Abstract
We report herein a biopolymer actuator with a modular design that allosterically transduces ligand binding into an aqueous demixing phase transition. The biopolymer actuator consists of two modular domains: a ligand binding protein domain, calmodulin (CaM) that is fused to a transducer domain, a stimulus responsive elastin-like polypeptide (ELP) that exhibits a reversible lower critical solution temperature (LCST) phase transition. We demonstrate that binding of calcium to CaM spontaneously triggers the phase transition of the attached ELP, leading to the formation of meso-micro scale particles depending on the chain length of the ELP. This behavior is reversible, as chelation of the bound calcium results in dissolution of the assembled particles; is selective for calcium as opposed to magnesium; and is abolished by the binding of a peptide ligand that is specific to calcium-bound CaM. These results are, to our knowledge, the first demonstration of biomolecular recognition triggered, allosteric regulation of the LCST phase transition of a polymer, and are significant because they expand the available triggers of the LCST transition of stimulus responsive polymers to biochemical ligand binding. The ability to allosterically trigger the LCST transition of ELPs by biomolecular recognition will be useful for developing “smart” polymer actuators that capitalize upon the myriad ligand-protein pairs that are available from biology, for application in the design of selective pull-down assays in proteomics, drug delivery, and in the design of nanoscale biomolecular devices.
Keywords: biopolymer, elastin-like polypeptide, actuator, allostery, ligand binding, inverse phase transition, LCST transition, calmodulin
INTRODUCTION
Polymers that exhibit a lower critical solution temperature (LCST) transition in response to changes in their solution environment are an interesting class of stimulus responsive –smart– polymers.1,2 Polymers in this class, such as poly(N-isopropyl acrylamide) and their biological analogs such as elastin-like polypeptides are soluble in aqueous solution below their LCST (also termed inverse transition temperature: Tt), and undergo an aqueous demixing phase transition above their LCST resulting in the formation of an insoluble, polymer-rich phase.2 This reversible aqueous demixing behavior can be triggered by small changes in the solution environment (pH, temperature, and ionic strength),3–7 and variants of these polymers can be designed to exhibit the LCST transition in response to redox triggers and light.8–10 The LCST transition of these polymers has also been exploited in protein-polymer conjugates where the LCST triggered conformational collapse of the polymer in the vicinity of the ligand binding site of the protein has been shown to control the activity of the protein.11–16 To our knowledge, however, there are no examples of engineered systems that exhibit the reverse behavior, wherein ligand binding to a protein can trigger the LCST phase transition of an appended polymer.
Motivated by these previous studies, we demonstrate proof-of-concept of a modular allosteric biopolymer actuator whose LCST behavior can be reversibly triggered by a biomolecular recognition event. The actuator consists of a ligand binding domain and a LCST transduction domain, as shown in Figure 1. The LCST transduction domain of the biopolymer actuator is composed of an elastin-like polypeptide (ELP), which is an artificial polypeptide composed of repeats of the pentapeptide Val-Pro-Gly-Xaa-Gly, where the “guest residue” Xaa is any amino acid except proline.2 We chose an ELP as the transduction domain for the following reasons: first, ELPs exhibit LCST behavior, as they are highly soluble in water below their LCST, but undergo a reversible, soluble-insoluble phase transition when the temperature is raised above their LCST, leading to aggregation of the polypeptide.2,17 Second, unlike synthetic polymers that exhibit LCST behavior, ELPs can be genetically encoded with a protein of interest,18–22 which allows site-specific, stoichiometric and monodisperse conjugates to be readily synthesized by recombinant DNA techniques. Third, we have observed that ELPs and their fusion proteins far more sensitive to changes in their local environment than poly(NIPAAm), 23, 24 which led us to hypothesize that the subtle perturbation of the immediate environment imposed by a ligand binding event was more likely to be transmitted to an appended ELP chain than a synthetic LCST polymer.
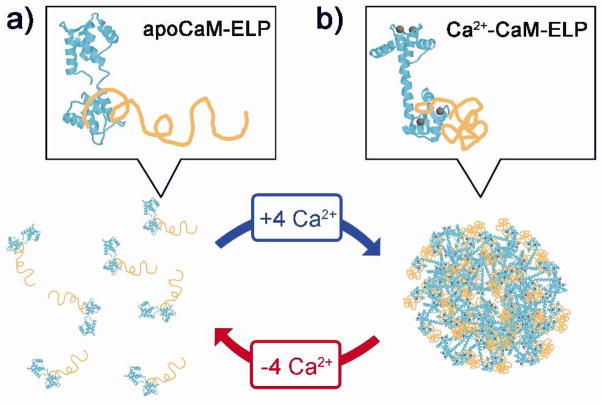
Figure 1.
Schematic representation of the allosteric, calcium triggered control of phase transition of CaM-ELP fusion proteins. Binding of Ca2+ to apoCaM allosterically triggers the LCST transition of the attached ELP, resulting in the formation of meso-microscale particles depending upon the chain length of the ELP. Chelation of the bound Ca2+ by EDTA reverses the LCST transition. Legend: CaM is colored cyan, ELPs are orange, and Ca2+ ions are gray. The N-terminal lobe of CaM is oriented to the top, and the C-terminal lobe to the bottom of each panel. a) Soluble monomer of apoCaM-ELP. b) Reversible calcium triggered formation of particles of Ca2+-CaM-ELP. Structural data of CaM were taken from the Protein Data Bank, accession codes: apoCaM(1CFD) and Ca2+-CaM(3CLN).
