The transcription factor XBP1 (X-box-binding protein 1) is essential for plasma cell differentiation and immunoglobulin secretion. This study indicates that the active form of XBP1, XBP1(S), provides a specific marker of advanced plasma differentiation, and in lymphoid malignancies is restricted to plasma cell-derived neoplasms and plasmablastic diffuse large B-cell lymphomas.
Keywords: plasma cell, lymphomas, XBPS1(s), monoclonal antibody
Abstract
The transcription factor XBP1 (X-box-binding protein 1) is essential for plasma cell (PC) differentiation and immunoglobulin secretion. XBP1 is widely expressed, but its activity is precisely controlled by mRNA splicing in response to endoplasmic reticulum (ER) stress. It is the active form of XBP1, XBP1(S), which is required for PC differentiation. The relationship between XBP1(S) expression and PC differentiation in human tissue and its expression in hematologic malignancies has eluded assessment. With a novel antibody, we now define XBP1(S) expression in a large series of normal and neoplastic lymphoid tissues. We establish that XBP1(S) provides a specific marker of advanced plasma differentiation and in lymphoid malignancies is restricted to PC-derived neoplasms and plasmablastic diffuse large B-cell lymphomas. XBP1(S) expression delineates heterogeneity amongst plasmablastic diffuse large B-cell lymphomas, identifying a distinct tumor sub-group. Furthermore, our results establish a direct and practical means of assessing ER stress in human tumors.
Introduction
Differentiation of B-cells to plasma cell (PC) requires reprogramming of gene expression, mediated by a transition in transcription factor network. B-cell lymphoproliferative disorders can be related to stages of this process.1 A key component which remains to be assessed is activation of the transcription factor X-box binding protein 1 (XBP1), a terminal event during differentiation.
An initiating event during differentiation is silencing of Paired box gene 5 (PAX5). Loss of PAX5 unravels B-cell identity2 and may facilitate high-level expression of XBP1.3,4 Repression of PAX5 is mediated by the transcriptional repressor B-lymphocyte induced maturation protein 1 (BLIMP1 also known as PRDM1).5 Both BLIMP1 and XBP1 are essential for PC differentiation6,7 and may act sequentially with Blimp1 required for induction of Xbp1.8 However a preplasmablast secretory stage of differentiation is observed in the presence of defective Blimp1 expression.9,10
XBP1 is a key component of the unfolded protein response (UPR).11 This stress response triggered by accumulation of unfolded protein in the ER, balances adaptive and apoptotic fates.12 During the UPR splicing of 26 nucleotides from XBP1 mRNA results in a reading frame shift, giving rise to an active form of XBP1 XBP1(S).13,14 The essential role for Xbp1 in PC differentiation, and immunoglobulin synthesis reflects a requirement for XBP1(S)15,16 and expansion of the secretory apparatus.8 XBP1(S) has eluded direct assessment in human tissue, a critical issue for our understanding of the UPR, humoral immunity and malignancies derived from differentiating B-cells and PCs.
Design and Methods
XBP1(S) monoclonal antibody
XBP1(S) cDNA was cloned into pIRES2-Myc-EGFP and XBP1(S) carboxy-terminus (amino-acids 165–367) was cloned into pGEX-6P1 (Promega). Anti-XBP1(S) monoclonal antibody (clone 143F) isotype IgG2a/κ was produced as described,17 with GST-XBP1(S)-carboxy-terminus as immunogen.
Tissue samples and tissue microarrays
TMAs were prepared containing samples of normal tissue and 679 pre-treatment lymphoma biopsies (CNIO Tissue Bank) diagnosed according to WHO classification criteria.18
B-cell tumors: chronic lymphocytic leukemias/small lymphocytic lymphoma (B-CLL/SLL) n=21, mantle cell lymphoma (MCL) n=14, follicular lymphoma (FL) n=29, Burkitt’s lymphoma (BL) n=18, marginal zone lymphoma (splenic, extranodal and nodal) (MZL) n=25, DLBCL n=268, plasmablastic DLBCL n=25, lymphoplasmacytic lymphoma (LL) n=9 and myeloma/plasmacytoma n=40.
T/NK-cell tumors: peripheral T-cell lymphoma (PTCL) n=21, anaplastic large cell lymphoma (ALCL) n=4, T-angioimmunoblastic lymphoma n=10, mycosis fungoides/Sézary syndrome n=5 and T-cell lymphoblastic lymphoma/leukemia n=3. Use of human tissue was approved by the CNIO and Research Ethics Committee (UK) reference number 07/Q1206/47.
