfection mix was adjusted to 3.0 μg by the addition of pcDNA. NF-κB promoter activation was analyzed by measurement of luciferase 48 h after transfection. Values obtained were normalized according to protein content, and the results are expressed in terms of the fc in comparison to the results for the negative control (pcDNA only, left bar in each panel; set to 1). The luciferase assays were performed at least three times; mean values and standard deviations of relative light units (RLU) are shown. −, absent; *, P ≤ 0.05; n.s., not significant.
DISCUSSION
RTCM is a novel method which can be used for high-throughput analyses of gene functions by either overexpressing or silencing genes of interest. Despite numerous advances achieved in this technology since its first description in 2001 (70), the number of published studies using RTCM is still quite limited (for a review, see reference 53). Most of the published studies addressed proof-of-concept topics. A major advantage of RTCM studies in comparison to classical high-throughput studies is that several slides can be printed with the same experimental setup, and slides can be stored for several months to reconfirm the obtained results at a later date. Ambitious studies performing genome-wide overexpression analyses of single-gene effects on NF-κB activation in HEK 293T cells with classical luciferase reporter assays in 96- or 384-well format were published recently (20, 31). We hypothesized that the analysis of the combination effects of a limited number of genes extracted from biologically meaningful samples, such as infectious agents (viruses and bacteria), or from pathogenetically relevant gene clusters found by transcriptome analysis to be coexpressed might be an interesting approach for the application of RTCM.
Based on these considerations, RTCM was used here for the first time to analyze combinatory gene effects of an infectious agent. We investigated NF-κB activation by all HHV-8 genes individually and by pairwise combinations of a selected set of genes. The NF-κB signal transduction pathway is considered to be a prime candidate triggering the pathogenesis of Kaposi's sarcoma and PEL (11, 24, 25, 44). Inhibition of NF-κB leads to lytic-protein synthesis and virus reactivation in infected B lymphocytes (5). This indicates that NF-κB plays an important role in the regulation of the balance between the latent and lytic life cycle phases of HHV-8.
We established a reporter gene approach to detect NF-κB activation in RTCM. This method was highly reproducible, as demonstrated in 38 replications with more than 14,000 transfections (Fig. 2B). Most importantly, the validity of the results obtained with RTCM was clearly demonstrated by the fact that two previously known activators (K13 and the LMP-1 gene [positive control]) and an inhibitor of NF-κB (K10.5) were confirmed in the screening.
The HHV-8 genes K13 and ORF75 were found to be the major activators of the NF-κB signaling pathway in the RTCM analysis. The sequence accuracy and successful expression of all HHV-8 genes used in our study were demonstrated recently (45). However, in the RTCM, 17% of the transfected HHV-8 gene-carrying plasmids were too weakly expressed to be detected by immunocytochemistry. Of note, K13 was also only weakly expressed (Fig. 2A), but it robustly activated NF-κB in RTCM. Nevertheless, we investigated in classical transfection experiments whether additional activators of NF-κB might have escaped our detection in the RTCM study. In fact, two further genes, K15 and ORF7, significantly (P ≤ 0.05) activated NF-κB when large amounts of plasmid DNA (2.5 μg) were transfected. K15 encodes a latency-associated membrane protein and has been previously described as an activator of NF-κB (4). ORF7 encodes a homologue of a processing and transport protein and as yet has not been associated with activation of NF-κB. In contrast to ORF75 and K13, both genes (K15 and ORF7) had no effect on NF-κB activation when smaller amounts (1.0 μg) of plasmid DNA were used. Therefore, it has to be determined whether the expression of K15 and ORF7 in HHV-8-infected cells may be sufficiently high to contribute to NF-κB activation. ORF74 has been described as an activator of NF-κB (48). However, we did not observe ORF74-mediated NF-κB activation in the RTCM analysis or in classical transfection experiments (data not shown). The reason for the negative result with ORF74 needs to be determined, as this gene was strongly expressed in the RTCM.
