Abstract
Protein functions require conformational motions. We show here that the dominant conformational motions are slaved by the hydration shell and the bulk solvent. The protein contributes the structure necessary for function. We formulate a model that is based on experiments, insights from the physics of glass-forming liquids, and the concepts of a hierarchically organized energy landscape. To explore the effect of external fluctuations on protein dynamics, we measure the fluctuations in the bulk solvent and the hydration shell with broadband dielectric spectroscopy and compare them with internal fluctuations measured with the Mössbauer effect and neutron scattering. The result is clear. Large-scale protein motions are slaved to the fluctuations in the bulk solvent. They are controlled by the solvent viscosity, and are absent in a solid environment. Internal protein motions are slaved to the beta fluctuations of the hydration shell, are controlled by hydration, and are absent in a dehydrated protein. The model quantitatively predicts the rapid increase of the mean-square displacement above ≈200 K, shows that the external beta fluctuations determine the temperature- and time-dependence of the passage of carbon monoxide through myoglobin, and explains the nonexponential time dependence of the protein relaxation after photodissociation.
Keywords: beta process, dielectric, hydration, solvent
Proteins are dynamic systems that interact strongly with their environment (1). Most texts and publications show proteins in unique conformations and naked, without hydration shell and bulk solvent, while fluctuations are rarely mentioned. The unified model of protein dynamics presented here is a radical departure from this picture. In this model, the protein provides the structure for the biological function, but it is dynamically passive. The fluctuations in the bulk solvent power and control the large-scale motions and shape changes of the protein in a diffusive manner (2–4), whereas the fluctuations in the hydration shell power and control the internal protein motions such as ligand migration (5, 6). The hydration shell consists of ≈2 layers of water that surround proteins as shown in Fig. 1(7–12). Protein functions depend on the degree of hydration, h, defined as the weight ratio of water to protein. Dehydrated proteins do not function. Some proteins begin to work at h ≈ 0.2 (11) but full function may require h > 1. The controls exerted by the bulk solvent and the hydration shell are possible because the protein interior is fluid-like (13); the intrinsic viscosity of a protein is small, about like water (14–16). The image of the protein being essentially passive and being slaved to the environment is not an idle speculation. It is based on experiments using myoglobin (Mb) that led to the seminal concepts that underlie the present work: (i) Proteins do not exist in a unique conformation; they can assume a very large number of conformational substates (CS) (17, 18). (ii) The CS can be described by an energy landscape (17). (iii) The landscape is organized in a hierarchy; there are energy valleys within energy valleys within energy valleys (19). The description of the effects of the bulk solvent and the hydration shell is based on these concepts. Because knowledge of the fluctuations in glass-forming liquids and of the energy landscape of proteins is essential for understanding these results, we discuss these topics first.
Fig. 1.
The hydration shell of myoglobin (Mb). Diagram of myoglobin (blue surface) with 1,911 water molecules (CPK model), the approximate number needed for optimal function (h = 2). The waters form a shell ≈5 Å thick around the protein. Approximately 200 water molecules are distinguishable from background with high-resolution X-ray crystallography.
The α and β Processes (20, 21)
Glass-forming liquids have two types of equilibrium fluctuations, α and β.* One tool to study these fluctuations is dielectric relaxation spectroscopy (21). The sample is placed in a capacitor, a sine-wave voltage U1(ν) of frequency ν is applied, and the resulting current is converted into a voltage U2(ν) that characterizes the dielectric spectrum. Our spectra exhibit two prominent peaks that characterize α, or primary, and β, or secondary, relaxations. The α process describes structural fluctuations. The mechanical Maxwell relation,
connects the rate coefficient kα(T) for the α fluctuations to the viscosity η(T). Here, G0 is the infinite-frequency shear modulus that depends only weakly on temperature and on the material. In a solid, where the viscosity is extremely large, Eq. 1 implies that the α fluctuations are essentially absent. The rate coefficient kα(T) can usually be approximated by the Vogel–Tammann–Fulcher (VTF) relation,
D, Aα, and T0 are experimentally determined coefficients. Theoretical models for the effect of the α process on protein dynamics exist (4, 22). In contrast, the β processes in supercooled liquids and glasses are essentially independent of the solvent viscosity and exist even in the glassy state (23, 24). Two classes of β fluctuations occur in molecular liquids (25–27). The Johari–Goldstein (J-G) fluctuations are related to the α fluctuations; at some temperature, the two merge. In contrast, the “statistically independent” β fluctuations do not merge; they are faster than α at temperatures below a crossing temperature Tcr, cross at Tcr, and are slower above Tcr. Their rate coefficient kβ(T) can be approximated by a standard Arrhenius relation, kβ(T) = Aβ exp[−Hβ/(kBT)].
