Abstract
Using Cre-loxP-mediated recombination, we established a highly efficient and reproducible system that generates autonomous HPV-18 genomes in primary human keratinocytes (PHKs), the organotypic raft cultures of which recapitulated a robust productive program. While E7 promoted S-phase re-entry in numerous suprabasal differentiated cells, HPV DNA unexpectedly amplified following a prolonged G2 arrest in mid- and upper spinous cells. As viral DNA levels intensified, E7 activity diminished and then extinguished. These cells then exited the cell cycle to undergo virion morphogenesis. High titers of progeny virus generated an indistinguishable productive infection in naïve PHK raft cultures as before, never before achieved until now. An immortalization-defective HPV-18 E6 mutant genome was also characterized for the first time. Numerous cells accumulated p53 protein, without inducing apoptosis, but the productive program was severely curtailed. Complementation of mutant genomes by E6-expressing retrovirus restored proper degradation of p53 as well as viral DNA amplification and L1 production. This system will be invaluable for HPV genetic dissection and serves as a faithful ex vivo model for investigating infections and interventions.
Keywords: Human papillomavirus infection program, organotypic cultures of primary human keratinocytes, Cre-loxP-mediated recombination in vivo, cell cycle regulation, immortalization- and replication-defective E6 mutant, p53 protein stabilization
HPVs are nonenveloped DNA viruses with a circular genome of ∼7.9 Kb that replicate as multicopy nuclear plasmids. Various HPV genotypes infect mucosal or cutaneous squamous epithelia, where they may cause benign hyperproliferation. A small fraction of infections by the mucosotropic high-risk (HR) HPVs (e.g., HPV-16, HPV-18, and related genotypes) can undergo neoplastic progression to high grade intraepithelial lesions and cancers, specifically cervical, penile, anal, and oropharyngeal carcinomas (for a review, see zur Hausen 2002). In contrast, infections by low-risk (LR) types (e.g., HPV-6 and HPV-11) rarely promote carcinogenesis.
Wounding of epithelium is necessary to establish infections in basal keratinocytes, but viral DNA amplification only occurs in mid- and upper spinous cells undergoing terminal squamous differentiation (Stoler and Broker 1986). Major and minor capsid proteins, L1 and L2, express in the superficial strata of the epithelia where progeny virions assemble and eventually shed as cornified envelopes desquamate (for review, see Chow and Broker 2007). HPV DNA replication depends on cellular replication machinery and substrates to complement the viral origin-binding protein E2 and the replicative helicase E1 (for review, see Chow and Broker 2006). Because spinous cells have withdrawn from the cell cycle, HPV E7 protein destabilizes p130 pocket protein, thereby disrupting the homeostasis of the squamous epithelium and promoting S-phase re-entry (Zhang et al. 2006; Genovese et al. 2008 and references therein). HPV E6 protein blocks p53 transactivating functions (Thomas and Chiang 2005), though its significance in productive infections remains unknown; E6 alone is neither necessary nor sufficient to induce S-phase re-entry in spinous cells (Cheng et al. 1995). In addition, HR HPV E7 and E6 can destabilize pRB and p53, respectively (for reviews, see Münger and Howley 2002; zur Hausen 2002). Thus their inappropriate overexpression in cycling basal cells can initiate neoplastic progression and, in vitro, immortalize primary human keratinocytes (PHKs).
Organotypic cultures of PHKs or selected epithelial cell lines have been a core format for HPV investigations. Briefly, keratinocytes stratify and differentiate into a squamous epithelium in 10 or more days when cultured on a dermal equivalent, consisting of collagen with embedded fibroblasts, and held at the medium:air interface. This “raft” culture system can support the viral productive program (Dollard et al. 1992; Meyers et al. 1992). Nonetheless, current methods for HPV genetic analyses in such cultures have severe limitations. Typically, linear or recircularized viral DNA, excised from a recombinant plasmid, is cotransfected into PHKs along with another plasmid encoding a selectable marker (for reviews, see McLaughlin-Drubin and Meyers 2005; Wilson and Laimins 2005). Since transfection of nonsupercoiled DNA is inefficient and PHKs senesce after only a few passages, selection and expansion of drug-resistant colonies depend on HR HPV oncogenes to immortalize PHKs. One alternative has been to use certain immortalized epithelial cell lines as recipient cells (Fang et al. 2006b; for review, see Lambert et al. 2005). In either case, only a small number of spinous cells in raft cultures of immortalized cells support viral DNA amplification and progeny virus production. Critically, there is no report that these low yields of virus can initiate a de novo productive infection in PHK raft cultures. Rather, infectious virus is demonstrated by detecting spliced early viral transcripts. Cell lines are typically used in these assays for their elevated susceptibility to infection over PHKs (Ozbun 2002). This inefficient productive program could explain the inconsistent results for E5 and E4 mutants regarding their roles in viral DNA amplification. Moreover, immortalization-defective E6 mutants cannot be studied, as the mutant plasmids were rapidly lost in transfected cells (Thomas et al. 1999; Oh et al. 2004). Thus, there is a critical need for an efficient strategy to obviate the need for immortalized cells and to conduct analyses in raft cultures of PHKs.
In this study, we developed just such a system. It is highly efficient, yet simple and reproducible. Importantly, high titers of progeny virions elicited, for the first time, equally productive infections in fresh PHK raft cultures. The productive program revealed novel virus–host interactions. The significance of this new methodology is highlighted by the successful characterization of an immortalization-defective E6 mutant in PHK raft cultures. This new system would be invaluable for definitive genetic analyses of HPVs and for investigating infection processes and validating potential antiviral agents for intervention.
Results
Excision and amplification of the HPV-18 genomic plasmids in PHKs
Upon PHKs transfection with pNeo-loxP HPV-18 parental vector alone or together with pCAGGS-nlsCre, >30% of the cells typically survived the acute G418 selection. Cre-mediated recombination should generate a circular 7.9-Kb HPV-18 genome and a pNeo vector, each now with a single loxP site (Fig. 1A). This 34-base-pair (bp) loxP sequence is placed in the upstream regulatory region (URR) between nucleotides 7473 and 7474 of HPV-18, a site downstream from the late poly-A to which no host or viral transcription or other regulatory factors are known to bind. For simplicity, excised HPV-18 DNA is hereafter referred to as wild type because, by all criteria, the patterns of its early and late gene expression are identical to genuine HPVs in productively infected patient specimens (for review, see Doorbar 2006). Most important, the virions generated were infectious in naïve PHK raft cultures, recapitulating the same productive program.
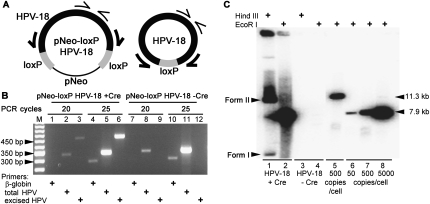
Figure 1.
HPV DNA excision and amplification. (A) Schematic diagrams depicting the parental pNeo-loxP HPV-18 and Cre-excised HPV-18 plasmids and PCR primers (thin and thick arrows) to detect total and excised HPV DNA. (B) Ethidium bromide-stained agarose gel of PCR amplification products after 20 and 25 cycles, revealing total HPV DNA (332 bp), excised HPV DNA (450 bp), or β-globin gene (275 bp) from submerged PHK cultures after transfection with pNeo-loxP HPV-18 plasmid with (lanes 1–6) or without (lanes 7–12) the nls-Cre expression plasmid. (M) DNA length markers. (C) Southern blot hybridization for HPV genomic plasmid in day-12 raft cultures. Total DNA from raft cultures of PHKs transfected with pNeo-loxP HPV-18 in the presence (lanes 1,2) or absence (lanes 3,4) of the nls-Cre expression plasmid. Length and copy number standards for EcoR I-linearized parental plasmid (lane 5) or HPV-18 genomic DNA (lanes 6–8).
