Abstract
Reconstructing the evolutionary history of Hox cluster origins will lead to insights into the developmental and evolutionary significance of Hox gene clusters in vertebrate phylogeny and to their role in the origins of various vertebrate body plans. We have isolated two Hox clusters from the horn shark, Heterodontus francisci. These have been sequenced and compared with one another and with other chordate Hox clusters. The results show that one of the horn shark clusters (HoxM) is orthologous to the mammalian HoxA cluster and shows a structural similarity to the amphioxus cluster, whereas the other shark cluster (HoxN) is orthologous to the mammalian HoxD cluster based on cluster organization and a comparison with noncoding and Hox gene-coding sequences. The persistence of an identifiable HoxA cluster over an 800-million-year divergence time demonstrates that the Hox gene clusters are highly integrated and structured genetic entities. The data presented herein identify many noncoding sequence motifs conserved over 800 million years that may function as genetic control motifs essential to the developmental process.
The Hox gene clusters, first described in Drosophila as the Antenapedia and Bithorax clusters, control pattern formation along the anterior–posterior axis in bilateral animals (1). There is a single Hox gene cluster in all invertebrate species reported to date (2–5). In contrast, multiple Hox clusters have been reported in all vertebrate species examined (6–9). Reconstructing the evolutionary history of vertebrate Hox cluster duplication is necessary to identify orthologous clusters in different vertebrate species, to establish phylogenetic relationships among and between clades, and to elucidate the role of Hox cluster duplication in the vertebrate radiation.
We have isolated two genomic clones containing Hox clusters from the primitive horn shark, Heterodontus francisci. One (Het1) is 81.2 kilobases (kb) in length, and the other (Het2) is 98.8 kb. Both have been sequenced completely and compared inter se and with other vertebrate Hox sequences. The Heterodontus Hox cluster data reported herein provide definitive map positions for the Hox genes within the shark clusters as well as extensive sequence information in coding and noncoding domains.
Materials and Methods
Genomic DNA Sequencing.
Nucleotide sequences were primarily determined by using the shotgun method, and unsequenced gaps were filled by the primer walking method (10). Two distinct dye termination kits (dRhodamine terminator cycle sequencing kit and BigDye terminator cycle sequencing kit, Perkin–Elmer) were used and analyzed by a 377 PRISM DNA sequencer (Perkin-Elmer). Regions covered with only one shotgun clone were resequenced with the other dye termination kit. The entire sequencing region was ensured by covering at least one highly reliable reading (<550-nucleotides-long from a sequencing primer). For regions covered by unidirectional readings or covered by less than three readings, all the sequence trace patterns were inspected to eliminate base calling errors such as band compressions. The obtained sequence was finally confirmed by comparing a restriction map deduced from genomic sequence with an experimentally constructed restriction map (10).
Cluster Alignments and Sequence Comparisons.
Alignments were computed by an experimental version of the blast program (11), with the following alignment scores: match = +1, transition = −0.7, transversion = −1.0, gap open = −4.0, and gap extension = −0.2. The regions that are “strongly conserved” between the human HoxA sequence and the Heterodontus HoxM sequence, indicated by orange, were computed with more stringent scores: match = +1, transition = −0.9, transversion = −1.1, gap open = −6.0, and gap extension = −0.2. The alignments are drawn as a percentage identity plot (12) by using an updated version of the pmps program (13). These programs compute and display alignments of genomic sequences and can be run on user-supplied data at http://globin.cse.psu.edu/pipmaker.
P1 Artificial Chromosome (PAC) Library.
We have constructed a horn shark PAC library (2× coverage; ref. 14). Multiple clones (n = 1,000) were arrayed in single wells to reduce the number of samples required because of very large genome size (8 × 109 bp). Genomic DNA pools from the horn shark PAC library were screened with two degenerate primer sets for amplifying homeodomains (15), and putative Hox genes were identified by hybridization to PAC colonies. End sequences of PAC clones having Hox genes were sequenced, and specific PCR primers were designed to identify overlapping clones. The Hox content of each positive PAC clone was determined by using PCR assays with primers to the cognate genes, by subsequent DNA sequencing, and by Southern hybridization to restriction digests of respective PAC clones. Subsequently, Hox cluster-containing PAC clones were subjected to clone fingerprinting (14, 16).