Proteins are subject to α and β processes (5, 28–30). These do not originate in the protein, but in the bulk solvent and the hydration shell. We have measured dielectric spectra of Mb from <140 to >300 K in different solvents. Fig. 2A displays the peak rate coefficients (k = 2πν) of α and β in a 50/50 (wt/wt) glycerol/water solvent as a function of 1,000 K/T. The two processes cross at Tcr ≈ 210 K. The α process behaves like a typical α relaxation in supercooled liquids. The β process behaves like the “statistically independent” process in molecular glass formers. β is easy to observe well below Tcr where α is extremely slow. In the crossing region, α and β are difficult to separate and we only show the continuation well above Tcr. We retain the name “β relaxation,” but label it with a subscript h. Fig. 2C shows βh relaxation spectra at 160 K in 50/50 (wt/wt) glycerol/water samples without and with Mb at different hydration levels. Already the sample without Mb is inhomogeneously broadened. Adding Mb broadens the spectrum, shifts the peak to lower frequencies, and adds substates at both the low and high part of the spectrum. We assign the additional substates and βh to the hydration shell of the protein. The fact that βh can be observed well below 200 K where the solvent becomes solid means that it does not depend on viscosity. Thus, α is eliminated if Mb is embedded in a solid. We therefore measured the dielectric relaxation of Mb embedded in solid poly(vinyl) alcohol (PVA) at different levels of hydration (Fig. 2B). The parameters for the βh relaxation depend on hydration, as shown in Table 1. At small values of hydration, the activation enthalpy is very large. With increasing hydration, the activation enthalpy moves toward typical values of β relaxations. For hydration levels above h = 0.5 a transition occurs at ≈270 K (not shown in Fig. 2B). It is likely due to the melting of small ice crystals. Ice crystals cannot form if water is closely confined. For h < 0.5, the water molecules are indeed closely confined to the hydration shell. At h > 0.5, there is enough water to permit the formation of small ice crystals.
Fig. 2.
Relaxation processes in Mb. (A) Arrhenius plot of the α and the βh processes of Mb embedded in a 50:50 (wt/wt) glycerol/water solvent with a water–protein weight ratio h = 1, measured with dielectric relaxation spectroscopy. The α process (blue) follows a VTF relation (Eq. 2). The βh process (red) approximately follows an Arrhenius law. (B) Arrhenius plot of the βh processes for Mb embedded in PVA for various values of the hydration h. (C) Dielectric spectra of 50:50 (wt/wt) glycerol/water samples at 160 K for h = 0.5, 2.5, and ∞ (no Mb).
Table 1.
Hβ, Aβ, and Δε of the β relaxations for Mb embedded in PVA at various h
| h | Hβ, kJ/mol | log(Aβ /s−1) | Δε* |
|---|---|---|---|
| 0.2 | 135.5 ± 7.0 | 28.9 ± 1.3 | 14.1 ± 2.5 |
| 0.4 | 79.2 ± 2.2 | 20.5 ± 0.4 | 15.0 ± 0.5 |
| 0.9 | 48.7 ± 1.3 | 16.3 ± 0.3 | 24.4 ± 17.8 |
| 2.1 | 41.2 ± 1.7 | 14.6 ± 0.4 | 31.0 ± 15.5 |
*Δε (Eq. 5) varies with T. The values given are at T = 265 K for h = 0.2, 0.4, and 2.1 and T = 260 K for h = 0.9.