Total DNA isolated from G418-resistant PHKs was analyzed using PCR and primer sets specific for total HPV-18 DNA, excised viral DNA (Fig. 1A), or β-globin gene as a positive control. In the absence of nls-Cre, only the total HPV DNA band was observed (Fig. 1B, lanes 7–12). In the presence of nls-Cre, the similar intensities of the total and the excised HPV-18 bands demonstrated efficient excision recombination (Fig. 1B, lanes 1–6). The excision-specific DNA band was confirmed by direct DNA sequence (data not shown). G418-resistant PHKs were also immediately developed into raft cultures. Total DNA from 12-d raft cultures was subjected to Southern blot analysis following digestion with a no-cut (Hind III) or one-cut (EcoR I) restriction enzyme (Fig. 1C). The 11.3- Kb parental plasmid was below detection in the presence or absence of nls-Cre (Fig. 1C, cf. lanes 2–4 and lane 5). In contrast, in the presence of nls-Cre, an HPV-18 plasmid of 7.9-Kb length amplified to ∼2000–5000 copies per diploid chromosome equivalent in different experiments (Fig. 1C, cf. lanes 1,2 and lanes 6–8; data not shown). These results demonstrate that excision from the parental plasmid is prerequisite to HPV-18 DNA amplification in the squamous epithelium. The productivity per cell is an approximation. Although many of the cells have gone through S phase and could very well be tetraploid, viral DNA amplified only in a fraction of the cells (see Fig. 2). On the other hand, the amount of host DNA remaining in the residual nuclei in the stratum corneum (see Fig. 2) is not known.
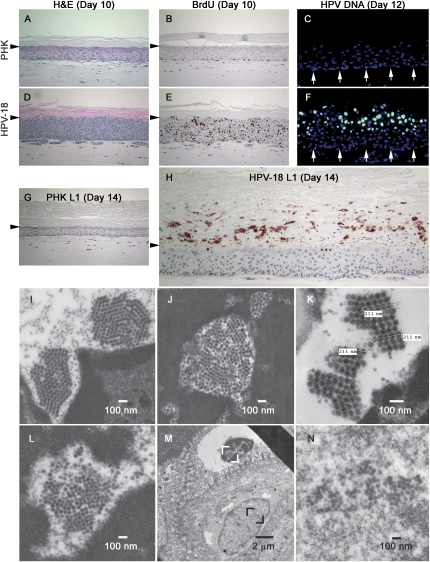
Figure 2.
Productive HPV-18-containing PHK raft cultures and visualization of HPV virions by TEM. (A–H) Four-micron sections of control (A–C,G) or HPV-18-containing PHK raft cultures (D–F,H) analyzed. Day-10 culture sections were stained with H&E (A,D) or probed for BrdU incorporation (reddish brown, B,E) after a 12-h exposure immediately prior to harvest. (C,F) DNA-FISH to reveal HPV-18 DNA (green) in a day-12 culture sections. Arrows point to the basal stratum. (G,H) IHC to detect the major capsid protein L1 (reddish brown) in a day-14 culture sections. Horizontal arrowheads point to the boundary between the upper cornified layer and live epithelium below. Under the epithelium is the collagen matrix. (I–N) TEM of ultrathin sections of day-14 cultures to visualize virions. (I,J) Paracrystalline arrays of mature virions in degenerated nuclei with condensed chromatins. (K) Three measurements, each across five virions in a closely packed ordered array, gave the same value of 211 nm, or 42.2 nm/virion diameter. (M) Two adjacent cells shown in low magnification. (L) The enlarged area (white brackets) in the upper cornified envelope with a degenerated nucleus. (N) The enlarged area (black brackets) in the lower, living cells with a normal nucleus.
Recapitulation of the HPV-18 productive program in raft cultures
Hematoxylin and eosin (H&E) staining showed that day-10 HPV-18 plasmid-containing raft cultures were mildly hyperproliferative when compared with untransfected PHK raft cultures (Fig. 2A,D). When probed with an antibody to bromodeoxyuridine (BrdU), positive cells were distributed stochastically in all cell strata of the HPV-18 raft cultures (Fig. 2E), similar to patient specimens and raft cultures expressing E7 gene from a proretrovirus (Cheng et al. 1995; Genovese et al. 2008 and references therein). In contrast, only basal keratinocytes in normal PHK raft cultures were BrdU-positive (Fig. 2B). DNA-fluorescence in situ hybridization (DNA-FISH) of day-12 cultures revealed intense HPV-18 DNA nuclear signals in a high percentage of mid- to upper spinous cells, with occasional punctate signals observed in basal and parabasal cells (Fig. 2F). This abrupt increase in viral DNA is identical to the pattern consistently observed in naturally occurring HPV lesions (Stoler and Broker 1986). By day 14, abundant L1 antigen was readily detected in numerous cornified envelopes and in some superficial live cells across the entire length of raft cultures, verifying a highly productive program (Fig. 2H). Cultures of control PHKs or PHKs transfected with pNeo-loxP HPV-18 alone remained negative for L1 and viral DNA signals (Fig. 2C,G; Supplemental Fig. S1C; data not shown). Multiple independent time course experiments showed that low L1 signals typically appeared on day 10 in a small number of superficial cells. Signals dramatically increased in the stratum corneum on day 12, and then leveled off by day 14 or 16, after which virtually all signals were restricted to cornified envelopes (Supplemental Fig. S1). This shutoff in late gene expression in superficial live cells coincided with diminished host DNA replication (see Fig. 4A, below; data not shown) and thinning of the raft cultures (Supplemental Fig. S1). Raft cultures of immortalized cells with HPV-31b genomes also had reduced late mRNA levels after day 14 (Ozbun and Meyers 1997).
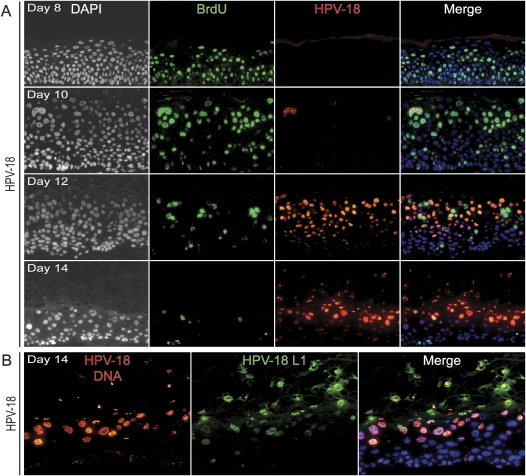
Figure 4.
(A) HPV-18 DNA amplification in differentiated keratinocytes lags behind cellular DNA replication. HPV-18-containing PHK raft cultures were harvested on days 8, 10, 12, and 14, each following a 12-h incubation with BrdU. Sections were probed for HPV-18 DNA (red) and incorporated BrdU (green). Cellular DNA was revealed by DAPI (blue). (Left column) For better visualization of tissue morphology, DAPI staining is also presented in black-and-white images. (B) Detection of viral DNA (red) and the major capsid protein L1 (green) in a 14-d-old culture.
Visualization of HPV-18 virions in organotypic cultures
Examination of day-14 raft culture sections by transmission electron microscopy (TEM) revealed numerous viral particles in the cornified envelopes, each with a degenerated, shrunken nucleus containing electron-opaque, presumably highly condensed chromatins (Fig. 2I,J). The persistence of residual nuclei in cornified envelopes (termed parakeratosis) is one the hallmarks of productive HPV infections. Viral particles were electron dense and icosahedral in shape and often formed paracrystalline arrays. Measurements with an internal scale of the TEM approximated the diameter of virions to be 42.2 nm (Fig. 2K), consistent with previous reports that used similar TEM techniques (Dollard et al. 1992 and references therein). Purified HPV virions are often referred to be 55 nm in diameter. The smaller size observed in tissue sections might be attributable to extensive dehydration procedures during sample preparation or to positive staining as opposed to the negative staining of purified virions.
We also observed what appeared to be slightly larger empty or immature particles, as they were of lower electron density or less distinct in overall shape and capsomere patterning. Paracrystalline arrays of mature virions were invariably disrupted by the inclusion of such immature or empty particles (Fig. 2I,J,L). Interestingly, live superficial cells rarely contained highly ordered arrays of mature virions. For instance, the more superficial of two adjacent cells in Figure 2M had a degenerated nucleus, in which paracrystalline arrays of mature virions had begun to form (Fig. 2L), while a live cell immediately below, with a normal-appearing nucleus, contained probably “immature” virions that were not yet organized into large ordered arrays (Fig. 2N).
