Abstract
Clinical trials have shown oncolytic adenoviruses to be tumor selective with minimal toxicity toward normal tissue. The virus ONYX-015, in which the gene encoding the early region 1B 55-kDa (E1B-55K) protein is deleted, has been most effective when used in combination with either chemotherapy or radiation therapy. Therefore, improving the oncolytic nature of tumor-selective adenoviruses remains an important objective for improving this form of cancer therapy. Cells infected during the G1 phase of the cell cycle with the E1B-55K deletion mutant virus exhibit a reduced rate of viral late protein synthesis, produce fewer viral progeny, and are less efficiently killed than cells infected during the S phase. Here we demonstrate that the G1 restriction imposed on the E1B-55K deletion mutant virus is due to the viral oncogene encoded by open reading frame 1 of early region 4 (E4orf1). E4orf1 has been reported to signal through the phosphatidylinositol 3′-kinase pathway leading to the activation of Akt, mTOR, and p70 S6K. Evidence presented here shows that E4orf1 may also induce the phosphorylation of Akt and p70 S6K in a manner that depends on Rac1 and its guanine nucleotide exchange factor Tiam1. Accordingly, agents that have been reported to disrupt the Tiam1-Rac1 interaction or to prevent phosphorylation of the ribosomal S6 kinase partially alleviated the E4orf1 restriction to late viral protein synthesis and enhanced tumor cell killing by the E1B-55K mutant virus. These results demonstrate that E4orf1 limits the oncolytic nature of a conditionally replicating adenovirus such as ONYX-015. The therapeutic value of similar oncolytic adenoviruses may be improved by abrogating E4orf1 function.
Conditionally replicating adenoviruses are a novel class of biological agents used to treat cancer (57). The E1B-55K deletion mutant virus ONYX-015, originally known as dl1520 (4), is one of the first of such agents (7). H101 is another E1B-55K deletion mutant adenovirus that is being used for tumor therapy in China (30, 78). We previously reported that cells infected during the G1 phase of the cell cycle with E1B-55K deletion mutant adenoviruses exhibit a reduced rate of viral late protein synthesis, produce fewer viral progeny, and are less effectively killed than cells infected during S phase (34, 35, 66). These observations indicated that the E1B-55K deletion mutant virus ONYX-015 is restricted in cells infected in G1. This restriction is significant because a large fraction of cells within a tumor exist in the G1 phase of the cell cycle (71). Here we show that the G1 restriction imposed on the E1B-55K deletion mutant virus is due to the viral oncogene encoded by open reading frame 1 of early region 4 (E4orf1).
The E4orf1-encoded protein is a small adapter molecule that associates with PDZ domain-containing proteins including MUPP1, PATJ, MAGI-1, ZO-2, and Dlg1 (46). PDZ domain-containing proteins often serve as scaffolds for the assembly of signaling complexes at the plasma membrane (64). Through its association with PDZ domain-containing proteins, the E4orf1-encoded protein promotes signaling through the phosphatidylinositol 3′-kinase (PI3-kinase) pathway to effectors such as protein kinase B (Akt), the mammalian target of rapamycin (mTOR), and the S6 ribosomal protein kinase (p70 S6K) (27, 54). Through these effectors, PI3-kinase alters protein synthesis and cell survival (21, 28). E4orf1 is the principal oncogenic determinant of species D adenovirus type 9 (42). The transforming ability of E4orf1 can be blocked by the PI3-kinase inhibitor LY249002 (27). However, phosphorylation of p70 S6K can also proceed by pathways that are independent of PI3-kinase or Akt. For example, the Rho-like GTPase Rac1 can activate p70 S6K (17). Rac1 is itself regulated by cellular factors to which it binds, including the Rac1-specific guanine nucleotide exchange factor T-cell lymphoma invasion and metastasis 1 protein (Tiam1). Tiam1 and the neural tissue-associated F-actin-binding protein neurabin II or spinophilin recruit p70 S6K into a complex containing Rac1, resulting in increased phosphorylation of p70 S6K (12, 36, 50). Interestingly, both Tiam1 and neurabin II are PDZ-containing proteins. These observations provided a potential basis by which E4orf1 may modulate protein synthesis and cell survival.
In this report, we show for the first time that E4orf1 restricts the abilities of the E1B-55K deletion mutant virus to produce viral progeny, to direct viral late protein synthesis, and to kill tumor cells. Drugs that are reported to prevent phosphorylation of p70 S6K or to disrupt the interaction between Tiam1 and Rac1 increase the cell-killing ability of the E1B-55K deletion mutant virus to nearly the same level observed for an E1B-55K/E4orf1 double mutant and the wild-type virus. By uncovering a role for E4orf1 in the course of a lytic adenovirus infection, this study presents novel genetic and pharmacological means by which the effectiveness of replicating oncolytic adenoviruses can be improved.
MATERIALS AND METHODS
Cell culture and cell viability.
Cervical carcinoma-derived HeLa and adenovirus E1-transformed 293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum. Non-small-cell lung carcinoma-derived H1299 cells and glioblastoma-derived U87 and U251 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were maintained and studied at 37°C in a humidified atmosphere with 5% CO2. Cell culture media, cell culture supplements, and serum were obtained from Life Technologies (Gaithersburg, MD). Because the metabolic activity of adenovirus-infected cells appears to vary with the genotype of the infecting virus (data not shown), surrogate measures of viability that monitor mitochondrial activity such as the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide or 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium assay were found to be inappropriate. For that reason, cell viability was measured by trypan blue dye exclusion (65).
Cell cycle synchronization.
Synchronously dividing cells were obtained by a combination of mitotic detachment and hydroxyurea block as originally described in reference 15 with the modifications described in references 35 and 53. Cells synchronized by this method displayed synchrony (>80% S-phase content at the appropriate time) for three cycles. Synchronously dividing cells were infected as the cells passed through the indicated stage of the cell cycle, during the first hour of S phase (early S phase) or during the first 3 h of G1 (early G1). S-phase-enriched populations of cells were produced by one of two means with comparable outcomes. First, cells were treated with 1 mM hydroxyurea (Sigma, St. Louis, MO) in normal growth medium for 24 h; upon replacement of the medium with normal medium or virus infection medium, the cells resumed cycling from the G1/S border. Alternatively, cells were exposed to uracil arabinoside, which was previously reported to increase the S-phase content of leukemic cells (16). Cells were cultured in normal medium with 0.2 to 0.4 mM uracil arabinoside (ICN/MP Biomedical, Solon, OH). After 48 h, the medium was replaced with virus in infection medium. The fraction of cells in S phase was determined at the time of infection by propidium iodide staining and DNA analysis by flow cytometry as described previously (35, 53). Each method yielded a population of viable HeLa cells with at least 80% of the cells in S phase.
Viruses.
The wild-type virus used for these studies was dl309 (44). E1B mutant viruses included dl338 (56a), dl1520 (4), and dl110 (2). The E4or1/E4orf2 double-mutant virus and the E1B-55K/E4orf1/E4orf2 triple-mutant virus dl1018 were described previously (9). Additional mutant viruses included E4orf1 and E4orf2 mutants in351 and in352, respectively (37). The E1B-55K/E4orf1 mutant virus MAT2 was created by recombination of the E1B-55K mutation of dl1520 with the E4orf1 mutation of in351 as described in reference 66. E1B-55K/E4orf2 mutant virus 223 was created by similar methods to include the E1B-55K deletion mutation of dl1520 and the E4orf2 mutation of in352. An E1 deletion mutant adenovirus vector expressing the T17N dominant-negative form of Rac1 (see, for example, reference 14) was purchased (Cell Biolabs, Inc., San Diego, CA). Viruses were propagated by using 293 cells (45). The titer of each viral preparation was determined by plaque assay and tested by a fluorescent focus assay for infectivity with antibodies to E1A or the E2A DNA-binding protein (33). Infections were performed at a true multiplicity of 20 to 30 (35).
Flow cytometry.
DNA content was determined by flow cytometry with a FACScalibur instrument (Becton Dickinson and Co., Franklin Lakes, NJ) and CellQuest (Becton Dickinson) or ModFit software (Verity Software House, Topsham, ME) as described in references 35 and 53.
Transmission electron microscopy to measure viral progeny.
