Abstract
Rationale: Secondhand tobacco smoke (SHS) and traffic-related air pollutants are associated with asthma and allergy. Diesel exhaust particles (DEPs) and SHS can interact with allergens in exacerbating allergic airway diseases through generation of reactive oxygen species. Glutathione S-transferases (GSTs) metabolize reactive oxygen species and detoxify electrophilic xenobiotics present in SHS and DEPs.
Objectives: We tested the hypotheses that functional GSTM1-null genotype and GSTP1 codon 105 variants (Ile105 and Val105) are determinants of allergic responses to SHS, and that responses to SHS and DEPs are correlated.
Methods and Measurements: In a randomized, placebo-controlled crossover trial, 19 ragweed allergen–sensitive subjects who had previously participated in a DEP trial were challenged intranasally with allergen after having been exposed to either clean air or SHS at separate visits. Nasal allergen–specific IgE, histamine, IL-4, and IFN-γ levels were measured before and after allergen challenge.
Main Results: Individuals with GSTM1-null or GSTP1 Ile105 genotypes showed larger nasal responses to allergens with SHS compared with clean air. GSTM1-null subjects had a larger increase in IgE than GSTM1-present subjects (median, 173.3 vs. 46.7 U/ml; p = 0.03), and the Ile105 GSTP1 genotype subjects had increased histamine (median, 10.2 vs. 4.6 nM; p = 0.01) after SHS plus allergen challenge. Responses to SHS and DEPs were correlated. Enhancement of IgE and histamine was greatest in the subjects with both the GSTM1-null and GSTP1 Ile/Ile genotypes.
Conclusions: GSTM1 and GSTP1 are important cytoprotective factors that reduce SHS and DEP exacerbation of allergic responses.
Keywords: GSTM1; GSTP1; histamine, IgE; tobacco smoke
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Secondhand smoke causes a wide variety of health effects. Glutathione S-transferases metabolize reactive oxygen species and may modify the effects of secondhand smoke.
What This Study Adds to the Field
We show that the glutathione S-transferase GSTM1 and GSTP1 Val105 variants reduce the adjuvant effects of secondhand smoke on allergic responses to common allergens.
Secondhand smoke (SHS) causes a wide spectrum of adverse health effects (1–5). Common allergic airway diseases, including asthma and allergic rhinitis, account for a substantial fraction of the huge burden of ill health that arises from this ubiquitous pollutant. This burden is amplified because individuals with asthma and other allergic airway diseases have greater sensitivity to SHS, with increased symptoms, medication use, medical care usage, allergic responses, and respiratory illnesses (1–5).
The explanation for the heightened sensitivity among those with allergic airway diseases is likely multifactorial, but involves synergistic effects of SHS and allergens in initiating and exacerbating allergic airway diseases (6–8). In conjunction with allergen, SHS can act to enhance IgE antibody responses, T-helper cell (Th) 2 cytokine production, and histamine release in vivo (6). Traffic-related air pollutants have the same synergistic effects on allergic response, as shown by studies of the model pollutant, such as diesel exhaust particles (DEPs) (9, 10). Although the mechanisms by which SHS and other air pollutants, such as DEPs, affect allergic symptoms are still uncertain, a growing body of evidence supports a role of oxidative stress in initiating and enhancing airway inflammation (11–14).
Cigarette smoke is an extremely potent oxidant mixture, with large amounts of oxidant radicals produced per puff (15). These oxidants, which include nitric oxide, superoxide, and semiquinone radicals, can react together to form peroxynitrite (ONOO−), hydroxyl radical, and hydrogen peroxide (16). Components of DEPs have also been shown to produce reactive oxygen species (ROS) and oxidative stress (13, 17, 18). Inflammatory cells, such as alveolar macrophages, stimulated by these oxidants can, in turn, produce ROS. These can directly injure the airway epithelium (19) and also activate redox-sensitive transcription factors, such as nuclear factor-κB and activation protein-1, which regulate expression of many cytokines involved in inflammation and allergy. In model systems, the enhancement of allergic airway responses by inhaled xenobiotics can be blocked by antioxidants (12, 13, 20). In humans, enzymatic antioxidants play a role in allergic airway responses to xenobiotics (9). The role of antioxidants in allergic responses to inhaled xenobiotics suggests that sensitivity to the effects of SHS and DEP may be related to variation in antioxidant defenses, and therefore, response to SHS and DEP may be correlated.
Airway antioxidant defenses are mediated, in part, by enzymatic antioxidants, including glutathione S-transferases (GSTs) (21). GSTs are a large family of proteins that participate in antioxidant defenses through a number of mechanisms, including ROS metabolism, repair of ROS damage, and detoxification of xenobiotics present in SHS (22, 23). We investigated GSTM1 and GSTP1 genotypes because these genes are expressed in the respiratory tract, are involved in detoxification of ROS and chemicals present in SHS and DEP, and have common functional variant alleles that result in either the total absence or a marked alteration in the enzyme's activity (24). A growing body of literature documents that GSTM1 and GSTP1 genetic variants influence allergic airway disease and might explain variation in SHS responsiveness and common responsiveness to SHS and DEPs (9, 25–27). Because the effects of SHS and DEPs on allergic responses are, in part, mediated by increased oxidative stress, we hypothesized that these GST genotypes contribute to both SHS and DEP enhancement of allergic responses, and that individual responses to both SHS and DEPs are correlated based on individual levels of oxidant defenses. To test the hypothesis that common functional genetic variants, the null genotypes for GSTM1 and GSTP1 codon 105 variants (Ile105 → Val105), modulate SHS enhancement of allergic responses, we conducted a randomized, placebo-controlled cross-over trial of the airway allergic response enhancement by SHS using an established human nasal challenge model. Nasal allergic responses were measured after challenge with allergen plus either clean air or SHS to determine whether functional variants in GSTs can account for the variation in interindividual responsiveness to SHS. The trial was conducted in subjects who had previously participated in a trial examining DEP responses using the same randomized, placebo-controlled crossover trial design (9). Conducting the trial in the same subjects allowed comparison of correlation of SHS and DEP responses.