Phylogenetic Analysis.
To determine the similarity of horn shark exon sequences for each paralogous gene and PCR fragments of homeoboxes to known Hox sequences, blastx searches of the GenBank database were done. Sequences were aligned with the clustal x program (17) for phylogenetic analyses. The regions difficult to align were excluded from data files. The gene phylogenetic trees were generated by maximum parsimony by using paup (version 3.1; ref. 18) and by neighbor-joining method (19) by using programs from the phylip package (version 3.572c; ref. 20) with 1,000 bootstrap replications. Nodes with low support (<70%) in all analyses have been collapsed to polychotomies. Reconstructing the phylogeny of genes in paralogous group 9, protein sequences of exons 1 and 2 from human A9 (GenBank accession no. P31269), human D9 (P28356), mouse B9 (P20615), mouse C9 (P09633), zebrafish A9a (AF071248), zebrafish A9b (AF071249), zebrafish B9a (AF071256), zebrafish C9a (AF071267), and zebrafish D9a (AF071268) were selected from GenBank and aligned with those of horn shark HoxM-9 and HoxN-9 by using clustal x. Nucleotide sequences of human Evx-1 (X60655), zebrafish Evx-1 (X71845), mouse Evx-2 (M93128), and zebrafish Evx-2 (X99290) were obtained from GenBank and aligned with those of horn shark EvxN.
Results and Discussion
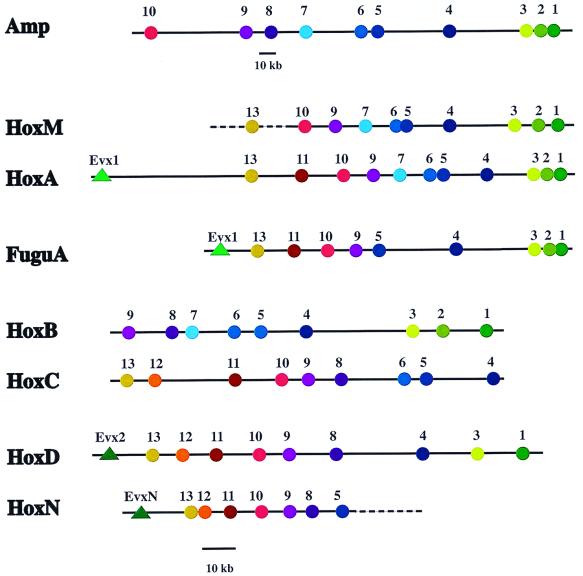
A Heterodontus PAC library was screened to isolate clones containing large segments of the genomic Hox clusters. The sequence and the contig map information of clones reveal the presence of two distinct Hox clusters in Heterodontus designated HoxM and HoxN. Two clones, Het1 with an insert of 81.2 kb and Het2 with an insert of 98.8 kb, have been completely sequenced. A complete cluster has been isolated for HoxM by the isolation of a clone that overlaps with Het1 shown by dashed extensions to HoxM in Fig. 1. The HoxN is still incomplete in its 3′ terminus (Fig. 1). The Hox gene composition and the spacing of genes are shown in Fig. 1.
Figure 1.
Comparisons of Hox gene spacing and gene dropouts between the amphioxus Hox cluster (Amp), human HoxA (HoxA), Fugu HoxA (FuguA; ref. 21), Heterodontus HoxM (HoxM), mouse HoxB (HoxB), mouse HoxC (HoxC), Heterodontus HoxN (HoxN), and mouse HoxD (HoxD) clusters. Spacing between gene coding regions is based on contig and sequence analysis. GenBank accession numbers for the human HoxA cluster are AC004079 and AC004080. Dotted lines indicate extensions not yet sequenced. The scale below Amp is for Amp only, and the scale below HoxN is for the other clusters.