At least 100 cells per experiment were scored for the presence or absence of progeny virus particles by transmission electron microscopy as described previously (35, 66).
Plaque assays for viral yields.
Virus yield was determined by plaque assay with 293 cells as described previously (35, 53).
Viral late protein synthesis.
Infected cells were pulse-labeled for 1 h with 35S-labeled amino acids (Tran35S-label; MP Biomedicals, Costa Mesa, CA) at various times after infection. Cellular proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the viral late proteins were quantified by phosphorescence imaging with a Molecular Dynamics PhosphorImager instrument and ImageQuant analysis software (Molecular Dynamics, Sunnyvale, CA).
Immunoblotting.
Protein from equivalent numbers of cells were separated by SDS-PAGE, transferred to a nitrocellulose support, and analyzed by immunoblotting as described previously (65). Monospecific primary antibodies (p70 S6K, phosphospecific p70 S6k [Thr389], Akt, phosphospecific Akt [S473], phosphospecific p70 S6K [S235/236], Rac1) were obtained from Cell Signaling Technology (Danvers, MA) and used according to the manufacturer's instructions. Polyclonal rabbit antiserum specific for adenovirus was kindly provided by Arnie Berk (University of California at Los Angeles) and used at a dilution of 1:10,000. Immune complexes were visualized with horseradish peroxidase-conjugated secondary antibody from Jackson ImmunoResearch Laboratories (West Grove, PA) and the SuperSignal chemiluminescent substrate from Pierce (Rockford, IL).
Statistics.
Log-transformed values were compared by the two-tailed t test with the Holm correction for multiple comparisons or Tukey's honest significant difference algorithm. Cell viability was analyzed by logistic regression with a quasibinomial model to determine the half-life and 95% confidence interval (CI). Values are presented with the mean and the standard deviation, standard error of the mean, or 95% CI as indicated. P values of less than 0.05 were considered significant.
RESULTS
The E1B-55K deletion mutant adenovirus ONYX-015/dl1520 is restricted in cells infected in G1.
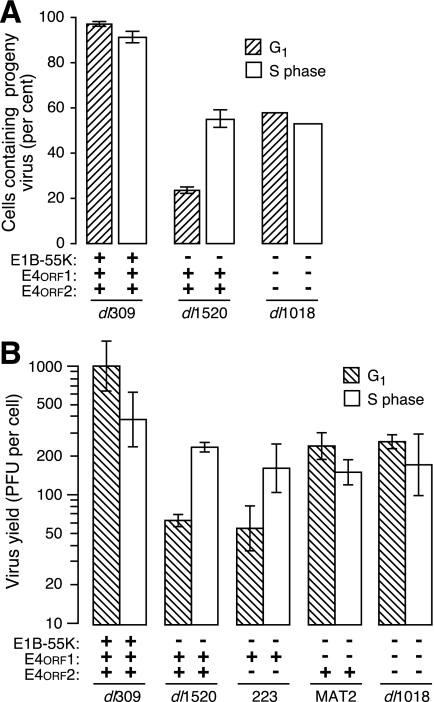
Cells infected during the G1 phase of the cell cycle with E1B-55K deletion mutant viruses produce fewer viral progeny, synthesize viral structural proteins at a reduced rate, and are less efficiently killed than cells infected during the S phase (34, 35, 66). Consequently, compared to cells infected in the S phase, E1B-55K deletion mutant viruses such as ONYX-015 can be viewed as G1 restricted. This restriction is significant because a large fraction of the cells within a tumor exist in the G1 phase of the cell cycle (71). To elucidate the basis for this restriction, synchronously dividing HeLa cells were infected with the wild-type virus dl309, the E1B-55K deletion mutant virus dl1520, or the E1B-55K, E4orf1, and E4orf2 triple-mutant virus dl1018 and evaluated by electron microscopy for viral progeny. Virtually all of the wild-type virus-infected cells contained progeny virus, whereas more S-phase cells infected with the E1B-55K single-mutant virus contained viral progeny than did cells infected during G1. Surprisingly, the G1 restriction was absent in cells infected with dl1018 (Fig. 1A). These results suggest that E4orf1, E4orf2, or both restrict the production of E1B-55K mutant viral progeny in cells infected in G1.
FIG. 1.
E4orf1 imposes a G1 restriction on virus production directed by E1B-55K deletion mutant adenoviruses. Synchronously dividing cultures of HeLa cells were generated by mitotic detachment and HU treatment as described in Materials and Methods. Cells were infected at early G1 (hatched bars) or early S phase (light bars) with the indicated viruses. (A) Infected cells were harvested after 32 h, and at least 100 infected cells from multiple samples were evaluated by transmission electron microscopy for the presence of progeny viral particles. The percentages of cells containing progeny viral particles from four (dl309 and dl1520) and two (dl1018) independent experiments are shown with the standard deviation indicated by error bars. (B) Synchronously dividing HeLa cells were generated and infected as in panel A with the viruses indicated in panel B. After 72 h, the yield of progeny virus was determined by plaque assay and is presented as the infectious virus per infected cell. Results from three independent experiments are represented as the mean with standard error of the mean. The status of the relevant viral gene is indicated below each bar.
To determine if E4orf1 or E4orf2 was responsible for the G1 restriction, additional E1B-55K deletion mutant viruses with mutations in the E4orf1 or E4orf2 gene were created and their replication was evaluated. The wild-type virus replicated to nearly equivalent levels irrespective of the phase of the cell cycle, whereas the E1B-55K deletion mutant virus dl1520 produced more viral progeny in cells infected during S phase than during G1 (Fig. 1B). Replication of the E1B-55K/E4orf2 double-mutant virus 223 was indistinguishable from dl1520; both viruses produced more progeny in cells infected during S phase than in those infected during G1. By contrast, the E1B-55K/E4orf1 double-mutant virus MAT2 and the E1B-55K/E4orf1/2 triple-mutant virus dl1018 produced statistically equivalent (P > 0.2) amounts of viral progeny from cells infected in the S phase and from cells infected in G1 (Fig. 1B). These results demonstrate that E4orf1 acts in G1 cells infected with the E1B-55K deletion mutant virus to restrict virus production.
E4orf1 restricts viral late protein synthesis.
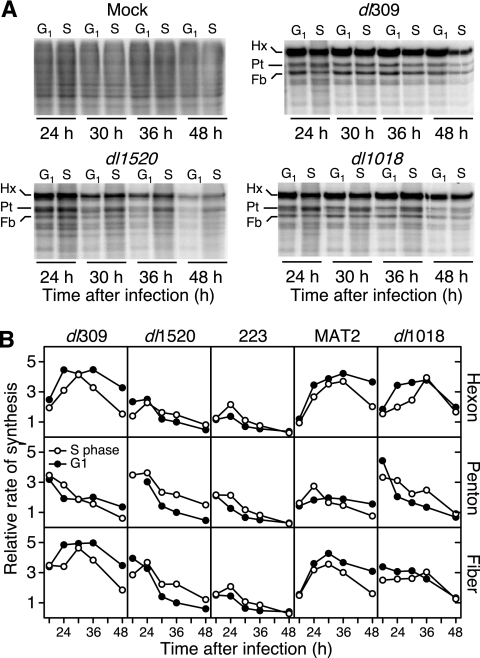
To determine if E4orf1 restricts viral late protein synthesis in cells infected in G1, synchronously dividing cells were infected at either G1 or S phase with the viruses indicated in Fig. 2. The infected cells were pulse-labeled with radioactive amino acids, and the newly synthesized viral late proteins were visualized by gel electrophoresis and phosphorescence imaging. Representative images are shown in Fig. 2A. Quantitative phosphorescence imaging confirmed that wild-type virus dl309 directed the synthesis of more viral late protein than E1B-55K single-mutant virus dl1520 or E1B-55K/E4orf2 double-mutant virus 223 (Fig. 2B). As reported previously, the wild-type virus directed more viral late protein synthesis in cells infected in G1 than in cells infected in the S phase, whereas dl1520 directed the synthesis of more viral late proteins in cells infected in the S phase than in cells infected in G1 even though dl1520 blocked host protein synthesis in cells infected in the S phase less effectively than in cells infected in G1 (35). The cell cycle dependence and temporal pattern of protein synthesis were the same for both 223 and dl1520. Strikingly, the time courses of viral late protein synthesis directed by wild-type virus dl309, E1B-55K/E4orf1/2 triple-mutant virus dl1018, and E1B-55K/E4orf1 double-mutant virus MAT2 were nearly identical (Fig. 2A and B). Protein synthesis directed by these mutant viruses resembled that of the wild-type virus in that the rate of viral late protein synthesis in cells infected in G1 nearly always exceeded the rate of synthesis in cells infected in the S phase. These results show for the first time that E4orf1 restricts viral late protein synthesis during an E1B-55K deletion mutant virus infection and suggest that the characteristic defect in viral late gene expression observed for E1B-55K single-mutant viruses such as ONYX-015 (see, for example, references 26 and 76) can be alleviated by abrogating E4orf1 function.