The Energy Landscape
Proteins can assume a very large number of conformational substates that are organized in the energy landscape (17, 18, 31). At any instant of time, the structure of a protein, including its hydration shell, is given by the set of coordinates of its N atoms, where N is of the order of 103. The set of coordinates is a point in a 3N-3-dimensional conformation space. The number of CS in a protein and its hydration shell is extremely large. Assume that each of 100 side chains and each of 200 water molecules in the hydration shell can assume two positions. The number of CS, or points in the conformation space, then is 21002200 or ≈10100. These points, together with the barriers between them, form the EL. A change of the protein structure in real space appears as the motion from one point to another in the conformation space. What has been gained by the introduction of the EL? If the EL were featureless, not much, apart from expressing the fact that proteins exist in a large number of conformations. However, the EL is structured into a number of tiers (19). The top three tiers are relevant for function. Lower tiers have been discovered in cryogenic experiments (32), but we do not consider them. Fig. 3A gives a 1-dimensional cross-section through the Mb energy landscape, drawn ≈20 years ago (33). The top tier, CS0, contains three taxonomic substates, so called because they can be fully characterized. Two of the taxonomic substates in Mb, A0 and A1, have been studied in detail. They have different infrared spectra (34, 35), different structures (36), and different functions (37), namely catalysis and dioxygen storage. Initially, the origins of the equilibrium fluctuations EF1 (now βh) and EF2 (now α) in Fig. 3A were obscure. The barriers between substates were assumed to be properties of the protein and the hierarchy was defined by the barrier heights. Experiments prove, however, that the α fluctuations originate in the bulk solvent, whereas the βh fluctuations originate in the hydration shell. Because the α fluctuations obey the VTF relation, and the βh fluctuations obey the Arrhenius law, it is not possible to use barrier heights to define the hierarchy. We define the hierarchy by the fluctuation rate coefficients at ambient temperature. In a 50:50 (wt/wt) glycerol/water solvent, the rate coefficients at 300 K are approximately kα ≈ 1011 s−1 and kβ ≈ 108 s−1. We therefore assign the α fluctuations to tier 2 and the βh fluctuations to tier 1. Transitions in tier 0 are slower and can involve both α and βh fluctuations. The resulting dynamics are shown in Fig. 3B, a projection of the high-dimensional conformation space on a 2-dimensional conformation plane. Each small circle in this plane corresponds to a CS, characterized by its 103 coordinates. The number of points is of the order of 10100. Fig. 3B shows only a very, very small fraction of the conformation plane. Consider now the life of a protein that starts out in the CS denoted by the black square. At ambient temperature the protein changes its conformation on average every 1/kα or ≈10−11 s. The changes are steps in the EL. Within the time 1/kβ or ≈10−8 s the protein visits a domain containing ≈103 CS. We call this region an α basin, shown purple in Fig. 3B. After an average time 1/kβ the protein makes a larger jump in the conformation space and traces out a new α basin. The process of forming new α basins continues. Together they form a β basin, shown in red. The walk in the EL forming α and β basins continues even if a reaction occurs, for instance, a transition from A0 to A1. Now, consider a second protein starting in a different CS. It makes a similar random walk in the conformation space. Because the total number of CS is many orders of magnitude >1010 it is unlikely that the two proteins will ever be in the same conformation, or that a protein returns twice to the same substate. No two proteins in a sample have exactly the same structure. In a protein ensemble, each protein will go its own way and experiments observe an average.
Fig. 3.
The energy landscape of myoglobin. (A) A 1-dimensional cross-section through the energy landscape of myoglobin as plotted in 1989 (33). It was assumed that the barriers between substates were a property of the protein and that the hierarchy could be defined by the height of the barriers. (B) Present view. Projection of the high-dimensional energy landscape (EL) on a 2-dimensional conformation plane. Each small circle in the plane represents a conformational substate, specified by a vector x(t) ≡ {x1, x2, …, xN} for N atoms. Only a very very small fraction of substates are shown. At the ambient temperature a protein starting in a specific CS makes a random walk in tier 2 with the average time for each step given by 1/kα and it carves out an α basin. In terms of A these fluctuations represent the “equilibrium fluctuations 2” or EF2. After an average time 1/kβ ≈ 10−8 s the protein jumps to another α basin. It continues the random walk (Lévy walk) and carves out a β basin. The random walk continues even if the protein undergoes a process, for instance by a jump from A0 to A1. At low temperatures, kβ ≫ kα and the protein hops between α basins faster than individual basins are explored.
Protein Dynamics
Protein motions that involve shape changes, such as folding or opening of a gate to permit the entry and exit of ligands, are slaved to the α fluctuations in the bulk solvent (2, 3). Here, we discuss the slaving of internal protein processes to the βh fluctuations in the hydration shell. The external fluctuations are measured with dielectric relaxation, the internal fluctuations by using the Mössbauer effect in 57Fe (38) and elastic neutron scattering. In the Mössbauer effect, a fraction f(T) of gamma rays is emitted without energy loss (39). The experimental data are unfortunately not usually given as the measured f(T), but by the mean-square displacement (msd), 〈 x2(T)〉, of the iron atom. The Lamb–Mössbauer factor f(T) and 〈 x2(T)〉 are related by
For 57Fe k02 = 53 Å−2. The data in Fig. 4Inset for 57Fe in a myoglobin crystal show that the msd is linear in T up to ≈200 K and then increases rapidly. To predict the increase we consider a Mössbauer emitter in a fluctuating environment (39). As long as the conformational fluctuations are slower than the lifetime τMö = 140 ns only vibrations decrease f(T). The vibrations lead to the approximately linear increase of 〈x2(T)〉 below 200 K. If the fluctuations are faster than τMö an additional fraction of the gamma rays becomes inelastic. We assume that the total 〈x2(T)〉 is composed of a vibrational and a conformational component (18),
〈x2〉v is found by linearly extrapolating the low-temperature data. Then, 〈x2〉c is given by 〈x2〉t − 〈x2〉v. Inserting 〈x2〉c into Eq. 3 yields fc(T), which is displayed in Fig. 4.
