Abstract
Müllerian inhibiting substance (MIS) is a key element required to complete mammalian male sex differentiation. The expression pattern of MIS is tightly regulated in fetal, neonatal, and prepubertal testes and adult ovaries and is well conserved among mammalian species. Although several factors have been shown to be essential to MIS expression, its regulatory mechanisms are not fully understood. We have examined MIS promoter activity in 2-day postnatal primary cultures of rat Sertoli cells that continue to express endogenous MIS mRNA. Using this system, we found that the region between human MIS−269 and −192 is necessary for full MIS promoter activity. We identified by DNase I footprint and electrophoretic mobility-shift analyses a distal steroidogenic factor-1 (SF-1)-binding site that is essential for full promoter activity. Mutational analysis of this new distal SF-1 site and the previously identified proximal SF-1 site showed that both are necessary for transcriptional activation. Moreover, the proximal promoter also contains multiple GATA-4-binding sites that are essential for functional promoter activity. Thus multiple SF-1- and GATA-4-binding sites in the MIS promoter are required for normal tissue-specific and developmental expression of MIS.
Keywords: Müllerian inhibiting substance promoter
Müllerian inhibiting substance (MIS), also called anti-Müllerian hormone, a glycoprotein homodimer belonging to the transforming growth factor β superfamily, is a critical component of sex differentiation responsible for regression in the male embryo of the Müllerian ducts, which in a normal female embryo become the uterus, fallopian tubes, and upper vagina (1). In the rat, MIS is expressed in fetal Sertoli cells from the time of testis differentiation (13 days postcoitum) (2). Both MIS mRNA and protein remain high after birth and fall precipitously after day 5 to a low level, where they remain throughout adult life. Conversely, MIS mRNA is undetectable in the fetal ovaries but becomes increasingly expressed after birth in the granulosa cells of developing follicles (3–5).
The complex expression pattern of MIS suggests that it is tightly regulated. The MIS genes from mouse (6), rat (7), bovine (8), porcine (9), chicken (10), and human (8, 11) have been cloned, and all mammalian proximal promoters show regions of evolutionary conservation (Fig. 1). The mouse MIS transcriptional start site is located only 328 bp downstream of the SAP62 gene, suggesting that the region conferring critical regulation of MIS expression is located within close proximity of the transcriptional start site (12).
Figure 1.
Comparison of rat, mouse, bovine, porcine, and human MIS proximal promoters. Boldface indicates regions conserved among species, with species designation to the left. Capital letters refer to nucleotides conserved among more than three species; underlines indicate gaps introduced to optimize alignment. The number from the transcription start site was derived from the human sequence; mouse−180 is also designated. +1 marks the transcriptional start sites.
Several factors important for sex determination have been proposed to regulate MIS expression, including SRY (13), SOX9 (14, 15), SF-1 (16, 17), WT-1 (18, 19), Dax-1 (19), and GATA-4 (20, 21), and all except Dax-1 and possibly WT-1 can bind to the −180 bp region upstream of the transcriptional start site of MIS. Moreover, evidence is convincing that transcriptional up-regulation of MIS requires coordinate interactions between SF-1 and SOX9 (15), GATA-4 (21), and WT-1 (19); Dax-1 is thought to down-regulate MIS transcription through an interaction with SF-1 (19). In addition, the developmental expression of each of these factors precedes that of MIS.
We have examined MIS promoter activity in early postnatal primary Sertoli cells, which have been shown to produce endogenous MIS for short periods of time in culture. Because the primary Sertoli cell culture exhibited very low transient transfection efficiencies, we developed an assay using adenoviral MIS promoter constructs engineered to drive luciferase expression to examine the DNA sequences important for functional MIS expression. The proteins responsible for the activity of these regions were investigated further by DNase I footprinting and electrophoretic mobility-shift assays (EMSA). These experiments identify a new distal SF-1 site that is necessary for transcription of human MIS in Sertoli cells at the perinatal stage of development. In addition, we show that multiple GATA-4-binding sites as well as both SF-1 sites are necessary for full promoter activity at this stage.
Materials and Methods
Plasmid Construction.
Deletion MIS promoter luciferase constructs, MIS−269/+8 (MIS−269-luc), MIS−106/+8 (13), and pA3 luciferase (pA3-luc) were used with new deletion constructs, MIS−192/+8 luciferase and MIS−38/+8 luciferase. The KpnI-BamHI fragments of MIS−269-luc and pA3-luc, which includes the luciferase coding region with or without the MIS promoter, respectively, were inserted between the KpnI and BamHI sites of pLEP3 to produce pLEP-MIS−269 and pLEP-pA3, respectively. The KpnI-HindIII fragments of MIS−192/+8, MIS−106/+8, and MIS−38/+8 were inserted between the KpnI and HindIII sites of pLEP-pA3.