The HHV-8 gene K13 was the strongest NF-κB activator. K13 encodes a viral FLICE-inhibitory protein (FLIP) (18, 21, 52, 59) and was well established as an NF-κB activator before (9, 32, 56). We were the first to show that K13-encoded viral FLIP is localized, exceptionally, both in the cytoplasm and the nucleus of the cell, whereas cellular FLIP molecules are exclusively cytoplasmic (45). This has subsequently been confirmed by others (33). NF-κB activation by K13 is involved in its antiapoptotic functions (15, 55, 60) and mediates the inhibition of lytic viral replication (67, 68). The fact that none of the pairwise HHV-8 gene combinations showed a statistically significant higher level of NF-κB activation than K13 alone in RTCM (Fig. 3C and Table 1) underlines the dominant role of K13 in NF-κB activation.
The HHV-8 gene ORF75 was identified by RTCM as a novel NF-κB activator. ORF75 encodes a homologue of phosphoribosylformylglycineamide amidotransferase (FGARAT), which is part of the viral tegument (3, 22, 42, 43, 69). The combination effect of K13 and ORF75 on NF-κB activation was not investigated in the RTCM analysis; therefore, the cooperation of the two genes was tested in classical transfection experiments using luciferase as a reporter. Both proteins cooperated more than additively in the activation of NF-κB (for K13, fc = 3.4; for ORF75, fc = 2.1; and for K13 plus ORF75, fc = 7.5) and, consequently, may cooperate in the establishment of NF-κB-mediated effects in infected cells.
The specific activation pathways used by ORF75 were analyzed by using different inhibitors of the classical (KD-TAK1 and IκBα S32/36A) and alternative (KD-NIK) NF-κB pathways. These experiments indicated that ORF75 activates NF-κB preferentially through the classical pathway, because it was strongly and selectively inhibited by KD-TAK1. Interestingly, IκBα S32/36A, which acts downstream of TAK1, only slightly inhibited the effects of ORF75 (Fig. 4B) but efficiently blocked NF-κB activation by K13 (56; data not shown). This suggested that ORF75 and K13 may use partly different routes of the classical pathway downstream of TAK-1, which may maintain NF-κB activation in different cellular and environmental conditions. ORF75 may deactivate other IκB molecules, compared to K13, or may deactivate IκBα S32/36A by phosphorylation of amino acids other than S32 and S36. In fact, it has been shown that phosphorylation of Y42 and Y305 can also lead to deactivation of IκBα (64).
In order to support the relevance of our findings, we investigated the impact of ORF75 on NF-κB activation in HHV-8-infected cells. It is a subject of controversy whether ORF75 is a latently or lytically expressed gene (22, 38, 46). However, we detected robust expression of ORF75 mRNA in HHV-8-infected HEK 293 cells in the absence of lytic stimulation (Fig. 4D), indicating that ORF75 is latently expressed. ORF75 expression in these cells could be efficiently inhibited (95%) with a specific siRNA. Most importantly, this resulted in significantly reduced NF-κB activation (∼40%) (Fig. 4E). On consideration of the results shown here and by others, the residual NF-κB activity most likely has to be attributed to K13 (2, 9, 19, 60). Altogether, these results showed that ORF75 contributes significantly to NF-κB activation in HHV-8-infected cells. This may be biologically meaningful, as the product of ORF75 is a viral tegument protein (3, 42, 43, 69). Accordingly, the ORF75 protein may activate NF-κB directly after viral entry without de novo protein synthesis and may regulate the early phases in the establishment of the latent infection. It is well in agreement with this hypothesis that K13 expression can only be detected after 2 h (26), whereas NF-κB activation is already observed 5 to 15 min after de novo infection of cells by HHV-8 (44).
K10.5 and K1 were identified as inhibitors of NF-κB in the RTCM. K10.5 encodes viral interferon regulatory factor 3. It is expressed in the latent phase of the viral life cycle (22, 41, 46) and has been shown to be required for the survival of HHV-8-infected PEL cells (65). Inhibition of NF-κB activation by K10.5 has also been observed by others (49), supporting the validity of the RTCM results. Certainly, it is contradictory that both NF-κB activation and K10.5 (an inhibitor of NF-κB) are required for the survival of PEL cells (24, 25, 49, 65). However, the detection of constitutively active NF-κB in HHV-8-infected PEL cells suggests that the inhibitory function of K10.5 is overruled by the cooperative activity of K13 and ORF75 in latently infected cells. This is further supported by our finding that K10.5 did not significantly inhibit NF-κB activation by ORF75.