FIG. 2.
E4orf1 imposes a G1 restriction on viral late protein synthesis directed by E1B-55K deletion mutant adenoviruses. (A) Synchronously dividing cultures of HeLa cells were generated by mitotic detachment and HU treatment and mock infected or infected with the indicated viruses at early G1 or early S phase. Infected cells were pulse-labeled for 1 h with 35S-labeled amino acids at 24, 30, 36, and 48 hpi. Labeled proteins were separated by SDS-PAGE, and the newly synthesized protein was visualized and quantified by phosphorescence imaging. Representative gels are shown with the viral late structural proteins hexon, penton, and fiber indicated as Hx, Pt, and Fb, respectively. (B) The relative rates of synthesis of the viral hexon, penton, and fiber proteins were determined by pulse-labeling as indicated for panel A. The rate of synthesis of each protein was normalized to the rate measured for dl1520 at 36 hpi in cells infected in G1. G1 rates are shown with closed circles, and S-phase rates are shown with open circles.
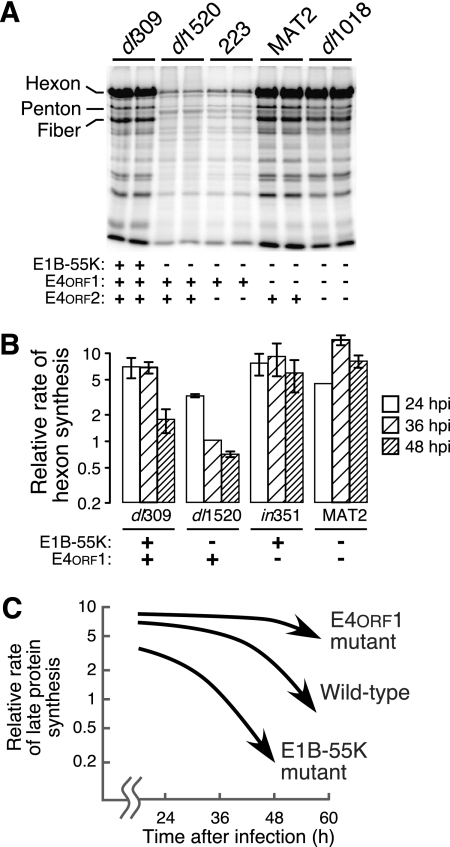
To determine if E4orf1 restricts viral late protein synthesis only in the absence of E1B-55K, additional viruses bearing mutations in the E1B-55K, E4orf1, or E4orf2 gene were evaluated with asynchronously dividing cells. Because we observed the greatest differences in rates of viral late translation around 36 h postinfection (hpi), the rate of synthesis of the hexon, penton, and fiber proteins measured in each experiment was normalized to the mean rate of synthesis measured at 36 hpi for the E1B-55K/E4orf2 double-mutant and E1B-55K single-mutant viruses. Differences among the rates of viral late protein synthesis were modest at 24 hpi (Fig. 3B). By contrast, at 36 hpi, the rates of viral late protein synthesis directed by the E1B-55K single-mutant-like viruses were substantially reduced (6-fold, range of 3- to 14-fold in 24 experiments) compared to other viruses. By 48 hpi, the rate of late protein synthesis directed by the wild-type virus had dropped to a level similar to that of the E1B-55K single-mutant-like viruses (P > 0.3). Strikingly, the rate of protein synthesis directed by E1B-55K/E4orf1 double-mutant virus MAT2 or E4orf1 single-mutant virus in351 remained significantly higher than those of the E1B-55K single-mutant-like viruses and the wild-type virus (P < 0.01 and P < 0.014, respectively). This trend was observed for infected cells evaluated as late as 72 hpi (data not shown) and is represented schematically in Fig. 3C.
FIG. 3.
E4orf1 restricts viral late protein synthesis in adenovirus-infected cells. (A) Duplicate cultures of asynchronously dividing HeLa cells were infected with the indicated viruses and pulse-labeled with 35S-labeled amino acids at 36 hpi. Labeled proteins were separated by electrophoresis and visualized by phosphorescence imaging. The late structural proteins hexon, penton, and fiber are indicated. The status of the relevant viral gene is indicated below each lane. (B) Asynchronously dividing HeLa cells were infected with the indicated viruses and pulse-labeled for 1 h with 35S-labeled amino acids at 24, 36, and 48 hpi. Radioactivity incorporated into the hexon protein was quantified by phosphorescence imaging. This value was normalized to the amount of radioactivity incorporated into hexon in dl1520 virus-infected cells at 36 hpi. The values shown are the mean of 9 to 23 independent infections with the standard error of the mean indicated. The status of the relevant viral gene is indicated below each virus. (C) The relative rate of late protein synthesis directed by representative viruses at late times after infection is schematically represented for an E4orf1 deletion mutant virus, an E1B-55K deletion mutant virus, and the wild-type virus. E4orf1 restricts viral late protein synthesis at all times of infection in the E1B-55K mutant background and at very late times in the context of a wild-type virus infection.
E4orf1 restricts the cell-killing potential of E1B-55K deletion mutant virus ONYX-015.
To determine if E4orf1 also restricts the cytolytic nature of the E1B-55K deletion mutant virus, HeLa cells were infected with the same viruses analyzed in Fig. 3 and cell viability was determined over the course of 7 days. Logistic regression was used to determine the time required to kill one-half of the infected cells (t1/2) and 95% CI. As expected, wild-type virus dl309 killed more quickly (t1/2 = 3.9 ± 0.3 days) than E1B-55K single-mutant virus dl1520 (t1/2 = 6.4 ± 0.4 days) or E1B-55K/E4orf2 double-mutant virus 223 (t1/2 = 6.2 ± 0.5 days). Surprisingly, both the E1B-55K/E4orf1 double-mutant virus and triple-mutant virus dl1018 killed cells more quickly (t1/2 = 4.9 ± 0.2 and 5.0 ± 0.2 days, respectively) than either single-mutant virus dl1520 or E1B-55K/E4orf2 double-mutant virus 223. These findings confirm that the presence of E4orf1 limits the cytolytic properties of the E1B-55K deletion mutant virus in HeLa cells.
HeLa cells express the human papillomavirus type 18 (HPV-18) E6 protein, which, like the E4orf1-encoded protein, targets several PDZ domain-containing proteins (31, 46). However, results obtained with HeLa cells were replicated with non-HPV-transformed cells, including the lung carcinoma-derived H1299 cell line and p53-positive and -negative glioblastoma-derived cell lines (data not shown). Thus, it seems unlikely that any shared targets of the E4orf1 and HPV-18 E6 proteins influenced the apparent restriction imposed by E4orf1 on virus production, viral late protein synthesis, and cell killing. These results led us to investigate possible E4orf1-related signals that are involved in the restriction of the E1B-55K mutant virus.
Akt and p70 S6K are phosphorylated in the presence of the PI3-kinase inhibitor LY294002 in adenovirus-infected cells.
