Abstract
Tumors with mutant BRAF and those with receptor tyrosine kinase (RTK) activation have similar levels of phosphorylated ERK, but only the former depend on ERK signaling for proliferation. The mitogen-activated protein kinase, extracellular signal-regulated kinase kinase (MEK)/ERK-dependent transcriptional output was defined as the genes whose expression changes significantly 8 h after MEK inhibition. In V600EBRAF cells, this output is comprised of 52 genes, including transcription factors that regulate transformation and members of the dual specificity phosphatase and Sprouty gene families, feedback inhibitors of ERK signaling. No such genes were identified in RTK tumor cells, suggesting that ERK pathway signaling output is selectively activated in BRAF mutant tumors. We find that RAF signaling is feedback down-regulated in RTK cells, but is insensitive to this feedback in BRAF mutant tumors. Physiologic feedback inhibition of RAF/MEK signaling down-regulates ERK output in RTK cells; evasion of this feedback in mutant BRAF cells is associated with increased transcriptional output and MEK/ERK-dependent transformation.
Keywords: BRAF mutation; dual specificity phosphatase; mitogen-activated protein kinase, extracellular signal-regulated kinase kinase inhibition
The RAF/mitogen-activated protein kinase, extracellular signal-regulated kinase kinase (MEK)/ERK signaling cascade regulates multiple processes required for the proliferation and survival of normal cells. Hyperactivation of the pathway results in dysregulated cell proliferation and malignant transformation in model systems and occurs commonly in human tumors. These findings have led to the concept that deregulation of proliferation in malignant cells is often mediated by activation of ERK signaling. In human tumors, ERK activation occurs as a result of receptor tyrosine kinase (RTK) activation or mutations in members of the RAS and RAF gene families (1, 2). RAS mutation is common in malignancy, including lung, pancreatic, and colon cancer, whereas BRAF mutation occurs in the majority of melanomas, with lower frequencies in thyroid cancers and other tumors (3–7). In some cancers, the pathway is driven by hyperactivated growth factor receptors, such as HER2 in breast cancer (8, 9) or EGFR in lung cancer and glioblastomas (10, 11).
These findings suggest that the RAF/MEK/ERK pathway is an attractive target for therapeutic intervention. The proliferation of tumors with BRAF mutation is sensitive to inhibition of ERK signaling with selective inhibitors of MEK (12). Such agents are now in clinical trial and have antitumor activity in patients with melanoma (13, 14). However, against expectation, the proliferation of tumors in which ERK signaling is driven by RTKs is insensitive to MEK inhibition (12).
These results suggest that the biologic consequences of activation of ERK signaling in tumors are poorly understood. It is possible that ERK output is elevated in both tumors with RTK activation and tumors with BRAF mutation, but that the pathway is necessary for proliferation only in the latter. Alternatively, the level of phosphorylated ERK (pERK), which has been used as a surrogate marker for pathway activation, may not be an accurate measure of pathway output. Thus, although pERK levels are similar in tumors with BRAF mutation and those with RTK activation, ERK pathway signaling output may be elevated only in the former.
To address these issues, we sought to identify the genes whose expression changes rapidly after MEK inhibition and that therefore comprise the transcriptional output of the MEK/ERK pathway. A program of 52 genes was found to be differentially regulated after treatment of V600EBRAF tumor cells with a selective inhibitor of MEK. In contrast, no such set of genes was identified in a group of tumor cells with RTK activation. These data are consistent with elevation of ERK signaling output in MEK/ERK-dependent tumors with V600EBRAF.
The profile of genes dependent on MEK/ERK signaling for expression in V600EBRAF tumor cells includes transcription factors associated with ERK-dependent transformation, such as members of the ETS family, FOS and MYC. The profile also includes feedback regulators of ERK signaling, several of which are markedly overexpressed compared with levels found in RTK-activated tumor cells. The increased ERK signaling output and overexpression of genes that feedback-inhibit the pathway suggest that feedback inhibition of RAF signaling is ineffective in V600EBRAF cells. Consistent with this idea, in RTK-driven cells, RAF/MEK signaling is feedback-inhibited by a MEK/ERK-dependent pathway. In contrast, there is no evidence for such feedback in tumors with V600EBRAF. Thus, activation of signaling output and induction of transformation by oncoproteins such as V600EBRAF may depend on their insensitivity to or evasion of normal feedback control.
Results
Identification of a Set of 52 Genes Dependent on MEK/ERK Activity in V600EBRAF Tumor Cells.