We compared the position of Hox genes within the horn shark clusters HoxM and HoxN, the amphioxus Hox cluster, the Fugu rubripes HoxA cluster, the human HoxA cluster, and the mouse HoxB, HoxC, and HoxD clusters (Fig. 1). The map positions of the human HoxA and the horn shark clusters HoxM and HoxN are based on full sequence data, whereas the location of the Hox genes of the amphioxus cluster (4), the Fugu HoxA cluster (21), and the mouse HoxB, HoxC, and HoxD clusters has been determined by contig mapping. The horn shark HoxM and HoxN clusters are clearly different, based on gene location (i.e., spacing) and gene presence/absence (i.e., gene dropout). HoxM shows a high degree of correspondence to the amphioxus and the human HoxA clusters where patterns of gene spacing and dropout are remarkably similar. Hox genes 1, 2, and 3 are closely spaced. Long intervals separate gene 4 from genes 1, 2, and 3 and 5 and 6. Genes 5 and 6 are closely spaced, and then moderate and regular spacings separate the remaining genes extending 5′. These spacing and dropout relationships are maintained between amphioxus and vertebrate HoxA clusters even though the amphioxus cluster is approximately two and half times longer than the human HoxA cluster (100 kb).
The HoxN cluster is more similar to the mouse HoxD cluster than to the mammalian HoxA cluster, based on gene spacing and dropout (Fig. 1). Hox genes 5 and 8–13, and EvxN and Evx-2 are present in both horn shark HoxN and mouse HoxD clusters, respectively. Hox5 is present in the horn shark HoxN cluster but absent in the mouse HoxD cluster, and Hox6 and Hox7 are absent in both clusters.
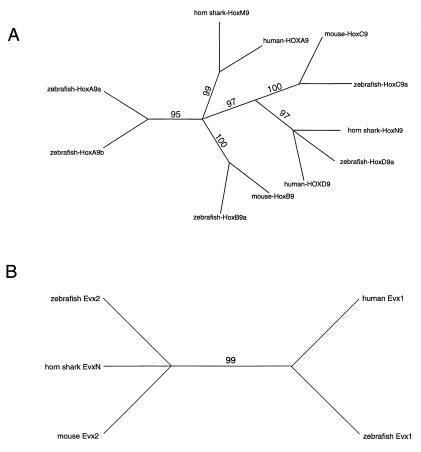
To determine the branching relationship of the Heterodontus cluster units with their mammalian and bony fish orthologs, we have carried out phylogenetic analyses based on amino acid sequences of Hox exon 1 and 2 coding regions (Fig. 2A). We restricted our analysis to Hox9 genes, because Hox9 is present in all four mammalian clusters and does not require any weighting adjustments. The branching pattern strongly supports the homology of shark HoxM and mammalian HoxA. It is interesting that zebrafish HoxAa and HoxAb genes separate independently, suggesting that the duplication of the HoxA cluster into HoxAa and HoxAb clusters in the ray-finned fishes may have been coupled with a sequence divergence of the paralogous Hox9 coding domains. The Hox9 branching pattern again reinforces the divergence between the shark HoxM and HoxN clusters and, in addition, supports a homology relationship between shark HoxN and mammalian and zebrafish HoxD genes. The zebrafish has only a single HoxD cluster.
Figure 2.
Phylogenetic relationships between Hox9 genes and Evx. Numbers above the lines are bootstrap values obtained in 1,000 replicates for maximum parsimony. (A) Phylogenetic analysis of amino acid sequences of exons 1 and 2 of Hox9 genes of human, mouse, zebrafish, and horn shark. (B) Phylogenetic analysis of nucleotide sequences of Evx-1 and Evx-2 of human, mouse, and zebrafish and EvxN of horn shark.