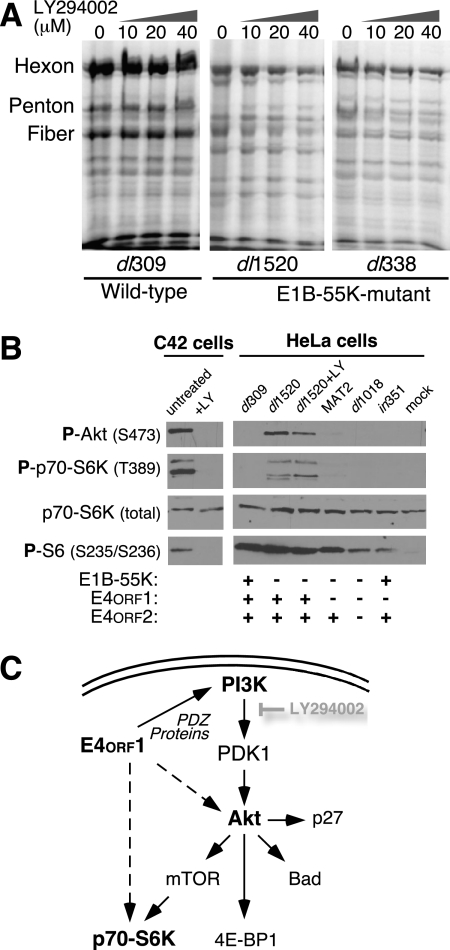
The E4orf1-encoded protein stimulates signaling through PI3-kinase to targets such as Akt, p70 S6K, and mTOR (27, 54). Because these PI3-kinase targets can regulate cellular translation (11, 21, 28), we first evaluated the contribution of the PI3-kinase pathway to viral late protein synthesis. The PI3-kinase inhibitor LY294002 has been reported to interfere with the ability of E4orf1 to activate the PI3-kinase pathway. We therefore expected that LY294002 would mimic the absence of E4orf1 during a dl1520 infection and increase the rate of viral late protein synthesis. Contrary to our expectation, viral late protein synthesis decreased in a concentration-dependent manner in E1B-55K deletion mutant virus-infected HeLa cells exposed to LY294002 (Fig. 4A), as well as in human cells of other origins, including lung carcinoma and glioblastoma (data not shown). Thus, it seems unlikely that E4orf1 restricts viral late protein synthesis by stimulating PI3-kinase activity.
FIG. 4.
The PI3-kinase inhibitor LY294002 diminishes viral late protein synthesis and does not reduce Akt and p70 S6K phosphorylation in adenovirus-infected cells. (A) Asynchronously dividing HeLa cells were infected with either the wild-type virus dl309 or the E1B-55K deletion mutant virus dl1520 or dl338. Four hours after infection, LY294002 was added at the indicated concentration. At 36 hpi, the cells were pulse-labeled with 35S-labeled amino acids and the labeled proteins were visualized by SDS-PAGE and phosphorescence imaging. (B) Akt and p70 S6K are phosphorylated in dl1520 virus-infected cells. The C4-2 prostatic cancer cell line with constitutive activation of the PI3-kinase pathway was left untreated or exposed to 50 μM LY294002 (LY) for 36 h before cellular proteins were separated by SDS-PAGE, transferred to nitrocellulose, and sequentially analyzed by blotting with antibodies specific for phospho-Akt (Ser473), phospho-p70-S6K (Thr389), total p70-S6K, and phosphoribosomal protein S6 (serines 235 and 236). Asynchronously dividing HeLa cells were mock infected or infected with the indicated viruses and exposed to 50 μM LY294002 at 4 h after infection. At 36 hpi, cells were harvested and protein was separated by SDS-PAGE, transferred to nitrocellulose, and analyzed with the indicated antibodies. The status of the relevant viral gene is indicated below each lane.
Phosphorylation of the PI3-kinase downstream effectors Akt and p70 S6K was evaluated with phosphospecific antibodies to serve as an indicator of activation (61). Phosphorylation of Akt, p70 S6K, and S6 in C4-2 cells, which contain a constitutively activated PI3-kinase pathway (77), was largely eliminated by 50 μM LY249002 (Fig. 4B). In contrast to C4-2 cells, HeLa cells do not contain measurably phosphorylated Akt, p70 S6K, and S6. Therefore, any increase in the phosphorylation of these proteins upon infection would be due to viral activity. Interestingly, HeLa cells infected with E1B-55K single-mutant virus dl1520 showed significant phosphorylation of Akt and p70 S6K at 18 to 24 hpi in comparison to the wild-type virus or other viruses lacking E4orf1 (Fig. 4B). The difference in Akt phosphorylation between dl1520 and wild-type virus-infected cells was less dramatic by 36 hpi (data not shown). Nonetheless, in dl1520 virus-infected cells, LY294002 reduced but did not eliminate the phosphorylation of Akt at Ser473. Additionally, and in contrast to C4-2 cells, LY294002 did not alter the total amount of phosphorylated p70 S6K in dl1520 virus-infected HeLa cells; rather, LY294002 appeared to alter the distribution of phosphorylated forms recognized by the antibody to phospho-Thr389 (Fig. 4B). Similar effects were seen in dl1520 virus-infected H1299 and U87 cells (data not shown). To verify that these observations were indeed due to E4orf1, we generated E4orf1-expressing H1299 cells. The constitutive level of Akt phosphorylation was elevated in the E4orf1-expressing cells. Treatment of these cells with LY294002 did not eliminate the phosphorylation of Akt, thus recapitulating the observation in dl1520 virus-infected cells (data not shown). These results led us to conclude that if E4orf1 contributes to the phosphorylation and activation of p70 S6K and Akt in dl1520 virus-infected cells, it may act through both PI3-kinase-dependent and -independent pathways.
Involvement of Rac1 in E4orf1-induced phosphorylation of p70 S6K and perhaps Akt.
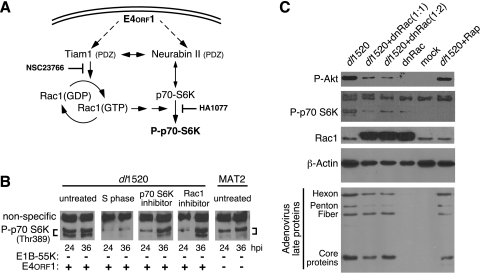
The small GTPase Rac1 can contribute to the phosphorylation of p70 S6K (17). The activity of Rac1 is governed by binding partners such as Tiam1 and neurabin II. Tiam1 is a widely distributed guanine nucleotide exchange factor that is specific for Rac1 (50). Neurabin II has been reported to bind both Tiam1 and p70 S6K (12, 13). Intriguingly, both Tiam1 and neurabin II contain PDZ domains. Because the E4orf1-encoded protein is able to bind to many PDZ domain-containing proteins, the E4orf1-encoded protein could possibly interact with Tiam1, neurabin II, or both and promote the phosphorylation of p70 S6K through Rac1 (Fig. 5A).
FIG. 5.
E4orf1-induced phosphorylation of p70 S6K and Akt may involve Rac1. (A) Schematic representation of a potential signaling pathway initiated by E4orf1 through the PDZ domain-containing proteins Tiam1 and neurabin II. Solid arrows identify known pathways and interactions. (B) HeLa cells were blocked at the G1/S border by exposure to HU for 24 h, released from the block, and infected 1 h later (S phase) or infected as an asynchronously dividing culture with the E1B-55K single-mutant virus dl1520 or the E1B-55K/E4orf1 double-mutant virus MAT2. At 4 hpi, the growth medium was replaced with fresh medium or with medium containing the p70 S6K inhibitor HA1077 at 50 mM or the Tiam1-Rac1 inhibitor NSC23766 at 40 μM. At 24 and 36 hpi, cellular proteins were analyzed by immunoblotting for phosphorylation of Thr389 on p70 S6K. A nonspecific cross-reacting protein is indicated which served as a loading control. The status of the relevant viral gene is indicated below each lane. (C) Asynchronously dividing HeLa cells were mock infected or infected with a nonreplicating adenovirus vector expressing dnRac1 (dnRac) at a multiplicity of 20 (1:1) or 40 (1:2). After 24 h, the cells were again mock infected or infected with the E1B-55K deletion mutant virus dl1520 at a multiplicity of 20. After 1 h, rapamycin (Rap) was added to 50 nM to one culture. Cells were harvested 36 h after infection with dl1520. Infected cell proteins were separated by SDS-PAGE and analyzed by immunoblotting with monospecific antibodies (phospho-Akt, phospho-P70 S6K, Rac1, β-actin) or polyclonal serum specific for the adenovirus late proteins.