METHODS
Study Design
The effects of SHS and GSTM1 and GSTP1 genotypes on allergic airway responses were studied by conducting a randomized, placebo-controlled crossover trial in 19 ragweed-allergic young adults who had previously participated in a trial of DEP effects using the same protocol (9). SHS exposure occurred at least several months after the DEP challenges. The genotyping was done in a blinded protocol after the DEP study was completed. The information about participation in previous studies was not provided to the team conducting the challenge assays.
Subjects were randomized to first receive either a 2-h exposure to clean air or SHS. Subjects and staff were blinded to allergen condition; however, it was impossible to blind to the SHS exposure condition. On completion of exposure, subjects performed a nasal lavage and were then randomly challenged with ragweed allergen or placebo (300 μl saline). Nasal lavages were performed at 10 min, 24 h, 4 d, and 7 d after allergen/placebo challenge. In each trial, volunteers then underwent the other exposure condition at least 6 wk after the initial exposure condition, and were again blindly challenged intranasally with allergen after either controlled, 2-h chamber exposures to SHS or clean air. This resulted in four challenge conditions in random order: (1) clean air plus saline challenge; (2) clean air plus allergen challenge; (3) SHS plus saline challenge; and (4) SHS and allergen challenge. Genotyping was conducted using a blinded protocol.
Study Population
We recruited nonsmoking volunteers from Los Angeles, California, who had a history of allergy consistent with allergic rhinitis, a positive epicutaneous skin test to short ragweed (an allergen not present in Southern California), and displayed immediate allergic symptoms when challenged with ragweed allergen Amb a 1. Although subjects had positive skin tests for other allergens, all were asymptomatic during the period of the trial. Characteristics for the study population are shown in Table 1. Based on interview responses, none of the volunteers had any atypical exposure to pollutants, none had used topical or systemic steroids in the 3 mo before the study or oral antihistamines for the previous week, none had ever received allergy immunotherapy, and none were exposed to household smokers. The volunteers did not take any medication for the 3 d before or during the duration of the study. The research was approved by the Human Subject Protection Committee of the University of California at Los Angeles. All subjects provided written consent.
TABLE 1.
SELECTED CHARACTERISTICS OF STUDY VOLUNTEERS
| n | % | |
|---|---|---|
| Total | 19 | 100.0 |
| Female | 12 | 63 |
| Male | 7 | 37 |
| Age, yr | ||
| 20–25 | 7 | 37 |
| 26–30 | 8 | 42 |
| 30–34 | 4 | 21 |
| Ethnicity | ||
| White | 8 | 42 |
| Hispanic | 5 | 26 |
| African American | 1 | 5 |
| Asian | 5 | 26 |
| Genotype | ||
| GSTM1 | ||
| Null | 14 | 74 |
| Present | 5 | 26 |
| GSTP1 Ile105Val (A→G) | ||
| Ile/Ile | 13 | 68 |
| Ile/Val | 6 | 32 |
| Val/Val | 0 | 0.0 |
Allergen Challenge and Nasal Lavage
The protocol for allergen challenge has been previously described (10, 28). Briefly, a positive allergen challenge level was determined using symptoms for each volunteer by step-wise exposure to increasing intranasal doses of ragweed extract. The severity of allergic symptoms in response to an allergen challenge was measured before and after nasal challenge using a similar system to that previously reported by Togias and colleagues (29, 30). The severity of nasal itching, nasal congestion, and rhinorrhea were rated by subjects on a scale ranging from 0 to 3 (no symptom, mild, moderate, or severe). In addition, the number of sneezes were enumerated and scored from 0 to 3 (none, 1–2, 3–4, or > 5). Thus, subjects could have a maximum symptom score of 12. A significant correlation has previously been shown between symptom scores and rhinoscopic evaluation (29). The allergen dose was increased until a symptom severity score (composed of sneezing, irritation, and congestion) of 5 of 12 was achieved. This dose of allergen was used in all subsequent challenges. We conducted a nasal lavage before challenges, and ragweed IgE levels were very low or undetectable.
Nasal lavage is a well-established procedure for studying the effects of air pollutants on the upper airway (31), and this procedure was performed as previously described (10, 28). Briefly, 5 ml normal saline was instilled into each nostril of the subjects, and wash fluid was collected after a 10-s residence time. The subject then performed four subsequent washes. The lavage fluid was centrifuged at 350 × g for 10 min at 4°C, and aqueous supernatant was stored at −20°C until assayed. Histamine was measured in nasal lavages performed before and immediately after SHS/clean air exposure, and 10 min after allergen exposure. All other measurements were conducted at 96 h after challenge.
Controlled SHS Exposure
The generation of SHS and the controlled exposure protocol has previously been described (6). Briefly, resting subjects were exposed in a 700-ft3 chamber with controlled temperature, humidity, and ventilation. SHS was generated from the sidestream smoke of 1R4F reference cigarettes (University of Kentucky Tobacco Health Research Institute, Lexington, KY) using an RM G1 Borgwaldt smoking machine (Hamburg, Germany) and the Federal Trade Commission guidelines for generation of sidestream smoke, which specify 1 inhalation every 55 s, with an inhalation/exhalation cycle of 5 s (a total of 5 cigarettes in a 2-h exposure). Chamber SHS exposures excluded mainstream smoke. Clean air exposures were performed using the same protocol without smoke generation. All exposures were done at the same time of day (between 9 and 11 a.m.) to avoid diurnal variation.
The 2-h SHS exposures were well tolerated by all subjects, and the only adverse symptoms were mild irritation of the eyes and throat lasting less than 5 min after exposure in 5 subjects. The mean level of particulate matter was 310 and 46 μg/m3 during SHS exposures and clean air exposures, respectively.