We chose calmodulin (CaM) as the ligand binding domain for proof-of-principle of this concept. Given the potentially vast number of ligand binding proteins that could potentially be harnessed for this application, we narrowed the choice to CaM, dictated by the hypothesis that ligand binding to a protein (or peptide) could isothermally trigger the phase transition of an appended ELP if one or more of the following conditions were met upon ligand binding: (1) a significant decrease in the surface charge of the protein; or (2) an increase in the fraction of solvent-exposed hydrophobic surface area. These two requirements are based upon our observation that the increase in the LCST of an ELP fusion protein relative to the same ELP is positively correlated with the fraction of solvent exposed ionizable residues on the protein and is negatively correlated with the solvent exposed hydrophobic surface area.24
These requirements suggested CaM as an obvious candidate for allosteric transduction of the ELP phase transition, because each apoCaM molecule binds 4 Ca2+ ions, which are coordinated by 11 Asp and 4 Glu residues,25 which leads to a decrease in the fraction of surface area attributed to ionizable groups in Ca2+ bound CaM relative to apoCaM. Structural studies of CaM have also shown that binding of Ca2+ triggers a conformational change in CaM,26 which results in the exposure of a significant amount of non-polar surface area to solvent.27–29 Because the effect of these Ca2+ triggered structural changes on the LCST behavior of CaM-ELP are additive, we hypothesized that they would reversibly modulate the LCST transition of CaM-ELP under isothermal conditions. Calcium itself is attractive as a ligand because it is a ubiquitous second messenger in cells, and exhibits significant gradients in its concentration in vivo,30,31 so that a calcium-triggered biopolymer could potentially be useful for in vivo application in drug delivery and imaging by triggering the release of a drug or imaging agent from a loaded particle. The conformational changes of CaM upon Ca2+ binding have been previously exploited in the design of biosensors,32 reversible biomaterials,33 hydrogels,34,35 calcium sensitive MRI contrast agents,36 and Ca2+ dependent biological motors.37 However, the interaction between calcium and calmodulin has not been previously used to allosterically trigger the phase transition of an appended LCST polymer, which is a significant omission, given the long-standing interest in the unique aqueous phase behavior of this class of polymers and the large number of technological applications of these materials.
RESULTS AND DISCUSSION
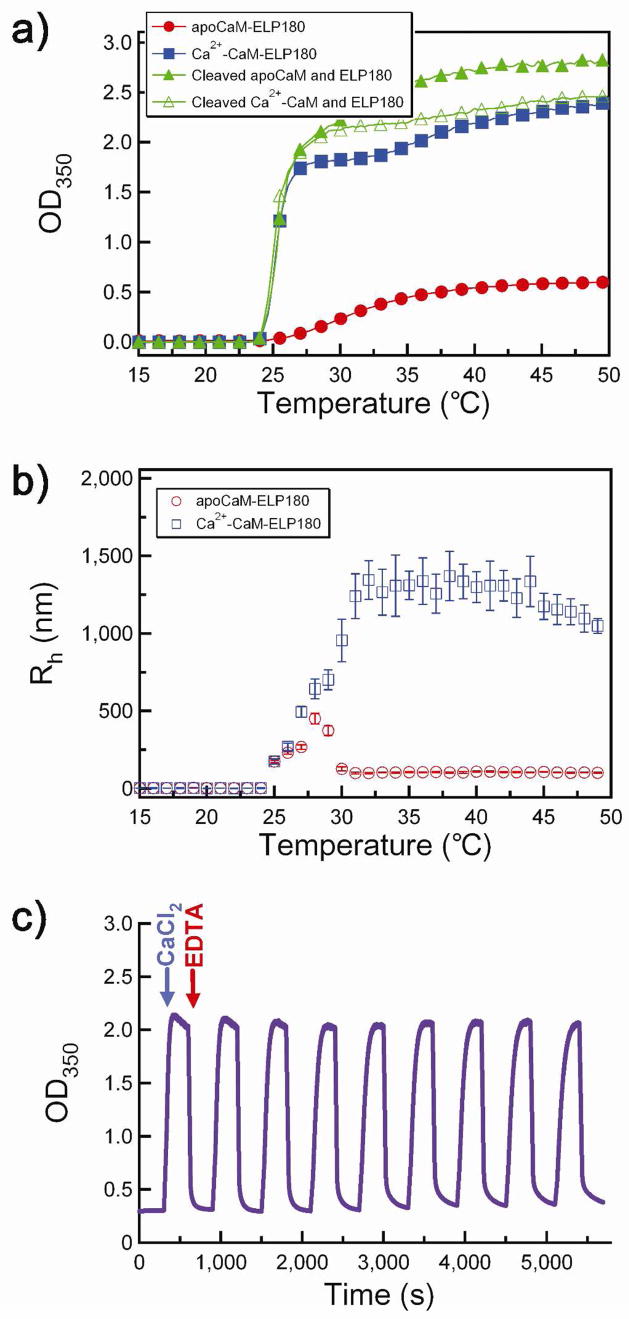
A CaM-ELP fusion with a 180 pentapeptide ELP repeat (CaM-ELP180) was expressed in E. coli from a synthetic gene and purified by inverse transition cycling.18 Figure 2 shows turbidity profiles by UV-vis spectrophotometry (Figure 2a) and particle size measurement by DLS (Figure 2b) as a function of solution temperature of a 25 μM solution of CaM-ELP180 without (apoCaM-ELP180) and with calcium (Ca2+-CaM-ELP180). A broad phase transition was observed for apoCaM-ELP, with a maximum OD350nm of ~0.6, which corresponds to nanoscale particles with an Rh of ~100 nm as determined by DLS (Figure 2b). This result suggests that significant electrostatic repulsion occurs during aggregation of apoCaM-ELP, which limits the terminal size of the aggregate. In contrast, Ca2+ bound CaM-ELP showed a change in the LCST behavior to lower temperatures and the formation of large aggregates with a quasi-steady state Rh of ~1.2 μm (Figure 2b). To examine whether the calcium sensitivity of the ELP LCST transition was allosteric, we liberated CaM from ELP via digestion by thrombin at a thrombin recognition site encoded between the two partners, and confirmed cleavage of the CaM-ELP fusion protein by SDS-PAGE (Figure S1 in the Supporting Information). Cleavage of CaM from its ELP partner completely abolished the calcium sensitivity of the phase transition (Figure 2a). Furthermore, the LCST behavior of the cleaved fusion protein was identical to that of an ELP of the same composition and chain length that had not been fused to CaM (results not shown). This finding is important because it demonstrates that allosteric transduction of Ca2+ binding to trigger the ELP phase transition requires the two domains to be physically connected, as a physical mixture of ELP and apoCaM does not spontaneously exhibit calcium dependent LCST transition behavior under identical solution conditions. Furthermore, we note that the calcium sensitivity of the CaM-ELP fusion protein occurs at concentrations that are at least 3-orders of magnitude lower than would be expected simply on the basis of the effect of salt on the LCST behavior of polymers.38
Figure 2.