The proximal SF-1 mutation (MIS−269-pSF-1-mut-luc) was produced by PCR by using MIS−269-luc as a template with primers CMH 106 and 317 and CMH 107 and 316 [see supplemental Table 1 in the supplemental data on the PNAS web site (www.pnas.org) for sequences of oligonucleotides]; PCR products were inserted into pGEM and pCR (TA vector kit, CLONTECH), respectively. The ApaI fragment of 316/107-pCR was inserted into the 317/106-pGEM ApaI site. Insert orientation was confirmed by sequencing. The distal SF-1 mutation and double SF-1 mutations were produced by using the Quick Change system (Stratagene) with oligonucleotides TC043 and TC044 and MIS−269-luc or MIS−269-pSF-1-mut-luc as templates, respectively. The proximal GATA mutation, MIS−269-pGATA-mut-luc, was constructed similarly to MIS−269-pSF-1-mut-luc with primers CMH 106 and 313 and CMH 107 and 312. The construct with mutations introduced into both the distal and reverse GATA sites for the distal GATA mutation and the construct containing all three GATA mutations were made by using the Quick Change system with oligonucleotides AHL01 and AHL02 and MIS−269-luc or MIS−269-pGATA-mut-luc, respectively. All mutations were digested by HindIII and inserted into the HindIII site of pLEP. The inserted sequences were confirmed by thermosequencing with primer CMH123. pCMV-GATA-4 was prepared by ligation of the 3.3-kb EcoRI fragment of PUC18-GATA-GT2 to the EcoRI site of pCMV6.
Recombinant Adenoviruses.
Adenovirus type 2-based recombinant viruses were generated by using a two-cosmid system developed in the MGH Gene Therapy Center (X.W., M. W. Freeman, and B. Seed, unpublished data). The recombinant adenoviral vectors contain the luciferase gene driven by various MIS promoter fragments or the green fluorescent protein (GFP) driven by a cytomegalovirus (CMV) early promoter. The adenovirus vectors were propagated to high-titer stocks and purified by using CsCl gradient centrifugation as described (22). The viral particle number was determined by UV absorbance at 260 nm and described as optical particle units (OPU). The virus solution was diluted in PBS to 1011 OPU/ml and stored until use at −70°C in PBS containing 10% glycerol and 0.2% BSA. All mutations were confirmed by directly sequencing the adenoviral vectors after PCR amplification with primers CMH 106 and 107. Multiple independent viral preparations were analyzed for each promoter construct.
Culture of Postnatal 2 (P2)-Day-Old Rat Sertoli Cells.
Sertoli cells were isolated from 2-day-old rats by using a sequential enzymatic procedure (23, 24). The animal protocol was approved by the Massachusetts General Hospital Internal Review Board. Briefly, testes were harvested from 10–18 male pups, decapsulated, and digested twice in 0.5 mg/ml collagenase D and 0.1 mg/ml DNase I in M199 Hepes solution (pH 7.2) at 32°C for 30–50 min and once in 0.5 mg/ml hyaluronidase in Hanks' balanced salt solution (HBSS) at 32°C for 30 min. Digested seminiferous tubule fragments were suspended at 1–6 × 105/ml in DMEM mixed 1:1 with F-12 medium containing 0.5 mg/ml insulin, 0.5 mg/ml transferrin, 0.1% BSA, and 5 mM l-glutamine and plated in extracellular matrix gel (Matrigel, Sigma) pretreated 24- (500 μl) or 6-well plates (2 ml). Cultured cells were incubated at 37°C with 5% CO2, then 12 h later washed two to three times with Ca2+, Mg2+ free HBSS to remove peritubular cells, thus producing a yield of highly purified Sertoli cells. Media were changed every 2 days; the cells reached 80–90% confluence in 3 days. Cells in one well of a 24-well plate from each culture were stained for alkaline phosphatase to detect peritubular cells (25) and 3β-hydroxysteroid dehydrogenase to detect Leydig cells (26) to confirm the purity of the Sertoli cell preparation.
Northern Blot.
mRNA was prepared from Sertoli cell cultures in 6-well plates by using Quickprep (Amersham) or from the tissue of P2 testes or spleen. poly(A)+ mRNA (0.8 μg) from each sample was electrophoresed on a 1.5% agarose-formaldehyde gel. The gel was blotted to nylon membrane (Hybond-N; Amersham) and the RNA UV crosslinked. A rat MIS EcoRI-NotI fragment from pCMV-rMIS that included the fifth exon of rat MIS (7), a rat GATA-4 EcoRI fragment from PUC18-GATA-GT2 (27), and a mouse SF-1 EcoRI fragment from pCMV-SF-1 (28), was used to prepare 32P-labeled probes by the random hexamer method (29). The blot was stripped and rehybridized with a β-actin cDNA probe as a loading control.
Adenoviral Infection and Luciferase Reporter Assay.