In tumors, ligand-dependent or mutational activation of RTKs or mutational activation of pathway intermediates such as RAS and BRAF lead to activation of ERK signaling, which is associated with demonstrably high intracellular levels of pERK (15–18). However, whereas the proliferation and growth of V600EBRAF tumors is sensitive to inhibition of MEK/ERK signaling, tumors with RTK activation are resistant (12). Thus, high levels of pERK do not correlate with dependence of the tumor cell on MEK/ERK for proliferation. It is also possible that pERK expression is a poor measure of activation of ERK pathway signaling and that, in reality, the output of the pathway varies as a function of the mechanism by which it is driven.
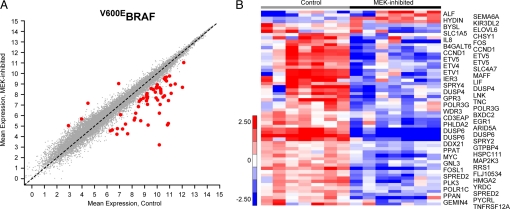
To address this question, we compared the ERK signaling output in a panel of V600E BRAF tumors (sensitive to pathway inhibition) with that found in a panel of tumor cell lines in which the pathway is driven by RTKs (uniformly insensitive; Fig. S1 and ref. 12). We operationally defined the transcriptional output of the pathway as comprising those genes whose expression changes significantly after MEK inhibition. PD0325901 is a selective inhibitor of MEK1 and MEK2 kinases that rapidly abrogates ERK signaling (19). In pilot studies in the melanoma cell line SkMel-28, the inhibitor caused significant changes in the expression of genes involved in signaling and transcription by 2 h, which increased by 8 h after treatment (SI Text, Fig. S2, and Table S1). Seven cell lines with V600EBRAF (melanoma, SkMel-1, SkMel-5, SkMel-19, SkMel-28, and Malme3M; colon cancer, Colo205 and HT29) were then treated with 50 nM PD0325901 for 8 h and a paired statistical analysis [significance analysis of microarrays (SAM); see Experimental Procedures and ref. 20], was used to identify the set of transcripts differentially expressed upon MEK inhibition in this group of cells (Fig. 1). This method identified 52 genes [false discovery rate (FDR) <2%, δ = 1.505], represented by 60 unique probe sets, that comprise, by our operational definition, the MEK-dependent transcriptional output in cells with V600EBRAF. Forty-eight of the genes were down-regulated, in a range from 2- to 124-fold; 4 of the genes were up-regulated, from 3- to 6-fold. At least 16 genes in the profile are established components of ERK signaling networks (Fig. 2). Among those genes most changed in response to MEK inhibition were ERK targets such as CCND1, multiple transcription factors, and genes involved in the feedback inhibition of MEK/ERK signaling [dual specificity phosphatase 6 (DUSP6), DUSP4, SPRY2, SPRY4, and SPRED2] (21, 22). Nine of the transcription factors (MYC, FOS, FOSL1, EGR1, IER3, ETV1, ETV5, ETV4, and MAFF) are previously described targets of ERK signaling. The remaining 36 genes in the profile have not been previously identified as components of ERK signaling networks in published databases (23, 24). The complete gene program is shown in Fig. 1B. In Table S2 the genes are listed in order of the magnitude of their change after MEK inhibition.
Fig. 1.
Identification of the transcriptional output of the MEK/ERK pathway in V600EBRAF tumor cells. (A) The plot identifies the 52 genes (probe sets in red) significantly changed in expression upon MEK inhibition, above background (probe sets for all other genes in gray) in V600EBRAF cells (n = 7), determined by SAM analysis. (B) Details of expression levels for the 52 genes (60 probe sets, rows), in each of the 7 cell lines (columns) with and without MEK inhibition (heatmap values: red = increased expression, blue = decreased expression). Parameters in SAM: final largest median FDR is 1.03% and δ cutoff is 1.505.
Fig. 2.
A molecular interaction map displaying the functional interaction of genes in the MEK/ERK output profile with established roles in ERK signaling. A molecular interaction map was constructed for major constituents of the RAS/RAF/MEK/ERK signaling pathway and downstream effectors. The genes in the output profile that have established functional relationships to ERK signaling are indicated by gray shading, with the nature of their relationship (i.e., direct phosphorylation, transcriptional activation) indicated by using the symbols described by Pommier and colleagues (43) and shown in the legend.
No ERK-Dependent Transcriptional Output Profile Was Identified in Tumor Cells with RTK Activation.