Fig. 4.
The temperature dependence of the dielectric relaxation area aβ(T) and the recoilless fraction fc(T) of the Mössbauer effect. The similar temperature dependence between the two independently measured functions demonstrates that the internal protein motions, characterized by fc(T), are determined by the external βh process, characterized by aβ(T). Note that fc(T) is difficult to measure at low temperatures because a small change in 〈x2〉v changes fc(T) strongly. It is also difficult to measure at high temperatures, because the elastic component becomes very small. (Inset) The temperature dependence of the mean-square displacement (〈x2(T)〉) (msd) of the heme iron in Mb. The msd (black circles) was determined by using the Mössbauer effect for 57Fe in a metmyoglobin crystal (38). At ≲200 K, the msd is due to vibrations and is nearly linear in T. At ≳200 K, the msd increases rapidly. The red diamonds are the msd calculated by assuming the conformational part of the msd is dominated by βh fluctuations.
We now postulate that the increase in the msd above ≈200 K is due to βh fluctuations that are faster than τMö. Fig. 5A shows the βh spectrum at 265 K in a water/myoglobin solution embedded in solid PVA, where the α fluctuations are absent. The vertical line at log(ν/s−1) = 6.06 represents kMö = 1/τMö. The area aβ(265 K) to the left of kMö gives the fraction of fluctuations that are too slow to reduce the Lamb–Mössbauer factor. We thus have fc(265 K) = aβ(265 K). Relaxation spectra measured at many temperatures and shown in Fig. 5B then yield aβ(T). The dielectric relaxation and the Mössbauer effect must be measured on samples with the same hydration. The value of h inside a myoglobin crystal has been computed to be 0.4 (40, 41). We therefore use relaxation spectra for h = 0.4 in Fig. 5B. The area aβ(T) can be determined by fitting the spectra in Fig. 5B with the Havriliak–Negami function (21),
where k = 2πυ, and b and c are fitting parameters. The total area of the spectrum is a″(T) = ∫−∞∞ ε″(k, T)d(log k). The fractional area aβ(T) is approximately a sigmoidal probability given by
 |
The resulting aβ(T) is given in Fig. 4. fc(T) and aβ(T) are close from 200 to 300 K, corresponding to a change of kβ(T) by more than a factor of a thousand. The result proves that the fluctuations seen by the heme iron follow aβ(T) in the hydration shell. Neutron-scattering experiments also show a rapid increase in the msd at ≈200 K (42, 43). The data evaluation for neutron scattering is more complicated, but the result is clear. The β fluctuations are responsible for the rapid increase of the msd (6). The conclusion is surprising. One would expect different parts of the protein's interior to fluctuate with different rates. That the environments of the heme iron and the interior protons fluctuate like the hydration shell suggests a different picture: Major internal protein motions are slaved to the βh fluctuations in the hydration shell. The similar temperature dependence of aβ(T) and fc(T) shown in Fig. 4 can be checked by calculating 〈x2(T)〉 with Eq. 3 by using fc(T) = aβ(T) as input. The result is shown as the red diamonds in Fig. 4 Inset. This curve is not a fit to the Mössbauer data; it is a prediction based on the dielectric relaxation data in the Mb hydration shell. This result shows that the rapid increase of the msd at ≈200 K is not a “dynamic transition”; it is caused by the βh fluctuations in the hydration shell.
Fig. 5.
The determination of the area fraction aβ(T) by using dielectric relaxation spectra. The Mb was embedded in solid PVA (h = 0.4) so that the α relaxation was eliminated. (A) The fitted dielectric relaxation spectrum of the βh relaxation in Mb at 265 K. The vertical line denoted by kMö gives the dephasing rate corresponding to τMö = 140 ns, the mean lifetime of 57Fe. The fractional area to the left of kMö is aβ(T). (B) Dielectric loss spectra ε″(T) at different temperatures. The symbols are from the measurement and the lines plot the fits to Eq. 5.