Infection was performed on day 3 of culture of the P2 Sertoli cells. The medium was changed before infection, and 32.5 μl of various dilutions of adenoviral constructs in PBS was added to fresh medium and incubated for 1 h at 37 C. After 1 h, cells were washed with HBSS, and 500 μl of fresh medium was added. Forty-eight hours later, the cells were extracted with 150 μl of passive lysis buffer for 15 min, scraped, and lysed by one round of freezing and thawing. Twenty to forty microliters of lysate was used for each luciferase assay (Promega) and 4–15 μl for each protein assay (Bio-Rad) to standardize the luciferase values. GFP-infected cells were trypsinized after 48 h incubation, resuspended with DMEM containing 10% female FBS (Aires Scientific, Montgomery, IL), mixed with 0.4% trypan blue, and GFP-positive- and trypan blue-excluded cells were counted to determine the efficiency of infection. Statistical analysis was performed by the unpaired t test.
Nuclear Extract.
Nuclear extracts were prepared according to the method of Dignam (30). Protein concentration was determined by using a protein assay kit (Bio-Rad) and stored at −70°C. COS cells were transfected with 10 μg CMV-GATA-4 per 10 cm plate by Fugene 6 (Roche Molecular Biochemicals).
DNA Footprint Analysis.
Radiolabeled DNA probes for footprinting analysis were constructed by PCR amplification as described previously (31). Briefly, the oligonucleotide CMH106, phosphorylated with [32P-γ] ATP and T4 polynucleotide kinase, was used as a 5′-primer for amplification by PCR to create the MIS−269/+8 promoter fragments with CMH107. The binding of protein to DNA was performed in a total volume of 100–120 μl, with 2 μg poly(dI⋅dC), 2 × 104 cpm labeled probe, and various quantities of nuclear extract. The volumes were adjusted with nuclear extract buffer (buffer C/buffer D = 1:6). After 30 min at 25°C, 60 μl of 5 mM CaCl2/10 mM MgCl2 solution was added. The DNA was digested with 0.1–1.0 unit DNase I at 25°C for 100 sec and terminated with 300 μl stop solution. The digestion products were analyzed on a 6% sequencing gel alongside sequencing reactions primed with the same oligonucleotide (CMH106).
EMSA.
Ten picomoles of double-stranded oligonucleotides with 5′ overhangs were 32P-labeled by using the fill-in method (29) and filtered through a G-50 Sephadex Quick spin column (Roche Molecular Biochemicals). One microliter of competitor or antibody was combined with 3–5 μg nuclear extract in a total volume of 14 μl containing 300 μg/ml BSA, 15% glycerol, 20 mM Hepes (pH 7.9), 70 mM NaCl, 30 mM KCl, 0.5 μg poly(dI⋅dC), 5 mM DTT, 1 mM EDTA, and 0.3 mM PMSF. After 10 min at 30°C, 2 × 104 cpm of probe in 1 μl was added and further incubated for 20 min at 30°C. The reactions were resolved on a 4% nondenaturing gel at 30 mA for 2 h at 4°C. The SF-1 and GATA-4 antibodies were from Upstate Biotechnology (Lake Placid, NY) and Santa Cruz Biotechnology, respectively.
Results and Discussion
P2 Rat Primary Sertoli Cell Cultures Express MIS, GATA-4, and SF-1 mRNA.
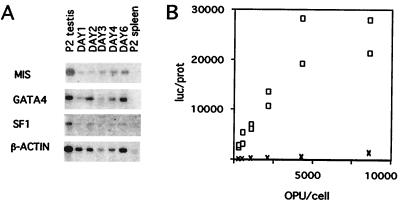
Purification of P2 Sertoli cells was confirmed by showing less than 5% alkaline phosphatase and 1% 3β-hydroxysteroid dehydrogenase staining, as previously described (23). MIS mRNA could be detected in increasing amounts in the cultured Sertoli cells on days 1, 2, 3, 4, and 6 of culture when compared with the β-actin signal (Fig. 2A). This pattern confirmed a recent report showing that the MIS mRNA level increased from day 2 to 4 in cultures of P2 Sertoli cells plated on Matrigel (23). GATA-4 and SF-1 RNA was also detected on all days of culture, suggesting that these important transcription factors could be functional at this time in development.
Figure 2.
(A) MIS mRNA expression during culture of P2 Sertoli cells. poly(A) RNA signals for MIS (2.0 kb), GATA-4 (3.1 kb), and SF-1 (3.8 kb) were present in P2 rat testes (positive control) and in isolated Sertoli cells from P2 rat testes after 1, 2, 3, 4, and 6 days in culture, but not in P2 rat spleen (negative control). The β-actin cDNA probe was hybridized on the same filter after stripping as a loading control. Exposure was 3 days for MIS and β-actin, 4 days for GATA-4, and 5 days for SF-1. (B) Luciferase response mediated by the MIS−269 promoter. The y-axis displays the relative luciferase (luc)/protein (prot) expression of the MIS−269 (o) and pA3 (X) vectors. (see Materials and Methods). Experiments were performed in duplicate at the same OPU/cell ratio.