To determine whether the transcriptional output of the ERK pathway is similar in tumors in which RTKs drive the pathway, this experiment was repeated with a set of 5 cell lines with WTBRAF and RTK activation: HER2-amplified breast cancers (BT474, SkBr3), EGF receptor (EGFR)-amplified breast cancers (MDA-468), squamous cell cancer (A431), and EGFR mutant lung cancer (NCI-H1650). No genes were identified that were significantly altered in expression in response to MEK inhibition in this group of cells, although pERK was effectively inhibited (Fig. S3A). No new genes were identified, nor did the genes identified in the BRAF output profile change significantly in the RTK group (Fig. S3B). This finding suggests that the transcriptional output of the ERK pathway is significantly elevated in V600EBRAF compared with RTK-activated tumor cells. This conclusion was confirmed when these data were reanalyzed by a nonparametric rank-order analysis (SI Text). In this analysis, the MEK output genes were randomly distributed among a ranked list of descending gene expression change (from highest magnitude change to lowest) in the RTK group of cells.
To determine whether the transcriptional output of the ERK pathway was similar in vivo, we assessed the effect of MEK inhibition on gene expression in SkMel-28 (V600EBRAF) and BT474 (HER2-amplified) xenograft tumors from mice treated with a single oral dose of 25 mg/kg PD0325901 or vehicle for 8 h. In each tumor from a treated mouse, ERK phosphorylation was completely inhibited (Fig. S4 A). In V600EBRAF SkMel-28 xenografts, the expression of the previously identified genes, including DUSP6, Sprouty, and several transcription factors, were among those most significantly altered (Fig. S3B). In contrast, in BT474, the majority of the ERK output profile genes did not change significantly in response to MEK inhibition. We performed rank-order analyses on these samples as well and found that the genes in the ERK output profile were nonrandomly distributed among the genes with the highest magnitude of change in response to MEK inhibition in the SkMel-28 xenografts (enrichment score 0.753). In contrast, the output genes were randomly distributed among all ranked genes in BT474 (enrichment score 0.216), consistent with the absence of an identifiable ERK output signature in these tumors in vivo.
To confirm that the genes identified by microarray were MEK-dependent, their expression before and after MEK inhibition was determined by immunoblotting and quantitative RT-PCR. The expression of several proteins decreased after drug treatment in all 7 V600EBRAF cell lines examined (representative immunoblots of SPRY2, SPRY4, ETV1, ETV5, and cyclin D1 are shown in Fig. 3A). Profound reductions in DUSP6 (98%) and SPRY2 (99%) mRNA levels were observed 4 h after MEK inhibition; maximal decreases (89–95%) in ETS family and CCND1 mRNAs occurred later (Fig. 3B). Loss of protein expression of Sprouty and ETS family members and cyclin D1 was also demonstrated in vivo in SkMel-28 xenografts (Fig. S4 B).
Fig. 3.
Expression of genes in the MEK/ERK output profile is down-regulated after MEK inhibition. The MEK dependence of genes identified as components of the MEK/ERK transcriptional output profile is confirmed by immunoblot and RT-PCR. (A) SkMel-5 cells in tissue culture were treated for the times indicated with 50 nM PD0325901 and lysates were subjected to immunoblotting with the indicated antibodies. (B) mRNA was isolated from SkMel-5 treated with drug as in A and analyzed by RT-PCR for levels of the indicated mRNAs. Values are relative mRNA level compared with those before treatment.
Effectors and Feedback Inhibitors of MEK/ERK Signaling Are Overexpressed in V600EBRAF Tumor Cells.
The expression of a set of genes comprised of effectors of RAF/MEK/ERK signaling and feedback regulators of the pathway is under ERK control in V600EBRAF tumor cells but not in RTK cells. These data suggest that the transcriptional output of ERK signaling is selectively elevated in V600EBRAF tumors. The differential expression of the output profile genes in V600EBRAF compared with RTK cells is shown in Table S2. Some of the genes are overexpressed in BRAF mutant compared with RTK cells, with differences varying from 2.2- to 54-fold. They include transcription factors downstream of ERK (ETV1, ETV4, ETV5, FOS), and feedback effectors (DUSP6, SPRY2, SPRY4, SPRED2; examples are shown in Fig. S5A). The expression of these genes requires ERK signaling and is thus elevated and under MEK control in V600EBRAF cells. Other genes are not differentially expressed in the 2 cell types (examples are shown in Fig. S5B) but are MEK-dependent only in BRAF cells. These genes include some that are important in many types of tumors (i.e., CCND1, MYC, IER3, EGR1) and regulated by multiple oncogenic pathways, so that activation of MEK/ERK signaling may be sufficient, but not necessary for their expression (12, 25). Quantification of the composite expression of the 52-gene, operationally-defined ERK output signature, calculated as the mean of Z-scores, showed that, by this measure as well, that aggregate ERK pathway output is greater in V600EBRAF, than in RTK-activated tumor cells (Fig. S5C).
Levels of pERK Are Similar in Tumor Cells with V600EBRAF and Those with RTK Activation.