Biological Roles of the Conformational Fluctuations
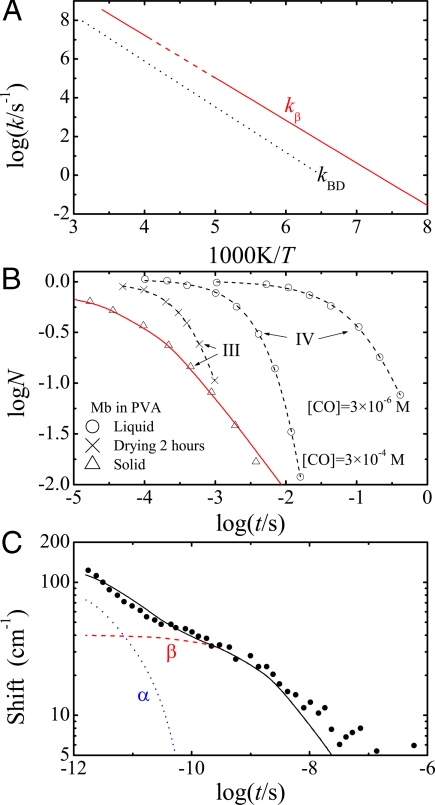
The best-studied function of Mb is binding of CO. Before the mid-1970s binding was assumed to be a simple 1-step process. Flash photolysis experiments extending over a broad range of temperature and time shattered this belief and showed that association and dissociation of CO are complex (17, 44, 45). Here, we demonstrate that our model predicts the temperature dependence and the nonexponential time dependence of the motion of the CO through the protein and of the relaxation of the protein structure after photodissociation. CO bound to the heme iron in the heme cavity (state A) is photodissociated by a laser flash and the CO moves into the heme cavity (state B). At temperatures ≲150 K the CO rebinds to the heme iron without leaving the heme cavity. Above 150 K, the CO also moves to other cavities, particularly to the Xe-1 pocket (state D), and returns from there to rebind (46). Above ≈200 K some CO can escape into the bulk solvent (state S). We expect that the internal motions are controlled by the βh fluctuations in the hydration shell. Consider the transition B→D. Doster and collaborators have measured the rate coefficient kBD(T) in different solvents (44). (kBD is called kBC in ref. 44.) The average coefficient is shown in Fig. 6A, together with kβ(T) from Fig. 2A. The two curves are parallel over more than a factor of 107 in rate, proving that the βh fluctuations permit the CO to move from the heme pocket to Xe-1. The fact that kBD(T) is smaller than kβ(T) is not surprising: CO may have to try more than once to find an open gate.
Fig. 6.
The βh relaxation predicts internal motions. (A) The rate coefficient kBD(T) for the passage of CO from the heme pocket to the Xe-1 cavity follows kβ(T) over nearly 8 orders of magnitude. (B) Rebinding of CO to Mb in PVA. The data are from figure 6 of ref. 17. The curves denoted by IV are obtained in a freshly prepared sample of Mb in PVA while the sample is still liquid. After drying for ≈2 h, process IV disappears and the exponential but [CO]-independent and faster process III appears. On complete drying, when PVA is solid, the nonexponential curve III is obtained. The red solid line is the time dependence as predicted by the βh fluctuation data. (C) The relaxation of the charge-transfer band III in Mb after photodissociation at room temperature (48). The shift of band III after photodissociation is plotted versus time. The highly nonexponential time dependence of the band position shift can be reproduced with the sum of an α process predicted by Eq. 8 and a βh process predicted by Eq. 9.
Next we consider in Fig. 6B the nonexponential time dependence of the CO binding (17). Mb was embedded in liquid PVA and rebinding of CO after flash photolysis was observed during the drying process. Initially, rebinding was exponential in time and proportional to the CO concentration, proving that rebinding originated in the solvent. After some drying, rebinding became faster and concentration-independent, but remained exponential. After further drying, rebinding became nonexponential. The behavior is related to α and βh. In the liquid solvent the viscosity is small, kα(η) is of the order of 1010 s−1, and CO can escape into the solvent and return. As the viscosity increases, kα(η) decreases to a value where CO can no longer escape, rebinding becomes a unimolecular reaction ruled by kβ. The internal rebinding remains exponential in time as long as the rebinding rate coefficient is smaller than kα(η). On further drying, the PVA becomes solid, and the kinetics become nonexponential in time. To compare N(t) for “process III” in Fig. 6B with the βh fluctuations in the hydration shell, we note that N(t) gives the fraction of CO molecules that have not rebound at time t. N(t) should be proportional to the area aβ(t), like that plotted in Fig. 4, but now calculated at a fixed temperature as a function of time. We further expect N(t) to be slower than aβ(t) because the CO visits the internal cavities many times before binding (17, 44). We therefore set
On the log-log scale in Fig. 6B the slowing corresponds to a shift of log(NIII(t)) by log(c). The result, shown as a red curve in Fig. 6B, gives log(c) = −4. The fact that Eq. 7 holds proves that the transit of CO through the protein is indeed controlled by the hydration shell fluctuations.