Antibody, Histamine, and Cytokine Determination
All samples were run in duplicate and repeated if there was more than 10% variation between the duplicates. Total and ragweed-specific IgE in nasal washes were measured by isotype-specific ELISAs, as previously described (10, 32, 33), with minor modifications. The cytokines interleukin (IL)-4 and IFN-γ were measured using commercial ELISA kits (BD Pharmingen, San Diego, CA) following 20-fold concentration of the samples using Amicon filters (Millipore, Billerica, MA), per the manufacturers' instructions. Histamine was measured in nasal lavages performed before and immediately after SHS/clean air exposure, as well as 10 min after allergen exposure. Histamine levels were determined using a commercial assay (Immunotech, Brea, CA), per the manufacturer's instructions. The sensitivity of the assay was 0.5 nM.
Genotyping
Buccal cells were collected from participants as a source of genomic DNA. Details of buccal cell processing and genotyping assays have been previously described (9, 26). Briefly, DNA was extracted using a Puregene DNA isolation kit (Gentra, Minneapolis, MN). GSTM1, GSTT1, and GSTP1 genotypes were determined by real-time polymerase chain reaction using a TaqMan 7700 (Applied Biosystems, Foster City, CA). The presence or absence of a fluorescent amplification signal was used as an indication of whether the GSTM1 and T1 alleles were present or absent in a particular genomic DNA sample. Analysis of the single nucleotide polymorphism at codon 105 in the GSTP1 gene (Ile105Val) was performed using allele-specific probes. Assays were repeated on a subset of the samples for quality control, and genotyping was successfully completed for all 19 subjects.
Statistical Analysis
The lower limit of detection for each assay was used in analyses for subjects with values below the limit of detection. The median levels and differences of allergen-specific IgE, IL-4, IFN- γ, and histamine after allergen challenge and after allergen plus SHS challenge were examined using Wilcoxon's matched-pairs signed ranks tests. The effect of GSTM1-null and GSTP1 105 variant genotypes on allergen-specific IgE, histamine, IL-4, and IFN-γ levels after allergen alone or SHS plus allergen were assessed by comparisons of median response levels between single locus and bilocus genotypes (for single gene or joint effects of two genes) using Wilcoxon's rank sums test. Correlations between responses to DEPs and SHS were computed using Pearson's correlation coefficients. All analyses were conducted using SAS software (SAS Institute, Cary, NC), and all reported p values are based on a two-sided alternative hypothesis.
RESULTS
Subject Population and SHS Enhancement of Allergic Responses
GSTM1-null genotype was present in 73.7% of subjects (Table 1). Most of the subjects were homozygous for the GSTP1 Ile105 wild-type allele (68.4%), and none of the subjects were homozygous for the GSTP1 Val105 variant allele. The population of subjects was selected based on nasal allergy status. This selection factor likely explains the genotype distribution that differs from that observed in general population studies (< 50%).
We have previously reported that SHS enhances allergen-driven IgE, histamine, and IL-4 responses, while decreasing IFN-γ production (34). As shown in Table 2, exposure to SHS plus allergen increased nasal allergen-specific IgE levels from a median of 12.2 U/ml to 101.5 U/ml; p < 0.0001) among the 19 subjects. Exposure to SHS plus allergen increased IL-4 and decreased IFN-γ levels compared with allergen challenge alone, consistent with a Th2 cytokine response characteristic of an allergic response (9). Histamine levels increased from a median of 3.6 to 12.5 nM (p < 0.0001) after SHS plus allergen exposure. Exposure to SHS in the absence of allergen did not result in the formation of a detectable increase in allergen-specific antibodies, histamine, IL-4, or IFN-γ (see Table E1 in the online supplement). We have also previously reported the same pattern of allergen-driven effects for DEP exposure in the same group of subjects (9).
TABLE 2.
NASAL RESPONSES IN 19 SUBJECTS AFTER EXPOSURE TO CLEAN AIR PLUS ALLERGEN OR SECONDHAND SMOKE EXPOSURE PLUS ALLERGEN, AND THE DIFFERENCES IN RESPONSE*
| Median (min∼max)
|
||||
|---|---|---|---|---|
| Response | Clean Air + Allergen | SHS + Allergen | Difference† | p Value‡ |
| IgE, U/ml | 12.2 (1.1∼27.5) | 101.5 (23.5∼746.5) | 95.0 (8.9∼725.5) | < 0.0001 |
| IL-4, U/ml | 0.2 (0.2∼0.7) | 3.5 (0.2∼13.3) | 3.2 (−0.4∼13.1) | < 0.0001 |
| IFN-γ, ng/L | 0.6 (0.2∼1.6) | 0.3 (0.1∼1.4) | −0.2 (−1.5∼0.1) | < 0.0001 |
| Histamine, nM | 3.6 (0.9∼6.8) | 12.5 (0.9∼24.7) | 9.1 (−0.9∼20.6) | < 0.0001 |
Definition of abbreviation: IL = interleukin; SHS = secondhand smoke.
Level after secondhand smoke exposure plus allergen minus level after clean air plus allergen.
Differences in response between clean air plus allergen or SHS exposure plus allergen (level after SHS exposure plus allergen minus level after clean air plus allergen). The minimum differences are not simple differences between the minimums, and the maximum differences are not simple differences between the maximums. Histamine values were collected in lavages at 10 min, and other responses at 4 d after challenge.
Two-sided Wilcoxon matched pairs signed-rank tests for difference medians.