Reversible change of LCST behavior of CaM-ELP180 by Ca2+ binding. a) Solution turbidity at 350 nm (OD350) as a function of solution temperature for apoCaM-ELP180 (filled circles, red), Ca2+-CaM-ELP180 (filled squares, blue), thrombin-cleaved apoCaM-ELP180 (filled triangles, green), and thrombin-cleaved Ca2+-CaM-ELP180 (open triangles, green). b) Hydrodynamic radius (Rh) of apo-CaM-ELP180 (open circles, red) and Ca2+-CaM-ELP180 (open squares, blue) as a function of temperature. c) Reversible control of self-assembly of CaM-ELP fusion as a function of Ca2+ binding. The OD350 was measured as a function of time after sequential injection of Ca2+ followed by EDTA, as indicated by the arrows. Nine cycles of addition of Ca2+ followed by EDTA are shown in the Figure.
The reversibility of the LCST transition behavior of CaM-ELP in response to calcium was investigated in an experiment where we monitored the turbidity at 350 nm (as a measure of the aggregate size) by sequential addition of Ca2+ and EDTA (Figure 2c) to a 25 μM solution of CaM-ELP in HEPES buffer at 32°C. At 32°C, apoCaM-ELP forms nanoparticles with an Rh of ~100 nm, as measured by DLS (Figure 2b). Addition of 0.5 mM CaCl2 led to an increase in the solution turbidity to ~2.2, which is consistent with the formation of microparticles with an Rh of ~1.2 μm (Figure 2b). Subsequent addition of 0.5 mM EDTA to chelate the calcium led to a structural transition in CaM-ELP, as seen by a decrease in turbidity to ~0.3, which corresponds to the formation of ~100 nm Rh nanoparticles (Figure 2b). This behavior was completely reversible as shown by repeated cycling of Ca2+/EDTA in the same experiment at 32°C, indicating that the fusion protein could be toggled back and forth between nanoparticles (−Ca2+) and micron-size aggregates (+Ca2+) (Figure 2c).
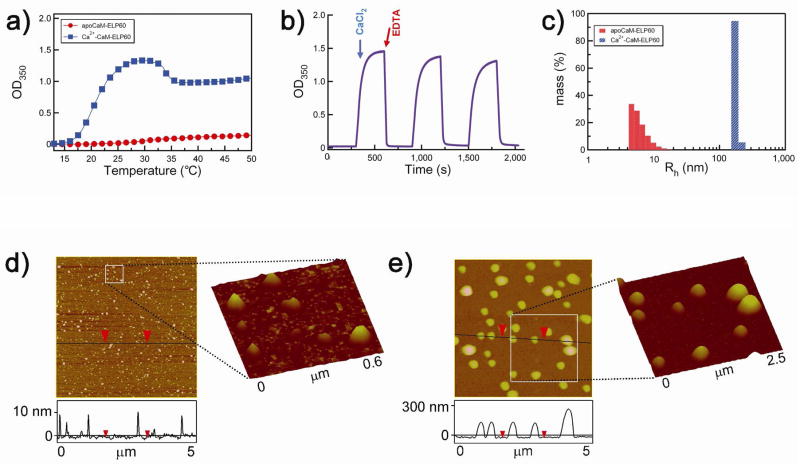
In order to demonstrate that this behavior is not restricted to a single CaM-ELP construct, we also investigated the Ca2+ triggered behavior of another ELP fused to CaM. ELP60 has the same guest residue composition as ELP180, but is a third in length of ELP180, as it consists of 60 VPGXG pentapeptides (see Experimental section for ELP sequences). The LCST transition behavior of CaM-ELP60 in 0.4 M (NH4)2SO4 is shown in Figure 3a. We chose 0.4 M (NH4)2SO4 for these experiments instead of 1 M NaCl (previously used with the longer CaM-ELP180) because the LCST of ELPs shows a 1/l (l = chain length) dependence,39 so that the LCST of ELP60 is significantly greater in 1 M NaCl than that of CaM-ELP180. In order to bring the LCST to close to room temperature, we chose 0.4 M (NH4)2SO4 because the dependence of the LCST on salt follows the Hofmeister series,38,40 so that (NH4)2SO4 is more effective than NaCl in reducing the LCST to an experimentally convenient temperature.