Adenovirus Infection of Early Postnatal Sertoli Cells.
Because primary Sertoli cells exhibited poor transfection efficiency in transient transfection assays, we infected the Sertoli cells with adenoviral vectors, which can achieve high titers, can infect a wide spectrum of cells, and exhibit a high efficiency of gene transfer. Because adenovirus is episomal, cis-acting influences should also be minimized (32). It has previously been shown that adenovirus can successfully infect Sertoli cells both in vitro and in vivo (33). Incubation of our primary Sertoli cell cultures for 1 h with 7,000 OPU/cell of a CMV-driven GFP adenovirus resulted in 30% GFP positive cells (data not shown).
The virus titer was optimized for each MIS deletion luciferase construct. The MIS−269 luciferase construct and the other deletion MIS promoter constructs showed peak luciferase values between 5,000 and 10,000 OPU/cell (Fig. 2B and data not shown). The relative luciferase units were expressed per microgram of protein (luc/prot) to normalize values by protein concentration, which should reflect the amount of cells in the well, because coinfection of two adenoviral constructs reduced the efficiency of infection (data not shown). To compare experiments, the highest duplicate luc/prot for each deletion or mutation construct in each experiment was divided by the average of the peak luc/prot value of MIS−269-luc in the same experiment and termed “relative luc/prot.” To confirm our results, multiple viral preparations were tested for each construct.
The Region Between Human MIS−269 and −192 Is Necessary for MIS Promoter Activity in P2 Sertoli Cells.
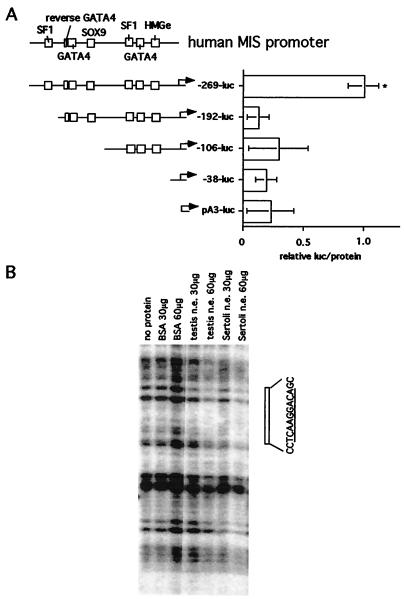
The relative luc/prot values exhibited by infection of the P2 Sertoli cell culture with the MIS−269 luciferase construct were highest compared with that of the other deletion MIS promoter constructs (Fig. 3A). Duplicate infection assays using different preparations of the same deletion adenoviral constructs reproduced these results (data not shown).
Figure 3.
(A) The region between human MIS−269 and −192 is important for human MIS transcription in the Sertoli cells from P2 rat testes. The conserved binding sites of the human MIS proximal promoter preserved in each deletion construct and the length of the human MIS promoter are shown in the schematic (Left). Relative luc/protein (see Materials and Methods) values (Right) were compared statistically by unpaired t test. Asterisk (*) indicates P-value < 0.01 compared with the −269+8 promoter. n = 8, 6, 6, 6, and 10, for pA3, MIS−38, MIS−106, MIS−192, and MIS−269, respectively. (B) DNase I footprinting analysis of the region between −269 and −192 of the human MIS promoter. The 5′ phosphorylated MIS−269 probe was incubated with 30 or 60 μg of BSA or with nuclear extract from P2 testes or Sertoli cells. As indicated, DNase I was added, and the resultant digests were electrophoresed. DNase I (0.1 units) was added in the lanes incubated without protein and 1.0 unit in the lanes incubated with nuclear extracts or BSA. The open box (Right) depicts the sequence of the footprinted region with its underlined distal SF-1- binding site.
The same MIS deletion constructs were used to infect monkey kidney-derived COS cells, which do not express MIS. Under these conditions, MIS−269 failed to enhance luciferase expression above basal pA3-luc luciferase values (data not shown), indicating that the greater expression caused by MIS−269 is specific in P2 Sertoli cells, and the region between −269 and −192 is important for regulation of MIS transcription at this developmental stage.
Sertoli Cell Nuclear Extract Protects an SF-1-Binding Site Between −269 and −192 by DNase I Footprint Assay.
To determine the sequence responsible for increased promoter activity between MIS−269 and −192, a footprinting assay was performed by using nuclear extracts from P2 rat Sertoli cells. We observed a footprint between −219 and −209 of the sequence “CTCAAGGACAG” (Fig. 3B); this site is similar to the consensus SF-1-binding site sequence “(T/C)CAAGG(T/C)C(A/G)” with a single mismatch “TCAAGGACA.” This site is conserved in the rat, mouse, bovine, porcine, and human promoter (Fig. 1). Footprinting at this distal SF-1 site was also observed by using nuclear extract from whole P2 rat testis (Fig. 3B).