Role of Genotype on SHS Adjuvancy
The enhancement of the allergic response by SHS was larger for the majority of individuals with either a null GSTM1 or a homozygous GSTP1 Ile105 genotype compared with those with GSTM1 or a GSTP1 Val105 variant (see Figure 1 and the online supplement). As shown in Table 3, subjects who were GSTM1 null had a significantly larger increase in antiragweed IgE (median, 173.3 vs. 46.7 U/ml; p = 0.03) after SHS plus allergen challenge compared with subjects with GSTM1-present genotype. Compared with subjects with a GSTP1 Val105 variant, subjects with the homozygous wild-type Ile105 GSTP1 genotype had a trend toward a larger increase in allergen-specific IgE after SHS plus allergen challenge (median, 162.2 vs. 51.1 U/ml; p = 0.07). The IFN-γ and IL-4 levels did not show consistent patterns by genotype. Neither GSTM1 nor GSTP1 genotypes modified the allergic response to allergen challenge alone. In addition, GSTT1 genotype was not associated with SHS-enhanced allergic responses (data not shown).
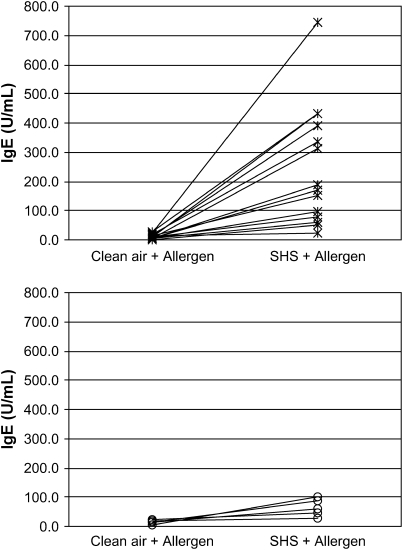
Figure 1.
Nasal allergen-specific IgE response 96 h after clean air plus allergen and secondhand smoke (SHS) plus allergen by GSTM1 genotype. Asterisks, GSTM1 null; open circles, GSTM1 present.
TABLE 3.
EFFECTS OF GSTM1 AND GSTP1 GENOTYPES ON MEDIAN DIFFERENCES IN NASAL ALLERGIC RESPONSES BETWEEN CLEAN AIR PLUS ALLERGEN AND SECONDHAND SMOKE EXPOSURE PLUS ALLERGEN*
| Median Difference (min∼max)
|
|||||
|---|---|---|---|---|---|
| Genotyping | n | IgE (U/ml) | IL-4 (U/ml) | IFN-γ (ng/L) | Histamine (nM) |
| GSTM1 | |||||
| Present | 5 | 46.7 (8.9∼95.0) | 3.2 (0.0∼5.4) | −0.2 (−1.0∼−0.1) | 8.4 (−0.9∼10.2) |
| Null | 14 | 173.3 (11.3∼725.5) | 4.0 (−0.4∼13.1) | −0.2 (−1.5∼0.1) | 9.4 (−0.9∼20.6) |
| p Value† | 0.03 | 0.33 | 0.61 | 0.43 | |
| GSTP1 | |||||
| Ile/Ile | 13 | 162.2 (24.6∼725.5) | 2.9 (0.0∼13.1) | −0.2 (−0.9∼0.1) | 10.2 (2.3∼20.6) |
| Ile/Val | 6 | 51.0 (8.9∼423.6) | 3.3 (−0.4∼12.2) | −0.5 (−1.5∼−0.1) | 4.6 (−0.9∼10.1) |
| p Value† | 0.07 | 0.93 | 0.20 | 0.03 | |
Level after secondhand smoke exposure plus allergen minus level after clean air plus allergen.
Two-sided Wilcoxon rank sum test. Histamine values were collected in lavages at 10 min, and other responses at 4 d, after challenge.
Parallel effects of the GSTP1 Ile105 genotypes were observed with SHS-enhanced histamine release at 10 min after challenge (Table E3). In subjects homozygous for GSTP1 Ile105, histamine levels were significantly higher after SHS plus allergen challenge than in subjects with the GSTP1 Val105 genotype (median, 10.2 vs. 4.6 nM; p = 0.03). In contrast, those with the GSTM1-null genotype showed no difference in histamine levels after SHS plus allergen challenge compared with subjects with GSTM1.
We found that the joint GSTM1 and GSTP1 genotype was an important determinant of response to SHS (Table 4); of the 14 allergic subjects with the GSTM1-null genotype, 11 had the homozygous GSTP1 Ile105 genotype. Subjects with the GSTM1-null and homozygous GSTP1 Ile105 genotype had the greatest enhancement of allergen-specific IgE (184.4 U/ml) compared with all other genotype groups. Similarly, subjects with this combined genotype had the greatest enhancement of allergen-specific histamine levels (10.3 nM) with SHS exposure.
TABLE 4.
JOINT EFFECTS OF GSTM1 AND GSTP1 GENOTYPES ON MEDIAN DIFFERENCES IN NASAL ALLERGIC RESPONSES BETWEEN CLEAN AIR PLUS ALLERGEN AND SECONDHAND SMOKE EXPOSURE PLUS ALLERGEN*
| Genotypes
|
Secondhand Smoke Exposure Response Median Difference (min∼max)
|
|||||
|---|---|---|---|---|---|---|
| GSTM1 | GSTP1 Ile105Val | n | IgE (U/ml) | IL-4 (U/ml) | IFN-γ (ng/L) | Histamine (nM) |
| + | Ile/Ile | 2 | 59.8 (24.6∼95.0) | 2.7 (0.0∼5.4) | −0.5 (−0.9∼−0.1) | 9.3 (8.4∼10.2) |
| + | Ile/Val | 3 | 46.7 (8.9∼74.7) | 3.2 (0.0∼3.3) | −0.2 (−1.0∼−0.2) | 3.2 (−0.9∼10.1) |
| − | Ile/Ile | 11 | 184.4 (47.6∼725.5)† | 2.9 (0.4∼13.1) | −0.2 (−0.8∼0.1) | 10.3 (2.3∼20.6)‡ |
| − | Ile/Val | 3 | 55.2 (11.3∼423.6) | 11.3 (−0.4∼12.2) | −0.9 (−1.5∼−0.1) | 6.0 (−0.9∼6.3) |
Level after secondhand smoke exposure plus allergen minus level after clean air plus allergen. Histamine values were collected in lavages at 10 min, and other responses at 4 d, after challenge.