Productive infections of PHK raft cultures by HPV-18 virus
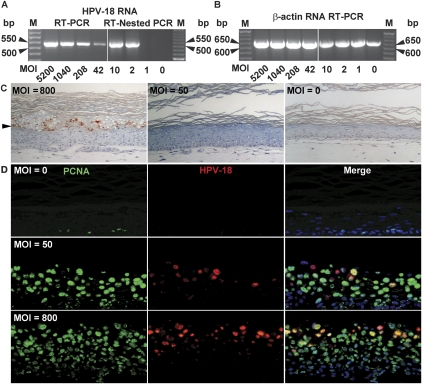
The titers of two independent virus preparations were determined by quantitative real-time PCR to be 3.2 × 108 and 4.9 × 108 per raft culture, each initiated from 2 × 105 to 2.5 × 105 PHKs. Virus infectivity was examined in submerged PHK cultures by detecting a spliced E6–E7–E1^E4 cDNA fragment using RT–PCR. The 521-bp cDNA was easily visualized in ethidium bromide-stained agarose gels at an multiplicity of infection (MOI) of 42 and as low as 5 in other experiments (our unpublished results). The band intensity increased with the MOI and was absent in uninfected PHKs (MOI = 0) (Fig. 3A, left). The identity of the cDNA was verified by direct DNA sequencing (data not shown). RT-nested PCR detected the same cDNA fragment at an MOI of 2 (Fig. 3A, right). In contrast, β-actin cDNA was detected in all samples (Fig. 3B). As a further test, infected PHKs were developed into raft cultures and then harvested on day 14. At MOI of 800 or higher, cultures contained widespread L1 antigen-positive cornified envelopes (Fig. 3C, left panel; data not shown). Unexpectedly, at MOIs of 400 and lower, the cultures had little or no L1 antigen (Fig. 3C, middle panel; data not shown).
Figure 3.
Infectivity assays of HPV-18 virions in PHKs. HPV virions were titered by quantitative real-time PCR. (A) HPV cDNA detection in submerged PHK cultures after infection at the indicated MOI. A 521-bp cDNA fragment of a spliced early viral mRNA was detected by ethidium bromide staining after RT–PCR (left panel) or RT-nested PCR (right panel). (B) A 642-bp-long β-actin cDNA served as an internal control. (M) 50-bp ladder. (C) Immunohistochemical detection of the HPV major capsid protein L1 (reddish brown) in 14-d raft cultures of PHKs infected with HPV virions.(Left panel) MOI of 800 or higher. (Middle panel) MOI of 50 up to 400. An uninfected culture is shown in the right panel. Arrowheads point to the boundary between the upper cornified strata and live epithelium below. (D) Four-micron sections from the raft cultures above were probed for PCNA (Alexa Fluor 488, green) and for viral DNA (Cy3, red). Cellular DNA was revealed by DAPI (blue).
Interestingly, regardless of L1 productivity, all infected cultures were mildly hyperplastic relative to uninfected ones (Fig. 3C, right panel), indicative of successful infection and early viral gene expression. To verify this interpretation, sections were probed for viral DNA and PCNA, a DNA polymerase δ processivity factor induced in suprabasal cells by HPV E7 (Cheng et al. 1995). In control PHK cultures, PCNA was only detected in basal cells (Fig. 3D). In contrast, in all infected cultures, down to an MOI of 50, the lowest MOI tested, most differentiated cells were positive for PCNA, indicative of successful infection in the majority if not all the cells. Further, occasional pockets of superficial cells in these nonproductive cultures had low levels of amplified viral DNA (Fig. 3D; data not shown). Accordingly, we suggest that HPV DNA must amplify to a high copy number for efficient late protein expression, contrary to a previous report (Spink and Laimins 2005). Rarely we detected faint L1 antigen in cells not also having high viral DNA. Interestingly, intense viral DNA signals detected at productive (high) MOIs were restricted to the differentiated strata, as in cultures initiated by DNA cotransfection. This latter observation indicates either that a majority of virions failed to enter cells under our conditions or that the preponderance of internalized viral DNA was lost.
Temporal order of host DNA replication and HPV-18 DNA amplification
Intriguingly, in productive raft cultures initiated by DNA transfection, the stochastic distribution of BrdU incorporation in cells of all strata did not reflect the pattern of amplified viral DNA in the more differentiated strata (Fig. 2E,F). Simultaneous visualization of BrdU and viral DNA revealed that the majority of cells intensely positive for HPV-18 DNA were negative for strong BrdU and vice versa. Only a very small fraction of cells had colocalized signals, regardless of the order in probe application or an additional denaturation step (data not shown). These results were inconsistent with the expectation that viral DNA amplifies concurrently with host DNA replication.
To investigate this dichotomy, HPV-18-containing PHK raft cultures were pulse-labeled with BrdU for the final 12 h and harvested on days 8, 10, 12, and 14 and analyzed as before (Fig. 4A). A large fraction of nuclei on days 8 and 10 were intensely positive for BrdU. There was virtually no detectable HPV DNA on day 8. A low level of amplified viral DNA was typically detected in a small percentage of superficial cells on day 10. On day 12, BrdU-positive spinous cells decreased while those with intense HPV DNA signals markedly increased. By day 14, few spinous cells incorporated BrdU, while the majority of mid- and upper spinous cells were strongly positive for viral DNA. These inverse patterns of host DNA replication and viral DNA amplification were reproducible in multiple experiments, with strong viral DNA signals appearing on day 12 ± 2. The fraction of spinous cells with high viral DNA significantly declined in older cultures, as did spinous cells in S phase (data not shown). Regardless of culture age, there was little colocalization of intense BrdU and strong HPV DNA signals. Occasionally, distinct HPV DNA foci were seen in spinous cells with intense BrdU signals, suggestive of initiation of viral DNA amplification. However, these cells were not in S phase, as they were negative for cyclin A (see Fig. 5). Thus, BrdU signals represent host DNA replication. As HPV DNA only amounts to ∼1% of host DNA by mass even at 5000 copies per cell, the amount of BrdU incorporated into HPV DNA is clearly not sufficient to generate strong signals.
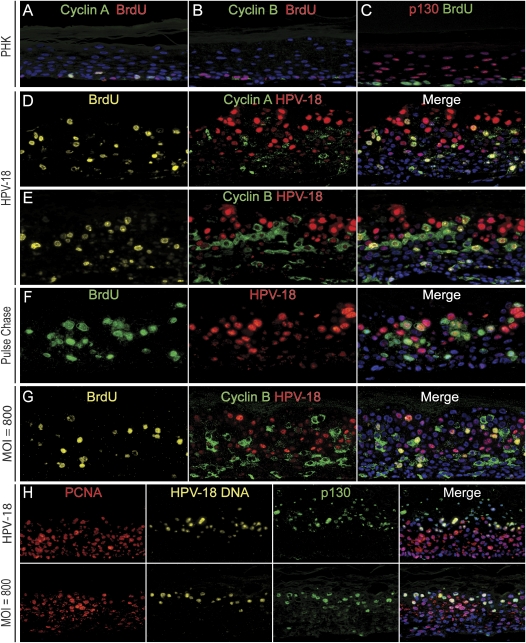
Figure 5.
HPV-18 plasmid DNA amplification as it relates to the cell cycle. (A–C) Day-10 raft cultures of normal PHK raft cultures. (D–F) Day-12 raft cultures initiated from PHKs transfected with HPV-18 DNA. Sections were analyzed for cyclin A (green in A,D) or cyclin B (green, B,E), HPV-DNA FISH (red, D–F), and for BrdU (red in A,B, green in C, or yellow Alexa Fluor 647 in D,E). (F) An HPV-18-containing raft culture was pulsed with BrdU for 6 h on day 10 and then chased for 48 h prior to harvest. (G) Day 14 of PHK raft cultures infected with HPV-18 virus at MOI of 800. The section was probed for viral DNA (red), cyclin B1 (green), and BrdU (gold). (H) Patterns of PCNA (red), HPV-18 DNA (gold), and p130 (green) in day-14 raft cultures of PHKs initiated after HPV-18 DNA transfection (top row) or HPV-18 virus infection at MOI of 800 (bottom row). Cellular DNA was revealed by DAPI (blue) in all panels.
Punctate dots or streaks of viral DNA signals were observed in the stratum corneum, consistent with DNA packaged in virions as observed in Figure 3. Indeed, some of the L1 signals colocalized with viral DNA (Fig. 4B). One of the factors attributed to the incomplete colocalization of these two signals is the difficulty in detecting packaged viral DNA and possibly a significant loss of virions during DNA denaturation for in situ hybridation. The latter is evident when one compares the reduction in DNA signals in the cornified strata relative to those in the live cells. Similarly, the L1 signals revealed in this double-immunofluorescence (IF) image were much reduced relative to those detected by immunohistochemistry (IHC) without the denaturation step (Fig. 2H; Supplemental Fig. S1).