Figure 3.
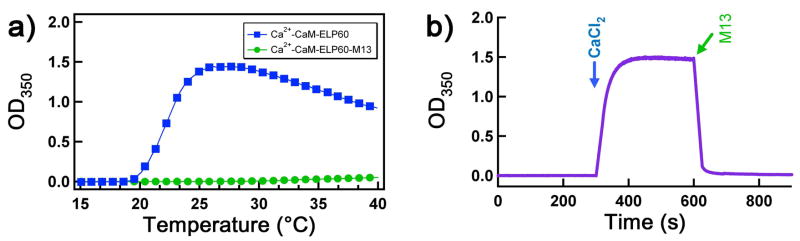
Effect of sequential addition of Ca2+ and EDTA on LCST behavior of CaM-ELP60. a) Turbidity at 350 nm (OD350) as a function of solution temperature for apoCaM-ELP60 (filled circles, red), Ca2+-CaM-ELP60 (filled squares, blue). b) Reversible control of self-assembly of CaM-ELP fusion as a function of Ca2+ binding. The OD350 was measured as a function of time after sequential injection of Ca2+ followed by EDTA as indicated by the arrows. Three cycles of addition of Ca2+ followed by EDTA are shown in the Figure. c) Rh measured by DLS of apoCaM-ELP60 and Ca2+-CaM-60, showing the existence of apoCaM-ELP60 as a soluble monomer and Ca2+-CaM-ELP60 as a monodisperse nanoparticle with a Rh of ~180 nm. Tapping mode AFM images of: d) apoCaM-ELP60 and e) Ca2+-CaM-ELP60. Vertical scale bar indicates height of the features in the 5×5 μm images. The squares demarcate regions of d, and e, whose magnified views are shown as 3-D reconstructions of the AFM images.
Figure 3a shows that in the absence of Ca2+, the baseline turbidity of apoCaM-ELP at room temperature was zero, which is consistent with the fact that apoCaM-ELP is in a soluble state with a Rh of 6.2 ± 2 nm (mean ± s.d.) (Figure 3c). In contrast, Ca2+ bound CaM-ELP showed a significant increase in the solution turbidity at room temperature (Figure 3a) due to the formation of particles with an Rh of 180 ± 31 nm (Figure 3c). This behavior was completely reversible, as shown by the repeated formation of meso-scale particles by addition of 0.5 mM Ca2+ and their dissolution rapidly back to soluble CaM-ELP molecules by the addition of 0.5 mM EDTA at room temperature (Figure 3b).
We directly confirmed the reversible formation of these meso-scale structures by tapping mode AFM of freeze-dried samples. As shown in Figure 3d, in the absence of Ca2+, apoCaM-ELP60 forms particles with a nominal height of 12 ± 2.5 nm (n = 50) and a lateral diameter of 69 ± 15 nm. Although these lateral sizes are significantly greater than the hydrated diameter of a single apoCaM-ELP (~12 nm), we believe they are caused by tip-induced broadening of the features41 for the following reasons: (1) in control AFM experiments, in which apoCaM was deposited onto a polyallyl-amine hydrochloride (PAH) coated Si wafer from a 1 μM solution in water at room temperature, conditions under which apoCaM exists as a soluble monomer (with a Rh of ~6 nm), we observed a sparse distribution of individual particles with a height of 10.4 ± 3.4 nm (n = 40) and a lateral diameter of 61.2 ± 2.2 nm, (Figure S2 in the Supporting Information); (2) in separate control experiments, we also carried out tapping mode AFM on 9.9 nm diameter gold nanoparticles in air and observed that their lateral diameter varied between 29.7 ± 3.5 nm to 35.6 ± 3.8 nm for scanning speed of 2 μm/s and 16 μm/s respectively (Figure S3 in the Supporting Information). Together, these control experiments showed that particles with a known size of ~10 nm are significantly broadened in AFM imaging, which strongly suggests that the features observed in Figure 2d are those of individual apoCaM-ELP60 molecules. Upon addition of 0.5 mM CaCl2, and subsequent imaging of freeze-dried CaM-ELP60, particles with a height of 276 ± 63 nm (n = 50) and a lateral diameter of 331 ± 68 nm were observed (Figure 3e), which are consistent with the formation of the mesoscale particles with a hydrated diameter of ~360 nm observed by DLS. The agreement between the DLS and AFM results is consistent with the notion that particles in this size range would not be expected to exhibit significant tip-induced broadening, as they are significantly larger than the AFM tip radius.
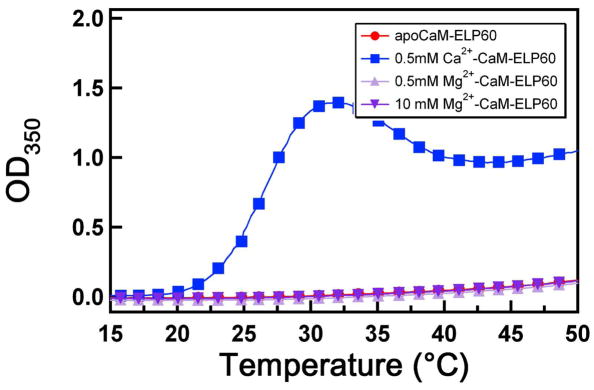
We also examined the sensitivity of the reversible phase transition of CaM-ELP to MgCl2, which is a physiologically relevant divalent cation.42 Figure 4 shows that a concentration of up to 10 mM MgCl2 had no appreciable effect on the phase transition of CaM-ELP60 and was similar to that of apoCaM-ELP60. In contrast, Ca2+ bound CaM showed a marked change in its phase behavior compared to apoCaM, even at a CaCl2 concentration as low as 0.5 mM. These results are consistent with a structural study of Mg2+ binding to CaM,43 that showed that the structure of Mg2+-CaM is nearly the same as apoCaM, as Mg2+ binding had no effect on the overall conformation of the protein. This experiment strongly supports our hypothesis that the key to Ca2+-triggered self-assembly and control of particle size in the CaM-ELP fusion rests in the ability of Ca2+ to trigger a large conformational change in the protein that exposes hydrophobic residues and consumes charge.