IgE p = 0.0232, the Wilcoxon rank sum test compared to other three M1-P1 genotype groups.
Histamine p = 0.0632, the Wilcoxon rank sum test compared with other three M1-P1 genotype groups.
Correlation of SHS and DEP Responses
Allergic responses to SHS and DEP challenge were significantly correlated (Table 5). The correlation of allergic responses to SHS and DEP challenges varied from 0.54 for histamine to 0.79 for IL-4. The correlations were generally stronger in the GSTM1-null group than in the GSTM1-present group—a trend that was not as apparent by GSTP1 genotype. Individual responses to both SHS and DEPs are shown for each endpoint and genotype in Figures E2 and E3.
TABLE 5.
CORRELATION OF DIFFERENCES IN NASAL ALLERGIC ENDPOINTS WHEN EXPOSED TO CLEAN AIR PLUS ALLERGEN OR SECONDHAND SMOKE EXPOSURE PLUS ALLERGEN VERSUS CLEAN AIR PLUS ALLERGEN OR DIESEL EXHAUST PARTICLES PLUS ALLERGEN
| Response*
|
||||
|---|---|---|---|---|
| IgE | IL-4 | Histamine | IFN-γ | |
| All (n = 19) | 0.62 (0.005) | 0.79 (< 0.0001) | 0.54 (0.02) | 0.63 (0.005) |
| By GSTM1 genotypes | ||||
| GSTM1 present (n = 5) | 0.87 (0.07) | 0.95 (0.02) | 0.34 (0.58) | 0.79 (0.11) |
| GSTM1 null (n = 14) | 0.54 (0.04) | 0.74 (0.003) | 0.51 (0.07) | 0.58 (0.03) |
| By GSTP1 genotypes | ||||
| GSTP1 Ile/Ile (n = 13) | 0.62 (0.03) | 0.73 (0.005) | 0.37 (0.22) | 0.75 (0.003) |
| GSTP1 Ile/Val (n = 6) | 0.45 (0.37) | 0.91 (0.01) | 0.36 (0.49) | 0.77 (0.07) |
Values presented are correlations (p values for H0: ρ = 0).
The data concerning diesel exhaust particles have been published previously (9). Histamine values were collected in lavages at 10 min, and other responses at 4 d, after challenge.
DISCUSSION
SHS causes a broad spectrum of highly prevalent adverse respiratory health outcomes (1, 2). A large body of evidence supports a causal role for SHS in allergic airway disease occurrence (1, 2). We have previously shown that SHS can interact with allergen to exacerbate acute and immediate allergic responses in both animal models and human exposure trials (6). Here, we demonstrate that susceptibility to SHS may be controlled by functional variation in airway antioxidant defenses. In the present randomized exposure trial, we show that common variants in GSTM1 and GSTP1, which have been associated with atopy (allergy, asthma, and atopic dermatitis), mediate the enhancement of allergic responses by SHS at levels commonly encountered in indoor environments. We also observed that individual responses to SHS and DEPs are correlated across the endpoints measured in this study. Our results provide further evidence that the severity of common allergic airway diseases, such as allergic rhinitis and asthma, is a consequence of the interplay between common genetic variants and environmental exposures (9, 35–37). The correlated responses to different exposures suggest that some individuals are at higher risk than others for allergic symptoms when exposed to a spectrum of environmental stressors.
The importance of these results, from a clinical and public health perspective, is reflected in the high frequency of variants in these genes in most populations, as well as the ubiquitous exposure to SHS. The null allele variant of GSTM1 and Ile/Ile genotype for GSTP1 are present in approximately 50 and 40% of individuals, respectively (38). Although expression of GSTM1 is highest in the liver, it has also been identified in lung and nasal tissue (22). Individuals who have the null genotype completely lack class μ GST isoenzyme activity. Similarly, GSTP1 is the major GST gene expressed in human airways, and the different GSTP1 codon 105 variants have different enzymatic activities. Based on our findings on the joint effects of GSTM1 and GSTP1, we estimate that 15–20% of the general population may be unprotected from the enhancement of allergic responses by SHS. Among allergic individuals, the proportion of the population that is highly susceptible to SHS enhancement is likely to be larger, because these variants are associated with increased risk for atopy (25). This increased risk may arise because these polymorphisms are directly involved in atopy per se, or because they modulate the role of pollutants on atopy development.
The findings on the role of GSTM1 and GSTP1 in allergic responsiveness in the present study are consistent with those of our previous report that GSTP1 and GSTM1 play important roles in susceptibility to the adjuvant effects of DEPs (9). In the presence of DEPs, individuals with GSTM1-null or the GSTP1 Ile105 wild-type genotypes showed enhanced nasal allergic responses. Compared with subjects with a functional GSTM1 genotype, subjects with the GSTM1-null genotype had a significantly greater increase in IgE and histamine after DEP plus allergen challenge. The Ile105 GSTP1 genotype was associated with an increase in IgE and histamine after DEP plus allergen challenge. As observed after SHS exposure, the DEP enhancement was greatest in the subjects with both the GSTM1-null and GSTP1 Ile/Ile genotypes. IgE production is regulated by IL-4 and the formation of a Th2 cytokine milieu. It is, therefore, somewhat surprising that we failed to find statistically significant differences in IL-4 and IFN-γ elicited by SHS. One likely explanation is that this is due to a type 2 error. Another is that very small changes in these cytokines can result in large changes in IgE. Alternatively, because oxidant pollutants, such as DEP, can directly target B cells to enhance class switching, it is possible that the effect of SHS on IgE production may be independent of IL-4. It is also possible that DEPs and tobacco smoke inherently differ in their allergy-modulating pathways. However, the correlation of responses for SHS and DEPs argues against different mechanisms.