K1 encodes a transmembrane glycoprotein with a functional immunoreceptor tyrosine-based activation motif (29). A consensus has not yet been reached in the literature about the effect of K1 on NF-κB activation. K1-mediated NF-κB activation (39), as well as inhibition of tetradecanoyl phorbol acetate-induced NF-κB activation (28), has been described. This indicates that the impact of K1 on NF-κB activation may depend on the kind of activation (tetradecanoyl phorbol acetate induced versus uninduced). We are the first to demonstrate that K1 inhibits both HHV-8 genes that are major activators of NF-κB (K13 and ORF75). The specificity of this activity was demonstrated by the fact that p65-induced promoter activity was also inhibited, while the activity of a control promoter (EF-1α) remained unaffected. An NF-κB-inhibitory function of K1 is in agreement with the biology of HHV-8. It has been shown that K1 is a lytic gene (54), augmenting lytic replication (27). On consideration that inhibition of NF-κB in latently infected cells is sufficient to induce the lytic cycle (5), K1 may significantly promote the lytic replication of HHV-8. Altogether, these results suggest that HHV-8 genes encode different molecules, such as the products of K1, K10.5, K13, and ORF75, which may contribute to the switch from latency to lytic replication by modulating NF-κB activity.
In summary, we have described the first successful application of RTCM for the systematic analysis of pathofunctions of an infectious agent. With this approach, two novel viral molecules (encoded by ORF75 and K1) that are regulators of the NF-κB signaling pathway were identified. Our findings underscore the strength of systems biology to provide new insights into pathogenesis and new molecular targets for the treatment of infectious diseases.
Acknowledgments
We thank Michael Bauer and Mahimaidos Manoharan for excellent technical support, Susanne Reed for help in writing, and Matthew Miller for critical reading of the manuscript (all from the Division of Molecular and Experimental Surgery). The generous support of Werner Hohenberger (Director of the Department of Surgery) is gratefully acknowledged.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG-SPP1130, DFG-GK1071, and STU317/2-1), German Cancer Aid (Deutsche Krebshilfe, Apoptose-Schwerpunktprogramm), and the Interdisciplinary Center for Clinical Research (IZKF) of the University of Erlangen-Nuremberg to M.S. Additional support was obtained by a tandem-project grant of the IZKF to M.S. and F.N. and a grant from the European Union project (TargetHerpes) to F.N.
The authors have no conflicting interests.
Footnotes
Published ahead of print on 7 January 2009.
REFERENCES
- 1.Algarté, M., H. Nguyen, C. Heylbroeck, R. Lin, and J. Hiscott. 1999. IκB-mediated inhibition of virus-induced beta interferon transcription. J. Virol. 732694-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagneris, C., A. V. Ageichik, N. Cronin, B. Wallace, M. Collins, C. Boshoff, G. Waksman, and T. Barrett. 2008. Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome. Mol. Cell 30620-631. [DOI] [PubMed] [Google Scholar]
- 3.Bechtel, J. T., R. C. Winant, and D. Ganem. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 794952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkmann, M. M., M. Glenn, L. Rainbow, A. Kieser, C. Henke-Gendo, and T. F. Schulz. 2003. Activation of mitogen-activated protein kinase and NF-κB pathways by a Kaposi's sarcoma-associated herpesvirus K15 membrane protein. J. Virol. 779346-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, H. J., M. J. Song, H. Deng, T. T. Wu, G. Cheng, and R. Sun. 2003. NF-κB inhibits gammaherpesvirus lytic replication. J. Virol. 778532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 3321186-1191. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman, E., and D. M. Knowles. 1999. The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Semin. Cancer Biol. 9165-174. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 2661865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhary, P. M., A. Jasmin, M. T. Eby, and L. Hood. 1999. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene 185738-5746. