Abstract
The success of plant reproduction depends on pollen-pistil interactions occurring at the stigma/style. These interactions vary depending on the stigma type: wet or dry. Tobacco (Nicotiana tabacum) represents a model of wet stigma, and its stigmas/styles express genes to accomplish the appropriate functions. For a large-scale study of gene expression during tobacco pistil development and preparation for pollination, we generated 11,216 high-quality expressed sequence tags (ESTs) from stigmas/styles and created the TOBEST database. These ESTs were assembled in 6,177 clusters, from which 52.1% are pistil transcripts/genes of unknown function. The 21 clusters with the highest number of ESTs (putative higher expression levels) correspond to genes associated with defense mechanisms or pollen-pistil interactions. The database analysis unraveled tobacco sequences homologous to the Arabidopsis (Arabidopsis thaliana) genes involved in specifying pistil identity or determining normal pistil morphology and function. Additionally, 782 independent clusters were examined by macroarray, revealing 46 stigma/style preferentially expressed genes. Real-time reverse transcription-polymerase chain reaction experiments validated the pistil-preferential expression for nine out of 10 genes tested. A search for these 46 genes in the Arabidopsis pistil data sets demonstrated that only 11 sequences, with putative equivalent molecular functions, are expressed in this dry stigma species. The reverse search for the Arabidopsis pistil genes in the TOBEST exposed a partial overlap between these dry and wet stigma transcriptomes. The TOBEST represents the most extensive survey of gene expression in the stigmas/styles of wet stigma plants, and our results indicate that wet and dry stigmas/styles express common as well as distinct genes in preparation for the pollination process.
The plant female reproductive organ, the pistil, has a dual function: the production of the female gametophytes in the ovary and the discrimination of the pollen grains that land on the stigma surface. Different pollen grains are carried to the stigma by insects, wind, water, or by direct contact between the open anther and the stigma, initiating the progamic phase. During this phase, intensive pollen-pistil interactions occur, including important recognition events that determine pollen fate. Ultimately, the success of the pollination process depends on a congruous and compatible recognition of the pollen and on appropriate conditions for pollen hydration, pollen tube germination, and directional growth toward the ovules, all taking place at the specialized tissues of the stigma/style.
Angiosperms have evolved two main strategies to perform pollen-pistil interactions. One consists of species containing dry stigmas, which at maturity have a hydrated proteinaceous extracuticular layer or pellicle but no free-flowing secretion. Generally, dry stigmas have a papillate receptive surface, and on the pollen side they are associated with trinucleate pollen grains (Hiscock and Allen, 2008). When the self-incompatibility system is present in species with dry stigmas, it is often of the sporophytic type (Heslop-Harrison and Shivanna, 1977). The model plants Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) have dry stigmas. Another strategy consists of wet stigmas, which at the receptive state exhibit a fluid secretion or exudate at the stigmatic surface. These stigmas usually have a smooth surface and are associated with binucleate (a vegetative cell and a generative cell) pollen grains (Hiscock and Allen, 2008). When the self-incompatibility system is present in species with wet stigmas, it is often of the gametophytic type (Heslop-Harrison and Shivanna, 1977). Solanaceous species such as tobacco (Nicotiana tabacum) have wet stigmas. In this species, the pollen grains first come into contact with the superficial stigmatic exudate that completely covers the grains, which in turn stimulates the pollen tubes to emerge. The exudate is also present at the intercellular spaces of the stigmatic secretory zone and transmitting tissue in mature pistils, where it is known as extracellular matrix, through which the pollen tubes grow toward the ovary. The importance of the exudate for the directional pollen tube growth in tobacco has already been shown (Goldman et al., 1994).