The Distal SF-1 Site as Well as the Proximal Site Can Bind SF-1.
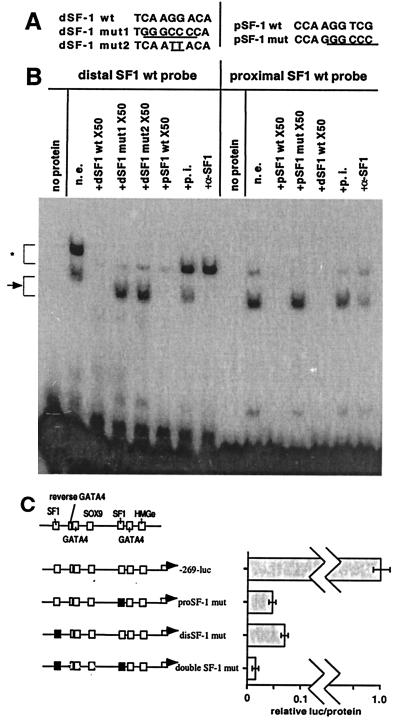
To further confirm that the footprinted sequence was able to bind SF-1 protein, EMSAs were done by using probes from both the distal and proximal SF-1 sites and nuclear extracts from the P2 Sertoli cells. Identical complexes were formed with both distal and proximal SF-1 probes (Fig. 4B) and were blocked by 50× excess unlabeled wild-type oligonucleotides of both sites but not by 50× excess mutated oligonucleotides [TC047 and 048, KW03, 04, 05, 06; see supplemental Table 1 (www.pnas.org)]. These complexes were also blocked or diminished by preincubation of the nuclear extract with an SF-1 antibody. Preincubation of nuclear extract with preimmune serum had no effect. The faint slower migrating complexes were considered nonspecific because they were competed by both excess unlabeled mutated and wild-type oligonucleotides and were not blocked by the SF-1 antibody.
Figure 4.
The distal as well as proximal SF-1 sites are necessary for MIS promoter activation. (A) Wild-type (wt) and mutated (mut) pSF-1 (proximal SF-1) or dSF-1 (distal SF-1) oligonucleotides were used in EMSAs; the mutated sequences are underlined. (B) Nuclear extract from P2 Sertoli cells was incubated with the distal or proximal SF-1 site wt probe alone (n.e.) or with (+) the designated double-stranded oligonucleotide competitor or antibody. The gel curves slightly upward on the left. The arrow indicates the SF-1 complexes, and the asterisk indicates slower migrating nonspecific bands. A normal rabbit serum was used as preimmune serum (p.i.), and α-SF-1 indicates the use of the anti-mouse SF-1 rabbit polyclonal IgG (Upstate Biotechnology). Exposure time was 4 h 30 min. (C) The black boxes indicate the mutated SF-1 site. Proximal or proSF-1, distal or disSF-1, and double SF-1 mutations are shown. The x-axis shows relative luciferase (luc)/protein (n = 5) expression.
Both SF-1 Sites Are Required for MIS−269 Luciferase Activity.
To study the functional significance of the upper SF-1 site, we constructed luciferase adenoviral vectors containing mutations in the distal SF-1 site (disSF-1 mut), proximal SF-1 site (proSF-1 mut), and both distal and proximal SF-1 sites (double SF-1 mut) of the MIS−269 promoter. Repeated infection studies showed that mutation of both SF-1 sites completely abolished MIS−269 luciferase activity, whereas mutation of either the proximal or distal SF-1 site alone exhibited very weak promoter activity that was nonetheless significantly greater than the SF-1 double mutation (P < 0.001), indicating that the distal SF-1 site, as well as the proximal SF-1 site, is necessary for functional induction of reporter expression by the MIS−269 promoter (Fig. 4C).
Proximal and Distal GATA-Binding Sites Are Necessary for Proper MIS Expression.
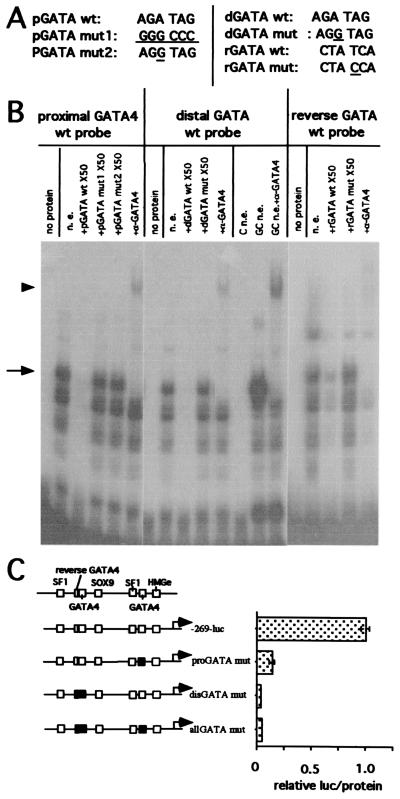
We found an upstream GATA consensus sequence located between −168 and −163 of human MIS (distal GATA), which was identical to the proximal GATA site sequence (AGATAG); we also observed a reversed GATA sequence site (TATC) just upstream of the distal GATA site (Fig. 1). To study the importance of these regions, which are conserved only in the bovine, porcine, and human MIS promoters, EMSAs were performed. The distal GATA site formed the same major protein–DNA complexes from P2 Sertoli cell nuclear extract as the proximal GATA site. The complexes with the reversed GATA site bound probe poorly, requiring a 2-day exposure for detection of complexes compared with a 6-h exposure for the proximal and distal GATA sites (Fig. 5B), hence suggesting that the contribution of the reverse GATA site is likely to be minimal.