To examine further the possible HoxD relationship with the shark HoxN cluster, we examined the branching relationships of the shark EvxN and mammalian and zebrafish Evx-1 and Evx-2 genes (Fig. 2B). This analysis is based on nucleotide sequence comparisons, because the boundaries of the Evx coding domains have not been clearly defined. The data support an orthologous relationship between shark EvxN and Evx-2. This result further strengthens the identity of shark HoxN as being a HoxD-like cluster, because Evx-2 is closely linked to the HoxD cluster in mammals and ray-finned fishes. We also compared the available amphioxus amino acid sequence data for Amphi-Hox genes 1–4 with those of Heterodontus and some vertebrates (data not shown). These results were ambiguous, giving weak associations of Amphi-Hox genes 1, 2, 3, and 4 with HoxA, HoxB, HoxC, and HoxC genes, respectively. We suspect that the divergence time between the amphioxus and the sharks is extreme and may account for this discrepancy. This suspicion is strengthened by recent findings that show that the Amphi-Hox genes 1–4 (particularly Amphi-Hox2) have distinctly different expression patterns than their corresponding genes in the vertebrates (22). In summary, the phylogenetic branching patterns of informative Hox genes supports a close relation of HoxA and the shark HoxM cluster and of HoxD and the shark HoxN cluster.
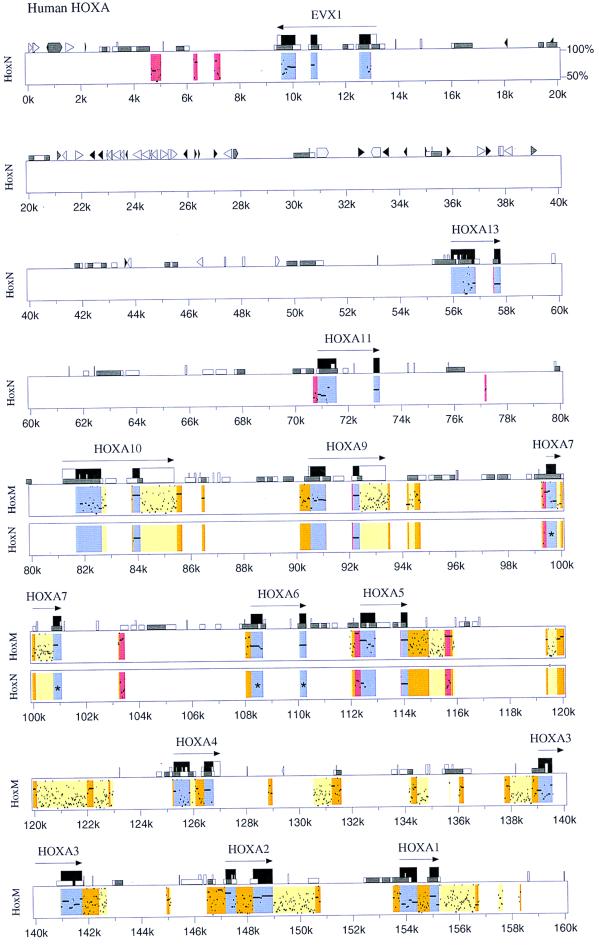
We are restricted to making extensive sequence comparisons between horn shark HoxM and HoxN and human HoxA clusters, because these are the only Hox clusters for which extensive and reliable sequence data are available at present. The horn shark HoxM cluster shows a high degree of sequence similarity to the human HoxA cluster in both the coding and noncoding regions (Fig. 3). There are in excess of 100 sites that show matches higher than 50% similarity and more than 30 that exceed 75%. Some of the conserved elements extend over 100 bp. Horn shark HoxN shows much less sequence similarity to human HoxA than does horn shark HoxM.
Figure 3.