To fulfill their unique functions, stigmas/styles should express genes encoding the proteins required for their appropriate development and accomplishment on pollen-pistil interactions. It is expected that many of these genes are stigma/style specific or preferential. Recent large-scale studies have identified stigma/style-specific genes in the dry stigma species Arabidopsis and rice (Swanson et al., 2005; Tung et al., 2005; Li et al., 2007). However, in wet stigma species, only a few individual genes have been characterized so far, and little is known about the stigma/style-specific genes important for successful pollination and reproduction. In order to identify these genes and have a general overview of the gene expression profile in wet stigmas, we constructed a tobacco stigma/style cDNA library and generated a database containing the sequences of these cDNA clones (TOBEST). A macroarray analysis of 782 independent transcripts/genes was performed and revealed 46 putative stigma/style-preferential genes. The expression of 10 of these genes was studied in the different organs of the tobacco plant by real-time quantitative reverse transcription (qRT)-PCR, confirming the pistil-preferential expression for nine of them. Most of the 46 tobacco putative stigma/style-preferential genes were not found in the Arabidopsis stigma/style data sets (Swanson et al., 2005; Tung et al., 2005) after a comparison using the BLAST algorithm. In addition, a search in the TOBEST database for the stigma/style-specific genes from this dry stigma species has demonstrated the absence of many of the Arabidopsis genes. Our results indicate that wet and dry stigmas/styles express common as well as distinct sets of genes in preparation for the pollination process.
RESULTS AND DISCUSSION
Production and Analysis of a Tobacco Stigma/Style EST Collection
As a way to identify genes involved in pistil development and the preparation of the pistil for pollination in a wet stigma species, we constructed and sequenced a tobacco cDNA library from a pool of stigmas/styles at stages 1 to 11 of flower development (Koltunow et al., 1990). Stigmas/styles of flowers with dehisced anthers (stage 12) were not included to avoid pollen contamination. A total of 12,480 cDNA clones were sequenced from their 5′ ends, generating 11,216 high-quality ESTs (with Phred quality ≥ 20). As shown at Supplemental Figure S1, the majority of the individual reads (58.3%) are longer than 500 bp, and the smallest one has 184 bases. The information was organized in a database denominated TOBEST (http://gbi.fmrp.usp.br/mhelena). In order to identify ESTs derived from the same transcripts/genes, we used the CAP3 program (Huang and Madan, 1999) to organize redundant ESTs into overlapping contigs. The analysis resulted in a total of 6,177 clusters (1,749 contigs [TOBC] and 4,428 singlets [TOBS]), representing putative different transcripts/genes.
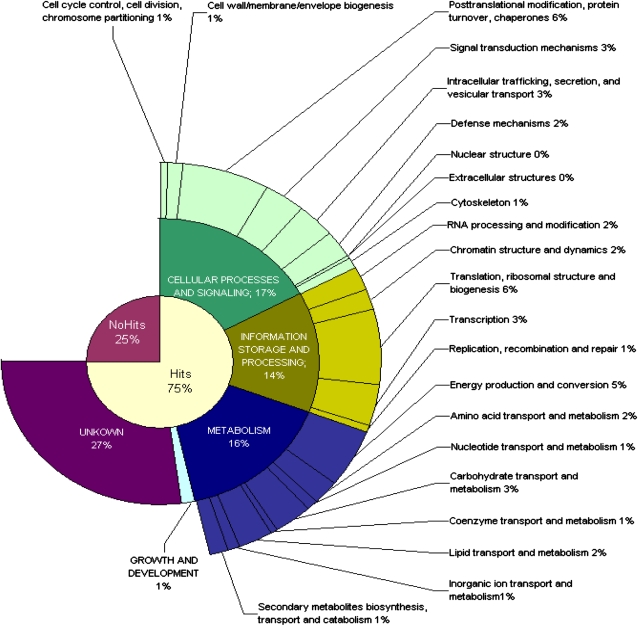
The clusters were analyzed for homology to other sequences using the BLASTX program (Altschul et al., 1997) in the National Center for Biotechnology Information (NCBI) nonredundant protein database. From the 6,177 clusters, a total of 1,552 (25.1%) showed no significant similarity (cutoff e-value of ≤10−5) to sequences present in the database and constitute the “No hits” category (Fig. 1). The corresponding amino acid sequences of the remaining 4,625 clusters were compared with the NCBI KOG database through the online KOGnitor tool and were categorized. A total of 2,894 clusters were automatically categorized by the KOGnitor and 1,731 remained as “No related KOG.” During this phase, expert categorization was introduced to examine the 605 sequences that were among the “No related KOG” but contained a putative annotation defined by the BLAST analysis (for details, see Supplemental Materials and Methods S1). The category “Function unknown” includes the clusters that matched to unknown proteins, expressed, unnamed, putative, or hypothetical proteins, that were initially categorized as “No related KOG,” as well as the ones categorized by KOGnitor as “General function prediction only” and “Function unknown,” with no indication of their corresponding functions. A complete overview of the categorization results of the tobacco stigma/style transcriptome is shown in Figure 1.