HPV-18 genome amplification occurs in G2-arrested cells
To verify and elaborate upon the above conclusions, we probed the raft cultures for the S-phase cyclin A, amplified viral DNA, and BrdU incorporation. Control PHK raft cultures demonstrated a few basal cells positive for both cyclin A and BrdU (Fig. 5A). Cyclin A was induced in many suprabasal cells in HPV-18 raft cultures in early times, but the fraction of cyclin A-positive cells progressively decreased by days 12 and 14 (Fig. 5D; Supplemental Fig. S2), as expected from the reduction in BrdU-positive cells as the cultures aged (Fig. 4A). At all time points, many nuclei positive for cyclin A were also positive for BrdU, indicative of cells well into S phase. Cells positive for nuclear cyclin A but negative for BrdU could represent very early S phase prior to extensive cellular DNA replication. Another small subset of cells was positive for BrdU but contained cytoplasmic cyclin A or no cyclin A. These cells may have been at the S/G2 transition or in G2. Most importantly, the great majority of cells with amplified HPV DNA were distinct from cells positive for both cyclin A and BrdU. These observations demonstrate that viral DNA amplification did not occur concurrently with host DNA replication in S phase.
If not in S phase, did HPV DNA amplify in G2? We next probed for the mitosis promoting factor (MPF) component cyclin B1, amplified viral DNA, and BrdU incorporation. Nuclear import of the cyclin B1/cdk1 complex is essential for the conclusion of G2 and initiation of mitosis. In control PHK raft cultures, weak cyclin B1 signals were detected in occasional basal cells positive for BrdU (Fig. 5B). In day-8 HPV-18 raft cultures, cytoplasmic cyclin B1 was detected in a small fraction of spinous cells, and there was a modest increase in day-10 cultures. The signals were typically observed in intense BrdU-positive cells (Supplemental Fig. S3). On day 12, there was a more dramatic increase in the signal strength and in the number of cytoplasmic cyclin B1-positive spinous cells (Fig. 5E; Supplemental Fig. S3). This increase coincided with the reduction in cyclin A- and BrdU-positive cells. On day 14, when amplified or packaged HPV DNA filled most of the mid- to upper differentiated nuclei and extended into the stratum corneum, only a few cyclin B1-positive cells were observed in the lower spinous cells. Critically, it was in the nuclei of many of the cyclin B1-positive midspinous cells that low to moderate HPV DNA signals first appeared (Fig. 5E; Supplemental Fig. S3).
To investigate further the relationship between BrdU-positive cells and those with amplified viral DNA, we probed raft cultures that had been pulsed with BrdU for 6 h on day 10 and harvested on day 12. There was an increased population of cells in which intense viral DNA colocalized with strong BrdU signals (Fig. 5F). Collectively, these observations demonstrate that HPV DNA amplification initiated after cellular DNA replication in cells that are subsequently arrested in G2.
When these experiments were conducted on day-14 raft cultures of PHKs productively infected with HPV-18 virions at MOI of 800, the same relative distributions of cyclin B1, amplified viral DNA, and BrdU were observed (Fig. 5G). These results verify that the delayed viral DNA amplification relative to S phase and the loss of cyclin B1 subsequent to viral DNA amplification are not an artifact of raft cultures containing transfected HPV-18 DNA.
Cessation of HPV-18 E7 activity in cells with abundant viral DNA
Intriguingly, spinous cells with high HPV DNA were negative for cyclin B1 signals in cultures of any age (Fig. 5E,G; Supplemental Fig. S3). We considered a number of possible explanations: (1) Cyclin B1 was degraded as cells transitioned through mitosis. This is unlikely because the enlarged nuclei in these cells are consistent with a greater than 2n DNA content (Chien et al. 2002). (2) Cyclin B was degraded to allow cellular DNA rereplication. This is also unlikely as cells with high viral DNA were negative for either nuclear cyclin A or intense BrdU (Fig. 5D; Supplemental Fig. S2). (3) Cells exit the cell cycle following viral DNA amplification due to the loss of E7 activity, resulting in cyclin B1 disappearance. Evidence is consistent with this last interpretation, as described below.
Our laboratory has recently shown that p130, not pRB, is readily detected in differentiated cells of normal squamous epithelia from diverse body sites and of PHK raft cultures. Expression of HR or LR HPV E7 proteins destabilizes p130 to enable S-phase re-entry by spinous cells (Genovese et al. 2008). Thus, the loss of p130 is an excellent indicator of E7 activity. As before, p130 was detected in the differentiated strata of normal PHK raft cultures, while BrdU signals were observed in cycling basal cells (Fig. 5C). The cells in the lower strata of HPV-18 DNA transfected raft cultures with no amplified viral DNA were positive for PCNA but lacked p130, indicative of functional E7 (Fig. 5H, top row). In contrast, moderate to high p130 signals were observed in cells with moderate to high HPV DNA signals. In addition, cells with high p130 were negative for PCNA. Both these latter observations are consistent with a reduction and eventual loss in E7 activity upon viral DNA amplification. This extinction of E7 activity explains why cells with amplified viral DNA do not re-enter additional rounds of S phase and thus cease to incorporate BrdU or to express cyclin A and cyclin B1. Similarly, this loss of PCNA and reappearance of strong p130 signals was also seen in cells with high viral DNA in PHK raft cultures productively infected with HPV-18 virus (Fig. 5H, bottom row).
An essential role of E6 in viral DNA amplification and virion production
To demonstrate that our experimental system indeed obviates a dependence on HPV E6 oncoprotein for long-term viral plasmid maintenance and PHK immortalization while permitting analyses in PHK raft cultures, we prepared an HPV-18 mutant in which the E6 gene (which spans nucleotides 105–579) was deleted from the E6*I intron coding sequence (nucleotides 234–415). The great majority of HR HPV E6 transcripts are alternatively spliced intragenically or intergenically, abrogating the ability to encode the E6 protein. The predominant HPV-18 E6*I mRNA contains a frameshift that terminates translation shortly after the splice. The resulting E6*I peptide has been reported to bind to and antagonize the E6 protein, preventing it from functioning as a ubiquitin ligase in conjunction with E6AP to degrade p53 and other E6-targeted host proteins (Pim and Banks 1999). Due to their low abundance, E6 and E6* transcripts and their encoded peptides have not been localized within the stratified epithelium in naturally infected patient specimens.
We harvested raft cultures containing either E6*I mutant genome or the wild-type HPV-18 genome over a time course extending to 22 d. The wild-type HPV-18 cultures were highly productive as before (Supplemental Fig. S1B) whereas the mutant HPV-18 cultures were virtually negative for L1 (Fig. 6A, left panel). The histology of the mutant-containing cultures was in between that of the control PHK raft cultures and those containing the HPV-18 plasmid, except in older cultures, when the epithelia were thin and similar to the control cultures. To examine whether the cells have lost the HPV-18 mutant DNA, we probed for PCNA by IHC. A great majority of the superbasal cells were positive. Relative to cultures containing the wild-type genome, a few pockets in the epithelium were negative for PCNA (Fig. 7, left panels). Thus the majority of the cells still harbored the mutant plasmid DNA. However, BrdU-positive cells were reduced relative to raft cultures harboring the wild-type HPV-18 (data not shown). Indeed, most of the spinous cells did not have the enlarged nuclei typical of tetraploid cells.
Figure 6.
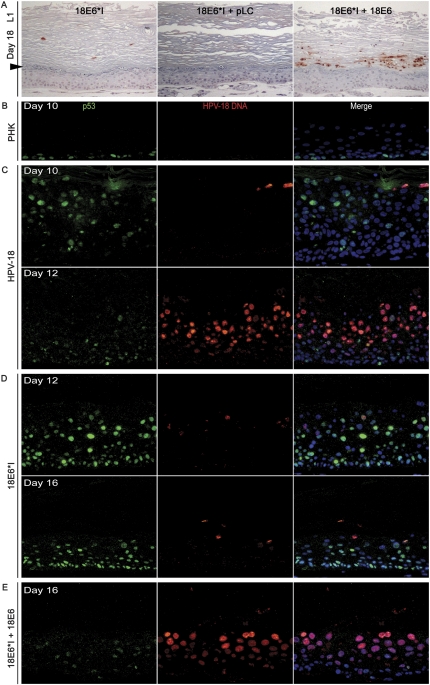
Complementation of HPV-18 E6*I genome by a retrovirus delivering HPV-18 URR-E6. Raft cultures were harvested over a time course. (A) IHC to detect L1 (reddish brown) on day-18 raft cultures. (Left panel) HPV-18 E6*I-containing cultures. (Middle panel) HPV-18 E6*I-containing PHKs infected with the empty pLC retrovirus. (Right panel) HPV-18 E6*I-containing PHKs trans-complemented with the pLJ HPV-18 URR-E6 retrovirus. Arrowhead points to the boundary between the upper cornified strata and live epithelium below. (B–E) Double-fluorescence detection of p53 (green) and HPV-18 DNA (red) in raft cultures. (B) Day-10 normal PHKs. (C) Day-10 and day-12 wild-type HPV-18-containing PHKs. (D) Day-12 and day-16 HPV-18 E6*I-containing PHKs. (E) Day-16 HPV-18 E6*I-containing PHKs trans-complemented with pLJ HPV-18 URR-E6. Cellular DNA was stained with DAPI (blue).