Comparisons of nucleotide sequence between human HoxA, Heterodontus HoxM, and HoxN Hox clusters. Sequence comparisons are based on the human HoxA cluster as a reference. Kilobase (k) markings relate to the human HoxA cluster. Actual spacings between genes and gene dropouts are shown in Fig. 1. Color codes and symbols are as follows. Blue signifies coding regions. Yellow indicates weakly conserved noncoding regions. Orange indicates strongly conserved noncoding regions. Red indicates noncoding sequences conserved in both Heterodontus HoxM and HoxN clusters at the same position. The long horizontal arrows indicate direction of transcription. Tall black boxes indicate protein coding regions. Tall open boxes show, where possible, untranslated regions of a gene as determined from mRNA sequences in GenBank (namely, Evx-1, HoxA10, HoxA9, and HoxA4). Medium size open boxes (e.g., at position 13.9k) denote simple repeats and low-complexity regions. Short open boxes (e.g., 4k) demarcate CpG islands, with open boxes indicating a CpG/GpC ratio between 0.6 and 0.75 and gray indicating a ratio above 0.75. Interspersed repeat elements are shown as triangles and pointed boxes, where black triangles signify MIR elements, light gray triangles represent SINES other than MIR, light gray pointed boxes designate LINE1, and dark gray represent all other repetitive elements. Asterisks (*) in the middle of exons of Hox6 and Hox7 genes of HoxN indicate gene dropouts of those Hox genes in HoxN cluster.
Sequence similarities are confined to the immediate Hox gene cluster regions. There is an abrupt drop off of sequence similarity 5′ of HoxA-13 with the exception of the Evx coding region itself and three noncoding regions immediately beyond Evx (Fig. 3, colored red between 4 kb and 8 kb). The conserved sequences beyond Evx identify conserved noncoding motifs common to Evx-1 (human HoxA) and Evx-2 (horn shark HoxN). This relationship predicts the presence of shared noncoding motifs 3′ of the Evx-1 and Evx-2 genes in vertebrate species generally. These conserved motifs may have a functional relationship with the Evx genes themselves and/or possibly with Hox genes in the adjacent Hox clusters. An abrupt reduction in sequence similarity is also seen in the region 3′ of HoxA1.
Middle repetitive elements are exceedingly rare within the HoxA cluster in agreement with our previous report (23). Only one is detected in the HoxA cluster (at around 129.5 kb; Fig. 3). Middle repetitive elements are abundant in the regions flanking the HoxA cluster. This exclusion property may have implications with respect to the structural and functional stability of the clusters. Middle repetitive elements have been shown to serve as sites for recombination leading to chromosomal rearrangements such as inversions, translocations, and excisions (24, 25). An additional possibility is that the introduction of repetitive elements affects gene spacing relationships within the clusters, which may perturb gene expression.
Many highly similar sequence motifs exist within the noncoding regions between the horn shark HoxM and the human HoxA clusters. These sequence similarities can be estimated conservatively to have been maintained over a period of about 800 million years, taking into account the diversification of clusters in both the horn shark and human lineages. Such remarkable stability suggests a conserved role for these sequences associated with vital biological function. These highly conserved elements are candidates for transcriptional control motifs such as promoters, enhancers, and insulators. Striking are concentrations of short conserved repeats seen in untranslated regions immediately 3′ to a number of the Hox coding regions, namely Hox1, Hox2, Hox3, Hox5, Hox9, and Hox10 in the human HoxA/horn shark HoxM comparisons (Fig. 3). These 3′ UTR sequences may play a role in Hox gene translational regulation and/or cytoplasmic targeting and, in a different context, have the potential to be used advantageously as signatures for the precise identification of Hox gene orthologs and paralogs.