Figure 5.
The proximal and distal GATA sites are essential for human MIS promoter activation. (A) The mutated oligonucleotide sequences of probes used in this EMSA are underlined. (B) Nuclear extract from P2 Sertoli cells (n.e.), untransfected COS cells (C n.e.), or COS cells transfected with pCMV-GATA-4 (GC n.e.) were incubated with the distal, proximal, or reverse GATA site oligonucleotide probes alone or with the specified competitor oligonucleotide or antibody. The arrow depicts the GATA-4 complexes, and the arrowhead shows the complex supershift by GATA-4 antibody (α-GATA4, goat polyclonal IgG, Santa Cruz Biotechnology). The exposure was for 5 h 45 min for the proximal and distal GATA probes and 42 h for the reverse GATA probe. (C) The mutated proximal GATA site and the combined mutations of the distal and reversed GATA sites, both alone and together, are indicated by the black boxes (Left). All mutation constructs of MIS−269 promoter (n = 4) show (Right) a decrease of luciferase (luc)/protein expression.
The complexes observed with the GATA site probes and P2 Sertoli cell nuclear extract are also seen with nuclear extract from COS cells transiently transfected with an expression construct for GATA-4, whereas nontransfected COS cell nuclear extract forms no complexes with these probes. The slower migrating complex observed with the Sertoli cell or GATA-4 expressing COS cell nuclear extract was supershifted by incubation with a GATA-4-specific antibody to the same position (Fig. 5B). Because the GATA-4 specific antibody is directed to a C-terminal sequence not conserved in other known GATA family members, these results demonstrate that P2 Sertoli cells express GATA-4 protein. Because the complexes observed with Sertoli cell nuclear extract appear identical to that observed in COS cells overexpressing GATA-4, it is likely that GATA-4 is the major or only GATA family member in the P2 Sertoli cell nuclear extracts. The faster migrating complexes that are not blocked or supershifted with the GATA-4 antibody are likely to be from GATA-4 molecules that are partially degraded and have lost the C-terminal epitope, because they are also observed by using nuclear extracts from the GATA-4 expressing COS cells but not from the untransfected COS cell nuclear extract.
When the GATA-4-binding sites were individually or multiply mutated in the MIS−269 luciferase construct, their functional importance was made clear because reporter expression was abolished (Fig. 5C). Whereas Tremblay and Viger found that GATA-4 strongly synergized with SF-1 through the zinc-finger region of GATA-4 even in the absence of a GATA-4-binding site in the mouse (21), our data suggest that in the context of the human (and possibly the bovine and porcine) MIS promoters, two sets of SF-1- and GATA-4-binding sites are required for MIS expression.
Conclusions
To understand the mechanisms that regulate MIS expression, we developed a cell culture assay using primary Sertoli cells that express MIS endogenously and therefore contain all the factors necessary for transcription.
Using an adenovirus-mediated promoter assay in primary P2 Sertoli cells, we showed the region between MIS−269 and −192 that contains a second distal SF-1 site is necessary for transcriptional activation of the human MIS promoter and that at this earlier developmental stage, human MIS−180 is not sufficient for MIS expression. In contrast, mouse MIS−180 was capable of driving MIS expression in vivo in postnatal day 15 rat Sertoli cells (17). Because MIS is expressed to varying degrees in fetal, neonatal, prepubertal, and adult testes and adult ovary, it is reasonable to assume that different mechanisms may drive MIS expression during development, between the sexes, and between different mammalian species.