Activation of the RAF/MEK/ERK signaling pathway results in phosphorylation of ERK by MEK, and the intracellular level of pERK is used as a measure of pathway activation in tumors (26). As expected, tumor cell lines with activated RTKs and those with BRAF mutation had detectable levels of pERK, which, although variable, did not differ significantly between the 2 groups (Fig. 4). However, ERK pathway output was elevated in tumors with V600EBRAF compared with those with activated RTKs. Levels of pERK thus do not correlate with pathway output or dependence on the pathway. Furthermore, despite similar levels of pERK, the expression of feedback regulators of ERK signaling was markedly higher in the V600EBRAF cells. We hypothesized that these findings could be explained by differential sensitivities of the ERK signaling pathway to feedback inhibition.
Fig. 4.
Quantitation of pERK and pMEK levels in tumor cell lines with mutant and WT BRAF. pERK and pMEK were quantitated by immunoblotting cell lysates from 13 tumor cell lines with WTBRAF or V600EBRAF. Cells grown in 10% FBS were harvested at 70–80% confluence, and lysates resolved by SDS/PAGE were probed for pERK (phosphorylated p44/ p42 MAPK), pMEK, total ERK, and total MEK 1/2. Band intensity was quantitatively measured by densitometry reading of the immunoblot [P value (one-sided Wilcoxon) is cited in text for difference in pMEK expression between groups].
RAF Kinase Activity Is Feedback-Inhibited in Cells with Activated RTKs but Not in Those with Mutated BRAF.
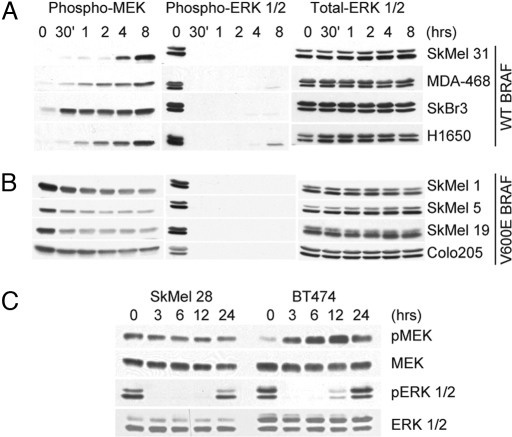
Physiologic activation of ERK is negatively feedback-regulated at multiple levels, both downstream and upstream of MEK. In RTK cells, pERK levels were elevated, but phosphorylated MEK (pMEK) levels were low or undetectable (Fig. 4), consistent with the possibility that RAF kinase or its activators are targets of negative feedback. This hypothesis is supported by the observation that MEK phosphorylation (but not total MEK protein) is rapidly induced in RTK cells exposed to PD0325901 (Fig. 5A), suggesting relief of feedback inhibition of RAF activity.
Fig. 5.
MEK phosphorylation is feedback inhibited in tumor cells with activated receptor tyrosine kinases, but not in those with V600EBRAF. Shown are immunoblots of pMEK and pERK 1/2 in WTBRAF cell lines (A), V600EBRAF cell lines (B), and SkMel-28 and BT474 xenograft tumors (C) treated with MEK inhibitor as a function of time. Cell lines were treated with 50 nM PD0325901, and mice were treated with a single oral dose of 25 mg/kg PD0325901. Total ERK and total MEK were unchanged.
In contrast, steady-state levels of pMEK were markedly elevated in V600EBRAF cells compared with RTK cells (Fig. 4; P = 0.004). Thus, pMEK, rather than pERK, levels are correlated with enhanced output of ERK signaling and sensitivity to MEK inhibition. The elevated levels of pMEK in V600EBRAF cells suggest that the ERK signaling pathway is insensitive to upstream feedback inhibition. This is the case: MEK kinase inhibition does not further induce pMEK in V600EBRAF cells, which decreases instead (Fig. 5B). In RTK-driven tumors, MEK inhibition relieves feedback inhibition of the pathway in vivo as well. In BT474 xenografts, pMEK levels were low, but induced after treatment with PD0325901, whereas SkMel-28 xenografts expressed high levels of pMEK, which decreased after MEK inhibition (Fig. 5C). Thus, the low levels of ERK output in the RTK cells occur in the setting of feedback inhibition of RAF phosphorylation of MEK. In contrast, feedback inhibition of RAF/MEK signaling is lost in V600EBRAF cells and is accompanied by high levels of MEK kinase activity and increased output of the pathway.