Figure 4.
Effect of Mg2+ ion binding to CaM on LCST behavior of CaM-ELP60. Solution turbidity at 350 nm (OD350) as a function of solution temperature for apoCaM-ELP60 (filled circles, red), Ca2+-CaM-ELP60 (filled squares, blue), Mg2+-CaM-ELP60 with 0.5 mM Mg2+ (filled triangles, light purple), Mg2+-CaM-ELP60 with 10 mM Mg2+(filled inverse triangles, dark purple).
Finally, it is well known that CaM bind to a number of peptide ligands and that this binding is calcium dependent.44 In order to assess the effect of the interaction of Ca2+-CaM with its peptide ligand on the LCST transition of the fusion protein, we chose a CaM binding peptide derived from the skeletal muscle Myosin Light Chain Kinase (termed M13 peptide).44 Figure 5a shows that in the presence of M13 peptide, the behavior of Ca2+ bound CaM-ELP-M13 as a function of temperature is similar to that of apoCaM. The altered temperature dependent aggregation observed is consistent with the burial of a large solvent exposed hydrophobic area in Ca2+ bound CaM upon binding of the M13 peptide and potentially the increase in overall charge,44 factors that are known to increase the LCST and decrease the size of aggregates of ELP fusion proteins.24 This behavior was also confirmed in an isothermal experiment, by sequential injection of Ca2+ followed by M13 peptide as indicated by the arrows at 25°C temperature (Figure 5b), which shows that the addition of M13 peptide at a temperature changes the LCST of Ca2+-CaM and disrupts the aggregated ELP.
Figure 5.
Effect of binding of M13 peptide with Ca2+-CaM-ELP60 on LCST behavior of the ELP fusion protein. a) Turbidity at 350 nm (OD350) as a function of solution temperature for Ca2+-CaM-ELP60 (filled squares, blue), Complex of Ca2+-CaM-ELP60 with M13 peptide, M13 (filled circles, green). b) Control of self-assembly of CaM-ELP fusion upon M13 peptide binding. The OD350 was measured as a function of time after sequential injection of Ca2+ followed by M13 peptide in a ratio of fusion protein:peptide (1:1) as indicated by the arrows.
We believe that these results are notable for the following reasons. First, the calcium triggered LCST behavior of CaM-ELP180 and CaM-ELP60 demonstrates that CaM-ELP fusions can be triggered to reversibly cycle between nanoparticles and microparticles (ELP180) or between soluble monomer and mesoscale particles (ELP60), and highlight the control over supramolecular organization available in this system by choice of the ELP and solution conditions. Second, to the best of our knowledge, these results are the first demonstration of an LCST transition in a polymer that is triggered by an allosteric ligand binding event. Although we do not have direct and unequivocal experimental evidence that the effect we see is allosteric in the conventional sense, we believe that this is indeed the case in that the binding of calcium to CaM triggers a conformational change in the protein which, we believe is further transmitted to the ELP via the collapse of the ELP chain. We believe that without collapse of the ELP chain, it is unlikely that the phase transition, as observed by the aggregation of the ensemble of ELP molecules would occur, as it has been shown by AFM studies45 and MD simulations46 that the release of bound water from the ELP with rising temperature leading to collapse of the ELP chain are the necessary molecular events that drive the phase transition of the ELP. We see this intramolecular collapse of the ELP triggered by calcium binding to the fusion partner, CaM, as the equivalent of an allosteric conformational change that would occur in the conventional case of a folded protein.
The ability to trigger an LCST transition by a ligand-binding event at a spatial location that is separate and distinct from the ELP is an important finding, because if it proves to be a general phenomenon, it will provide a simple, convenient and modular approach to design biopolymer actuators by splicing together ligand binding domains with ELPs. The closest analog to the present study are previous reports from the Stayton and Hoffman groups on the design of functional protein switches via conjugation of a protein to a synthetic, stimulus responsive polymer, poly(NIPAAm).11–15 In contrast, herein we have demonstrated the reverse behavior, that ligand binding to a protein can drive the LCST phase transition of an appended stimulus responsive polymer, a phenomenon that to our knowledge has not been previously reported.
CONCLUSIONS
The ability to modulate the phase transition behavior of CaM-ELP in response to Ca2+ binding demonstrated herein is a new form of allosteric actuation in an artificial protein-biopolymer construct. In the short term, we foresee two complementary lines of investigation to expand upon these studies: first, it will be important to dissect the mechanism of allosteric actuation by investigation of the calcium triggered behavior of CaM-ELP variants in which the calcium binding residues in CaM are mutated and the chain lengths and molecular composition of the attached ELPs are systematically varied. Second, we hope to expand this concept of allosteric transduction of the LCST transition to other ligand binding proteins and peptide sequences that display the requisite behavior upon ligand binding, namely a decrease in the solvent-accessible, charged surface area and a concomitant increase in surface hydrophobicity. Expanding the repertoire of protein-ligand binding pairs via which the LCST transition of ELPs and potentially other stimulus responsive polymers can be triggered is an important goal, because it would provide a new class of modular biopolymer actuators that can be driven by the enormous number of biochemical binding events that are available. We anticipate that the ability to reversibly control supramolecular organization of CaM-ELP in response to calcium binding demonstrated here will lead to new applications that exploit this unique actuation behavior in the design of biosensors, drug and gene delivery vehicles and nansocale devices.