The relaxation of Mb after photodissociation is a third example of the role of α and βh. The structures of deoxyMb and MbCO differ. After a laser flash dissociates the CO from the heme iron, Mb is still in the MbCO structure, but it relaxes to the deoxyMb structure. The relaxation can be followed by monitoring the wavenumber of the heme-charge transfer band III (47). Anfinrud and collaborators have measured the shift of the peak wavenumber as a function of time after photodissociation (48). A close look at their data in Fig. 6C suggests that two distinct processes are involved. We postulate that the fast process is α, which changes the protein shape to the deoxyMb form, and that the slow process is βh, which relaxes the internal structure. These two processes can be measured separately. Consider α first. Its time dependence is usually described by a stretched exponential (49),
For the glycerol-water solvent used by Anfinrud et al. the parameters are approximately kα = 1011 s−1 (25) and p = 0.65 (49). For βh we set
To calculate aβ(T) we use the spectrum for h = 2.5 in Fig. 2 C and kβ ≈ 108.7 s−1 as estimated from Fig. 2A for the peak at T ≈ 300 K. The spectrum shifted to this peak value then permits the calculation of aβ(t). Eqs. 8 and 9 predict the time dependence of the fast and the slow component in Fig. 6C and only Nα(0) and Nβ(0) must be taken from the data. A crude evaluation gives Nα(0) ≈ 100 cm−1 and Nβ(0) ≈ 40 cm−1. With these values and Eqs. 8 and 9 the predicted relaxation functions and their sum can be calculated and plotted in Fig. 6C. The sum agrees remarkably well with the experimentally determined relaxation function. We repeat that the solid curve in Fig. 6C is a prediction, and not a fit.
The unified model justifies the picture of proteins as picomachines (50). The protein provides the exquisite structural machinery that moves nearly without friction. The α fluctuations in the bulk power and control the shape of the protein. The βh fluctuations in the hydration shell power and control protein internal motions. This model implies that βh permits a broader range of control of protein function than α. The latter is essentially determined by the viscosity of the bulk solvent, which in cells probably does not vary much. The former, however, can be changed in many ways because the hydration shell is not uniform; it is highly structured. Protein surfaces have been designed by evolution and are craggy, contain outcrops and pockets, and have both charged and neutral residues (51, 52). Pockets and channels open and close (53). Addition of salts, sugars, or nucleic acids may change βh. Many molecules attach covalently to protein surfaces. Allostery, phosphorylation, and palmitoylation (54) are examples of candidates for the study of the effect of external factors on βh. The possibilities are endless.
Finally a caveat. Although the data used here can be explained with the α and the βh fluctuations, other motions exist and are important for protein function (51). The famous boson peak fluctuations may also play a role (55).
Acknowledgments.
We thank Albert Migliori and Jon Betts for experimental assistance and discussions, and Angel Garcia, S. Gnanakaran, Vas Lubchenko, Feri Mezei, Stephen Sligar, and Ranko Richert for discussions and comments on the manuscript. This work was supported by Department of Energy Contract DE-AC5206NA25396. J.S. and H.J. were supported by the Swedish Foundation for Strategic Research.
Footnotes
The authors declare no conflict of interest.
Systems undergo thermal fluctuations even in equilibrium. When moved out of equilibrium, the system relaxes toward equilibrium. So long as detailed balance is obeyed, relaxations and fluctuations are related by the fluctuation–dissipation theorem.