To determine if Rac1 contributes to the phosphorylation of p70 S6K in adenovirus-infected cells, phosphorylation at Thr389 was compared among dl1520- and MAT2-infected cells following exposure to the broad-spectrum inhibitor of p70 S6K phosphorylation HA1077 (22) or the specific inhibitor of the Tiam1-Rac1 complex NSC23766 (29). S-phase cells infected with dl1520, which do not exhibit the characteristic G1 restriction, contained dramatically less phosphorylated p70 S6K at both 24 and 36 hpi than did untreated dl1520 virus-infected cells (Fig. 5B). Similarly, MAT2-infected cells, which show no evidence of the G1 restriction, contained less phosphorylated p70 S6K than did untreated dl1520 virus-infected cells. In dl1520 virus-infected G1 cells, the inhibitor of p70 S6K phosphorylation HA1077 reduced the amount of phosphorylated p70 S6K detected at 24 hpi, with less of an impact at 36 hpi. A similar pattern was observed for infected cells exposed to the Tiam1/Rac1 inhibitor NSC23766 at 40 μM. In these studies, the pattern of p70 S6K phosphorylation observed in MAT2-infected cells was more similar to that observed in drug-treated dl1520 virus-infected cells than in nontreated dl1520 virus-infected cells. These results support the notion that p70 S6K may be differentially phosphorylated by a Tiam1/Rac1-related pathway in E1B-55K deletion mutant virus-infected cells.
To further investigate the contribution of Rac1 to the phosphorylation of Akt and p70 S6K, as well as to viral late protein synthesis, dl1520 virus-infected cells were coinfected with an adenovirus vector expressing dominant-negative Rac1 (dnRac1). Levels of β-actin were reduced by dnRac1 (Fig. 5C). A comparable effect was observed in THP1 cells expressing a similar construct (70), most likely reflecting a role for Rac1 in modulating the actin cytoskeleton. Phosphorylation of both Akt and p70 S6K was reduced by dnRac1 (Fig. 5C). HeLa cells infected with dl1520 were also treated with rapamycin to determine if the rapamycin-sensitive form of mTOR was responsible for the increase in Akt and p70 S6K phosphorylation. In agreement with the work of Sarbassov and associates (60), prolonged exposure of HeLa cells to rapamycin eliminated p70 S6K phosphorylation but had no effect on Akt phosphorylation (Fig. 5C).
Notably, even though both dnRac1 and rapamycin reduced or eliminated p70 S6K phosphorylation, neither substantially affected the accumulation of viral late proteins in cells infected with the E1B-55K deletion mutant virus dl1520 (Fig. 5C). In contrast to dnRac1 or rapamycin, HA1077 and NSC23766 produced a slight increase in viral late protein synthesis directed by the E1B-55K mutant virus. HA1077 elicited a 1.4-fold increase (P < 0.001, n = 9) in the rate of hexon synthesis, while the Tiam1/Rac1 inhibitor NSC23766 increased hexon synthesis by 1.7-fold over that in nontreated cells (P < 0.0001, n = 11). Neither drug affected the rate of hexon synthesis in MAT2-infected cells (data not shown), nor were these inhibitors able to recapitulate the high levels of hexon synthesis observed in MAT2-infected cells at 36 h (1.7-fold compared to 10-fold). These results provide additional support for the idea that E4orf1 promotes the phosphorylation of p70 S6K and Akt in a Rac1-dependent manner but that this is not likely the mechanism by which E4orf1 restricts late protein synthesis.
The PI3-kinase/Tiam1-Rac1 signals initiated by E4orf1 promote survival in adenovirus-infected cells.
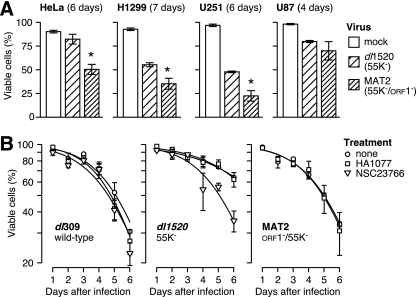
The results shown in Fig. 5 led us to infer that the regulation of viral late translation is largely independent of the phosphorylation status of p70 S6K and signaling through PI3-kinase. However, signaling through the p70 S6K (38), PI3-kinase (21, 28), and Rac1 (23, 43) pathways also promotes cell survival. Consequently, we tested the possibility that E4orf1 restricts the cell-killing potential of the E1B-55K deletion mutant virus through the aforementioned pathways. We first measured the impact of E4orf1 on the cell-killing potential of the E1B-55K mutant virus in four cell lines derived from tumors of three origins. Cell viability was measured after infection with either the E1B-55K single-mutant virus dl1520 or the E1B-55K/E4orf1 double-mutant virus MAT2. The double-mutant virus MAT2 killed these tumor cells more effectively than did the E1B-55K single-mutant virus (Fig. 6A). This result indicates that E4orf1 indeed restricts the cell-killing potential of the E1B-55K deletion mutant virus.
FIG. 6.
E4orf1 restricts the tumor cell-killing potential of E1B-55K mutant virus dl1520 (ONYX-015) in a Tiam1/Rac1-dependent manner. (A) Cell lines derived from human cervical carcinoma (HeLa), large-cell lung carcinoma (H1299), and malignant glioblastoma (U251, U87) were mock infected or infected with the indicated viruses at a multiplicity sufficient to infect all of the cells. The viability of the cells was determined by trypan blue dye exclusion on the indicated day after infection. Values represent the mean of three independent infections with the standard error of the mean. The asterisk identifies values for MAT2-infected cells that are significantly (P < 0.05) less than the viability of dl1520 virus-infected cells. (B) HeLa cells were infected with the wild-type virus (dl309), the E1B-55K mutant virus (dl1520), or the E1B-55K/E4orf1 double-mutant virus (MAT2) at a multiplicity of 20 and left untreated (circles) or were adjusted to 50 mM HA1077 (squares) or 40 μM NSC2366 (triangles) at 4 hpi. The fraction of viable cells was determined daily for 6 days after infection by trypan blue dye exclusion. The plotted values represent the mean of three independent infections with the standard error of the mean indicated by error bars. The lines represent best-fit curves determined by logistic regression with a quasibinomial model.
Since Fig. 5 indicates that E4orf1 promotes the phosphorylation of p70 S6K through Tiam1/Rac1, we tested the hypothesis that inhibiting the phosphorylation of p70 S6K and/or disrupting the Tiam1-Rac1 interaction would increase the cell-killing potential of the E1B-55K single-mutant adenovirus. Cells were infected with the wild-type virus, the E1B-55K single-mutant virus, or the E1B-55K/E4orf1 double-mutant virus and treated with the vehicle control, HA1077, or NSC232766. The fraction of viable cells was determined by trypan blue dye exclusion, and the half-life of the infected cells was estimated by regression analysis. The viability of mock-infected cells was unaffected by the drugs used in these experiments (data not shown). HeLa cells infected with the wild-type virus died at comparable rates, irrespective of drug treatment (t1/2 = 4.6 to 5.1 days). Untreated cells infected with the E1B-55K single-mutant virus died more slowly (t1/2 = 7.0 ± 0.6 days) than wild-type virus-infected cells (t1/2 = 5.1 ± 0.4 days), as reported previously (33, 65). Treatment of dl1520 virus-infected cells with HA1077 had no significant effect on the rate of HeLa cell death (t1/2 = 7.2 ± 1.1 days), although HA1077 did increase the rate at which dl1520 virus-infected H1299 or U87 cells died (data not shown). The Tiam1/Rac1 inhibitor NSC232766 accelerated the rate of cell killing mediated by the E1B-55K single-mutant virus (t1/2 = 5.1 ± 0.4 days) to that measured for the E1B-55K/E4orf1 double-mutant virus MAT2 (t1/2 = 5.1 ± 0.4 days) and the wild-type virus. Cell killing by the E1B-55K/E4orf1 double-mutant virus was not affected by treatment with HA1077 (t1/2 = 7.2 ± 1.1 days) or NSC232766 (data not shown). From these results, we inferred that E4orf1 acts through Tiam1-Rac1 to promote the survival of adenovirus-infected cells. These signals limit the cytolytic potential of the E1B-55K single-mutant adenovirus ONYX-015 (dl1520) and are expected to impose similar restrictions on other replication-competent oncolytic adenoviruses such as H101 as well.