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z. J. 2005. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 7758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chugh, P., H. Matta, S. Schamus, S. Zachariah, A. Kumar, J. A. Richardson, A. L. Smith, and P. M. Chaudhary. 2005. Constitutive NF-kappaB activation, normal Fas-induced apoptosis, and increased incidence of lymphoma in human herpes virus 8 K13 transgenic mice. Proc. Natl. Acad. Sci. USA 10212885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas, J. L., J. K. Gustin, B. Dezube, J. L. Pantanowitz, and A. V. Moses. 2007. Kaposi's sarcoma: a model of both malignancy and chronic inflammation. Panminerva Med. 49119-138. [PubMed] [Google Scholar]
- 13.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 964546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efklidou, S., R. Bailey, N. Field, M. Noursadeghi, and M. K. Collins. 2008. vFLIP from KSHV inhibits anoikis of primary endothelial cells. J. Cell Sci. 121450-457. [DOI] [PubMed] [Google Scholar]
- 16.Glenn, M., L. Rainbow, F. Aurade, A. Davison, and T. F. Schulz. 1999. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J. Virol. 736953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm, T., S. Schneider, E. Naschberger, J. Huber, E. Guenzi, A. Kieser, P. Reitmeir, T. F. Schulz, C. A. Morris, and M. Stürzl. 2005. EBV latent membrane protein-1 protects B cells from apoptosis by inhibition of BAX. Blood 1053263-3269. [DOI] [PubMed] [Google Scholar]
- 18.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 751857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guasparri, I., S. A. Keller, and E. Cesarman. 2004. KSHV vFLIP is essential for the survival of infected lymphoma cells. J. Exp. Med. 199993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Halsey, T. A., L. Yang, J. R. Walker, J. B. Hogenesch, and R. S. Thomas. 2007. A functional map of NFkappaB signaling identifies novel modulators and multiple system controls. Genome Biol. 8R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, S., C. Vincenz, M. Buller, and V. M. Dixit. 1997. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J. Biol. Chem. 2729621-9624. [DOI] [PubMed] [Google Scholar]
- 22.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenner, R. G., and C. Boshoff. 2002. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim. Biophys. Acta 16021-22. [DOI] [PubMed] [Google Scholar]
- 24.Keller, S. A., D. Hernandez-Hopkins, J. Vider, V. Ponomarev, E. Hyjek, E. J. Schattner, and E. Cesarman. 2006. NF-kappaB is essential for the progression of KSHV- and EBV-infected lymphomas in vivo. Blood 1073295-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller, S. A., E. J. Schattner, and E. Cesarman. 2000. Inhibition of NF-kappaB induces apoptosis of KSHV-infected primary effusion lymphoma cells. Blood 962537-2542. [PubMed] [Google Scholar]
- 26.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 783601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagunoff, M., D. M. Lukac, and D. Ganem. 2001. Immunoreceptor tyrosine-based activation motif-dependent signaling by Kaposi's sarcoma-associated herpesvirus K1 protein: effects on lytic viral replication. J. Virol. 755891-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, B. S., M. Paulose-Murphy, Y. H. Chung, M. Connlole, S. Zeichner, and J. U. Jung. 2002. Suppression of tetradecanoyl phorbol acetate-induced lytic reactivation of Kaposi's sarcoma-associated herpesvirus by K1 signal transduction. J. Virol. 7612185-12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, H., J. Guo, M. Li, J.-K. Choi, M. DeMaria, M. Rosenzweig, and J. U. Jung. 1998. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi's sarcoma-associated herpesvirus. Mol. Cell. Biol. 185219-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malinin, N. L., M. P. Boldin, A. V. Kovalenko, and D. Wallach. 1997. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature 385540-544. [DOI] [PubMed] [Google Scholar]
- 31.Matsuda, A., Y. Suzuki, G. Honda, S. Muramatsu, O. Matsuzaki, Y. Nagano, T. Doi, K. Shimotohno, T. Harada, E. Nishida, H. Hayashi, and S. Sugano. 2003. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene 223307-3318. [DOI] [PubMed] [Google Scholar]