Figure 7.
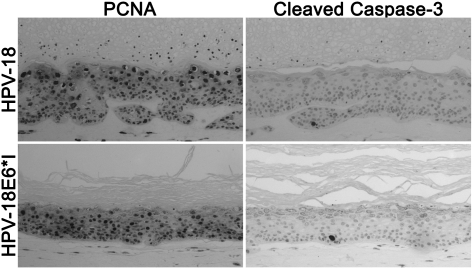
Widespread induction of PCNA and virtual absence of cleaved caspase 3 in raft cultures harboring the wild-type or E6*I mutant plasmid. IHC was conducted to detect PCNA (left column) or the cleaved caspase 3 (right column) in day-16 raft cultures of PHK transfected with the wild-type HPV-18 (top row) or the HPV-18 E6*I mutant (bottom row). The small dots in cornified layers represent parakeratosis.
Raft cultures harboring E6*I mutant accumulate high levels of p53 protein without experiencing apoptosis
To determine the defects of E6*I, we probed for viral DNA amplification and p53 protein. We demonstrated previously that p53 is induced by E7 in a fraction of spinous cells in PHK raft cultures, but it is degraded when E6 is also expressed (Jian et al. 1998). The induction of p53 in cultures with E6*I mutant genomes would therefore provide yet another marker of E7 activity. Further, a comparison of p53 distribution between raft cultures containing the wild-type and E6*I mutant genomes would shed light on the normal localization for E6 and E6* peptides.
In untransfected PHK cultures, p53 was detected in a few BrdU-positive and BrdU-negative basal cells but not in suprabasal cells (Fig. 6B). Day-10 cultures harboring wild-type HPV-18 genomes had low viral DNA, which increased by day 12, as described previously. Weak p53 signals were detected in some basal as well as mid- and upper spinous cells on day 10. On days 12 and 14, weak p53 was also detected in a fraction of cells in the lower strata, but not in mid- and upper spinous cells with high viral DNA (Fig. 6C; data not shown). In contrast, numerous basal and suprabasal cells in raft cultures harboring the E6*I mutant genomes were moderately or strongly positive for nuclear p53 at all time points. Importantly, only a small number of differentiated cells were positive for very low levels of amplified viral DNA, and these cells were negative for strong p53 (Fig. 6D). This pattern did not change out to 22 d. Poor amplification of HPV-18 E6*I mutant genomes would account for the scarce L1 signals.
Despite the high p53 protein, the cells did not undergo apoptosis based on histology of the tissues. Had extensive apoptosis taken place, we would not have been able to have raft cultures for >3 wk. To verify the absence of apoptosis, we probed raft cultures harboring E6*I mutant or the wild-type genome for one of the effector caspases, the cleaved caspase 3 (Taylor et al. 2008). There was little or no signal in either culture (Fig. 7, right panels). In contrast, the antibody yielded widespread signals in drug-treated tissues with apoptotic histology (E.-Y. Kho, H.-K. Wang, T.R, Broker, and L.T. Chow, unpubl.).
To ascertain that the defective phenotypes were not attributable to unintended mutations in E6*I genomes, we infected the E6*I DNA-containing PHKs with a retrovirus expressing HPV-18 E6 or E6–E7 controlled by the contiguous viral URR, which harbors transcription enhancers, the E6 promoter, and the HPV origin of replication. With either retrovirus, ∼30% of the cornified envelopes across the day-18 and older rafts were abundantly positive for L1 (Fig. 6A, right panel; data not shown). Empty vector-only retrovirus had no effect on L1 synthesis (Fig. 6A, middle panel). Areas of URR-E6-complemented day-16 HPV-18 E6*I cultures exhibited a pattern of p53 loss and HPV DNA amplification similar to day-12 wild-type cultures (Fig. 6E). The slower kinetics of L1 synthesis can be attributed to the delay in E6 expression from the complementing retrovirus versus wild-type HPV-18 cultures. In regions where complementation was not observed, the provirus may have integrated in chromosomal locations from which E6 expression was inadequate to overcome the dominant-negative E6*I suppression of the E6 function. Indeed, raft cultures of PHKs transduced with a retrovirus harboring HPV-18 or HPV-11 URR-βgal also exhibited patchy patterns of reporter gene expression (Parker et al. 1997). However, we cannot rule out that the mutant plasmid might have been lost in some of the cells, as suggested by the pockets of PCNA-negative cells. E6 trans-complementation was reproducible with three batches of PHKs. These results demonstrate that E6 plays a critical role in viral DNA amplification and virion production, possibly implicating p53 in inhibiting viral DNA amplification.
Discussion
By using the Cre-loxP recombination in vivo, we developed an efficient strategy to generate HPV genomic plasmids in PHKs suitable for organotypic raft cultures without extensive passages. The cultures support an unprecedented productive program that yields high titers of infectious HPV (Figs. 1–4) with little or no effects on squamous differentiation (our unpublished observations). Critically, the ability to examine an immortalization-defective E6 mutant phenotype (Fig. 6), which could not be investigated in PHK raft cultures until now, clearly demonstrates that our approach obviates the need for immortalized host cells. In so doing, we overcame the inefficient amplification of the wild-type viral DNA in immortalized cells, a limitation on HPV genetic analyses.
Lee et al. (2004) similarly used Cre-loxP recombination to generate an HPV-16 genomic plasmid in PHKs from chimeric adenoviruses, but the immortalized cells had a very low yield of virus. Construction of such a virus is time-consuming. Given that adenoviruses themselves are highly infectious to humans, the chimeric virus requires greater care in handling. In contrast, our method is simple, expedient, efficient, and reproducible. The critical factors are a high efficiency of cotransfection of supercoiled plasmids into PHKs, a rapid and effective acute drug selection for cells harboring the parental plasmid that expresses the drug resistance gene, and a highly efficient excision of HPV genomes from the parental plasmids by nls-Cre recombinase in vivo (Fig. 1). Relative to the wild-type genome, the HPV-18 plasmid generated has a 34-bp loxP insertion downstream from the late poly-A site. We believe this insertion exerts no adverse effects on the viral productive program. A mildly hyperproliferative tissue morphology, differentiation-dependent viral DNA amplification, and patterns of E7 activity and L1 protein synthesis (Figs. 2, 4, 5; Supplemental Fig. S1) all recapitulate those observed in productively infected patient tissues (Stoler and Broker 1986; Doorbar 2006). Moreover, the exuberant viral DNA amplification and robust virus production (Figs. 1–3) resemble those observed in HPV-11 infected foreskin xenografts in nude mice deficient in immune surveillance (Stoler et al. 1990).
Virion production and maturation
Taking into account the significant fraction of spinous cells with amplified viral DNA, the high numbers of amplified viral genomes per cell, and the numerous L1-positive cornified envelopes (Figs. 1–5), the titers of virus produced in our PHK rafts are orders of magnitude higher than viruses recovered from immortalized cells (see Frattini et al. 1997). A highly productive infection of naive PHKs in raft cultures by a human papillomavirus has finally been achieved (Fig. 3). Our TEM studies of tissue sections revealed that, in degenerated nuclei of cornified envelopes, HPV particles undergo a maturation that alters the particle surface to enable the tightly packed paracrystalline arrays (Fig. 2I–N), similar to those observed in highly productive canine papillomas (Campo 2002). Notably, disulfide bond formation between neighboring L1 capsomeres is required for virion maturation (Buck et al. 2005b and references therein) and apparently this process does not take place in the reducing environment of living cells, further linking the viral productive program to the successive stages of epithelial differentiation.
HPV-16 pseudovirions must undergo a maturation process in vitro to gain stability (Buck et al. 2005b). High titers of HPV pseudovirions are assembled in 293T or 293TT cells from L1/L2 expressed from vectors and cotransfected, recircularized HPV DNA (Buck et al. 2005a; Pyeon et al. 2005). However, no productive infection in PHK raft cultures has been reported despite efficient infection of immortalized or transformed cells. Given the wide use of HPV pseudovirions in diverse studies, a side-by-side comparison between pseudovirions against our genuine HPV virions will be particularly relevant in examining infection processes and outcomes.