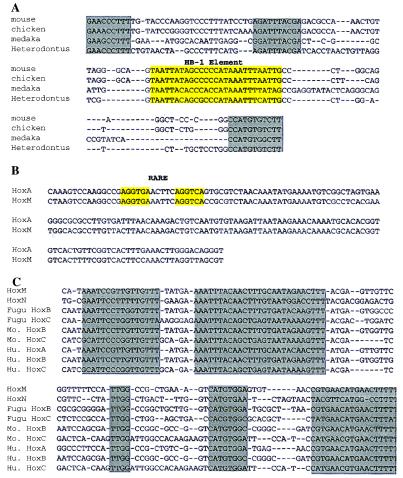
A number of the conserved sequences in the noncoding regions have been reported as control (i.e., enhancer) motifs (Fig. 4). This fact is the case for the HB-1 element in human HoxA4 between exons 1 and 2 (Fig. 3 at 126 kb and Fig. 4A). This homeodomain binding element has been described in Drosophila, bony fishes, birds, and mammals. HB-1 has been mapped to the introns of Ultrabithorax (Ubx), decapentaplegic (dpp), and Deformed (Dfd) in Drosophila species; to HoxA-4 and HoxB-4 in mouse, medaka, Fugu, and chicken; to HoxD-9 in mouse, human, and chicken; and to HoxA-7 in mouse. HB-1 is believed to be responsive to a number of homeobox gene products (26, 27). We also found a conserved motif located 5′ of the HB-1 motif. This element shows similarity to a consensus site found within the Pax-6 paired domain (Fig. 4A; refs. 27 and 28).
Figure 4.
Alignments of HB-1, retinoic acid response elements (RAREs), and the Hox8/Hox7–Hox6 four cluster sequences (H8/7–6 FCS). (A) Alignment of HB-1 elements located in the intron of the HoxA4 gene. The yellow box indicates the HB-1 element, and the gray box represents the other conserved motifs. The second gray box located upstream of the HB-1 element shows similarity to a consensus site found within the Pax-6 paired domain. (B) RAREs previously reported (29, 30) were detected and mapped to 122 kb. (C) Alignment of the H8/7–6 FCS. These sequences exist in all four clusters downstream of HoxA7, HoxB7, HoxD8, and HoxC8. Gray boxes indicate conserved regions in FCS. Mo., mouse; Hu., human.
Several RAREs previously reported were detected in our study (Fig. 4B). One showing a functional response to retinoic acid (29, 30) maps to 122 kb (Fig. 3). Other RARE sites have been described, termed CE-1 and CE-2 (31), which map downstream of human HoxA-1. These sequences were not observed in our sequence comparisons, suggesting that they have been lost in the independent evolution of the horn shark lineage or evolved after the shark/human split. The presence and absence of such motifs can prove valuable as binary markers for the establishment of phylogenetic relationships. Many more binary markers can be expected to emerge when Hox clusters are extensively sequenced and compared in vertebrate clades. The detection of known control motifs in our sequence comparisons as reported above suggests that many of the other conserved sequences we have detected may also possess functional properties.
A number of conserved sequences can be seen to exist in both the horn shark HoxM and HoxN clusters at the same relative position (see red coded regions in HoxM and HoxN; Fig. 3). We suspect these motifs will be present in many of the HoxA and HoxD orthologs. One of particular interest maps to a region between 103 kb and 104 kb (Figs. 3 and 4C). Additional searches and sequence alignments have shown that these sequences exist in all four clusters downstream of HoxA7, HoxB7, HoxD8, and HoxC8. Hox7 is absent in the mouse HoxC and HoxD clusters (Fig. 4C). This report is the first reported instance, to our knowledge, of noncoding sequence conservation at the same cluster position in all four clusters. The Hox genes at this position affect brachial development at the intersection of thoracic and cervical pattern formation domains. It should be noted that these sequences occur at a site similar to the separation of the Drosophila Ubx and Antp complexes.
Conclusions
Hox cluster duplication is a major feature of the vertebrate radiation. Amphioxus has a single Hox cluster containing most probably a full set of 13 Hox genes, although the presence of Hox13 is not yet confirmed. Moreover, it is likely that other protovertebrate clades in the deuterostome lineage such as the acorn worm (Saccoglossus kowalevskii) and the sea urchin (Strongylocentrotus purpuratus) have single clusters (5, 32). The lamprey (Petromyzon marinus and Lampetra planeri) has been reported to have at least three Hox clusters (33, 34). In this report, we show that the horn shark (H. francisci) has two clusters, minimally—additional clusters cannot be ruled out as yet. The bony fishes as represented by the zebrafish (Danio rerio) may have as many as seven to eight clusters (9), whereas representatives of the mammals, such as the mouse and human, have four well characterized clusters (33). Cluster duplication is invariably associated with the loss of Hox genes, although, in all instances studied, including Heterodontus as reported herein, 13 Hox genes are represented by at least one paralog among multiple clusters (33).