- 32.Matta, H., and P. M. Chaudhary. 2004. Activation of alternative NF-kappa B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1 beta-converting enzyme inhibitory protein (vFLIP). Proc. Natl. Acad. Sci. USA 1019399-9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matta, H., V. Punj, S. Schamus, L. Mazzacurati, A. M. Chen, R. Song, T. Yang, and P. M. Chaudhary. 2008. A nuclear role for Kaposi's sarcoma-associated herpesvirus-encoded K13 protein in gene regulation. Oncogene 275243-5253. [DOI] [PubMed] [Google Scholar]
- 34.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 3321181-1185. [DOI] [PubMed] [Google Scholar]
- 35.Naschberger, E., T. Werner, A. B. Vicente, E. Guenzi, K. Töpolt, R. Leubert, C. Lubeseder-Martellato, P. J. Nelson, and M. Stürzl. 2004. Nuclear factor-kappaB motif and interferon-alpha-stimulated response element co-operate in the activation of guanylate-binding protein-1 expression by inflammatory cytokines in endothelial cells. Biochem. J. 379409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 714187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ninomiya-Tsuji, J., K. Kishimoto, A. Hiyama, J. Inoue, Z. Cao, and K. Matsumoto. 1999. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398252-256. [DOI] [PubMed] [Google Scholar]
- 38.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 754843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prakash, O., O. R. Swamy, X. Peng, Z. Y. Tang, L. Li, J. E. Larson, J. C. Cohen, J. Gill, G. Farr, S. Wang, and F. Samaniego. 2005. Activation of Src kinase Lyn by the Kaposi sarcoma-associated herpesvirus K1 protein: implications for lymphomagenesis. Blood 1053987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rezaee, S. A., C. Cunningham, A. J. Davison, and D. J. Blackbourn. 2006. Kaposi's sarcoma-associated herpesvirus immune modulation: an overview. J. Gen. Virol. 871781-1804. [DOI] [PubMed] [Google Scholar]
- 41.Rivas, C., A. E. Thlick, C. Parravicini, P. S. Moore, and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozen, R., N. Sathish, Y. Li, and Y. Yuan. 2008. Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J. Virol. 824742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 9314862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadagopan, S., N. Sharma-Walia, M. V. Veettil, H. Raghu, R. Sivakumar, V. Bottero, and B. Chandran. 2007. Kaposi's sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J. Virol. 813949-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sander, G., A. Konrad, M. Thurau, E. Wies, R. Leubert, E. Kremmer, H. Dinkel, T. Schulz, F. Neipel, and M. Stürzl. 2008. Intracellular localization map of human herpesvirus 8 proteins. J. Virol. 821908-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 721005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulz, T. F. 2006. The pleiotropic effects of Kaposi's sarcoma herpesvirus. J. Pathol. 208187-198. [DOI] [PubMed] [Google Scholar]
- 48.Schwarz, M., and P. M. Murphy. 2001. Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor constitutively activates NF-kappa B and induces proinflammatory cytokine and chemokine production via a C-terminal signaling determinant. J. Immunol. 167505-513. [DOI] [PubMed] [Google Scholar]
- 49.Seo, T., J. Park, C. Lim, and J. Choe. 2004. Inhibition of nuclear factor kappaB activity by viral interferon regulatory factor 3 of Kaposi's sarcoma-associated herpesvirus. Oncogene 236146-6155. [DOI] [PubMed] [Google Scholar]
- 50.Sgarbanti, M., M. Arguello, B. R. tenOever, A. Battistini, R. Lin, and J. Hiscott. 2004. A requirement for NF-kappaB induction in the production of replication-competent HHV-8 virions. Oncogene 235770-5780. [DOI] [PubMed] [Google Scholar]
- 51.Soboleski, M. R., J. Oaks, and W. P. Halford. 2005. Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. FASEB J. 19440-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stürzl, M., C. Hohenadl, C. Zietz, E. Castanos-Velez, A. Wunderlich, G. Ascherl, P. Biberfeld, P. Monini, P. J. Browning, and B. Ensoli. 1999. Expression of K13/v-FLIP gene of human herpesvirus 8 and apoptosis in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 911725-1733. [DOI] [PubMed] [Google Scholar]