Viral DNA amplification in G2 arrested cells
Most intriguing is our finding that, in the great majority of the cells, HPV DNA amplification did not occur in cells in S phase. Rather, it follows host DNA replication. Significant colocalization of cellular DNA replication and amplified viral DNA occurs only upon a BrdU pulse followed by a long chase (Figs. 4, 5; Supplemental Figs. S2, S3). Furthermore, viral DNA first appears in cells strongly positive for cytoplasmic cyclin B1. Collectively, these observations strongly suggest that viral DNA amplification initiated in G2-arrested cells. For small DNA tumor viruses, the assumption has been that, after inducing an S-phase milieu by one of the viral “oncoproteins” (HPV E7, adenovirus E1A, and SV40/polyomavirus T-antigen), viral DNA then amplifies concurrently with host DNA replication in S phase. Nevertheless, the possibility that the two events might not exactly coincide has been suggested for HPV (Swindle et al. 1999). Others have observed that sporadic HPV DNA signals were separated spatially from S-phase markers (for review, see Davy and Doorbar 2007). However, in the absence of simultaneous detections for viral DNA, BrdU incorporation, and markers of cell cycle status in time course studies, as presented here, no definitive interpretations had been offered.
Why would HPV DNA amplification lag behind host DNA replication? The simplest and most straightforward explanation is that HPV is not able to hijack host replication machinery while it is engaged in replicating host DNA. A virus-induced G2 arrest would then create an uncontested window of opportunity for a highly efficient and rapid viral genomic amplification. A number of DNA and RNA viruses are known to arrest cells in G2 phase for reasons not well understood (for review, see Davy and Doorbar 2007). Perhaps viral replication in G2-arrested cells is not an uncommon occurrence, just not widely appreciated.
G2 arrest of proliferating cell lines caused by ectopic overexpression of the cytoplasmic HPV E1^E4 protein is well documented (Davy and Doorbar 2007). However, mutational analyses of the E4 gene have not resolved its role in viral DNA amplification (Nakahara et al. 2005; Wilson et al. 2005; Fang et al. 2006a). In our productive raft cultures, the E1^E4 protein did not colocalize with cyclin B1 and thus is unlikely to be responsible for G2 arrest (A. Duffy, H.-K. Wang, T. Broker, and L. Chow, unpubl.). The experimental system reported here is ideal for investigating its possible roles in the viral infection cycle. In contrast, in PHK raft cultures expressing HPV-18 E6 and E7 genes, there is an elevated transcription of cdk1, cyclin B1, other G2/M genes, and Wee1, a negative regulator of cyclin B1/cdk1 activation at G2/M (Garner-Hamrick et al. 2004). Indeed, our recent results in PHK raft cultures demonstrate that E7 alone causes cytoplasmic accumulation of cytoplasmic cyclin B1 in BrdU-positive cells (N. Banerjee, T. Broker, and L. Chow, unpubl.).
A critical function of E6 in viral DNA amplification
We show that the full-length E6 protein is critical for viral DNA amplification and hence capsid protein synthesis (Fig. 6). The defect of the E6*I mutant cannot be attributed to a loss of the plasmid, as E7-induced PCNA was detected in the great majority of the superbasal cells (Fig. 7). This interpretation is supported by the very high levels of p53 protein, which is induced in response to E7, in a great majority of cells (Fig. 6). The elevated p53 protein level may have trigger checkpoint control to inhibit cellular DNA replication. Also, in wild-type and E6*I raft cultures, viral DNA signals did not colocalize with p53 signals, suggesting that the high levels of p53 protein also inhibited viral DNA amplification. This interpretation would agree with transient HPV DNA replication assays in which ectopic p53 protein inhibits HPV DNA replication (Lepik et al. 1998). The ability to trans-complement the E6 deletion mutation in the context of an autonomous viral plasmid (Fig. 6) will facilitate future analyses of site-specific E6 mutations to verify the role of p53 and to identify additional E6 functions relevant to the viral productive program.
Comparative results between wild-type and E6*I mutant genome-containing raft cultures have several additional implications: (1) The function of E6 is not to counter E7-induced apoptosis from elevated levels of p53. In cervical carcinoma-derived HeLa cells, expression of E7 in the absence of E6 leads to apoptosis (DeFilippis et al. 2003). Overexpression of HPV-18 E6*I in CaSki cells, another cervical carcinoma cell line, antagonized E6 function and triggered p53-dependent apoptosis (Pim and Banks 1999). In addition, HPV-31 E6 and E7 have been reported to activate caspase 3, important for HPV-31 DNA amplification in raft cultures of immortalized cells (Moody et al. 2007). We did not observe apoptotic cells by histology in raft cultures with the wild type or the HPV E6*I mutant. Furthermore, we hardly ever detect cleaved caspase 3 in either raft cultures (Fig. 7). (2) The patterns of p53 detection suggest that E6 is expressed in both basal and in suprabasal cells in raft cultures. This could account for an increase in p53-positive cells in the basal stratum of E6*I-containing raft cultures, resulting in a less hyperproliferative morphology than in the wild-type HPV-18-containing cultures. Also revealing is the observation that, in cultures containing the wild-type HPV, the increased p53 signals in spinous cells relative to the basal cells would suggest an increase in spliced E6* transcripts during squamous differentiation, reducing but not abolishing the synthesis of the E6 protein. It is possible that, because of the anti-recombinational properties of p53 (Bertrand et al. 2004), the low levels of p53 remaining in the spinous cells could limit recombination of HPV DNA into oligomers that cannot be packaged into HPV capsids. An increase in E6* spliced RNAs could bolster translation of downstream early ORFs from polycistronic mRNA to support viral DNA amplification (Hubert and Laimins 2002; Tang et al. 2006). The absence of p53 in cell with high HPV DNA is consistent with the loss of E7 activity in these cells.
A switch from early to late phase
The temporal and spatial patterns of cyclin A and cyclin B, BrdU incorporation, viral DNA amplification, destabilization, and subsequent reappearance of p130 pocket protein (Fig. 5; Supplemental Figs. S2, S3), and the synthesis of the L1 protein in superficial cells following viral DNA synthesis (Fig. 2; Supplemental Fig. S1) in both DNA-transfected and HPV-infected PHK raft cultures all point to a transition from a state of viral DNA amplification to late protein synthesis for virion morphogenesis. What then inactivates E7 as viral DNA amplification progresses? The viral E6 promoter might be down-regulated by increasing levels of viral E2 protein expressed from the increasing viral DNA templates (Chow and Broker 2007 and references therein). Alternatively, elevated E2 protein has been reported to directly bind and inactivate E7 (Gammoh et al. 2006).
We propose the following scenario for the sequence of events in the productive phase. The E6 protein is expressed in the lower strata to increase the parabasal, transit-amplifying cell population. Elevated E7 expression, attributable to increased E6* mRNA in suprabasal differentiated cells, leads to p130 destabilization and S-phase re-entry. Following host chromosome duplication, spinous cells are arrested in a prolonged G2 phase, during which viral DNA amplification occurs, a process that appears to require E6 to degrade most of the p53 induced by E7. High levels of viral DNA amplification are accompanied by a reduction in and eventual loss of E7 activity, leading to a switch to late protein synthesis. The late events are characterized by a dramatic increase in E1^E4 protein (Doorbar 2006; Duffy et al., unpubl.) and the expression of capsid proteins in superficial cells (Figs. 2, 4; Supplemental Fig. S1) for virion morphogenesis. The assembled virions then mature in the cornified envelopes. Viral DNA amplification in the live tissue is then attenuated after the productive phase. The cause of this shutdown remains to be investigated.
In summary, we developed a simple and reproducible method to recapitulate a complete and highly productive infection cycle in organotypic cultures of PHKs in which viral genomic plasmids were generated by in vivo recombination. A detailed molecular portrait of virus–host interactions were revealed and further verified using raft cultures infected with the progeny virus. Our system is ideal for genetic and molecular analyses of mutants of HR as well as LR HPV types in PHK raft cultures. The recapitulation of the full infection cycle will finally enable realistic ex vivo evaluation of potential agents to prevent HPV infection or reproduction.