Discussion
Activation of the RAS/RAF/MEK/ERK pathway is a common feature of tumors and results from mutations in pathway intermediates, such RAS, BRAF, or NF1, or unregulated activation of upstream receptors, including hyperactivation of RTKs. There are several important unanswered questions about the pathway in malignancy, including how to quantitate its activity. ERK must be phosphorylated by MEK on threonine 202 and tyrosine 204 to be activated, so pERK levels are used as a measure of pathway activation in cells. How steady-state levels of pERK are related to information flow through the pathway is unknown, however. Elevated ERK activity is thought to subserve multiple aspects of the transformed phenotype, especially dysregulation of proliferation. Whereas MEK kinase inhibition in tumors with mutant BRAF has the expected consequences, loss of D-cyclin expression, inhibition of G1/S progression and tumor cell proliferation, and suppression of tumor growth in animals, it has none of these effects in tumors with activated RTKs (12).
Despite the insensitivity of RTK cells to MEK inhibition, their pERK levels are not significantly different from those detected in V600EBRAF cells. It is possible that the pathway is activated in both types of tumors, but is not necessary for the proliferation and transformation of cells in which RTKs drive the pathway. Alternatively, the pathway may be much more active in the BRAF cells. To resolve this question and obtain another and perhaps more accurate measure of ERK pathway activity, we measured the transcriptional “output” of the pathway in tumors in which it is driven by V600EBRAF or RTKs. In doing so, we found that, despite similar pERK levels in the 2 types of tumor, ERK pathway output was elevated only in the former. A set of 52 genes, the expression of which changes rapidly in response to MEK/ERK inhibition, was identified in tumor cells with V600EBRAF. The expression of these genes depends on MEK/ERK activation and comprises a measure of increased flux through the pathway. It is reasonable to assume that at least a portion of the biologic effects of V600EBRAF are mediated by the expression of these genes. The importance of these genes for the biology of V600EBRAF tumors is supported by their overlap with genes identified as overexpressed in metastatic melanoma or mutant BRAF melanoma (27–29) (Table S3 and Fig. S6).
Contrary to expectations, no ERK output profile could be identified in a panel of tumor cells in which the pathway is driven by activated HER family receptor tyrosine kinases. Neither the gene set identified in BRAF tumors nor any new set of genes was identified as significantly regulated by ERK signaling in these tumors. This result may be caused by the different cell lineages: BRAF mutations in melanomas and colon cancers, EGFR mutations in lung cancers, and HER2 amplification in breast cancers. However, this is unlikely to be the case, because the genes identified in the BRAF panel encode proteins that have general function as effectors (ETS family, MYC, FOS) and feedback regulators (Sprouty, DUSPs) of the ERK signaling pathway.
The results suggest another explanation, that the magnitude of the transcriptional output of MEK/ERK signaling varies as a function of the mechanism of activation of the pathway: it is markedly elevated in tumors with BRAF mutation and only marginally increased, if at all, in tumors with mutational activation or amplification of HER kinases. Thus, it would seem that the proliferation of RTK-driven tumors is resistant to MEK/ERK inhibition because ERK signaling output is not very elevated. The data do not rule out that ERK signaling plays other roles in the biology of these tumors that are mediated by direct phosphorylation of substrates or modest increases in transcriptional output, including effects on invasion and angiogenesis (30, 31).
The expression of a subset of the genes in the V600EBRAF-defined ERK output profile is not significantly different in BRAF and RTK tumors (examples, see Fig. S5B). However, their expression is MEK-dependent only in BRAF tumors and may be induced by other activated pathways in tumors in which ERK signaling output is not elevated. This idea is supported by the overlap between genes identified as ERK output in our study and those genes identified as down-regulated upon PI3K inhibition in PIK3CA or PTEN mutant cell lines (ref. 32, SI Text, Table S3, and Fig. S6). Such genes, exemplified in our panel by CCND1, IER3, and EGR1, may represent downstream targets important in mediating induction of transformation by multiple pathways. Thus, in BRAF tumors, cyclin D1 expression is MEK-dependent and PI3K-independent, whereas the reverse is true in HER2-dependent breast cancers (12, 25).
This explanation of the data presents us with 2 paradoxes: First, the levels of pERK in these tumors are not significantly different, yet the output of the pathway is much higher in the V600EBRAF cells. Second, the BRAF tumors express high levels of feedback inhibitors of the pathway, yet they have higher pathway output than the RTK cells. These apparent contradictions may be resolved by considering the implications of feedback down-regulation of the pathway in normal and transformed cells. Receptor activation of RAS/MEK/ERK signaling in normal cells is accompanied by rapid and complex feedback inhibition, resulting in a pulse of pathway activation followed by rapid down-regulation (33, 34). Feedback is mediated by induction of expression of proteins such as Sprouty (22) and the MAPK phosphatases [DUSPs (21)] and by direct phosphorylation and inhibition of proteins like RAF-1 (35).