- 53.Stürzl, M., A. Konrad, G. Sander, E. Wies, F. Neipel, E. Naschberger, S. Reipschläger, N. Gonin-Laurent, R. E. Horch, U. Kneser, W. Hohenberger, H. Erfle, and M. Thurau. 2008. High throughput screening of gene functions in mammalian cells using reversely transfected cell arrays: review and protocol. Comb. Chem. High Throughput Screen. 11159-172. [DOI] [PubMed] [Google Scholar]
- 54.Stürzl, M., C. Zietz, P. Monini, and B. Ensoli. 2001. Human herpesvirus-8 and Kaposi's sarcoma: relationship with the multistep concept of tumorigenesis. Adv. Cancer Res. 81125-159. [DOI] [PubMed] [Google Scholar]
- 55.Sun, Q., H. Matta, and P. M. Chaudhary. 2003. The human herpes virus 8-encoded viral FLICE inhibitory protein protects against growth factor withdrawal-induced apoptosis via NF-kappa B activation. Blood 1011956-1961. [DOI] [PubMed] [Google Scholar]
- 56.Sun, Q., H. Matta, and P. M. Chaudhary. 2005. Kaposi's sarcoma associated herpes virus-encoded viral FLICE inhibitory protein activates transcription from HIV-1 long terminal repeat via the classical NF-kappaB pathway and functionally cooperates with Tat. Retrovirology 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun, Q., H. Matta, G. Lu, and P. M. Chaudhary. 2006. Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-kappaB activation. Oncogene 252717-2726. [DOI] [PubMed] [Google Scholar]
- 58.Sylla, B. S., S. C. Hung, D. M. Davidson, E. Hatzivassiliou, N. L. Malinin, D. Wallach, T. D. Gilmore, E. Kieff, and G. Mosialos. 1998. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-kappaB through a pathway that includes the NF-kappaB-inducing kinase and the IkappaB kinases IKKalpha and IKKbeta. Proc. Natl. Acad. Sci. USA 9510106-10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386517-521. [DOI] [PubMed] [Google Scholar]
- 60.Thurau, M., G. Marquardt, N. Gonin-Laurent, K. Weinländer, E. Naschberger, R. Jochmann, K. R. Alkharsah, T. F. Schulz, M. Thome, F. Neipel, and M. Stürzl. 2009. Viral inhibitor of apoptosis vFLIP/K13 protects endothelial cells against superoxide-induced cell death. J. Virol. 83598-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Traenckner, E. B., H. L. Pahl, T. Henkel, K. N. Schmidt, S. Wilk, and P. A. Baeuerle. 1995. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. EMBO J. 142876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vieira, J., and P. M. O'Hearn. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325225-240. [DOI] [PubMed] [Google Scholar]
- 63.Wang, C., L. Deng, M. Hong, G. R. Akkaraju, J. Inoue, and Z. J. Chen. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412346-351. [DOI] [PubMed] [Google Scholar]
- 64.Waris, G., A. Livolsi, V. Imbert, J. F. Peyron, and A. Siddiqui. 2003. Hepatitis C virus NS5A and subgenomic replicon activate NF-kappaB via tyrosine phosphorylation of IkappaBalpha and its degradation by calpain protease. J. Biol. Chem. 27840778-40787. [DOI] [PubMed] [Google Scholar]
- 65.Wies, E., Y. Mori, A. Hahn, E. Kremmer, M. Stürzl, B. Fleckenstein, and F. Neipel. 2008. The viral interferon-regulatory factor-3 is required for the survival of KSHV-infected primary effusion lymphoma cells. Blood 111320-327. [DOI] [PubMed] [Google Scholar]
- 66.Xiao, G., E. W. Harhaj, and S. C. Sun. 2001. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell 7401-409. [DOI] [PubMed] [Google Scholar]
- 67.Ye, F. C., F. C. Zhou, J. P. Xie, T. Kang, W. Greene, K. Kuhne, X. F. Lei, Q. H. Li, and S. J. Gao. 2008. Kaposi's sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-kappaB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J. Virol. 824235-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao, J., V. Punj, H. Matta, L. Mazzacurati, S. Schamus, Y. Yang, T. Yang, Y. Hong, and P. M. Chaudhary. 2007. K13 blocks KSHV lytic replication and deregulates vIL6 and hIL6 expression: a model of lytic replication induced clonal selection in viral oncogenesis. PLoS ONE 2e1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ziauddin, J., and D. M. Sabatini. 2001. Microarrays of cells expressing defined cDNAs. Nature 411107-110. [DOI] [PubMed] [Google Scholar]