Figure 1.
Pie chart representing the tobacco stigma/style gene function categories. The “No hits” category is composed of the clusters/transcripts/genes that had no significant similar sequences (cutoff e-value of ≤10−5) in the NCBI nonredundant protein database after BLASTX analysis. The remaining sequences were automatically categorized by the KOGnitor tool, with a curator revision of the clusters initially categorized as “No related KOG.” The category “Function unknown” includes the clusters that matched to unknown proteins, expressed, unnamed, putative, or hypothetical proteins that were initially categorized as “No related KOG,” as well as those categorized by KOGnitor as “General function prediction only” and “Function unknown,” with no indication of their corresponding function. The slices indicate the number of clusters assigned to each category as a percentage of the total number of clusters (6,177).
The total number of clusters to which a molecular function could be assigned was 2,957 (47.9%). The most abundant categories are “Posttranslational modification, protein turnover, and chaperones” (6.4%), “Translation, ribosomal structure, and biogenesis” (6.2%), and “Energy production and conversion” (4.8%). These results suggest that the stigmas/styles are composed of highly active metabolic cells, with a high level of protein synthesis and degradation, in which energy availability is needed. They also indicate that the correct protein folding, modification, and complex formation should be important to guarantee appropriate pistil development and function. However, this is a groundwork assumption, since, at this moment, we are unable to compare the distribution of protein categories in the stigmas/styles with the representation of each category in the whole tobacco genome.
The cluster TOBS025G12 of the “Posttranslational modification, protein turnover, and chaperones” category, similar to the Arabidopsis SHEPHERD gene, which encodes an endoplasmic reticulum-resident HSP90-like protein (Ishiguro et al., 2002), is a good example of the importance of proteins belonging to this category for plant development. The Arabidopsis shepherd mutant shows expanded shoot apical meristems and floral meristems, disorganized root apical meristems, and defects in pollen tube elongation (Ishiguro et al., 2002). The cluster TOBS053A05, homologous to the Arabidopsis PINHEAD/ZWILLE, a translation initiation factor that has a role in the relative organization of central zone and peripheral zone cells in plant apical meristems (Lynn et al., 1999), shows the significance of proteins of the “Translation, ribosomal structure, and biogenesis” category. Our results are consistent with what was found at the Arabidopsis stigma (Swanson et al., 2005), in which most stigma genes functioned in protein modification and degradation.
The next most numerous category is “Transcription” (3.1%), which includes the transcription factors and is coherent with a stringent transcription regulation program for the pistil genes. The “Signal transduction mechanisms” category, corresponding to 2.9% of the TOBEST transcripts/genes, contains several predicted protein kinases and protein phosphatases that may have roles in cell-cell communication, a fundamental aspect during pollen-pistil interaction. The “Intracellular trafficking, secretion, and vesicular transport” (2.8%), “Carbohydrate transport and metabolism” (2.6%), and “Lipid transport and metabolism” (2.3%) categories contain the gene functions important for the roles exerted by the stigmatic secretory zone and the stylar transmitting tissue in secreting the exudate, which is rich in carbohydrates and lipids (Cresti et al., 1986). The “Defense mechanisms” (2.3%) category is also well represented, consistent with what was observed for the Arabidopsis stigma and transmitting tract gene sets (Tung et al., 2005) and for the rice stigma gene set (Li et al., 2007). Our data corroborate the notion of an overlap of genetic programs regulating reproduction processes and stress/defense responses, as proposed by Li et al. (2007). Similar to what was found in Arabidopsis pistils (Tung et al., 2005), in which there were few genes predicted to function in cell cycle and nucleic acid biosynthesis, in the TOBEST database the categories “Nucleotide transport and metabolism” (0.84%), “Replication, recombination, and repair” (0.68%), “Cell cycle control, cell division, and chromosome partitioning” (0.52%), and “Nuclear structure” (0.05%) are poorly represented.