Our data also indicate that the GATA site that is present in the human, porcine, and bovine promoter between the distal SF-1 site and the SOX9 site is required for expression as well, because mutation in either or both the proximal and distal GATA sites abolished MIS−269 expression. Recently targeted mutagenesis of the endogenous mouse MIS promoter in which mutations were “knocked in” the SOX9 site or the proximal SF-1 site showed that the promoter with a mutated SOX9 site was unable to initiate MIS transcription, whereas the promoter with a mutated proximal SF-1 site could express MIS but at significantly reduced levels (34). The present results could explain the failure of the proximal SF-1 site mutation to produce retained Müllerian ducts (34). Perhaps if both SF-1 sites were inactivated, one might observe a failure in Müllerian duct regression as seen with the SOX9 site mutation. It is possible that the distal SF-1 site supplemented the lost function of the proximal SF-1 site, because our data showed that the double SF-1 site mutation decreased the luciferase activity to a greater degree than did mutation of either SF-1 site alone. SOX9 expression appears to be essential for the higher levels of MIS transcriptional activation observed in fetal testis when SOX9 is maximally expressed. However, it may not be required for lower levels of MIS expression, as observed in the postnatal testis and adult granulosa cells, because SOX9 has not been observed in granulosa cells (35). Interestingly, because the SOX9-binding site is located between the distal and proximal SF-1- and GATA-4-binding sites, it is possible that the DNA bending ability of SOX9 could enhance the activation of the MIS promoter (36).
P2 Sertoli cells are capable of expressing endogenous MIS for a limited time when cultured on extracellular matrix, suggesting that interaction with other types of cells may be needed for more prolonged expression. To resolve this question, it may be necessary to perform infection of Sertoli cells in vivo or in organ culture in the context of the whole testis. Although use of the adenoviral system precludes coinfection of multiple constructs in a single experiment, it does, however, permit high infectability of a broad range of dividing and nondividing tissue that can be used to advantage to explore tissue interactions in fetal testicular and adult ovarian and testicular tissues.
Supplementary Material
Acknowledgments
We are indebted to Dr. Mason Freeman, Director of the Massachusetts General Hospital (MGH) Gene Therapy Center for invaluable advice and expertise. K.W. is funded by a fellowship from the W. Gerald Austen Fund at MGH and the GAR Foundation, T.C. is funded by a fellowship from the American Cancer Society, and P.K.D. is partially funded by Grant CA17393 from the National Institutes of Health. We thank Dr. Keith Parker, University of Texas Southwestern Medical Center, Dallas, TX, for pCMV-SF-1, Dr. Masamitsu Futai, Osaka University, Japan, for PUC18-GATA-GT2, and Dr. Chris Haqq for the promoter oligonucleotide probes. We thank Drs. Larry Jameson, Joel Habener, Pascal de Santa Barbara, Liz Perkins, and Jose Teixeira for critically reviewing the manuscript.
Abbreviations
- MIS
Müllerian inhibiting substance
- SF-1
steroidogenic factor-1, P2, postnatal day 2
- GFP
green fluorescent protein
- EMSA
electrophoretic mobility-shift assay
- CMV
cytomegalovirus
- OPU
optical particle unit
References
- 1.Lee M M, Donahoe P K. Endocrine Rev. 1993;14:152–164. doi: 10.1210/edrv-14-2-152. [DOI] [PubMed] [Google Scholar]
- 2.Donahoe P K, Ito Y, Marfatia S, Hendren W H., III Biol Reprod. 1976;15:329–334. doi: 10.1095/biolreprod15.3.329. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi M, Hayashi M, Manganaro T F, Donahoe P K. Biol Reprod. 1986;35:447–453. doi: 10.1095/biolreprod35.2.447. [DOI] [PubMed] [Google Scholar]
- 4.Hirobe S, He W-W, Lee M M, Donahoe P K. Endocrinology. 1992;131:854–862. doi: 10.1210/endo.131.2.1639028. [DOI] [PubMed] [Google Scholar]
- 5.Lee M M, Cate R L, Donahoe P K, Waneck G L. Endocrinology. 1992;130:847–853. doi: 10.1210/endo.130.2.1346380. [DOI] [PubMed] [Google Scholar]
- 6.Münsterberg A, Lovell-Badge R. Development (Cambridge, UK) 1991;113:613–624. doi: 10.1242/dev.113.2.613. [DOI] [PubMed] [Google Scholar]
- 7.Haqq C, Lee M M, Tizard R, Wysk M, DeMarinis J, Donahoe P K, Cate R L. Genomics. 1992;12:665–669. doi: 10.1016/0888-7543(92)90291-y. [DOI] [PubMed] [Google Scholar]