EXPERIMENTAL
Synthesis of CaM-ELP fusion gene
Synthetic genes encoding for the two different lengths of the ELP (ELP60 and ELP180) were synthesized by recursive directional ligation in pUC-19.47 ELP60 and ELP180 refer to an ELP composed of 60 and 180 VPGXG pentapeptides respectively, where the X is Val (V), Ala (A) and Gly (G) in a 5:2:3 ratio. The DNA sequence for this gene has been published previously.18 CaM-ELP genes (Invitrogen) were synthesized by inserting the CaM gene 5′ to the ELP gene in a modified pET25b vector. The modified pET25b vector was produced by replacement of the NotI to AvaI segment of pET25b (Novagen) with an oligonucleotide cassette (Integrated DNA Technologies) encoding for a thrombin cleavage site and an SfiI restriction site. The ELP gene was inserted into the SfiI restriction site as previously described.18
The CaM gene was retrieved from its plasmid (provided by Invitrogen) by PCR and TA cloning (Invitrogen), in which NdeI and SalI restriction sites were also incorporated 5′ and 3′ to the gene, respectively. The gene was excised from the TA vector using NdeI and SalI, separated from the linearized vector by gel electrophoresis and isolated from the agarose gel after electrophoresis (Qiaex II Gel Extraction kit; Qiagen). The modified pET25b vector was digested with NdeI and SalI, and the fusion protein was assembled by ligation of the CaM gene with the restricted pET25b vector containing the ELP gene. Correct assembly of the fusion protein gene was confirmed by DNA sequencing.
CaM-ELP expression and purification
The CaM-ELP fusions were expressed in BLR(DE3) E. coli (Novagen). The proteins were expressed as 1 L cultures of E. coli in CircleGrow™ media (Qbiogene, Carlsbad, Ca, USA), supplemented with 100 μg/ml ampicillin. One-liter cultures were inoculated with cells from 20 mL of a starter culture that was inoculated from frozen (−80°C) DMSO stocks and grown overnight. After inoculation, the culture was incubated for 24 h at 37°C and CaM-ELP was purified from other soluble E. coli proteins using a nonchromatographic separation method for recombinant proteins, inverse transition cycling,18 as follows: the ionic strength of the soluble lysate was increased by the addition of (NH4)2SO4 to cause aggregation of the CaM-ELP fusion protein in the cell lysate, and the aggregated CaM-ELP fusion protein in the cell lysate was separated from soluble E. coli proteins by centrifugation at ~20,000 rcf for 15 min at moderate temperatures (25–37°C). Purification of CaM-ELP180 typically required 0.4 M (NH4)2SO4, while that of CaM-ELP60 required 1 M (NH4)2SO4. The pellet containing the CaM-ELP fusion protein coacervate was dissolved in cold 10 mM HEPES buffer and centrifuged at 4°C to remove any particulate contaminants. This aggregation and dissolution process was repeated 3–4 times until the CaM-ELP fusion protein was determined to be approximately 95% pure of E. coli proteins by visualization on Coomassie blue or copper-stained SDS-PAGE gels. In a control experiment (Figure S1), CaM was liberated from its ELP fusion partner by enzymatic cleavage using thrombin as follows: CaM-ELP was cleaved overnight at 4°C from solutions containing approximately 100 μM CaM using 10 units of thrombin per μmole of fusion protein. Protein concentrations were determined by UV-visible spectroscopy using extinction coefficients at 280 nm calculated from the primary amino acid sequence with the software program Protean (DNA Star, Madison, WI).
Temperature and time dependent turbidity measurement
The temperature-dependent LCST behavior of the CaM-ELP fusion proteins was characterized by measuring the optical density at 350 nm (OD350) as a function of solution temperature. Solutions of the CaM-ELP fusion protein (25 μM) were heated at 1°C/min in a Cary 300 UV-visible spectrophotometer equipped with a multi-cell thermoelectric temperature controller (Varian Instruments, Walnut Creek, Ca) between 10–60°C. The experiments were performed in 10 mM HEPES with an additional 1 M NaCl (CaM-ELP180) or 0.4 M (NH4)2SO4 (CaM-ELP60). The LCST of CaM-ELP was defined as the temperature at which the OD reached 50% of its maximum value. The effect of reversible Ca2+-triggered changes of CaM-ELP turbidity was measured by their OD350 as a function of time at constant temperature as follows: prior to turbidity measurements, samples were equilibrated at a temperature of interest for 10 min. CaCl2 and EDTA (both at 0.5 mM final concentration) was alternately added every 5 min and the OD350 was recorded continuously as a function of time. The M13 skeletal muscle myosin light chain kinase peptide (M13 peptide), with the sequence KRRWKKNFIAVSAANRFKKISSSGAL (Anaspec Inc., San Jose, CA), was used to investigate the effect of the binding of a peptide ligand on the LCST transition of Ca2+-CaM. The OD350 was measured as a function of time after sequential injection of Ca 2+ (0.5 mM final concentration) into a solution of apoCaM-ELP60 (25 μM) followed by the addition of an equimolar concentration of the M13 peptide.