References
- 1.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 2.Fenimore PW, Frauenfelder H, McMahon BH, Parak FG. Slaving: Solvent fluctuations dominate protein dynamics and functions. Proc Natl Acad Sci USA. 2002;99:16047–16051. doi: 10.1073/pnas.212637899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frauenfelder H, Fenimore PW, Chen G, McMahon BH. Protein folding is slaved to solvent motions. Proc Natl Acad Sci USA. 2006;103:15469–15472. doi: 10.1073/pnas.0607168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lubchenko V, Wolynes PG, Frauenfelder H. Mosaic energy landscapes of liquids and the control of protein conformational dynamics by glass-forming solvents. J Phys Chem B. 2005;109:7488–7499. doi: 10.1021/jp045205z. [DOI] [PubMed] [Google Scholar]
- 5.Fenimore PW, Frauenfelder H, McMahon BH, Young RD. Bulk-solvent and hydration-shell fluctuations, similar to α- and β-fluctuations in glasses, control protein motions and functions. Proc Natl Acad Sci USA. 2004;101:14408–14413. doi: 10.1073/pnas.0405573101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, et al. Protein fluctuations explored by inelastic neutron scattering and dielectric relaxation spectroscopy. Phil Mag. 2008;88:3877–3883. [Google Scholar]
- 7.Rupley JA, Careri G. Protein hydration and function. Adv Protein Chem. 1991;41:37–172. doi: 10.1016/s0065-3233(08)60197-7. [DOI] [PubMed] [Google Scholar]
- 8.Halle B. Protein hydration dynamics in solution: A critical survey. Philos Trans R Soc Lond B. 2004;359:1207–1224. doi: 10.1098/rstb.2004.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy Y, Onuchic JN. Water mediation in protein folding and molecular recognition. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 10.Helms V. Protein dynamics tightly connected to the dynamics of surrounding and internal water molecules. Chem Phys Chem. 2007;8:23–33. doi: 10.1002/cphc.200600298. [DOI] [PubMed] [Google Scholar]
- 11.Careri G, Gratton E, Yang PH, Rupley JA. Correlation of IR spectroscopic, heat capacity, diamagnetic susceptibility and enzymatic measurements on lysozyme powder. Nature. 1980;284:572–573. doi: 10.1038/284572a0. [DOI] [PubMed] [Google Scholar]
- 12.Brovchenko I, Oleinikova A. Which properties of a spanning network of hydration water enable biological functions? Chem Phys Chem. 2008;9:2695–2702. doi: 10.1002/cphc.200800662. [DOI] [PubMed] [Google Scholar]
- 13.McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 14.Ansari A, Jones CM, Henry ER, Hofrichter J, Eaton WA. The role of solvent viscosity in the dynamics of protein conformational changes. Science. 1992;256:1796–1798. doi: 10.1126/science.1615323. [DOI] [PubMed] [Google Scholar]
- 15.Shibata Y, Kurita A, Kushida T. Real-time observation of conformational fluctuations in Zn-substituted myoglobin by time-resolved transient hole-burning spectroscopy. Biophys J. 1998;75:521–527. doi: 10.1016/S0006-3495(98)77541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansari A. Langevin modes analysis of myoglobin. J Chem Phys. 1999;110:1774–1780. [Google Scholar]
- 17.Austin RH, et al. Dynamics of ligand binding to myoglobin. Biochemistry. 1975;14:5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- 18.Frauenfelder H, Petsko GA, Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979;280:558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- 19.Ansari A, et al. Protein states and protein quakes. Proc Natl Acad Sci USA. 1985;82:5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donth E. The Glass Transition. Heidelberg: Springer; 2001. [Google Scholar]
- 21.Kremer F, Schönhals A. Broadband Dielectric Spectroscopy. Heidelberg: Springer; 2002. [Google Scholar]
- 22.Lubchenko V. Quantitative theory of structural relaxation in supercooled liquids and folded proteins. J Non-Cryst Solids. 2006;352:4400–4409. [Google Scholar]
- 23.Richert R. Heterogeneous dynamics in liquids: Fluctuations in space and time. J Phys Condens Matter. 2002;14:R703–R738. [Google Scholar]
- 24.Ngai KL, Paluch M. Classification of secondary relaxation in glass-formers based on dynamic properties. J Chem Phys. 2004;120:857–873. doi: 10.1063/1.1630295. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi Y, Puzenko A, Feldman Y. Slow and fast dynamics in glycerol-water mixtures. J Non-Cryst Solids. 2006;352:4696–4703. [Google Scholar]
- 26.Capaccioli S, Ngai KL, Shinyashiki N. The Johari-Goldstein beta relaxation of water. J Phys Chem B. 2007;111:8197–8209. doi: 10.1021/jp071857m. [DOI] [PubMed] [Google Scholar]
- 27.Mandanici A, Huang W, Cutroni M, Richert R. Dynamics of glass forming liquids. XII. Dielectric study of primary and secondary relaxations in ethylcyclohexane. J Chem Phys. 2008;128:124505. doi: 10.1063/1.2844797. [DOI] [PubMed] [Google Scholar]
- 28.Swenson J, Jansson H, Hedström J, Bergman R. Properties of hydration water and its role in protein dynamics. J Phys Condens Matter. 2007;19:205109. [Google Scholar]