Although tobacco is a self-compatible species, transcripts related to the gametophytic self-incompatible system, described in Solanaceae species, were identified in the TOBEST database. The singlet TOBS043D10 encodes a protein similar to the RNase NGR2 from Nicotiana glutinosa, which belongs to the RNase T2 family that includes the S-RNases. The clusters TOBC008H09, TOBC071A02, TOBC103B01, and TOBS116E01 encode proteins similar to the HT protein, an additional pistil factor necessary for the self-incompatible reaction (McClure et al., 1999), and TOBC017H10 encodes a putative S-RNase-binding protein p11 precursor (Cruz-Garcia et al., 2005). If these transcripts are capable of translating functional proteins, whether they are just relic genes from the self-incompatible response or have a genuine function in the self-compatible pollen-pistil interaction remains to be established.
It is interesting that at least 33 clusters (five contigs and 28 singlets) encode proteins (e.g. reverse transcriptase, transposase, RNase H, and integrase) related to (retro)transposable elements, indicating that these elements are active in the tobacco stigmas/styles. It is generally accepted that transposable elements are transcriptionally silent in most plant tissues under stress-free conditions. The fact that they are present as cDNAs in the TOBEST database suggests that (retro)transposable elements may be active in plant tissues related to reproduction, as observed for the P element in Drosophila, and/or that reproduction represents a developmental stress condition capable of activating the mobilization of transposable elements. The fact that several stress-response transcripts were identified in the stigmas/styles of tobacco plants cultivated in normal greenhouse conditions strengthens the hypothesis that flowering and reproducing is a stressful condition. As the stigmas/styles do not genetically contribute to the next generation, the activity of these elements would have no long-term instability consequence. However, the situation may be completely different if the mobilization of (retro)transposable elements also occurs within the tobacco ovaries and in the megaspore mother cell, which were not the focus of our study. In any case, as tobacco is an allotetraploid species, it would be more tolerant to (retro)transposable element mobility (Leitch and Leitch, 2008) than would diploid species. It is interesting that among the candidate stigma-preferentially expressed genes of rice there are three retrotransposon proteins: Os07g24100, Os03g63124, and Os12g01900 (Li et al., 2007).
Despite the fact that four plant genomes have already been sequenced (Arabidopsis, rice, poplar [Populus spp.], and Vitis), 25.1% of the sequences still show no statistically significant match (“No hits”) after BLASTX analysis. These sequences may correspond to rapidly evolving sequences whose homology is not easily detectable by computer programs. Alternatively, they may represent RNAs that do not code for proteins and have other functions, like regulating pistil development at the RNA level. Considering the “No hits” (25.1%) and the “Function unknown” (27%) categories, the TOBEST database contains 52.1% of novel pistil transcripts/genes for which no information is available, except for their expression in an organ with a very defined role in plants.
Genes Highly Expressed in the Tobacco Stigmas/Styles
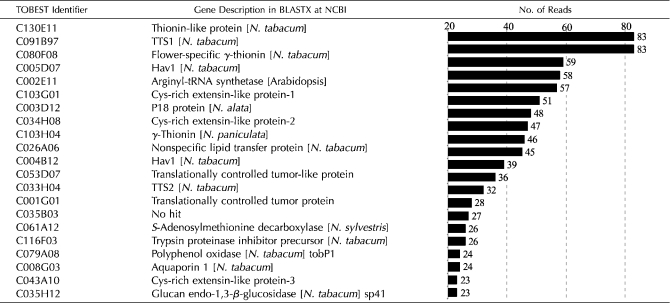
The fact that the sequence data are derived from a nonnormalized cDNA library allows us to infer gene expression levels based on the number of reads contained in the contigs. From 11,216 reads present in the TOBEST, a total of 885 reads (7.9%) belong to only 21 transcripts (Table I), probably corresponding to the highly expressed genes in the tobacco stigmas/styles. Two genes are expressed at the highest levels and account for 83 reads each. A second group of highly expressed genes is formed by 11 genes, which produce 59 to 32 transcripts each. A third group is formed by eight genes, each of them corresponding to 28 to 23 transcripts.
Table I.
Digital northern blotting providing estimates of transcript abundance of the 21 genes with higher expression levels in the tobacco stigma/style
The level of expression was based on analysis of the number of EST reads corresponding to each cluster in the nonnormalized stigma/style cDNA library used to generate the TOBEST database.