Dynamic light scattering
The particle size distribution of CaM-ELP fusions was measured as a function of temperature by dynamic light scattering. Samples were prepared at the protein and solvent compositions used in the turbidity measurements described above. Prior to particle size measurement, samples were centrifuged at 16,000×g for 10 min at 4°C remove insoluble debris and were then filtered through a 20 nm Whatman Anodisc filter at a temperature below the LCST. Autocorrelation functions were collected using DynaPro-LSR dynamic light scattering instrument (Protein Solutions, Charlottesville, VA) equipped with a Peltier temperature control unit. Light scattering data were collected at regular temperature intervals (1°C) as solutions were heated from 10 to 60°C. Data were obtained at each temperature by heating the cell to the temperature of interest, allowing the sample to equilibrate for 1 min, and then collecting 6 measurements, each with a 10 s collection time. The autocorrelation function was analyzed using the algorithm provided by the manufacturer to determine the hydrodynamic radius and mass faction of the ~10 nm to the μm scale particles.
Atomic force microscopy
The particle size of CaM-ELP was measured by tapping mode atomic force microscopy (AFM) (MultiMode, Veeco Digital Instruments, Woodbury, New York) in air using Si cantilevers with a force constant of ~42 N/m. Silicon substrates were cleaned with piranha solution (a 3:1 v/v mixture of sulfuric acid and hydrogen peroxide) at 80°C for 30 min (Caution: piranha is a vigorous oxidant and should be used with extreme caution!). In order to enhance the adsorption of CaM-ELP, cleaned silicon substrates were immersed in a 0.003 monomol/L (monomer mole/liter) solution of positively charged poly(allylamine) hydrochloride (PAH, MW 70000) in 1 M NaCl for 30 min. After deposition of PAH, the silicon substrates were rinsed with purified water and dried with a stream of nitrogen gas. The RMS roughness of the surface was < 1 nm as measured by AFM over a scan size of 5 μm. A drop of apoCaM-ELP60 (25 μM concentration unless otherwise specified) or Ca2+-CaM-ELP60 in HEPES, 0.4 M (NH4)2SO4, pH 7.4 at room temperature were deposited onto a PAH coated silicon substrate and plunged into liquid nitrogen. The sample was held under liquid nitrogen until it freeze-dried completely for 48 h. To obtain a low enough surface coverage to visualize individual protein molecules or aggregated particles, the deposition time varied between 5 and 60 s. To ensure a flat background before image analysis, the images were flattened and plane-fitted by using the microscope’s analysis software (Digital Instruments, Version 5.12r3). The height and lateral size of particles was determined from cross-sectional analysis of AFM images.
Supplementary Material
Supporting information contains: (1) SDS-PAGE gel showing thrombin cleavage of CaM-ELP180, and tapping mode AFM images in air of (2) freeze-dried apoCaM-ELP60 (from a1 μm concentration solution in water), and (3) 10 nm diameter gold nanoparticles adsorbed on a substrate.
Acknowledgments
We thank Dr. Wookyung Lee for assistance with the AFM imaging. This work was funded by National Institutes of Health Grant GM-061232.
References
- 1.Schild HG. Prog Polym Sci. 1992;17:163–249. [Google Scholar]
- 2.Urry DW. J Prot Chem. 1988;7:1–34. doi: 10.1007/BF01025411. [DOI] [PubMed] [Google Scholar]
- 3.Yamaoka T, Tamura T, Seto Y, Tada T, Kunugi S, Tirrell DA. Biomacromolecules. 2003;4:1680–1685. doi: 10.1021/bm034120l. [DOI] [PubMed] [Google Scholar]
- 4.Mao HB, Li CM, Zhang YJ, Furyk S, Cremer PS, Bergbreiter DE. Macromolecules. 2004;37:1031–1036. [Google Scholar]
- 5.Zhang YJ, Furyk S, Bergbreiter DE, Cremer PS. J Am Chem Soc. 2005;127:14505–14510. doi: 10.1021/ja0546424. [DOI] [PubMed] [Google Scholar]
- 6.Furyk S, Zhang YJ, Ortiz-Acosta D, Cremer PS, Bergbreiter DE. J Polym Sci Part A: Polym Chem. 2006;44:1492–1501. [Google Scholar]
- 7.Zhang Y, Furyk S, Sagle LB, Cho Y, Bergbreiter DE, Cremer PS. J Phys Chem C. 2007;111:8916–8924. doi: 10.1021/jp0690603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urry DW, Hayes LC, Gowda DC, Harris CM, Harris RD. Biochem Biophys Res Commun. 1992;188:611–617. doi: 10.1016/0006-291x(92)91100-5. [DOI] [PubMed] [Google Scholar]
- 9.Irie M. Adv Polym Sci. 1993;110:49–65. [Google Scholar]
- 10.Strzegowski LA, Martinez MB, Gowda DC, Urry DW, Tirrell DA. J Am Chem Soc. 1994;116:813–814. [Google Scholar]
- 11.Stayton PS, Shimoboji T, Long C, Chilkoti A, Chen GH, Harris JM, Hoffman AS. Nature. 1995;378:472–474. doi: 10.1038/378472a0. [DOI] [PubMed] [Google Scholar]
- 12.Ding ZL, Fong RB, Long CJ, Stayton PS, Hoffman AS. Nature. 2001;411:59–62. doi: 10.1038/35075028. [DOI] [PubMed] [Google Scholar]