We have previously shown that SHS acts as an adjuvant by interacting with allergens to affect the immune system and enhance allergic responses (6). In mice, SHS exposure has an adjuvant effect, characterized by an increase in antigen-specific IgE, elevated Th2 responses, and influx of eosinophils into the lungs (7, 8). It is unclear which of the more than 6,000 compounds in sidestream tobacco smoke act as adjuvants; however, other particulate pollutants resulting from incomplete combustion of organic materials, such as ambient particulate matter and diesel exhaust, contain many of the constituents of SHS, such as polyaromatic hydrocarbons, suggesting that a spectrum of xenobiotics in the environment can contribute to enhancement of responses to allergens (9, 17, 39–41). Human and murine in vitro and in vivo studies have demonstrated that DEP and polyaromatic hydrocarbons can also induce IgE production, increase Th2 cytokine production, select against Th1 cytokines, and augment histamine release, and that these effects are blocked by GSTM1 and GSTP1 (reviewed in References 42 and 43). Many of the proximal mechanisms underlying the effects of SHS, such as enhanced inflammation, formation of a Th2 cytokine environment, and the production of allergen-specific IgE antibodies, are believed to be driven by oxidative stress responses (10, 11, 28, 44). Inflammation itself is an oxidative event; SHS, DEPs, and a variety of other air pollutants can increase oxidative stress levels by stimulating the production of ROS directly and indirectly through enhanced airway inflammation. Small molecular and enzymatic antioxidants, such as the GSTs, can reduce the formation and effects of ROS. Here, we suggest that members of the GST family may play a key role in controlling the responses to a wide range of xenobiotics, in addition to SHS and DEPs, by detoxifying xenobiotic-derived ROS and oxidation products. We speculate that combustion products, as a class of complex mixtures, are likely to enhance allergic responses to allergens, and that variants in genes, including GSTM1 and GSTP1, may play an important role in determining who is protected from the adverse effects of these mixtures.
A growing number of epidemiologic studies have shown that GSTP1 and GSTM1 variants are associated with airway hyperresponsiveness and asthma, especially in relation to secondhand smoke or other xenobiotic exposure (25–27, 36, 37, 45). Although we investigated inflammatory responses in the upper airway, the genetic susceptibility to SHS and DEPs is likely to apply to the lower airway, as exposures, such as to SHS, can enhance airway hyperreactivity in humans and murine models. The GSTP1 gene product provides more than 90% of the GST activity in the lung, suggesting that lower airway as well as upper airway effects of SHS may be strongly modulated by GSTP1 function. Further support for common genetic effects over the upper and lower airways is provided by evidence that the GSTP1 Ile105/Ile105 genotype is associated with IgE levels and increased risk for atopy, which affects both the upper and lower airways (25).
Because the respiratory tract has multiple mechanisms to defend against insults from airborne toxins, it is unlikely that GSTM1 and GSTP1 are the only loci that confer protection from airborne pollutants, such as SHS and DEP, or that they have a unique protective mechanism (21). The variation in responses among subjects who are GSTM1 null suggests that other antioxidant genes with common functional polymorphisms exist that could contribute to susceptibility to more severe allergic responses on exposure to SHS and DEPs. For example, reduced nicotinamide adenine dinucleotide phosphate:quinone oxidoreductase, a flavoenzyme that can detoxify quinones and other oxidant chemicals, has also been reported to be protective in pollutant-induced asthma. Small molecule antioxidants and dietary intake may also contribute to antioxidant defenses directly, or by increasing expression of antioxidant genes (21, 46). Additional studies in larger populations are needed to discover additional protective loci with lower frequency variants, as well as to explore the role of dietary intake of antioxidants. Including genetic characterization of subjects in the design and analysis of inhalation challenge studies has great potential to advance our understanding of the role of the environment and susceptibility factors in allergic responses and other respiratory conditions.
In summary, SHS exposure is common in human populations, with estimates that up to 40% of children in the United States are exposed daily to SHS. Secondhand smoke causes a huge public health and clinical burden of disease. Our results provide important evidence that common genetic variants may determine who is protected and who is at risk for the adverse effects of SHS. Individuals with allergic airway diseases are recognized to be sensitive to the effects of SHS. We show that GSTM1 and the GSTP1 Val105 variant reduce the adjuvant effects of SHS on allergic responses to a common allergen. The elevated genetic susceptibility for a large number of individuals indicates great potential benefits of a public health intervention that targets reduction in SHS exposure. Although reductions in exposure are being achieved, more effective treatment of allergic airway disease may be possible via approaches that induce expression of GSTs.
Supplementary Material
Acknowledgments
The authors thank Krissy Nielsen and Christine Tidwell for assisting in the production and format of this manuscript.
Supported by the Southern California Environmental Health Sciences Center (grant 5P30ES007048), which was funded by the National Institute of Environmental Health Sciences; the Children's Environmental Health Center (grants 5P01ES009581, R826708-01, and RD831861-01), which was funded by the National Institute of Environmental Health Sciences and the Environmental Protection Agency; the University of California at Los Angeles, Allergy, Asthma, and Immunologic Disease Center (grant 5P01AI050495), which was funded by the National Institute of Allergy and Infectious Diseases and the National Institute of Environmental Health Sciences; the National Institute of Environmental Health Sciences (grant 5P01ES011627); the National Heart, Lung, and Blood Institute (grants 5R01HL61768 and 5R01HL076647); and the Hastings Foundation.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200509-1424OC on October 5, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Public Health Service. The health consequences of involuntary smoking. Report of the Surgeon General. Washington, DC: U.S. Public Health Service; 1986. DHHS Publication No. (PHS) 87-8398.
- 2.U.S. Environmental Protection Agency. Respiratory health effects of passive smoking: lung cancer and other disorders. Washington, DC: U.S. Environmental Protection Agency; 1992. EPA/600/6-90/006F.
- 3.California Environmental Protection Agency. Health effects of exposure to environmental tobacco smoke. Sacramento, CA: California Environmental Protection Agency; 1997. [DOI] [PMC free article] [PubMed]
- 4.Committee on the Assessment of Asthma and Indoor Air. Clearing the air: asthma and indoor exposures. Washington, DC: National Academy of Sciences; 2000.