In the steady state, mutational activation or overexpression of RTKs may cause only marginal activation of pathway output because activation of ERK above a threshold level is associated with phosphorylation and inhibition of RAF kinase by activated ERK and with induction of the expression of DUSPs. Moreover, any increase in ERK transcriptional output leads to increased DUSP activity and feedback dephosphorylation of ERK. This model accurately predicts our finding that levels of pMEK are quite low in HER2 and EGFR cells, probably caused by both direct feedback inhibition of RAF kinase and, perhaps, its upstream activators. This interpretation is consistent with our finding that pharmacologic inhibition of MEK rapidly abrogates this feedback and induces phosphorylation of MEK. Thus, the observed levels of pERK in these cells in the steady state are likely caused by a balance of low levels of MEK activity and the low levels of DUSP expression associated with marginal transcriptional output of the pathway (Fig. 6A).
Fig. 6.
Models of RAF/MEK/ERK signaling in tumor cells with RTK activation and with V600EBRAF. (A) RTK-activated tumor cells: feedback inhibition occurs at multiple levels of the pathway, down-regulating both RAF/MEK activation and ERK phosphorylation levels. Steady-state levels of pERK are caused by low levels of both MEK activity and ERK dephosphorylation by DUSPs. Output of the pathway is low. (B) V600EBRAF cells: RAF kinase is active and insusceptible to negative feedback. Increased pathway output leads to increased DUSP6 expression. Steady-state levels of pERK are caused by high levels of MEK and DUSP6 dephosphorylation of ERK.
In contrast, the elevation of ERK pathway output in V600EBRAF cells is associated with insensitivity of mutant BRAF activity to feedback inhibition. BRAF kinase activity is elevated in these cells and phosphorylation of its substrate MEK is significantly elevated compared with the almost undetectable levels found in tumors with activated RTKs. Furthermore, MEK inhibition does not reveal residual feedback inhibition of RAF; that is, it does not result in further elevation of pMEK in these cells. Elevation of RAF/MEK kinase activity is associated with increased output of the pathway, leading to the enhanced expression of a variety of transcription factors and DUSPs that dephosphorylate ERK. Thus, in BRAF tumor cells, elevated steady-state levels of pERK are caused by a balance between much higher levels of RAF kinase activity and DUSP6 expression than are found in RTK cells (Fig. 6B).
As MEK/ERK signaling output increases, DUSP6 expression increases, so that levels of ERK phosphorylation should not vary very much above a threshold level. The model predicts that, as reported here, DUSP6 expression and MEK phosphorylation, but not ERK phosphorylation, should be markers of elevated output of the RAF/MEK/ERK pathway. The steady state of pERK is maintained by increased DUSP transcription, but is accompanied by increased expression of other ERK effectors, ETS family members, MYC, and FOS transcription factors that may contribute to transformation.
This model has significant implications for the understanding of transformation. Constitutive activation of signaling by activated oncoproteins, such as RAS and RAF, is likely to result in constitutive negative feedback. Activation of signaling output by these oncoproteins will therefore require abrogation of some aspect of this feedback, either caused by insensitivity of the mutant protein to normal regulation or second hits that directly affect components of the feedback system. Mutant BRAF is insensitive to feedback, and the resultant increased output of the pathways is associated with elevated DUSP and Sprouty protein expression. Other oncogenic mutants may require loss of these proteins to effect transformation. In fact, DUSP and SPRY gene expression have been reported to be elevated in V600EBRAF melanomas (36, 37), but significantly reduced in a variety of other tumors (38–41). Evasion of pathway feedback may be a fundamental requirement for oncogenic transformation.
In this work, we have defined a set of genes that represents the transcriptional output of BRAF/MEK/ERK signaling in tumors with V600EBRAF, including genes that encode negative feedback regulators of the pathway and those that encode known effectors of RAS-dependent transformation in model systems. The latter class includes 3 members of the ETS transcription factor family and MYC, FOS, EGR1, and IER3. It is likely that future work will reveal that these transcription factors mediate important aspects of mutant BRAF-dependent transformation and constitute potential new therapeutic targets for melanoma and other tumors with the mutation.
Experimental Procedures
Cell Lines, Antibodies, and Reagents.
SkMel cell lines were obtained from A. Houghton (Memorial Sloan-Kettering Cancer Center) and maintained in RPMI medium 1640. All other cell lines were obtained from ATCC, maintained in the recommended medium, supplemented with 2 mM glutamine, 50 units/mL penicillin, 50 units/mL streptomycin, and 10% FBS, and incubated at 37 °C in 5% CO2. Antibodies against p42/44 MAPK, phospho-p42/44 MAPK, phospho-MEK, and MEK 1/2 were obtained from Cell Signaling. Anti-MKP-3 (DUSP6), MKP-2 (DUSP4), SPRY4, and cyclin D1 antibodies were obtained from Santa Cruz. Anti-SPRY2 antibody was obtained from Upstate. Anti-ETV1 and anti-ETV5 antibodies were obtained from Abnova. PD0325901 was a gift of Pfizer. Cells were treated with PD0325901 to a final concentration of 50 nM.