- 13.Shimoboji T, Ding ZL, Stayton PS, Hoffman AS. Bioconj Chem. 2002;13:915–919. doi: 10.1021/bc010057q. [DOI] [PubMed] [Google Scholar]
- 14.Shimoboji T, Larenas E, Fowler T, Kulkarni S, Hoffman AS, Stayton PS. Proc Natl Acad Sci USA. 2002;99:16592–16596. doi: 10.1073/pnas.262427799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimoboji T, Larenas E, Fowler T, Hoffman AS, Stayton PS. Bioconj Chem. 2003;14:517–525. doi: 10.1021/bc025615v. [DOI] [PubMed] [Google Scholar]
- 16.Gil ES, Hudson SA. Prog Polym Sci. 2004;29:1173–1222. [Google Scholar]
- 17.Urry DW. Prog Biophys Mol Biol. 1992;57:23–57. doi: 10.1016/0079-6107(92)90003-o. [DOI] [PubMed] [Google Scholar]
- 18.Meyer DE, Chilkoti A. Nature Biotech. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 19.Trabbic-Carlson K, Liu L, Kim B, Chilkoti A. Prot Sci. 2004;13:3274–3284. doi: 10.1110/ps.04931604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banki MR, Feng LA, Wood DW. Nature Methods. 2005;2:659–661. doi: 10.1038/nmeth787. [DOI] [PubMed] [Google Scholar]
- 21.Diehl MR, Zhang KC, Lee HJ, Tirrell DA. Science. 2006;311:1468–1471. doi: 10.1126/science.1122125. [DOI] [PubMed] [Google Scholar]
- 22.Jenikova G, Lao UL, Gao D, Mulchandani A, Chen W. Langmuir. 2007;23:2277–2279. doi: 10.1021/la0626151. [DOI] [PubMed] [Google Scholar]
- 23.Meyer DE, Shin BC, Kong GA, Dewhirst MW, Chilkoti A. J Controlled Release. 2001;74:213–224. doi: 10.1016/s0168-3659(01)00319-4. [DOI] [PubMed] [Google Scholar]
- 24.Trabbic-Carlson K, Meyer DE, Liu L, Piervincenzi R, Nath N, LaBean T, Chilkoti A. Prot Engin Design Selection. 2004;17:57–66. doi: 10.1093/protein/gzh006. [DOI] [PubMed] [Google Scholar]
- 25.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. J Mol Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 26.Babu YS, Bugg CE, Cook WJ. J Mol Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- 27.Finn BE, Evenas J, Drakenberg T, Waltho JP, Thulin E, Forsen S. Nature Struct Biol. 1995;2:777–783. doi: 10.1038/nsb0995-777. [DOI] [PubMed] [Google Scholar]
- 28.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Nature Struct Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Tanaka T, Ikura M. Nature Struct Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 30.Evenas J, Malmendal A, Forsen S. Curr Opin Chem Biol. 1998;2:293–302. doi: 10.1016/s1367-5931(98)80072-0. [DOI] [PubMed] [Google Scholar]
- 31.Brini M, Carafoli E. Cell Molec Life Sci. 2000;57:354–370. doi: 10.1007/PL00000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, Baker D, Tsien RY. Chem Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Topp S, Prasad V, Cianci GC, Weeks ER, Gallivan JP. J Am Chem Soc. 2006;128:13994–13995. doi: 10.1021/ja064546i. [DOI] [PubMed] [Google Scholar]
- 34.Ehrick JD, Deo SK, Browning TW, Bachas LG, Madou MJ, Daunert S. Nature Materials. 2005;4:298–302. doi: 10.1038/nmat1352. [DOI] [PubMed] [Google Scholar]
- 35.Murphy WL, Dillmore WS, Modica J, Mrksich M. Angew Chem Int Ed. 2007;46:3066–3069. doi: 10.1002/anie.200604808. [DOI] [PubMed] [Google Scholar]
- 36.Atanasijevic T, Shusteff M, Fam P, Jasanoff A. Proc Natl Acad Sci USA. 2006;103:14707–14712. doi: 10.1073/pnas.0606749103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konishi K, Uyeda TQP, Kubo T. FEBS Lett. 2006;580:3589–3594. doi: 10.1016/j.febslet.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 38.Zhang YJ, Trabbic-Carlson K, Albertorio F, Chilkoti A, Cremer PS. Biomacromolecules. 2006;7:2192–2199. doi: 10.1021/bm060254y. [DOI] [PubMed] [Google Scholar]
- 39.Meyer DE, Chilkoti A. Biomacromolecules. 2004;5:846–851. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 40.Zhang YJ, Cremer PS. Curr Opin Chem Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 41.Markiewicz P, Goh MC. Rev Sci Instrum. 1995;66:3186–3190. [Google Scholar]
- 42.Martin RB. Bioinorganic Chemistry of Magnesium. In: Sigel H, editor. Metal ions in Biological Systems. New York: Marcel Dekker Inc; 1990. pp. 1–13. [Google Scholar]
- 43.Ohki S, Ikura M, Zhang M. Biochem. 1997;36:4309–4316. doi: 10.1021/bi962759m. [DOI] [PubMed] [Google Scholar]
- 44.Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, Bax A. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 45.Valiaev A, Lim DW, Schmidler S, Chilkoti A, Zauscher S. J Am Chem Soc. 2008;130:10939–10946. doi: 10.1021/ja800502h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, Alonso DOV, Daggett V. J Mol Biol. 2001;305:581–592. doi: 10.1006/jmbi.2000.4306. [DOI] [PubMed] [Google Scholar]
- 47.Meyer DE, Chilkoti A. Biomacromolecules. 2002;2:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information contains: (1) SDS-PAGE gel showing thrombin cleavage of CaM-ELP180, and tapping mode AFM images in air of (2) freeze-dried apoCaM-ELP60 (from a1 μm concentration solution in water), and (3) 10 nm diameter gold nanoparticles adsorbed on a substrate.