- 5.Cook DG, Strachan DP. Health effects of passive smoking-10: Summary of effects of parental smoking on the respiratory health of children and implications for research. Thorax 1999;54:357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz-Sanchez D, Rumold R, Gong H Jr. Challenge with environmental tobacco smoke exacerbates allergic airway diseases in humans. J Allergy Clin Immunol 2006;118:441–446. [DOI] [PubMed] [Google Scholar]
- 7.Rumold R, Jyrala M, Diaz-Sanchez D. Secondhand smoke induces allergic sensitization in mice. J Immunol 2001;167:4765–4770. [DOI] [PubMed] [Google Scholar]
- 8.Seymour BW, Pinkerton KE, Friebertshauser KE, Coffman RL, Gershwin LJ. Second-hand smoke is an adjuvant for T helper-2 responses in a murine model of allergy. J Immunol 1997;159:6169–6175. [PubMed] [Google Scholar]
- 9.Gilliland FD, Li YF, Saxon A, Diaz-Sanchez D. Effect of glutathione-S-transferase M1 and P1 genotypes on xenobiotic enhancement of allergic responses: randomised, placebo-controlled crossover study. Lancet 2004;363:119–125. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Sanchez D, Tsien A, Fleming J, Saxon A. Combined diesel exhaust particulate and ragweed allergen challenge markedly enhanced in vivo nasal ragweed–specific IgE and skews cytokine production to a Th2-type pattern. J Immunol 1997;158:2406–2413. [PubMed] [Google Scholar]
- 11.Nel AE, Diaz-Sanchez D, Li N. The role of particulate pollutants in pulmonary inflammation and asthma: evidence for the involvement of organic chemicals and oxidative stress. Curr Opin Pulm Med 2001;7:20–26. [DOI] [PubMed] [Google Scholar]
- 12.Xiao GG, Wang M, Li N, Loo JA, Nel AE. Use of proteomics to demonstrate a hierarchical oxidative stress response to diesel exhaust particle chemicals in a macrophage cell line. J Biol Chem 2003;278:50781–50790. [DOI] [PubMed] [Google Scholar]
- 13.Li N, Hao M, Phalen RF, Hinds WC, Nel AE. Particulate air pollutants and asthma: a paradigm for the role of oxidative stress in PM-induced adverse health effects. Clin Immunol 2003;109:250–265. [DOI] [PubMed] [Google Scholar]
- 14.Howard DJ, Ota RB, Briggs LA, Hampton M, Pritsos CA. Environmental tobacco smoke in the workplace induces oxidative stress in employees, including increased production of 8-hydroxy-2′-deoxyguanosine. Cancer Epidemiol Biomarkers Prev 1998;7:141–146. [PubMed] [Google Scholar]
- 15.Church DF, Pryor WA. Free-radical chemistry of cigarette smoke and its toxicological implications. Environ Health Perspect 1985;64:111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacNee W. Oxidative stress and lung inflammation in airways disease. Eur J Pharmacol 2001;429:195–207. [DOI] [PubMed] [Google Scholar]
- 17.Hiura TS, Kaszubowski MP, Li N, Nel AE. Chemicals in diesel exhaust particles generate reactive oxygen radicals and induce apoptosis in macrophages. J Immunol 1999;163:5582–5591. [PubMed] [Google Scholar]
- 18.Li N, Kim S, Wang M, Froines J, Sioutas C, Nel A. Use of a stratified oxidative stress model to study the biological effects of ambient concentrated and diesel exhaust particulate matter. Inhal Toxicol 2002;14:459–486. [DOI] [PubMed] [Google Scholar]
- 19.Pettersen CA, Adler KB. Airways inflammation and COPD: epithelial–neutrophil interactions. Chest 2002;121:142S–150S. [DOI] [PubMed] [Google Scholar]
- 20.Whitekus MJ, Li N, Zhang M, Wang M, Horwitz MA, Nelson SK, Horwitz LD, Brechun N, Diaz-Sanchez D, Nel AE. Thiol antioxidants inhibit the adjuvant effects of aerosolized diesel exhaust particles in a murine model for ovalbumin sensitization. J Immunol 2002;168:2560–2567. [DOI] [PubMed] [Google Scholar]
- 21.Gilliland FD, McConnell R, Peters J, Gong H Jr. A theoretical basis for investigating ambient air pollution and children's respiratory health. Environ Health Perspect 1999;107(Suppl 3):403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes J, Pulford D. The glutathione-S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol 1995;30:445–600. [DOI] [PubMed] [Google Scholar]
- 23.Strange RC, Jones PW, Fryer AA. Glutathione S-transferase: genetics and role in toxicology. Toxicol Lett 2000;112:357–363. [DOI] [PubMed] [Google Scholar]
- 24.Talalay P, Fahey JW, Holtzclaw WD, Prestera T, Zhang Y. Chemoprotection against cancer by phase 2 enzyme induction. Toxicol Lett 1995;82–83:173–179. [DOI] [PubMed] [Google Scholar]
- 25.Fryer AA, Bianco A, Hepple M, Jones PW, Strange RC, Spiteri MA. Polymorphism at the glutathione S-transferase GSTP1 locus: a new marker for bronchial hyperresponsiveness and asthma. Am J Respir Cell Mol Biol 2000;161:1437–1442. [DOI] [PubMed] [Google Scholar]
- 26.Gilliland FD, Li YF, Dubeau L, Berhane K, Avol E, McConnell R, Gauderman WJ, Peters JM. Effects of glutathione S-transferase M1, maternal smoking during pregnancy, and environmental tobacco smoke on asthma and wheezing in children. Am J Respir Crit Care Med 2002;166:457–463. [DOI] [PubMed] [Google Scholar]