DISCUSSION
The E1B-55K deletion mutant adenovirus ONYX-015 and, more recently, H101 have been used to treat cancer. ONYX-015 is more effective in combination with other therapies than as a single agent (30, 78). Therefore, improving the oncolytic nature of this single agent should improve its value as a cancer therapy. We previously reported that E1B-55K deletion mutant adenoviruses replicated more effectively in cells infected during the S phase than in cells infected during G1 and that the mutant virus killed cells infected in the S phase more effectively than cells infected in G1 (33, 66). Because most of the cells in a tumor are in G1 (71), we chose to examine more closely the basis for the relative G1 restriction imposed on the E1B-55K deletion mutant virus. In this report, we show that E4orf1 limits viral progeny production, viral late protein synthesis, and cell killing. Consequently, conditionally replicating viruses such as ONYX-015 that contain an intact E4orf1 gene suffer from an intrinsic limitation to their oncolytic nature. This report also highlights the possibility that other viral genes contribute to the oncolytic nature of replication-competent adenoviruses in ways that have not been recognized previously.
Among many activities attributed to the E1B-55K protein, promoting viral late gene expression is key to a productive infection (reviewed in references 6 and 26). In the infected cell, the E1B-55K and E4orf6 proteins form an E3 ubiquitin ligase that includes cullin 5 (Cul5), elongins B and C, and Rbx1 (39, 58). Because viral late gene expression was reduced by proteasome inhibitors (19) or by expression of a dominant-negative Cul5 variant (76), the E1B-55K protein appears to promote late gene expression by the targeted degradation of a protein. The identity of this protein (or proteins) remains elusive. Cellular proteins known to be targeted by the adenovirus ubiquitin ligase are central to the cellular response to DNA damage. These targets include p53 (39, 58), Mre11 (67), and DNA ligase IV (3). Woo and Berk have noted that components of the nonsense-mediated mRNA decay pathway, which also participate in mRNA export, are targets of the DNA damage response (76). These investigators suggested that the inhibition of viral late mRNA export observed in cells infected with the E1B-55K mutant may stem from a failure to suppress signaling from cellular kinases that sense DNA damage. However, E4orf6 mutant viruses that failed to direct the degradation of Mre11 were not defective for late gene expression (8), indicating that targets other than Mre11 may be important in this regard. Recently, additional targets of the adenovirus ubiquitin ligase with no obvious role in the cellular DNA damage response have been identified. These include the nonstructural Rep52 and capsid proteins of adeno-associated virus type 5 (52). In view of the diverse requirements for the recruitment of specific substrates by the viral ubiquitin ligase (49, 63), it would not be surprising that additional cellular targets of the adenovirus ubiquitin ligase remain to be identified and that one of these may be critical for viral late gene expression.
Like the adenovirus E1B-55K- and E4orf6-encoded proteins, the human immunodeficiency virus type 1 (HIV-1) Vif-encoded protein forms an E3 ubiquitin ligase with Cul5, elongins B and C, and Rbx1. The HIV ubiquitin ligase directs the degradation of apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G, or APOBEC3G (5). APOBEC3G serves as an antiviral effector toward a wide variety of viruses and retroelements (40, 41). Several investigators have suggested that the Cul5-based adenovirus ubiquitin ligase targets antiviral effectors to promote efficient adenovirus replication (8, 74, 76). Further support for this notion may be inferred from the overlapping functions of the E1B-55K/E4orf6-encoded protein complex and the E4orf3-encoded protein. Like the E1B-55K/E4orf6-encoded protein complex, the E4orf3-encoded protein suppresses the DNA damage response, albeit by mislocalizing components of the MRN complex (1, 24, 68). Recently, the E4orf3-encoded protein has been shown to explicitly disable the interferon-mediated antiviral response by mislocalizing the cellular proteins Daxx and PML (72, 73). For these reasons, we favor the idea that cellular effectors directly engaged in the antiviral response are targeted by the E1B-55K/E4orf6-encoded protein complex.
It is tempting to speculate about possible connections among the cellular DNA damage response, the postulated antiviral activity targeted by adenovirus ubiquitin ligase, and G1 restriction. We reported that E1B-55K and E4orf6 deletion mutant adenoviruses replicate less effectively in cells infected during G1 than in S phase (34, 35). If the E1B-55K/E4orf6-encoded ubiquitin ligase must suppress an antiviral activity for efficient virus replication, it stands to reason that this antiviral activity is less pronounced in cells infected in the S phase. Because cells engaged in DNA synthesis must tolerate partially replicated DNA, single-stranded DNA, and transient DNA strand breaks, sensors that elicit an antiviral state in response to aberrant DNA may be subdued in S-phase cells compared to G1 cells. Consequently, viruses that are crippled in the ability to suppress the DNA damage response are predicted to be less restricted following infection of an S-phase cell. Because the E4orf3 single-mutant virus is not G1 restricted (34, 66), we anticipate that the E4orf3 targets Daxx and PML do not impose the G1 restriction observed in E1B-55K mutant virus-infected cells.
In this report, we show that disruption of E4orf1 alleviates the apparent G1 restriction imposed on E1B-55K mutant viruses (Fig. 1). E4orf1 may contribute to the development of an antiviral state that must be suppressed by the adenovirus ubiquitin ligase. Because cells infected with the E1B-55K/E4orf1 double-mutant virus synthesize wild-type levels of viral DNA (M. Thomas and D. Ornelles, unpublished observations), it seems unlikely that the act of viral DNA synthesis could serve as the only trigger of the antiviral response. E1B-55K/E4orf1 double-mutant viruses also direct the synthesis of wild-type levels of viral proteins (Fig. 2 and 3). Since the cytoplasmic levels of viral late mRNA remained the same in single- and double-mutant virus-infected cells (unpublished observations), we concluded that this effect was due to increased translational efficiency of viral late mRNA. Perhaps the postulated antiviral activity that is elicited by E4orf1 and suppressed by the adenovirus ubiquitin ligase limits viral progeny production at a posttranslational step. Finally, because neither of the two E4orf1/E1B-55K double-mutant viruses replicated to the same level as the wild-type virus (Fig. 1), it seems likely that the E1B-55K-encoded protein may contribute to the production of viral progeny independently of its role in promoting viral late gene expression.
The role of E4orf1 during a productive adenovirus infection remains poorly understood. E4orf1 mutant viruses were reported to be indistinguishable from the wild-type virus when studied in established tumor cell lines (10, 37). We also observed that in351 replicated in HeLa cells to levels equivalent to that of the wild-type virus dl309 (data not shown). However, E4orf1 may be important to virus replication in quiescent human cells, where it enables adenovirus to mobilize the translational machinery in nutrient-deprived cells (54, 55). Somewhat at odds with this notion, we show that E4orf1 limits viral late translation. Our findings indicate that E4orf1 counteracts the ability of the E1B-55K-encoded protein to promote viral late gene expression. Other ostensibly opposing activities among adenovirus products exist. For example, the E1A-encoded proteins stabilize p53 (47, 48) while the E1B-55K- and E4orf6-encoded proteins promote p53 degradation (51, 59). Some of these opposing activities may reflect a switch in the viral replication program, as is the case with the E1B-19K-encoded protein, which promotes cell survival at early times, while the E3 adenovirus death protein promotes death at late times (75). It appears that E4orf1 and E1B-55K exert opposing effects at the same time. Systems that incorporate opposing activities to maintain homeostasis are poised to respond rapidly to changes in the environment. Perhaps the integration of E4orf1 into a signal transduction cascade impinging on translation permits a more rapid response than could be achieved by changes at the level of transcription.
The E4orf1-encoded protein is a small polypeptide that occurs as monomers or trimers with distinct functions and binding partners associated with each form (18). The transforming ability of E4orf1 maps to the PDZ domain-binding element at the carboxy terminus of the protein. The ability to bind cellular PDZ domain-containing proteins is necessary for the E4orf1-encoded protein to transform cultured cells in a PI3-kinase-dependent manner (27, 32). One consequence of increased signaling through the PI3-kinase pathway by E4orf1 is the activation of Akt, mTOR, and p70 S6K. Our results suggest that E4orf1 may also elicit phosphorylation of both Akt and p70 S6K in a manner that depends on Rho-like GTPase Rac1. Because neither the PI3-kinase inhibitor LY294002 nor the suppression of Rac1 activity overcame the restriction to viral late protein synthesis, E4orf1 may regulate late protein synthesis through a pathway that has not yet been described. Intriguingly, the E4orf1-encoded protein was reported to bind a 70-kDa cellular phosphoprotein independently of its PDZ domain-binding element (18). It will be of interest to determine if this 70-kDa protein contributes to the ability of E4orf1 to restrict viral late protein synthesis.