Cell lines
Myeloma cell lines (RPMI-8226, SK-MM-2, KARPAS-640, NCI-H929 and LP-1), DLBCL cell line (OCI-LY3) and U937 human histiocytic lymphoma were from DSMZ, Germany. HEK 293T cells were transfected with pIRES2-MycXBP1(S) as described.6
Antibodies
BCL6 (clone GI191E/A8, CNIO), BLIMP1 (clone ROS6, CNIO or rabbit-polyclonal19), MUM-1/IRF4 (Santa Cruz Biotechnology), CD138 (Dako), CD38 (Dako) PAX5 (Santa Cruz Biotechnology), GAPDH (clone 26A, CNIO) and ACTIN (clone AC15, Sigma).
RT-PCR, Western blot, and immunostaining
RT-PCR for XBP1 splicing and Western blotting were as described.19,20 A Bond automated system (Leica) was used for XBP1(S) immunostaining of TMA sections. Double immunoenzymatic labeling was as described.6 In all immunostained paraffin sections, PCs provided an internal positive control. Multi-color immunofluoresence (MCIF) was performed on human tonsil tissue as described21 (see also Online Supplementary Appendix).
XBP1(S) scoring
Cases were scored semi-quantitatively by two independent observers (MC and SMM): negative (< 10% positive tumor cells), weak (10% to 50% positive tumor cells) and positive (>50% positive tumor cells).
Results and Discussion
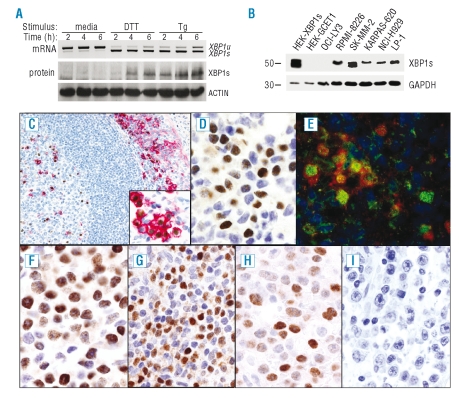
To track ER stress responses and address the relationship between XBP1 activation and PC differentiation in human tissue and lymphoid malignancies, we have raised an XBP1(S) specific monoclonal antibody which works in paraffin immunohistochemistry. To confirm specificity of this antibody we examined XBP1(S) protein expression and XBP1 mRNA splicing in U937 cells undergoing an UPR after treatment with dithiothreitol or thapsigargin.19 The expected correlation was observed with detection of a specific band at 54 kDa by Western blot following XBP-1 mRNA splicing (Figure 1A). Specificity was further confirmed by detection of a specific band in cells transfected with XBP1(S) expression vector and myeloma cell lines (Figure 1B). The OCI-LY3 cell line was used as a negative control.
Figure 1.
Characterization of anti-XBP1(S) monoclonal antibody and XBP1(S) expression patterns in normal tissue. (A) XBP1(S) protein is detected during the UPR following induced XBP1 mRNA splicing. U937 cells were left untreated or subject to an UPR with dithiothreitol (DTT) or thapsigargin (Tg) for indicated times, RT-PCR for XBP1 mRNA splicing (top) and Western blot with anti-XBP1(S) or anti-Actin monoclonal antibodies (bottom). In addition to the specific band at 54kDa, a non-specific band at 50kDa was detected in U937 cells (not shown). (B) Specificity of the XBP1(S) monoclonal antibody is further demonstrated by detection of a single 54 kDa band in transfectants and myeloma cell lines. Western blot of total protein extract from HEK cells transfected with XBP1(S) or GCET1 expression vector as indicated as well as myeloma cell lines and one DLBCL (OCI-LY3) cell line. (C) In tonsil XBP1(S) protein is strongly expressed by plasma cells present in germinal centers, and in subepithelial areas. Double immunoenzymatic staining reveals that the majority of XBP1(S) positive cells (brown) co-express CD138 (red). (Insert) Higher magnification of plasma cells in subepithelial area. (D) At higher magnification varied levels of XBP1(S) expression are evident amongst individual plasma cell. (E) Simultaneous detection of PAX5, BLIMP1 and XBP1(S) in human tonsil by MCIF demonstrates predominant co-expression of XBP1(S) (green) and BLIMP1 (red) in the absence of PAX5 (blue). (F, G, H, I) Immunostaining of lymphoid neoplasms for XBP1(S). Myeloma/plasmacytoma (F), lymphoplasmacytic lymphomas (G) and representative examples of XBP1(S) positive (H) and negative (I) plasmablastic DLBCL.