Materials and methods
Plasmids
The nls-Cre expression plasmid pCAGGS-nlsCre was a gift (Hardouin and Nagy 2000). All PCR primers and the construction of pNeo-loxP HPV-18 and pNeo-loxP HPV-18 E6*I plasmids are provided in Supplemental Table 1 and the Supplemental Material. For both plasmids, the 34-bp loxP sites flank the linear HPV-18 sequence upstream of nucleotide 7474 and downstream from nucleotide 7473. The vector carries the Neomycin resistance marker gene selectable in bacteria and in mammalian cells. In the HPV-18 E6*I mutant, the intron coding sequence (nucleotides 234–415) in the predominant E6*I mRNA was deleted. For trans-complementation experiments, the empty vector-only retrovirus pLC and pLJ HPV-18 URR-E6 or URR-E6/E7 retroviruses were used. Each expresses the Neomycin resistance gene (Cheng et al. 1995; Chien et al. 2002). All plasmids were purified by banding in CsCl-ethidium bromide equilibrium density gradients.
DNA transfection and organotypic raft cultures of PHKs
PHKs recovered from neonatal foreskins collected from UAB Hospital Well Baby Nursery with IRB approval were grown in keratinocyte serum-free medium (K-SFM) (Invitrogen). We seeded 2 × 105 PHKs at the second passage onto six-well plates, incubated overnight in K-SFM, and then cotransfected with 1 μg of pCAGGS-nlsCre and 5 μg of pNeo-loxP HPV-18 or pNeo-loxP HPV-18 E6*I plasmids using FuGENE 6 (Roche). After selection with 100 μg/mL of G418 (Invitrogen) for 4 d, the cells were cultured in fresh K-SFM for an additional 1 or 2 d. Each raft culture was then initiated from 2 × 105 to 2.5 × 105 PHKs (Banerjee et al. 2005 and references therein). As specified in each experiment, the cultures were grown for different numbers of days after being raised to the medium:air interface. To mark cells in S phase, BrdU was added to the medium prior to harvest as indicated in the figure legends. The cultures were fixed in 10% buffered formalin and embedded in paraffin. For trans-complementation, the G418-selected PHKs were transduced with various retroviruses. Raft cultures were initiated with or without a reselection using 300 μg/mL G418 for 2 d. BrdU labeling and tissue processing were similarly performed.
PCR and Southern blot analyses for HPV-18 DNA excision and amplification
Fifty nanograms of total DNA from submerged PHKs were examined by PCR amplification for Cre-mediated HPV DNA excision. The primer sets G, H, and I (Supplemental Table 1) detected total HPV-18 DNA, excised HPV-18 DNA, or the control β-globin gene, respectively, after electrophoresis in a 2% agarose gel and staining with ethidium bromide. For Southern blot hybridization, 15 μg of total epithelial DNA from day-12 raft cultures were digested overnight with Hind III (no cut in the parental plasmid or the excised HPV DNA) or with EcoR I (one cut in each). Copy number and length standards were prepared by mixing the parental plasmid or pGEM2-HPV-18 with 15 μg of cellular DNA from control PHK raft cultures to yield 50, 500, or 5000 copies of the HPV genome/diploid cell. The mixture was then digested with EcoR I, which linearizes the parental plasmid or excises the 7.9-Kb HPV-18 DNA from the pGEM2 vector. DNA samples were electrophoretically separated in 0.8% neutral agarose gels, and Southern-blotted membranes were hybridized to [α-32P]-labeled, genomic HPV-18 DNA probes. Blots were documented using a Storm 820 PhosphorImager (GE Healthcare).
Histology and in situ analyses
Four-micron sections of raft cultures were deparaffinized and stained with H&E for histology or by IHC to detect PCNA induction, BrdU, and cleaved caspase 3, and the L1 capsid antigen. DNA-FISH and indirect IF coupled with tyramide signal enhancement were conducted as described previously, with minor modifications (Van Tine et al. 2005). Additional details are provided in the Supplemental Material.
TEM of raft culture sections
Day-14 raft cultures were fixed with 2.5% glutaraldehyde in phosphate-buffered saline followed by 2% osmium tetroxide and embedded in Spurr resin (Electron Microscopy Sciences). One-hundred--nanometer sections were cut and stained with 1% uranyl acetate in 50% ethanol, followed with Reynold's lead citrate (EMS). Grids were scanned at 60 kV in a FEI T12 Spirit electron microscope (Advanced Microscopy Techniques). Images were electronically captured and measured using an AMT XR 60B digital camera and its software.
HPV-18 virion recovery and titer determination
HPV-18 virions were recovered from day-14 or day-16 epithelia as described (Favre 1975). To titer the virus, aliquots of the virus stocks were digested with DNase I (Invitrogen), which was then inactivated by heating for 5 min at 100°C. Packaged viral DNA was then purified by digestion with Proteinase K and phenol/chloroform extractions. Serial dilutions of viral DNA were analyzed by real-time quantitative PCR using SYBR GreenER qPCR SuperMix (Invitrogen) and primers J and K (Supplemental Table 1). As standards, purified pNeo-LoxP HPV-18 plasmid DNA was serially diluted to ∼40 to 4 × 108 copies per well. Forty cycle PCR amplification reactions in triplicate were performed in 384-well plates using the ABI 7900HT. Data were processed with SDS2.1 software (Applied Biosystems).
HPV-18 infectivity assays
Approximately 1 × 105 PHKs were inoculated with various amounts of virus stock, corresponding to an MOI of 5200, 1040, 208, 42, 10, 2, 1, or 0 in 1 mL of K-SFM and incubated overnight. The medium was changed and the cells were cultured for four more days. Total RNA was then extracted by Trizol (Invitrogen). Reverse transcription was conducted in a 50-μL reaction on 10 μg of RNA. One microliter of RT reaction was then subjected to 30 cycles of PCR or nested PCR amplification (30 cycles each) in a 35-μL reaction mixture to generate a cDNA fragment of the spliced HPV-18 E6–E7–E1^E4, RNA, or the β-actin mRNA, as described (Meyers et al. 2002). Fifteen microliters of each reaction were resolved by electrophoresis in a 2% agarose gel and visualized by ethidium bromide staining. PHKs were also infected with various MOIs in K-SFM overnight and developed into raft cultures, fixed on day 14, and processed as described.
Acknowledgments
We thank Leigh Millican of the UAB Cancer Center Core for assistance in TEM and the staff in the UAB Well Baby Nursery for collecting circumcised neonatal foreskins. DNA sequencing was provided by the UAB Center for AIDS Research Core Facility. This work was supported by USPHS grants CA83679 and CA107338. H.K.W. developed the system, prepared the raft cultures, and performed biochemical, IHC, and infection assays. A.A.D. conducted in situ hybridization and IF assays.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1735109.
Supplemental material is available at http://www.genesdev.org.