Although signaling through the PI3-kinase pathway did not appear to be the means by which E4orf1 restricts late protein synthesis, this activity may restrict virus-mediated cell killing. Accordingly, exposure to the PI3-kinase inhibitor LY294002 increased the cytolytic nature of the E1B-55K deletion mutant virus (data not shown). Moreover, pharmacological agents known to disrupt the Tiam1-Rac1 interaction increased the cytolytic nature of the E1B-55K deletion mutant virus to that of the E1B-55K/E4orf1 double-mutant virus (Fig. 6). Thus, E4orf1 signaling to Tiam1/Rac1 may limit the cytolytic nature of replication-competent oncolytic adenoviruses such as ONYX-015. Another limitation of ONYX-015 as a single agent for cancer therapy includes the limited spread of this virus throughout the tumor (57). Because E4orf1 restricts both virus production (Fig. 1) and the synthesis of viral structural proteins (Fig. 2 and 3), E4orf1 may further limit the oncolytic potential of the E1B-55K deletion mutant virus by reducing the yield of progeny virus in the tumor. Although it remains to be determined if the E1B-55K/E4orf1 double-mutant virus spreads more effectively in a tumor model than the E1B-55K deletion mutant virus, this seems likely in view of the pivotal role E4orf1 plays in determining so many of the characteristics of this virus.
Other viruses initiate signaling cascades through the Tiam1/Rac1 pathway. The EBNA3C latent protein of Epstein-Barr virus is an oncoprotein that targets the cellular protein Nm23-H1, which in turn prevents Nm23-H1 from limiting tumor metastasis (69). Nm23-H1 negatively regulates Tiam1 and inhibits Rac1 activation in vivo (56). The HIV-1 Nef protein stimulates activation of Vav1 (25), which is a Tiam1-like guanine nucleotide exchange factor (20, 62). Nef activation of Vav1 increases levels of virus transcription and replication (25). Because viruses as diverse as Epstein-Barr virus and HIV target pathways involving Tiam1/Rac1, it seems likely that this represents an important cellular target for additional viruses. In the case of the E1B-55K mutant adenovirus, disabling this cellular target appears to increase virus replication, increase the potential for virus spread, and increase virus-mediated cell death. Targeting the Tiam1/Rac1 pathway during oncolytic virus therapy may be especially effective because elevated Tiam1 and Rac1 activities have been associated with more aggressive cancers and a poor clinical outcome (29). Thus, targeting the Tiam1/Rac1 pathway in conjunction with oncolytic adenovirus therapy may improve the cancer cell-killing potential of the E1B-55K deletion mutant virus and be an effective multipronged approach to cancer therapy.
Acknowledgments
We thank Gary Ketner (Johns Hopkins University) and Tom Shenk (Princeton University) for providing viruses used in this study. We thank Arnie Berk (University of California at Los Angeles) for providing dl1520 and for the generous gift of adenovirus-specific polyclonal antiserum. We thank Ron Javier (Baylor College) for providing E4orf1 plasmids, as well as for valuable discussions. We also acknowledge the anonymous reviewers of the manuscript for their insightful comments. We thank Ken Grant of the Micromed facility of the Comprehensive Cancer Center of Wake Forest University for assistance with electron microscopy.
Cell culture reagents were provided by the Cell and Virus Vector Core Laboratory of the Comprehensive Cancer Center of Wake Forest University, which is supported in part by National Cancer Institute grant CA12197. Michael A. Thomas was supported in part by National Research Service award F31 CA11020 from the National Institutes of Health. This work was supported by Public Health Service grants CA77342 and CA77342S1 from the National Cancer Institute to David Ornelles.
Footnotes
Published ahead of print on 7 January 2009.
REFERENCES
- 1.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 7911382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss, L. E., and H. S. Ginsberg. 1984. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J. Virol. 50202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 817034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156107-121. [DOI] [PubMed] [Google Scholar]
- 5.Barraud, P., J. C. Paillart, R. Marquet, and C. Tisne. 2008. Advances in the structural understanding of Vif proteins. Curr. HIV Res. 691-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 247673-7685. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff, J. R., D. H. Kirn, A. Williams, C. Heise, S. Horn, M. Muna, L. Ng, J. A. Nye, A. Sampson-Johannes, A. Fattaey, and F. McCormick. 1996. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 274373-376. [DOI] [PubMed] [Google Scholar]
- 8.Blanchette, P., K. Kindsmuller, P. Groitl, F. Dallaire, T. Speiseder, P. E. Branton, and T. Dobner. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 822642-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174345-353. [DOI] [PubMed] [Google Scholar]
- 10.Bridge, E., S. Medghalchi, S. Ubol, M. Leesong, and G. Ketner. 1993. Adenovirus early region 4 and viral DNA synthesis. Virology 193794-801. [DOI] [PubMed] [Google Scholar]
- 11.Buchkovich, N. J., Y. Yu, C. A. Zampieri, and J. C. Alwine. 2008. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 6266-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchsbaum, R. J., B. A. Connolly, and L. A. Feig. 2003. Regulation of p70 S6 kinase by complex formation between the Rac guanine nucleotide exchange factor (Rac-GEF) Tiam1 and the scaffold spinophilin. J. Biol. Chem. 27818833-18841. [DOI] [PubMed] [Google Scholar]
- 13.Burnett, P. E., S. Blackshaw, M. M. Lai, I. A. Qureshi, A. F. Burnett, D. M. Sabatini, and S. H. Snyder. 1998. Neurabin is a synaptic protein linking p70 S6 kinase and the neuronal cytoskeleton. Proc. Natl. Acad. Sci. USA 958351-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burstein, E. S., D. J. Hesterberg, J. S. Gutkind, M. R. Brann, E. A. Currier, and T. L. Messier. 1998. The ras-related GTPase rac1 regulates a proliferative pathway selectively utilized by G-protein coupled receptors. Oncogene 171617-1623. [DOI] [PubMed] [Google Scholar]
- 15.Cao, G., L. M. Liu, and S. F. Cleary. 1991. Modified method of mammalian cell synchronization improves yield and degree of synchronization. Exp. Cell Res. 193405-410. [DOI] [PubMed] [Google Scholar]
- 16.Chandrasekaran, B., R. L. Capizzi, T. E. Kute, T. Morgan, and J. Dimling. 1989. Modulation of the metabolism and pharmacokinetics of 1-β-d-arabinofuranosylcytosine by 1-β-d-arabinofuranosyluracil in leukemic mice. Cancer Res. 493259-3266. [PubMed] [Google Scholar]
- 17.Chou, M. M., and J. Blenis. 1996. The 70 kDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell 85573-583. [DOI] [PubMed] [Google Scholar]
- 18.Chung, S. H., R. S. Weiss, K. K. Frese, B. V. Prasad, and R. T. Javier. 2008. Functionally distinct monomers and trimers produced by a viral oncoprotein. Oncogene 271412-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315234-244. [DOI] [PubMed] [Google Scholar]
- 20.Crespo, P., K. E. Schuebel, A. A. Ostrom, J. S. Gutkind, and X. R. Bustelo. 1997. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature 385169-172. [DOI] [PubMed] [Google Scholar]
- 21.Datta, S. R., A. Brunet, and M. E. Greenberg. 1999. Cellular survival: a play in three Akts. Genes Dev. 132905-2927. [DOI] [PubMed] [Google Scholar]
- 22.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 35195-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshpande, S. S., P. Angkeow, J. Huang, M. Ozaki, and K. Irani. 2000. Rac1 inhibits TNF-α-induced endothelial cell apoptosis: dual regulation by reactive oxygen species. FASEB J. 141705-1714. [DOI] [PubMed] [Google Scholar]
- 24.Evans, J. D., and P. Hearing. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 796207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fackler, O. T., W. Luo, M. Geyer, A. S. Alberts, and B. M. Peterlin. 1999. Activation of Vav by Nef induces cytoskeletal rearrangements and downstream effector functions. Mol. Cell 3729-739. [DOI] [PubMed] [Google Scholar]
- 26.Flint, S. J., and R. A. Gonzalez. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272287-330. [DOI] [PubMed] [Google Scholar]