The data reported herein show a strong similarity between the horn shark HoxM cluster, the amphioxus cluster, the Fugu HoxA cluster, and the human HoxA cluster on the basis of Hox gene number (dropout), gene spacing within the cluster, and sequence similarity in both coding and noncoding regions. We also show convincingly that the horn shark HoxM and HoxN clusters are divergent based on the same characteristics described above and that HoxM is orthologous to the mammalian HoxA cluster, whereas HoxN is an ortholog of HoxD. This finding provides evidence for the differentiation of HoxA- and HoxD-like clusters before or concomitant with the origin of the gnathostome vertebrates. It will be of considerable interest to characterize the Hox clusters in the jawless fishes to determine their cluster affinities to better resolve their position in vertebrate evolution. It is of interest that, although the horn shark HoxA-like cluster (HoxM) and the HoxD-like cluster (HoxN) differ significantly in terms of noncoding sequence, they retain a high degree of similarity in the Hox coding regions. This fact is consistent with the view that functional divergence in duplicated clusters has been mediated by a large-scale modification of noncoding control motifs. Hox gene swap experiments involving coding domains between organisms as different as Drosophila and the mouse also support this concept (35, 36). Finally, it should be emphasized that sequence similarity in noncoding DNA only suggests a functional role for conserved motifs. However, we and others have shown that a functional analysis is possible with a transgenic approach (37, 38).
Acknowledgments
We thank Chi-hua Chiu and Steve Irvine for critical reading, Dashzeveg Bayarsaihan for technical support, and Dana Weiss for illustrations. This work was supported by National Institutes of Health Grants GM09966 to F.R. and AI39008-01 to C.A. and National Science Foundation Grants IBN 9630567 to F.R. and G.W. and IBN 9614940 to C.A.
Abbreviations
- kb
kilobase
- PAC
P1 artificial chromosome
- RARE
retinoic acid response element
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF224262 and AF224263).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.030539697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.030539697
References
- 1.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 2.Akam M, Averof M, Castelli-Gair J, Dawes R, Falciani F, Ferrier D. Dev. Suppl. 1994. , 209–215. [PubMed] [Google Scholar]
- 3.Wang B B, Muller-Immergluck M M, Austin J, Robinson N T, Chisholm A, Kenyon C. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Fernandez J, Holland P W. Nature (London) 1994;370:563–566. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- 5.Martinez P, Rast J P, Arenas-Mena C, Davidson E H. Proc Natl Acad Sci USA. 1999;96:1469–1474. doi: 10.1073/pnas.96.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acampora D, D'Esposito M, Faiella A, Pannese M, Migliaccio E, Morelli F, Stornaiuolo A, Nigro V, Simeone A, Boncinelli E. Nucleic Acids Res. 1989;17:10385–10402. doi: 10.1093/nar/17.24.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham A, Papalopulu N, Krumlauf R. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 8.Harvey R P, Tabin C J, Melton D A. EMBO J. 1986;5:1237–1244. doi: 10.1002/j.1460-2075.1986.tb04352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amores A, Force A, Yan Y, Joly L, Amemiya C, Fritz A, Ho R K, Langeland J, Prince V, Wang Y, et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits J L, Wang J, Shimizu N. Genome Res. 1997;7:250–261. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz S, Miller W, Yang C M, Hardison R C. Nucleic Acids Res. 1991;19:4663–4667. doi: 10.1093/nar/19.17.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amemiya C T, Ota T, Litman G W. In: Analysis of Nonmammalian Genomes. Birren B, Lai E, editors. New York: Academic; 1996. pp. 223–256. [Google Scholar]