References
- Banerjee N.S., Chow L.T., Broker T.R. Retrovirus-mediated gene transfer to analyze HPV gene regulation and protein functions in organotypic ‘raft’ cultures. Methods Mol. Med. 2005;119:187–202. doi: 10.1385/1-59259-982-6:187. [DOI] [PubMed] [Google Scholar]
- Bertrand P., Saintigny Y., Lopez B.S. p53's double life: Transactivation-independent repression of homologous recombination. Trends Genet. 2004;20:235–243. doi: 10.1016/j.tig.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Buck C.B., Pastrana D.V., Lowy D.R., Schiller J.T. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 2005a;119:445–462. doi: 10.1385/1-59259-982-6:445. [DOI] [PubMed] [Google Scholar]
- Buck C.B., Thompson C.D., Pang Y.Y., Lowy D.R., Schiller J.T. Maturation of papillomavirus capsids. J. Virol. 2005b;79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo M.S. Animal models of papillomavirus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- Cheng S., Schmidt-Grimminger D.C., Murant T., Broker T.R., Chow L.T. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes & Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- Chien W.M., Noya F., Benedict-Hamilton H.M., Broker T.R., Chow L.T. Alternative fates of keratinocytes transduced by human papillomavirus type 18 E7 during squamous differentiation. J. Virol. 2002;76:2964–2972. doi: 10.1128/JVI.76.6.2964-2972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L.T., Broker T.R. Mechanisms and regulation of papillomavirus DNA replication. In: Campo M.S., editor. Papillomavirus research: From natural history to vaccines and beyond. Caister Academic Press; 2006. pp. 53–71. [Google Scholar]
- Chow L.T., Broker T.R. Human papillomavirus RNA transcription. In: DiMaio D., Garcea R.L., editors. The papillomaviruses. Springer; New York: 2007. pp. 109–131. [Google Scholar]
- Davy C., Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology. 2007;368:219–226. doi: 10.1016/j.virol.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippis R.A., Goodwin E.C., Wu L., DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 2003;77:1551–1563. doi: 10.1128/JVI.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollard S.C., Wilson J.L., Demeter L.M., Bonnez W., Reichman R.C., Broker T.R., Chow L.T. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. Genes & Dev. 1992;6:1131–1142. doi: 10.1101/gad.6.7.1131. [DOI] [PubMed] [Google Scholar]
- Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. (Lond.) 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- Fang L., Budgeon L.R., Doorbar J., Briggs E.R., Howett M.K. The human papillomavirus type 11 E1/\E4 protein is not essential for viral genome amplification. Virology. 2006a;351:271–279. doi: 10.1016/j.virol.2006.01.051. [DOI] [PubMed] [Google Scholar]
- Fang L., Meyers C., Budgeon L.R., Howett M.K. Induction of productive human papillomavirus type 11 life cycle in epithelial cells grown in organotypic raft cultures. Virology. 2006b;347:28–35. doi: 10.1016/j.virol.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Favre M. Structural polypeptides of rabbit, bovine, and human papillomaviruses. J. Virol. 1975;15:1239–1247. doi: 10.1128/jvi.15.5.1239-1247.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini M.G., Lim H.B., Doorbar J., Laimins L.A. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 1997;71:7068–7072. doi: 10.1128/jvi.71.9.7068-7072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammoh N., Grm H.S., Massimi P., Banks L. Regulation of human papillomavirus type 16 E7 activity through direct protein interaction with the E2 transcriptional activator. J. Virol. 2006;80:1787–1797. doi: 10.1128/JVI.80.4.1787-1797.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner-Hamrick P.A., Fostel J.M., Chien W.M., Banerjee N.S., Chow L.T., Broker T.R., Fisher C. Global effects of human papillomavirus type 18 E6/E7 in an organotypic keratinocyte culture system. J. Virol. 2004;78:9041–9050. doi: 10.1128/JVI.78.17.9041-9050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese N.J., Banerjee N.S., Broker T.R., Chow L.T. Casein kinase II motif-dependent phosphorylation of human papillomavirus E7 protein promotes p130 degradation and S-phase induction in differentiated human keratinocytes. J. Virol. 2008;82:4862–4873. doi: 10.1128/JVI.01202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardouin N., Nagy A. Gene-trap-based target site for cre-mediated transgenic insertion. Genesis. 2000;26:245–252. doi: 10.1002/(sici)1526-968x(200004)26:4<245::aid-gene50>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Hubert W.G., Laimins L.A. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 2002;76:2263–2273. doi: 10.1128/jvi.76.5.2263-2273.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian Y., Schmidt-Grimminger D.C., Chien W.M., Wu X., Broker T.R., Chow L.T. Post-transcriptional induction of p21cip1 protein by human papillomavirus E7 inhibits unscheduled DNA synthesis reactivated in differentiated keratinocytes. Oncogene. 1998;17:2027–2038. doi: 10.1038/sj.onc.1202142. [DOI] [PubMed] [Google Scholar]
- Lambert P.F., Ozbun M.A., Collins A., Holmgren S., Lee D., Nakahara T. Using an immortalized cell line to study the HPV life cycle in organotypic ‘raft’ cultures. Methods Mol. Med. 2005;119:141–155. doi: 10.1385/1-59259-982-6:141. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Yi S.M.P., Anderson M.E., Berger K.L., Welsh M.J., Klingelhutz A.J., Ozbun M.A. Propagation of infectious human papillomavirus type 16 by using an adenovirus and Cre/LoxP mechanism. Proc. Natl. Acad. Sci. 2004;101:2094–2099. doi: 10.1073/pnas.0308615100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepik D., Ilves I., Kristjuhan A., Maimets T., Ustav M. p53 protein is a suppressor of papillomavirus DNA amplificational replication. J. Virol. 1998;72:6822–6831. doi: 10.1128/jvi.72.8.6822-6831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin M.E., Meyers C. Propagation of infectious, high-risk HPV in organotypic ‘raft’ culture. Methods Mol. Med. 2005;119:171–186. doi: 10.1385/1-59259-982-6:171. [DOI] [PubMed] [Google Scholar]
- Meyers C., Frattini M.G., Hudson J.B., Laimins L.A. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- Meyers C., Bromberg-White J.L., Zhang J., Kaupas M.E., Bryan J.T., Lowe R.S., Jansen K.U. Infectious virions produced from a human papillomavirus type 18/16 genomic DNA chimera. J. Virol. 2002;76:4723–4733. doi: 10.1128/JVI.76.10.4723-4733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody C.A., Fradet-Turcotte A., Archambault J., Laimins L.A. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc. Natl. Acad. Sci. 2007;104:19541–19546. doi: 10.1073/pnas.0707947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münger K., Howley P.M. Human papillomavirus immortalization and transformation functions. Virus Res. 2002;89:213–228. doi: 10.1016/s0168-1702(02)00190-9. [DOI] [PubMed] [Google Scholar]
- Nakahara T., Peh W.L., Doorbar J., Lee D., Lambert P.F. Human papillomavirus type 16 E1^E4 contributes to multiple facets of the papillomavirus life cycle. J. Virol. 2005;79:13150–13165. doi: 10.1128/JVI.79.20.13150-13165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.T., Longworth M.S., Laimins L.A. Roles of the E6 and E7 proteins in the life cycle of low-risk human papillomavirus type 11. J. Virol. 2004;78:2620–2626. doi: 10.1128/JVI.78.5.2620-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun M.A. Human papillomavirus type 31b infection of human keratinocytes and the onset of early transcription. J. Virol. 2002;76:11291–11300. doi: 10.1128/JVI.76.22.11291-11300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun M.A., Meyers C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 1997;71:5161–5172. doi: 10.1128/jvi.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.N., Zhao W., Askins K.J., Broker T.R., Chow L.T. Mutational analyses of differentiation-dependent human papillomavirus type 18 enhancer elements in epithelial raft cultures of neonatal foreskin keratinocytes. Cell Growth Differ. 1997;8:751–762. [PubMed] [Google Scholar]
- Pim D., Banks L. HPV-18 E6*I protein modulates the E6-directed degradation of p53 by binding to full-length HPV-18 E6. Oncogene. 1999;18:7403–7408. doi: 10.1038/sj.onc.1203134. [DOI] [PubMed] [Google Scholar]
- Pyeon D., Lambert P.F., Ahlquist P. Production of infectious human papillomavirus independently of viral replication and epithelial cell differentiation. Proc. Natl. Acad. Sci. 2005;102:9311–9316. doi: 10.1073/pnas.0504020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink K.M., Laimins L.A. Induction of the human papillomavirus type 31 late promoter requires differentiation but not DNA amplification. J. Virol. 2005;79:4918–4926. doi: 10.1128/JVI.79.8.4918-4926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler M.H., Broker T.R. In situ hybridization detection of human papillomavirus DNAs and messenger RNAs in genital condylomas and a cervical carcinoma. Hum. Pathol. 1986;17:1250–1258. doi: 10.1016/s0046-8177(86)80569-x. [DOI] [PubMed] [Google Scholar]
- Stoler M.H., Whitbeck A., Wolinsky S.M., Broker T.R., Chow L.T., Howett M.K., Kreider J.W. Infectious cycle of human papillomavirus type 11 in human foreskin xenografts in nude mice. J. Virol. 1990;64:3310–3318. doi: 10.1128/jvi.64.7.3310-3318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindle C.S., Zou N., Van Tine B.A., Shaw G.M., Engler J.A., Chow L.T. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 1999;73:1001–1009. doi: 10.1128/jvi.73.2.1001-1009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Tao M., McCoy J.P., Jr, Zheng Z.M. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 2006;80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.C., Cullen S.P., Martin S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- Thomas M.C., Chiang C.M. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol. Cell. 2005;17:251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Thomas J.T., Hubert W.G., Ruesch M.N., Laimins L.A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tine B.A., Broker T.R., Chow L.T. Simultaneous in situ detection of RNA, DNA, and protein using tyramide-coupled immunofluorescence. Methods Mol. Biol. 2005;292:215–230. doi: 10.1385/1-59259-848-x:215. [DOI] [PubMed] [Google Scholar]
- Wilson R., Laimins L.A. Differentiation of HPV-containing cells using organotypic ‘raft’ culture or methylcellulose. Methods Mol. Med. 2005;119:157–169. doi: 10.1385/1-59259-982-6:157. [DOI] [PubMed] [Google Scholar]
- Wilson R., Fehrmann F., Laimins L.A. Role of the E1–E4 protein in the differentiation-dependent life cycle of human papillomavirus type 31. J. Virol. 2005;79:6732–6740. doi: 10.1128/JVI.79.11.6732-6740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Chen W., Roman A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc. Natl. Acad. Sci. 2006;103:437–442. doi: 10.1073/pnas.0510012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]