- 29.Cerveny S, Schwartz GA, Bergman R, Swenson J. Glass transition and relaxation processes in supercooled water. Phys Rev Lett. 2004;93:245702–245705. doi: 10.1103/PhysRevLett.93.245702. [DOI] [PubMed] [Google Scholar]
- 30.Swenson J, Jansson H, Bergman R. Relaxation processes in supercooled confined water and implications for protein dynamics. Phys Rev Lett. 2006;96:247802. doi: 10.1103/PhysRevLett.96.247802. [DOI] [PubMed] [Google Scholar]
- 31.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscape and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 32.Leeson DT, Wiersma DA, Fritsch K, Friedrich J. Energy landscape of myoglobin: An optical study. J Phys Chem B. 1997;101:6331–6340. [Google Scholar]
- 33.Frauenfelder H, Steinbach PJ, Young RD. Conformational relaxation in proteins. Chem Scripta. 1989;29A:145–150. [Google Scholar]
- 34.Alben JO, et al. Infrared spectroscopy of photodissociated carboxymyoglobin at low temperatures. Proc Natl Acad Sci USA. 1982;79:3744–3748. doi: 10.1073/pnas.79.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansari A, et al. Rebinding and relaxation in the myoglobin pocket. Biophys Chem. 1987;26:337–355. doi: 10.1016/0301-4622(87)80034-0. [DOI] [PubMed] [Google Scholar]
- 36.Yang F, Phillips GN., Jr Crystal structures of CO-, deoxy- and met-myoglobins at various pH values. J Mol Biol. 1996;256:762–774. doi: 10.1006/jmbi.1996.0123. [DOI] [PubMed] [Google Scholar]
- 37.Frauenfelder H, McMahon BH, Austin RH, Chu K, Groves JT. The role of structure, energy landscape, dynamics, and allostery in the enzymatic function of myoglobin. Proc Natl Acad Sci USA. 2001;98:2370–2374. doi: 10.1073/pnas.041614298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parak FG, Achterhold K. Protein dynamics on different timescales. J Phys Chem Solids. 2005;66:2257–2262. [Google Scholar]
- 39.Parak FG, Achterhold K, Croci S, Schmidt M. A physical picture of protein dynamics and conformational changes. J Biol Phys. 2007;33:371–387. doi: 10.1007/s10867-008-9102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong S-H, et al. Dynamical transition of myoglobin in a crystal: Comparative studies of X-ray crystallography and Mössbauer spectroscopy. Eur Biophys J. 2001;30:319–329. doi: 10.1007/s002490100152. [DOI] [PubMed] [Google Scholar]
- 41.Parak F, Hartmann H, Schmidt M, Corongiu G, Clementi E. The hydration shell of myoglobin. Eur Biophys J. 1992;21:313–320. doi: 10.1007/BF00188343. [DOI] [PubMed] [Google Scholar]
- 42.Doster W, Cusack S, Petry W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature. 1989;337:754–756. doi: 10.1038/337754a0. [DOI] [PubMed] [Google Scholar]
- 43.Engler N, Ostermann A, Niimura N, Parak FG. Hydrogen atoms in proteins: Positions and dynamics. Proc Natl Acad Sci USA. 2003;100:10243–10248. doi: 10.1073/pnas.1834279100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinert T, et al. Solvent composition and viscosity effects on the kinetics of CO binding to horse myoglobin. Biochemistry. 1998;37:717–733. doi: 10.1021/bi971508q. [DOI] [PubMed] [Google Scholar]
- 45.Tetreau C, Lavalette D. Dominant features of protein reaction dynamics: Conformational relaxation and ligand migration. Biochim Biophys Acta. 2005;1724:411–424. doi: 10.1016/j.bbagen.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Schotte F, et al. Watching a protein as it functions with 150-ps time-resolved x-ray crystallography. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 47.Stavrov SS. Optical absorption band III of deoxyheme proteins as a probe of their structure and dynamics. Chem Phys. 2001;271:145–154. [Google Scholar]
- 48.Jackson TA, Lim M, Anfinrud PA. Complex nonexponential relaxation in myoglobin after photodissociation of MbCO: Measurement and analysis from 2ps to 56 μs. Chem Phys. 1994;180:131–140. [Google Scholar]
- 49.Böhmer R, Ngai KL, Angell CA, Plazek DJ. Nonexponential relaxations in strong and fragile glass formers. J Chem Phys. 1993;99:4201–4209. [Google Scholar]
- 50.Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- 51.Pal SK, Zewail AH. Dynamics of water in biological recognition. Chem Rev. 2004;104:2099–2123. doi: 10.1021/cr020689l. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, et al. Mapping hydration dynamics around a protein surface. Proc Natl Acad Sci USA. 2007;104:18461–18466. doi: 10.1073/pnas.0707647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eyrisch S, Helms V. Transient pockets on protein surfaces involved in protein-protein interaction. J Med Chem. 2007;50:3457–3464. doi: 10.1021/jm070095g. [DOI] [PubMed] [Google Scholar]
- 54.Kang R. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lubchenko V, Wolynes PG. The origin of the boson peak and thermal conductivity plateau in low-temperature glasses. Proc Natl Acad Sci USA. 2003;100:1515–1518. doi: 10.1073/pnas.252786999. [DOI] [PMC free article] [PubMed] [Google Scholar]