- 27.Frese, K. K., S. S. Lee, D. L. Thomas, I. J. Latorre, R. S. Weiss, B. A. Glaunsinger, and R. T. Javier. 2003. Selective PDZ protein-dependent stimulation of phosphatidylinositol 3-kinase by the adenovirus E4-ORF1 oncoprotein. Oncogene 22710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fruman, D. A., R. E. Meyers, and L. C. Cantley. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67481-507. [DOI] [PubMed] [Google Scholar]
- 29.Gao, Y., J. B. Dickerson, F. Guo, J. Zheng, and Y. Zheng. 2004. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA 1017618-7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garber, K. 2006. China approves world's first oncolytic virus therapy for cancer treatment. J. Natl. Cancer Inst. 98298-300. [DOI] [PubMed] [Google Scholar]
- 31.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 195270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaunsinger, B. A., R. S. Weiss, S. S. Lee, and R. Javier. 2001. Link of the unique oncogenic properties of adenovirus type 9 E4-ORF1 to a select interaction with the candidate tumor suppressor protein ZO-2. EMBO J. 205578-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodrum, F. D., and D. A. Ornelles. 1998. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J. Virol. 729479-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodrum, F. D., and D. A. Ornelles. 1999. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 737474-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodrum, F. D., and D. A. Ornelles. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J. Virol. 71548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habets, G. G., E. H. Scholtes, D. Zuydgeest, R. A. van der Kammen, J. C. Stam, A. Berns, and J. G. Collard. 1994. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell 77537-549. [DOI] [PubMed] [Google Scholar]
- 37.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada, H., J. S. Andersen, M. Mann, N. Terada, and S. J. Korsmeyer. 2001. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc. Natl. Acad. Sci. USA 989666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 769194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes, R. K., M. H. Malim, and K. N. Bishop. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32118-128. [DOI] [PubMed] [Google Scholar]
- 41.Izumi, T., K. Shirakawa, and A. Takaori-Kondo. 2008. Cytidine deaminases as a weapon against retroviruses and a new target for antiviral therapy. Mini-Rev. Med. Chem. 8231-238. [DOI] [PubMed] [Google Scholar]
- 42.Javier, R. T. 1994. Adenovirus type 9 E4 open reading frame 1 encodes a transforming protein required for the production of mammary tumors in rats. J. Virol. 683917-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeong, H. G., H. J. Cho, I. Y. Chang, S. P. Yoon, Y. J. Jeon, M. H. Chung, and H. J. You. 2002. Rac1 prevents cisplatin-induced apoptosis through down-regulation of p38 activation in NIH3T3 cells. FEBS Lett. 518129-134. [DOI] [PubMed] [Google Scholar]
- 44.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17683-689. [DOI] [PubMed] [Google Scholar]
- 45.Jones, N., and T. Shenk. 1978. Isolation of deletion and substitution mutants of adenovirus type 5. Cell 13181-188. [DOI] [PubMed] [Google Scholar]
- 46.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 749680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, Z., C. P. Day, J. Y. Yang, W. B. Tsai, G. Lozano, H. M. Shih, and M. C. Hung. 2004. Adenoviral E1A targets Mdm4 to stabilize tumor suppressor p53. Cancer Res. 649080-9085. [DOI] [PubMed] [Google Scholar]
- 48.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7535-545. [DOI] [PubMed] [Google Scholar]
- 49.Luo, K., E. Ehrlich, Z. Xiao, W. Zhang, G. Ketner, and X.-F. Yu. 2007. Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J. 211742-1750. [DOI] [PubMed] [Google Scholar]
- 50.Mertens, A. E., R. C. Roovers, and J. G. Collard. 2003. Regulation of Tiam1-Rac signalling. FEBS Lett. 54611-16. [DOI] [PubMed] [Google Scholar]
- 51.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 9311295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nayak, R., K. D. Farris, and D. J. Pintel. 2008. E4Orf6-E1B-55k-dependent degradation of de novo-generated adeno-associated virus type 5 Rep52 and capsid proteins employs a cullin 5-containing E3 ligase complex. J. Virol. 823803-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ornelles, D. A., R. N. Broughton-Shepard, and F. D. Goodrum. 2007. Analysis of adenovirus infections in synchronized cells. Methods Mol. Med. 13183-101. [DOI] [PubMed] [Google Scholar]
- 54.O'Shea, C., K. Klupsch, S. Choi, B. Bagus, C. Soria, J. Shen, F. McCormick, and D. Stokoe. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J. 241211-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Shea, C. C., S. Choi, F. McCormick, and D. Stokoe. 2005. Adenovirus overrides cellular checkpoints for protein translation. Cell Cycle 4883-888. [DOI] [PubMed] [Google Scholar]
- 56.Otsuki, Y., M. Tanaka, S. Yoshii, N. Kawazoe, K. Nakaya, and H. Sugimura. 2001. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc. Natl. Acad. Sci. USA 984385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Post, D. E., F. R. Khuri, J. W. Simons, and E. G. Van Meir. 2003. Replicative oncolytic adenoviruses in multimodal cancer regimens. Hum. Gene Ther. 14933-946. [DOI] [PubMed] [Google Scholar]
- 58.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 153104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 713788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarbassov, D. D., S. M. Ali, S. Sengupta, J. H. Sheen, P. P. Hsu, A. F. Bagley, A. L. Markhard, and D. M. Sabatini. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22159-168. [DOI] [PubMed] [Google Scholar]
- 61.Sarbassov, D. D., D. A. Guertin, S. M. Ali, and D. M. Sabatini. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 3071098-1101. [DOI] [PubMed] [Google Scholar]
- 62.Schuebel, K. E., N. Movilla, J. L. Rosa, and X. R. Bustelo. 1998. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 176608-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwartz, R. A., S. S. Lakdawala, H. D. Eshleman, M. R. Russell, C. T. Carson, and M. D. Weitzman. 2008. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 829043-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sheng, M., and C. Sala. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 241-29. [DOI] [PubMed] [Google Scholar]
- 65.Shepard, R. N., and D. A. Ornelles. 2004. Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background. J. Virol. 789924-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shepard, R. N., and D. A. Ornelles. 2003. E4orf3 is necessary for enhanced S-phase replication of cell cycle-restricted subgroup C adenoviruses. J. Virol. 778593-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 68.Stracker, T. H., D. V. Lee, C. T. Carson, F. D. Araujo, D. A. Ornelles, and M. D. Weitzman. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 796664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7350-355. [DOI] [PubMed] [Google Scholar]
- 70.Sumita, C., M. Yamane, T. Matsuda, M. Maeda, T. Nariai, Y. Fujio, and J. Azuma. 2005. Platelet activating factor induces cytoskeletal reorganization through Rho family pathway in THP-1 macrophages. FEBS Lett. 5794038-4042. [DOI] [PubMed] [Google Scholar]
- 71.Tay, D. L., P. S. Bhathal, and R. M. Fox. 1991. Quantitation of G0 and G1 phase cells in primary carcinomas. Antibody to M1 subunit of ribonucleotide reductase shows G1 phase restriction point block. J. Clin. Investig. 87519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ullman, A. J., and P. Hearing. 2008. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 827325-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ullman, A. J., N. C. Reich, and P. Hearing. 2007. Adenovirus E4 ORF3 protein inhibits the interferon-mediated antiviral response. J. Virol. 814744-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 247686-7696. [DOI] [PubMed] [Google Scholar]
- 75.Wold, W. S., A. E. Tollefson, and T. W. Hermiston. 1995. E3 transcription unit of adenovirus. Curr. Top. Microbiol. Immunol. 199(Pt. 1)237-274. [DOI] [PubMed] [Google Scholar]
- 76.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, H. C., J. T. Hsieh, M. E. Gleave, N. M. Brown, S. Pathak, and L. W. Chung. 1994. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int. J. Cancer 57406-412. [DOI] [PubMed] [Google Scholar]
- 78.Yu, W., and H. Fang. 2007. Clinical trials with oncolytic adenovirus in China. Curr. Cancer Drug Targets 7141-148. [DOI] [PubMed] [Google Scholar]