Western Blot Analysis.
Cellular lysis was performed with Nonidet P-40 buffer, and lysates were resolved by SDS/PAGE as described (12). Linear quantitative determination of band intensity for both p44 and p42 MAPK forms (ERK1 and ERK2) was performed with Science Lab 2003 Image Gauge (Fujifilm).
Animal Studies.
Four- to 6-week-old nu/nu athymic female mice were obtained from the National Cancer Institute-Frederick Cancer Center. Experiments were carried out pursuant to institutional guidelines for the proper and humane use of animals in research and under an Institutional Animal Care and Use Committee-approved protocol. Tumors were generated by injecting 0.5–1.5 × 107 tumor cells together with reconstituted basement membrane (Matrigel; Collaborative Research). Before BT474 cell inoculation, 0.72 mg/day 17ß-estradiol pellets (Innovative Research of America) were inserted s.c. Mice were randomized to receive 25 mg/kg PD0325901 (n = 5) or vehicle only (n = 5). PD0325901 was formulated in 0.5% hydroxypropyl methylcellulose and 0.2% Tween 80 and administered by oral gavage. Mice were killed by CO2 euthanasia. To prepare lysates, tumor tissue was homogenized in 2% SDS lysis buffer and then processed as described (12). Two tumors from each treatment group were chosen at random for microarray studies.
Oligonucleotide Expression Array Analysis.
Cells were exposed to 50 nM PD0325901 for 2, 8, or 24 h or DMSO 0.1% as control. After TRIzol extraction, total RNA quality control was performed. Twenty-five to 50 ng of total RNA was run on an RNA 6000 Nano Assay (Agilent) using a Bioanalyzer 2100. Two micrograms of good-quality total RNA was labeled according to manufacturers' protocols. After reverse-transcription with oligo(dT)-T7 polymerase (Affymetrix), double-stranded cDNA was generated with the SuperScript double-stranded cDNA synthesis custom kit (Invitrogen Life Technologies). In an in vitro transcription step with T7 RNA polymerase (MessageAmp RNA kit; Ambion) the cDNA was linearly amplified and labeled with biotinylated nucleotides (Enzo Diagnostics). Ten micrograms of labeled and fragmented cRNA was then hybridized to the U133A2.0 expression array (Affymetrix). After hybridization staining, chips were processed according to the manufacturer and scanned with a high-numerical aperture and flying objective lens in the GS3000 scanner (Affymetrix). The image was quantified by using GCOS version 1.4 (GeneChip Operating System; Affymetrix).
PCR Analysis.
Total cellular RNA extraction was performed with the Qiagen RNeasy kit. Double-stranded cDNA was generated by using the SuperScript III First Strand Synthesis System (Invitrogen) for RT-PCR. Equal amounts of input RNA were used in a real-time PCR by using an ABI-7500 System and analyzed with Applied Biosystems 7500 System Sequence Detection Software version 1.3.1. Differences in expression were calculated by using the ΔΔCT method.
Data Analysis.
Primary analysis was executed with the Affymetrix GeneChip Operating Software for expression quantification and array normalization. Array data were subsequently log base-2 transformed for parametric analysis. SAM was performed on normalized data divided into multiple 2-group comparisons by using a 2-class t test to identify differentially expressed genes (20). Two-way average-linkage hierarchical clustering of an uncentered Pearson correlation similarity matrix was executed with Cluster and visualized with TreeView (42). All statistical analyses were performed with R 2.3.1.
Supplementary Material
Acknowledgments.
We thank E. Halilovic and O. Grbovic for valuable discussions; H. Zhao, Y. Zhao, and W. Wong for technical assistance; and A. Koff, C. Sawyers, and S. Chandarlapaty for careful review of the manuscript. This work was supported by grants from the National Institutes of Health (to N.R.), the William H. Goodwin and Alice Goodwin Foundation for Cancer Research, the Memorial Sloan-Kettering Cancer Center Experimental Therapeutics Program (to D.B.S. and N.R.), the Starr Foundation (to N.R.), and Pfizer Oncology (to N.R.). B.S.T. is a fellow of the Geoffrey Beene Cancer Research Center at Memorial Sloan-Kettering Cancer Center.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE10086).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900780106/DCSupplemental.