- 8.Cate R L, Mattaliano R J, Hession C, Tizard R, Farber N M, Cheung A, Ninfa E G, Frey A Z, Gash D J, Chow E P, et al. Cell. 1986;45:685–698. doi: 10.1016/0092-8674(86)90783-x. [DOI] [PubMed] [Google Scholar]
- 9.Pilon N, Behdjani R, Daneau I, Lussier J G, Silversides D W. Endocrinology. 1998;139:3803–3812. doi: 10.1210/endo.139.9.6193. [DOI] [PubMed] [Google Scholar]
- 10.Eusèbe D C, Clemente N d, Rey R, Pieau C, Vigier B, Josso N, Picard J-Y. J Biol Chem. 1996;271:4798–4804. doi: 10.1074/jbc.271.9.4798. [DOI] [PubMed] [Google Scholar]
- 11.Guerrier D, Boussin L, Mader S, Josso N, Kahn A, Picard J-Y. J Reprod Fertil. 1990;88:695–706. doi: 10.1530/jrf.0.0880695. [DOI] [PubMed] [Google Scholar]
- 12.Dresser D W, Hacker A, Lovell-Badge R, Guerrier D. Hum Mol Genet. 1995;4:1613–1618. doi: 10.1093/hmg/4.9.1613. [DOI] [PubMed] [Google Scholar]
- 13.Haqq C M, King C-Y, Ukiyama E, Falsafi S, Haqq T N, Donahoe P K, Weiss M A. Science. 1994;266:1494–1500. doi: 10.1126/science.7985018. [DOI] [PubMed] [Google Scholar]
- 14.da Silva S M, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R. Nat Genet. 1996;14:62–68. doi: 10.1038/ng0996-62. [DOI] [PubMed] [Google Scholar]
- 15.de Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, Scherer G, Poulat F, Berta P. Mol Cell Biol. 1998;18:6653–6665. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen W-H, Moore C C D, Ikeda Y, Parker K L, Ingraham H A. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 17.Giuili G, Shen W H, Ingraham H A. Development (Cambridge, UK) 1997;124:1799–1807. doi: 10.1242/dev.124.9.1799. [DOI] [PubMed] [Google Scholar]
- 18.Shimamura R, Fraizer G C, Trapman J, Lau Y-F C, Saunders G F. Clin Cancer Res. 1997;3:2571–2580. [PubMed] [Google Scholar]
- 19.Nachtigal M W, Hirokawa Y, Enyeart-VanHouten D l, Flanagan J N, Hammer G D, Ingraham H A. Cell. 1998;93:445–454. doi: 10.1016/s0092-8674(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 20.Viger R S, Mertineit C, Trasler J M, Nemer M. Development (Cambridge, UK) 1998;125:2665–2675. doi: 10.1242/dev.125.14.2665. [DOI] [PubMed] [Google Scholar]
- 21.Tremblay J J, Viger R S. Mol Endocrinol. 1999;13:1388–1401. doi: 10.1210/mend.13.8.0330. [DOI] [PubMed] [Google Scholar]
- 22.Gramham F L, Prevec L. In: Methods in Molecular Biology. Murray E J, editor. Clifton, NJ: Humana; 1991. [Google Scholar]
- 23.Arambepola N K, Bunick D, Cooke P S. Endocrinology. 1998;139:4489–4495. doi: 10.1210/endo.139.11.6315. [DOI] [PubMed] [Google Scholar]
- 24.LaQuaglia, M., Shima, H., Hudson, P., Takahashi, M. & Donahoe, P. K. (1986) J. Urol.136. [DOI] [PubMed]
- 25.Chapin R E, Phelps J L, Miller B E, Gray T J B. J Androl. 1987;8:155–161. doi: 10.1002/j.1939-4640.1987.tb02427.x. [DOI] [PubMed] [Google Scholar]
- 26.Steinberger E, Steinberger A, Vilar O. Endcrinology. 1966;79:406–410. doi: 10.1210/endo-79-2-406. [DOI] [PubMed] [Google Scholar]
- 27.Tamura S, Wang X-H, Maeda M, Futai M. Proc Natl Acad Sci USA. 1993;90:10876–10880. doi: 10.1073/pnas.90.22.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda Y, Lala D S, Luo X, Kim E, Moisan M-P, Parker K. Mol Endocrinol. 1993;7:852–860. doi: 10.1210/mend.7.7.8413309. [DOI] [PubMed] [Google Scholar]
- 29.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 30.Dignam J D, Lebovitz R M, Roeder R. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teixeira J, Kehas D J, Antun R, Donahoe P K. Proc Natl Acad Sci USA. 1999;96:13831–13838. doi: 10.1073/pnas.96.24.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robert J-J, Geoffroy M-C, Finiels F, Mallet J. J Neurochem. 1997;68:2152–2160. doi: 10.1046/j.1471-4159.1997.68052152.x. [DOI] [PubMed] [Google Scholar]
- 33.Blanchard K T, Boekelheide K. Biol Reprod. 1997;56:495–500. doi: 10.1095/biolreprod56.2.495. [DOI] [PubMed] [Google Scholar]
- 34.Arango N A, Lovell-Badge R, Behringer R R. Cell. 1999;99:409–419. doi: 10.1016/s0092-8674(00)81527-5. [DOI] [PubMed] [Google Scholar]
- 35.Heikinheimo m, Ermolaeva M, Bielinska M, Rahman N A, Narita N, Huhtaniemi I T, Tapanainen J S, Wilson D B. Endocrinology. 1997;138:3505–3514. doi: 10.1210/endo.138.8.5350. [DOI] [PubMed] [Google Scholar]
- 36.McDowall S, Argentaro A, Ranganathan S, Weller P, Mertin S, Mansour S, Tolmie J, Harley V. J Biol Chem. 1999;274:24023–30. doi: 10.1074/jbc.274.34.24023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.