In human tonsil, XBP1(S) protein was detected in nuclei of PCs within germinal centers (GC) and subepithelial areas, and in a minor percentage of small lymphocytes lacking PC phenotype (Figure 1C). Double immunoenzymatic staining showed that XBP1(S) was expressed in CD138 positive PCs (Figure 1C). A small number of XBP1(S) positive and CD138 negative cells, with centrocyte-like morphology, were observed in GC light zones. There was no evidence of XBP1(S) expression in T cells (data not shown). These observations are in agreement with previous studies of mRNA expression that identified unspliced XBP1 in all B-cell subsets, while XBP1(S) mRNA was restricted to PCs and centrocytes.22
XBP1(S) was expressed in all reactive PC populations in other organs, but XBP1(S) varied amongst individual PCs. While most expressed substantial levels of XBP1(S), occasional PCs had low expression or were negative (Figure 1D). The fact that XBP1(S) expression is not uniform is consistent with the adaptive role XBP1(S) plays, the short half-life of XBP1(S),13 and a close correlation between XBP1 splicing and active ER stress.
Next the relationship of XBP1(S) to PAX5 and BLIMP1 expression was directly examined. As expected, XBP1(S) was predominantly co-expressed with BLIMP1 in the absence of PAX5. Occasional cells weakly co-expressed PAX5 with both BLIMP1 and XBP1(S). Significantly, a rare but distinct population of cells co-expressed XBP1(S) and PAX5 in the absence of BLIMP1 (Figure 1E and Online Supplementary Figure S1).
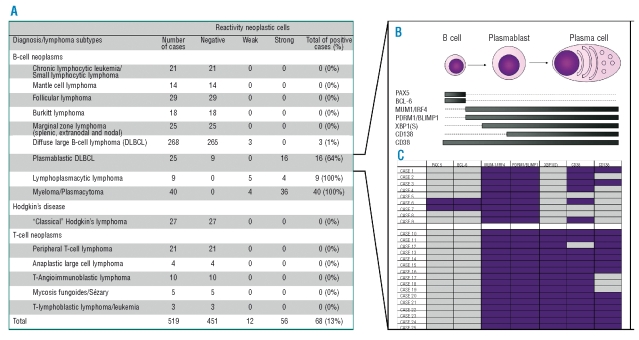
To establish the pattern of expression of XBP1(S) protein in human lymphomas we analyzed TMAs representative of B- and T-cell lymphomas (Figures 1F-I and Figure 2A). XBP1(S) protein was preferentially expressed in diseases characterized by plasmacytic differentiation such as previously described23 in myeloma/plasmacytomas (40/40 positive cases) (Figure 1F), and in lymphoplasmacytic lymphomas (9/9 positive cases) (Figure 1G). Amongst DLBCLs, expression was confined to the plasmablastic sub-type (16/25) (Figures 1H and I). Extended immunophenotyping further defined the relationship of XBP1(S) expression to differentiation in plasmablastic DLBCLs (Figures 2B and C). All cases were BLIMP1 and IRF4/MUM-1 positive and most were PAX5 and BCL6 negative. The 9 XBP1(S) negative cases were only partially CD38 positive and were CD138 negative. In contrast, XBP1(S), positive plasmablastic DLBCL were uniformly CD38 and CD138 positive.
Figure 2.
XBP1(S) expression in lymphoproliferative disorders. (A) Immunohistochemical analysis of XBP1(S) protein expression in lymphoma subtypes. (B) Schema illustrating distribution of markers across B cell to plasma cell differentiation. (C) Immunohistochemical analysis of plasmablastic DLBCLs, purple indicates positive expression. The first nine rows represent XBP1(S) negative cases characterized predominantly by absence of PAX5, BCL6 and CD138 and expression of MUM1/IRF4, BLIMP1 and CD38. The next 16 cases represent XBP1(S) positive plasmablastic DLBCLs. Note the predominace of CD38 and CD138 expression in these cases compared with XBP1(S) negative cases.
Thus our data demonstrate that in the B-cell lineage UPR activation is predominantly restricted to cells expressing BLIMP1 with repressed PAX5. However neither the presence of BLIMP1 nor loss of PAX5 is essential to allow XBP1(S) expression in B-cells in vivo. This may be consistent with a proposed phase of PAX5 functional inactivation observed in the effective absence of Blimp1 expression.9 Whether such XBP1(S) expressing B cells survive to give rise to functional PCs is uncertain. These patterns are paralleled in DLBCL in which XBP1(S) is restricted to the plasmablastic sub-type. Moreover, our results further delineate heterogeneity amongst these neoplasms. XBP1(S) expression identifies disease with advanced PC differentiation, which may be more closely related to myeloma than DLBCL,24 raising the question of alternate treatment choices for this sub-group.