References
- 1.Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 2.Hoshino R, et al. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene. 1999;18:813–822. doi: 10.1038/sj.onc.1202367. [DOI] [PubMed] [Google Scholar]
- 3.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Brose MS, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 5.Gorden A, et al. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer Res. 2003;63:3955–3957. [PubMed] [Google Scholar]
- 6.Cohen Y, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 7.Rajagopalan H, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 8.Ross JS, Fletcher JA. The HER-2/neu oncogene: Prognostic factor, predictive factor, and target for therapy. Semin Cancer Biol. 1999;9:125–138. doi: 10.1006/scbi.1998.0083. [DOI] [PubMed] [Google Scholar]
- 9.Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 10.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of nonsmall-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 12.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adjei AA, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26(13):2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LoRusso PM, et al. A phase 1–2 clinical study of a second generation oral MEK inhibitor PD 0325901 in patients with advanced cancer. J Clin Oncol. 2005;23 doi: 10.1200/JCO.2005.14.415. (abstr) [DOI] [PubMed] [Google Scholar]
- 15.Lewis TS, Shapiro PS, Ahn NG. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 16.Cobb MH, Goldsmith EJ. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 17.Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 18.Allen LF, Sebolt-Leopold J, Meyer MB. CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK) Semin Oncol. 2003;30:105–116. doi: 10.1053/j.seminoncol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Rinehart J, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced nonsmall-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens DM, Keyse SM. Differential regulation of MAP kinase signaling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, Bar-Sagi D. Modulation of signaling by Sprouty: A developing story. Nat Rev. Mol Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: Protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muise-Helmericks RC, et al. Cyclin D expression is controlled posttranscriptionally via a phosphatidylinositol 3-kinase/Akt-dependent pathway. J Biol Chem. 1998;273:29864–29872. doi: 10.1074/jbc.273.45.29864. [DOI] [PubMed] [Google Scholar]
- 26.Crews CM, Erikson RL. Purification of a murine protein-tyrosine/threonine kinase that phosphorylates and activates the Erk-1 gene product: Relationship to the fission yeast byr1 gene product. Proc Natl Acad Sci USA. 1992;89:8205–8209. doi: 10.1073/pnas.89.17.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoek KS, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 28.Smith AP, Hoek K, Becker D. Whole-genome expression profiling of the melanoma progression pathway reveals marked molecular differences between nevi/melanoma in situ and advanced-stage melanomas. Cancer Biol Ther. 2005;4:1018–1029. doi: 10.4161/cbt.4.9.2165. [DOI] [PubMed] [Google Scholar]
- 29.Haqq C, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irie HY, et al. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seton-Rogers SE, et al. Cooperation of the ErbB2 receptor and transforming growth factor β in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci USA. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamb J, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 33.Traverse S, Gomez N, Paterson H, Marshall C, Cohen P. Sustained activation of the mitogen-activated protein (MAP) kinase cascade may be required for differentiation of PC12 cells. Comparison of the effects of nerve growth factor and epidermal growth factor. Biochem J. 1992;288:351–355. doi: 10.1042/bj2880351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amit I, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39(4):503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 35.Dougherty MK, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 36.Bloethner S, et al. Effect of common B-RAF and N-RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis. 2005;26:1224–1232. doi: 10.1093/carcin/bgi066. [DOI] [PubMed] [Google Scholar]
- 37.Tsavachidou D, et al. SPRY2 is an inhibitor of the ras/extracellular signal-regulated kinase pathway in melanocytes and melanoma cells with wild-type BRAF but not with the V599E mutant. Cancer Res. 2004;64:5556–5559. doi: 10.1158/0008-5472.CAN-04-1669. [DOI] [PubMed] [Google Scholar]
- 38.Furukawa T, Sunamura M, Motoi F, Matsuno S, Horii A. Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am J Pathol. 2003;162:1807–1815. doi: 10.1016/S0002-9440(10)64315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo TL, et al. The ras/mitogen-activated protein kinase pathway inhibitor and likely tumor suppressor proteins, sprouty 1 and sprouty 2, are deregulated in breast cancer. Cancer Res. 2004;64:6127–6136. doi: 10.1158/0008-5472.CAN-04-1207. [DOI] [PubMed] [Google Scholar]
- 40.Kwabi-Addo B, et al. The expression of Sprouty1, an inhibitor of fibroblast growth factor signal transduction, is decreased in human prostate cancer. Cancer Res. 2004;64:4728–4735. doi: 10.1158/0008-5472.CAN-03-3759. [DOI] [PubMed] [Google Scholar]
- 41.Sutterluty H, et al. Down-regulation of Sprouty2 in nonsmall cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and -independent mechanisms. Mol Cancer Res. 2007;5:509–520. doi: 10.1158/1541-7786.MCR-06-0273. [DOI] [PubMed] [Google Scholar]
- 42.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genomewide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohn KW, Aladjem MI, Weinstein JN, Pommier Y. Molecular interaction maps of bioregulatory networks: A general rubric for systems biology. Mol Biol Cell. 2006;17:1–13. doi: 10.1091/mbc.E05-09-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.