- 15.Misof B Y, Wagner G P. Mol Phylogenet Evol. 1996;5:309–322. doi: 10.1006/mpev.1996.0026. [DOI] [PubMed] [Google Scholar]
- 16.Ota T, Amemiya C T. Genet Anal. 1996;12:173–178. [PubMed] [Google Scholar]
- 17.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swofford D L. paup, Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer; 1993. , Version 3.1. [Google Scholar]
- 19.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. phylip Phylogenetic Inference Package. University of Washington, Seattle: Department of Genetics; 1995. , Version 3.572c. [Google Scholar]
- 21.Aparicio S, Hawker K, Cottage A, Mikawa Y, Zuo L, Venkatesh B, Chen E, Krumlauf R, Brenner S. Nat Genet. 1997;16:79–83. doi: 10.1038/ng0597-79. [DOI] [PubMed] [Google Scholar]
- 22.Wada H, Garcia-Fernandez J, Holland P W. Dev Biol. 1999;213:131–141. doi: 10.1006/dbio.1999.9369. [DOI] [PubMed] [Google Scholar]
- 23.Hart C P, Bogarad L D, Fainsod A, Ruddle F H. Nucleic Acids Res. 1987;15:5495. doi: 10.1093/nar/15.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomilin N V. Int Rev Cytol. 1999;186:1–48. doi: 10.1016/s0074-7696(08)61050-5. [DOI] [PubMed] [Google Scholar]
- 25.Moran J V, DeBerardinis R J, Kazazian H H., Jr Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 26.Haerry T E, Gehring W J. Proc Natl Acad Sci USA. 1996;93:13884–13889. doi: 10.1073/pnas.93.24.13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haerry T E, Gehring W J. Dev Biol. 1997;186:1–15. doi: 10.1006/dbio.1997.8582. [DOI] [PubMed] [Google Scholar]
- 28.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 29.Packer A I, Crotty D A, Elwell V A, Wolgemuth D J. Development (Cambridge, UK) 1998;125:1991–1998. doi: 10.1242/dev.125.11.1991. [DOI] [PubMed] [Google Scholar]
- 30.Doerksen L F, Bhattacharya A, Kannan P, Pratt D, Tainsky M A. Nucleic Acids Res. 1996;24:2849–2856. doi: 10.1093/nar/24.14.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langston A W, Thompson J R, Gudas L J. J Biol Chem. 1997;272:2167–2175. doi: 10.1074/jbc.272.4.2167. [DOI] [PubMed] [Google Scholar]
- 32.Pendleton J W, Nagai B K, Ruddle F H. Proc Natl Acad Sci USA. 1993;90:6300–6304. doi: 10.1073/pnas.90.13.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruddle F H, Amemiya C T, Carr J L, Kim C B, Ledje C, Shashikant C S, Wagner G P. Ann NY Acad Sci. 1999;870:238–248. doi: 10.1111/j.1749-6632.1999.tb08884.x. [DOI] [PubMed] [Google Scholar]
- 34.Sharman A C, Holland P W. Int J Dev Biol. 1998;42:617–620. [PubMed] [Google Scholar]
- 35.McGinnis N, Kuziora M A, McGinnis W. Cell. 1990;63:969–976. doi: 10.1016/0092-8674(90)90500-e. [DOI] [PubMed] [Google Scholar]
- 36.Malicki J, Bogarad L D, Martin M M, Ruddle F H, McGinnis W. Mech Dev. 1993;42:139–150. doi: 10.1016/0925-4773(93)90003-g. [DOI] [PubMed] [Google Scholar]
- 37.Belting H G, Shashikant C S, Ruddle F H. Proc Natl Acad Sci USA. 1998;95:2355–2360. doi: 10.1073/pnas.95.5.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shashikant C S, Kim C B, Borbely M, Wang W, Ruddle F H. Proc Natl Acad Sci USA. 1998;95:15446–15451. doi: 10.1073/pnas.95.26.15446. [DOI] [PMC free article] [PubMed] [Google Scholar]