In the UPR, adaptive and apoptotic pathways are finely balanced.12 XBP1 splicing mediates a major adaptive pathway and identifies cells undergoing an active stress response. Our antibody provides a direct practical tool for assessing this in patient samples. Existing and novel treatment strategies aimed at myeloma and other secretory tumors in part act through destabilizing the balance of the UPR.25 We propose that application of our antibody in the diagnostic process may help predict response to such treatments.
Supplementary Material
Acknowledgments
the authors express their gratitude to all members of the Tumor Bank Network of the CNIO for their technical contribution and assistance. Manuscript received September 25, 2008. Revised version arrived on November 5, 2008. Manuscript accepted on November 10, 2008.
Footnotes
Funding: this work was supported by Ministerio de Sanidad y Consumo (G03/179, PI051623, PI052800, CIBER-ER) and Ministerio de Ciencia y Tecnología. (SAF2005- 00221, BIO 2005-01078). RT is a CRUK Senior Clinical Research Fellow.
The online version of this article contains a supplementary appendix.
Authorship and Disclosures
LM and GR designed research, performed experiments, analyzed data and wrote the paper. RT designed research, performed experiments, analyzed data, and wrote the paper. GD performed experiments, analyzed data, and wrote the paper. MAP designed research. MB and SB performed experiments and analyzed data. MC and SM analyzed data. RR and SB performed experiments.
The authors reported no conflicts of interest.
References
- 1.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 2.Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–81. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Reimold AM, Ponath PD, Li YS, Hardy RR, David CS, Strominger JL, et al. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nera KP, Kohonen P, Narvi E, Peippo A, Mustonen L, Terho P, et al. Loss of Pax5 promotes plasma cell differentiation. Immunity. 2006;24:283–93. doi: 10.1016/j.immuni.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–80. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia JF, Roncador G, García JF, Sánz AI, Maestre L, Lucas E, et al. PRDM1/BLIMP-1 expression in multiple B and T-cell lymphoma. Haematologica. 2006;91:467–74. [PubMed] [Google Scholar]
- 7.Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 8.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Kallies A, Hasbold J, Fairfax K, Pridans C, Emslie D, McKenzie BS, et al. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555–66. doi: 10.1016/j.immuni.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–69. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 11.Iwakoshi NN, Lee AH, Glimcher LH. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev. 2003;194:29–38. doi: 10.1034/j.1600-065x.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- 12.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–9. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–6. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 15.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–7. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 16.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–9. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 17.Roncador G, Brown PJ, Maestre L, Hue S, Martínez-Torrecuadrada JL, Ling KL, et al. Analysis of FOXP3 protein expression in human CD4+CD25+ regulatory T cells at the single-cell level. Eur J Immunol. 2005;35:1681–91. doi: 10.1002/eji.200526189. [DOI] [PubMed] [Google Scholar]
- 18.Jaffe ES, Harris NL, Stein H, Vardiman JW. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press; 2001. [Google Scholar]
- 19.Doody GM, Stephenson S, Tooze RM. BLIMP-1 is a target of cellular stress and downstream of the unfolded protein response. Eur J Immunol. 2006;36:1572–82. doi: 10.1002/eji.200535646. [DOI] [PubMed] [Google Scholar]
- 20.Maestre L, Fontan L, Martinez-Climent JA, Garcia JF, Cigudosa JC, Roncador G. Generation of a new monoclonal antibody against MALT1 by genetic immunization. Hybridoma (Larchmt) 2007;26:86–91. doi: 10.1089/hyb.2006.044. [DOI] [PubMed] [Google Scholar]
- 21.Doody GM, Stephenson S, McManamy C, Tooze RM. PRDM1/BLIMP-1 modulates IFN-gamma dependent control of the MHC class I antigen processing and peptide loading pathway. J Immunol. 2007;179:7614–23. doi: 10.4049/jimmunol.179.11.7614. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama Y, Stabach P, Maher SE, Mahajan MC, Masiar P, Liao C, et al. A limited number of genes are involved in the differentiation of germinal center B cells. J Cell Biochem. 2006;99:1308–25. doi: 10.1002/jcb.20952. [DOI] [PubMed] [Google Scholar]
- 23.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–60. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colomo L, Loong F, Rives S, Pittaluga S, Martínez A, López-Guillermo A, et al. Diffuse large B-cell lymphomas with plasmablastic differentiation represent a heterogeneous group of disease entities. Am J Surg Pathol. 2004;28:736–47. doi: 10.1097/01.pas.0000126781.87158.e3. [DOI] [PubMed] [Google Scholar]
- 25.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA. 2003;100:9946–51. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.