Among the highly expressed genes are the following: three contigs encoding γ-thionins, like the flower-specific γ-thionin, FST (Gu et al., 1992); two contigs for transmitting tissue-specific proteins, TTS1 and TTS2 (Cheung et al., 1993); two contigs encoding the lectin nictaba Hav1 (Chen et al., 2002); three contigs for the flower-predominant Cys-rich extensin-like protein, CELP (Wu et al., 1993); two contigs for the translationally controlled tumor protein; one contig encoding the embryo-defective ATP-binding/Arg-tRNA ligase; a contig for the homolog of the Nicotiana alata flower-specific P18 protein, which contains a domain that inhibits pectin methylesterases, PMEI; and one contig for a nonspecific lipid-transfer protein, LTP. One contig corresponding to the homolog of the N. alata stigma/style trypsin proteinase inhibitor precursor (Atkinson et al., 1993), a contig for an S-adenosylmethionine decarboxylase, one contig for the flower-specific polyphenol oxidase tobP1 (Goldman et al., 1998), one contig encoding an aquaporin, and one contig for the (1-3)-β-glucosidase sp41 (Ori et al., 1990) are also among the most abundantly expressed. There is still another highly expressed gene, corresponding to contig TOBC035B03, that has no significant similarity according to the analysis performed at NCBI (BLASTX) and The Arabidopsis Information Resource (TAIR; TBLASTX).
To test the assumption that the number of reads in a cluster approximately represents gene transcript levels in the tobacco stigma/style, we performed a semiquantitative RT-PCR experiment with two genes among the 21 highly expressed ones mentioned above: TOBC091B07 (TTS1), which is a contig composed of 83 clones, and TOBC026A06 (for a nonspecific LTP), which is composed of 45 clones. For comparison, TOBC110E12 (for β-actin), which contains three independent clones in the TOBEST cDNA library, and TOBS008D12 (for the bZIP transcription factor BZI-3), which is a singlet, were also included in the analysis. The results (Supplemental Fig. S2) showed that the number of cDNA clones in a cluster can be considered a good representation of the actual expression level of the corresponding gene.
At least five of these protein functions [γ-thionin, trypsin proteinase inhibitor, the lectin nictaba, polyphenol oxidase, and (1-3)-β-glucosidase] are associated with defense mechanisms and may protect the stigma/style from potential pathogen attack. As pistils are rich in nutrient and are very important for the plant life cycle, it is possible that, unlike the normal plant defense mechanisms that are turned on by pathogen attack, this organ is a priori protected by the expression of developmentally regulated pistil-specific genes encoding defense response proteins.
The other proteins are probably related to pistil development and/or pollen-pistil interactions. Their involvement in pollen-pistil interactions have been demonstrated for TTS (Cheung et al., 1995) and suggested for CELP (Wu et al., 1993). TTS is an arabinogalactan protein of the pistil extracellular matrix, which has been shown to attract pollen tubes and stimulates their growth (Cheung et al., 1995). Concerning the aquaporin, LTP, and PMEI similar sequences, there are reports about the importance of proteins with equivalent functions in the reproductive process (O'Brien et al., 2002; Park and Lord, 2003; Wolf et al., 2003, respectively). In Solanum chacoense, a fertilization-induced and developmentally regulated plasma membrane aquaporin (ScPIP2a) expressed in reproductive tissues has been identified (O'Brien et al., 2002). The maximal expression levels of ScPIP2a correlated with phases of rapid style elongation and papilla formation on stylar cortical cells, which is consistent with a role in cell elongation and expansion during pistil development. The possible roles of LTP and a pectin methylesterase inhibitor (PMEI) will be discussed below. It is noteworthy that many of these genes, corresponding to the highest transcript levels, were identified at the beginning of the 1990s by conventional methods like differential screening.