- 27.Mapp CE, Fryer AA, De Marzo N, Pozzato V, Padoan M, Boschetto P, Strange RC, Hemmingsen A, Spiteri MA. Glutathione S-transferase GSTP1 is a susceptibility gene for occupational asthma induced by isocyanates. J Allergy Clin Immunol 2002;109:867–872. [DOI] [PubMed] [Google Scholar]
- 28.Diaz-Sanchez D, Penichet-Garcia M, Saxon A. Diesel exhaust particles directly induce activated mast cells to degranulate and increase histamine levels and symptom severity. J Allergy Clin Immunol 2001;106:1140–1146. [DOI] [PubMed] [Google Scholar]
- 29.Togias A, Proud D, Kagey-Sobotka A, Norman P, Lichtenstein L, Naclerio R. The effect of a topical tricyclic antihistamine on the response of the nasal mucosa to challenge with cold, dry air and histamine. J Allergy Clin Immunol 1987;79:599–604. [DOI] [PubMed] [Google Scholar]
- 30.Togias A, Lykens K, Kagey-Sobotka A, Eggleston PA, Proud D, Lichtenstein LM, Naclerio RM. Studies on the relationships between sensitivity to cold, dry air, hyperosmolal solutions, and histamine in the adult nose. Am Rev Respir Dis 1990;141:1428–1433. [DOI] [PubMed] [Google Scholar]
- 31.Koren HS, Hatch GE, Graham DE. Nasal lavage as a tool in assessing acute inflammation in response to inhaled pollutants. Toxicology 1990;60:15–25. [DOI] [PubMed] [Google Scholar]
- 32.Diaz-Sanchez D, Dotson AR, Takenaka H, Saxon A. Diesel exhaust particles induce local IgE production in vivo and alter the pattern of IgE messenger RNA isoforms. J Clin Invest 1994;94:1417–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Sanchez D, Tsien A, Casillas A, Dotson AR, Saxon A. Enhanced nasal cytokine production in human beings after in vivo challenge with diesel exhaust particles. J Allergy Clin Immunol 1996;98:114–123. [DOI] [PubMed] [Google Scholar]
- 34.Diaz-Sanchez D, Riedl M. Diesel effects on human health: a question of stress? Am J Physiol Lung Cell Mol Physiol 2005;289:L722–L723. [DOI] [PubMed] [Google Scholar]
- 35.Holgate ST. Genetic and environmental interaction in allergy and asthma. J Allergy Clin Immunol 1999;104:1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YL, Lin YC, Lee YC, Wang JY, Hsiue TR, Guo YL. Glutathione S-transferase P1 gene polymorphism and air pollution as interactive risk factors for childhood asthma. Clin Exp Allergy 2004;34:1707–1713. [DOI] [PubMed] [Google Scholar]
- 37.Kabesch M, Hoefler C, Carr D, Leupold W, Weiland SK, von Mutius E. Glutathione S transferase deficiency and passive smoking increase childhood asthma. Thorax 2004;59:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.To-Figueras J, Gene M, Gomez-Catalan J, Galan MC, Fuentes M, Ramon JM, Rodamilans M, Huguet E, Corbella J. Glutathione S-transferase M1 (GSTM1) and T1 (GSTT1) polymorphisms and lung cancer risk among northwestern Mediterraneans. Carcinogenesis 1997;18:1529–1533. [DOI] [PubMed] [Google Scholar]
- 39.Barfknecht TR, Hites RA, Cavaliers EL, Thilly WG. Human cell mutagenicity of polycyclic aromatic hydrocarbon components of diesel emissions. Dev Toxicol Environ Sci 1982;10:277–294. [PubMed] [Google Scholar]
- 40.Kumagai Y, Arimoto T, Shinyashiki M, Shimojo N, Nakai Y, Yoshikawa T, Sagai M. Generation of reactive oxygen species during interaction of diesel exhaust particle components with NADPH-cytochrome p450 reductase and involvement of the bioactivation in the DNA damage. Free Radic Biol Med 1997;22:479–487. [DOI] [PubMed] [Google Scholar]
- 41.Soontjens CD, Holmberg K, Westerholm RN, Rafter JJ. Characterization of polycyclic aromatic compounds in diesel exhaust particulate extract responsible for aryl hydrocarbon activity. Atmos Environ 1997;31:219–225. [Google Scholar]
- 42.Kagawa J. Health effects of diesel exhaust emissions—a mixture of air pollutants of worldwide concern. Toxicology 2002;181–182:349–353. [DOI] [PubMed] [Google Scholar]
- 43.Diaz-Sanchez D, Proietti L, Polosa R. Diesel fumes and the rising prevalence of atopy: an urban legend? Curr Allergy Asthma Rep 2003;3:146–152. [DOI] [PubMed] [Google Scholar]
- 44.Li N, Alam J, Venkatesan MI, Eiguren-Fernandez A, Schmitz D, Di Stefano E, Slaughter N, Killeen E, Wang X, Huang A, et al. Nrf2 is a key transcription factor that regulates antioxidant defense in macrophages and epithelial cells: protecting against the proinflammatory and oxidizing effects of diesel exhaust chemicals. J Immunol 2004;173:3467–3481. [DOI] [PubMed] [Google Scholar]
- 45.Piirila P, Wikman H, Luukkonen R, Kaaria K, Rosenberg C, Nordman H, Norppa H, Vainio H, Hirvonen A. Glutathione S-transferase genotypes and allergic responses to diisocyanate exposure. Pharmacogenetics 2001;11:437–445. [DOI] [PubMed] [Google Scholar]
- 46.Romieu I, Sienra-Monge JJ, Ramirez-Aguilar M, Moreno-Macias H, Reyes-Ruiz NI, Estela del Rio-Navarro B, Hernandez-Avila M, London SJ. Genetic polymorphism of GSTM1 and antioxidant supplementation influence lung function in relation to ozone exposure in asthmatic children in Mexico City. Thorax 2004;59:8–10. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.