Genes Involved in Specifying Arabidopsis Pistil Identity Can Be Recognized in the TOBEST
The TOBEST database provides a resource for recognizing genes previously described as involved in specifying pistil identity as well as those that are necessary for the normal pistil morphology and function in other species, like Arabidopsis and snapdragon (Antirrhinum majus), two model species for the study of flower development. A TBLASTN similarity search was performed on the protein sequences of some genes previously described as crucial in Arabidopsis pistil development (Ferrándiz et al., 1999), comparing them with the TOBEST database. As shown in Table II, we found TOBEST sequences significantly similar (cutoff e-value of ≤10−5) to AGAMOUS (AG), FRUITFUL (FUL), SPATULA (SPT), CRABS CLAW (CRC), ETTIN (ETT), PINOID (PID), TOUSLED (TSL), CLAVATA1 (CLV1), CLV2, ERECTA (ER), LEUNIG (LUG), SUPERMAN (SUP), PERIANTHIA (PAN), FIDDLEHEAD (FDH), and WUSCHEL (WUS). To confirm the homology of the identified TOBEST sequences to these Arabidopsis genes, we performed a reverse analysis, a TBLASTX similarity search on the TOBEST sequences listed in Table II, comparing them with the TAIR database. The tobacco sequences for which the best hits in the Arabidopsis genome were the same Arabidopsis sequences initially used were considered their “true homologs” and, therefore, possibly responsible for the same biological process. In doing so, the assumption was made that functionality is transferable based on sequence conservation, to which, of course, there are many exceptions (Van der Hoeven et al., 2002). According to this assumption, the TOBEST database provided sequence information concerning important genes involved in pistil development, like the Nicotiana true homologs for AG, CRC, PID, ER, LUG, SUP, and FDH, from which only AG and CRC were previously identified in Nicotiana. The proof that the tobacco sequences revealed here are functionally equivalent and participate in the same biological processes as the Arabidopsis pistil development genes will depend on future experiments.
Table II.
Putative tobacco genes involved in specifying pistil identity or normal pistil morphology and function
These tobacco sequences were identified by the TBLASTN algorithm at the TOBEST database using the Arabidopsis sequences as query.
| Arabidopsis Genes Involved in Pistil Development | Accession No.a | TOBEST Identifier | BLAST e-Value/% Identity/% Similarity | Previously Identified in Nicotiana |
|---|---|---|---|---|
| AG | NP_567569 | C025G07 | 2e-50/69/81 | Yes |
| FUL | NP_568929 | C067C08b | 1e-59/70/87 | Yes |
| SPT | AAG33640 | S061H09c | 7e-14/52/86 | |
| CRC | AAD30526 | C029A08 | 3e-52/62/70 | Yes |
| ETT | AAC23589 | C075G08d | 2e-15/53/67 | |
| PID | AAF40202 | S107E07 | 2e-43/64/72 | |
| TSL | Q39238 | S028G11e | 4e-11/32/46 | |
| CLV1 | AAB58929 | S013B04f | 3e-29/42/60 | |
| CLV2 | AF177674 | S092C11g | 5e-22/41/59 | |
| ER | NP_180201 | C025H08 | 9e-69/84/92 | |
| LUG | AF277458 | C024G04 | 5e-46/59/74 | |
| SUP | Q38895 | S121D07 | 5e-25/80/85 | |
| PAN | AAD19660 | S001F08h | 7e-05/32/60 | Yes |
| FDH | AJ010713 | C125A09 | 4e-70/82/95 | |
| WUS | AJ012310 | C087E09i | 3e-13/47/73 |
Accession numbers of the Arabidopsis sequences used to search the TOBEST database.
Encodes a MADS box protein, but the best hit by TBLASTX at TAIR is to APETALA1 (At1g69120).
Has similarity to transcription factors of the bHLH family, but the best hit by TBLASTX at TAIR is to At4g02590, which encodes UNE12 (for UNFERTILIZED EMBRYO SAC12).
ETT is an auxin response factor (ARF3). The TOBC075G08 encodes an ARF, but the best hit by TBLASTX at TAIR is to ARF8 (At5g37020).
Encodes a protein kinase, but the best hit by TBLASTX at TAIR is to ATPDK1 (At5g04510).
Encodes a Leu-rich repeat protein kinase, but the best hit by TBLASTX at TAIR is to At1g67720 and not to At1g08590 (CLV1 receptor kinase).
Encodes a Leu-rich repeat transmembrane protein kinase, but the best hit by TBLASTX at TAIR is to At5g01890 and not to At1g65380 (CLV2).
Encodes a bZIP transcription factor, but the best hit by TBLASTX at TAIR is to ATBZIP42 (At3g30530).
Encodes a WUS-related homeobox protein. The best hit by TBLASTX at TAIR is to At2g33880 (WOX9) and not to At2g17950 (WUS).
We also searched the TOBEST database for the presence of HECATE1 (HEC1), HEC2, and HEC3 (Gremski et al., 2007) and No Transmitting Tract (NTT; Crawford et al., 2007). The HEC genes encode basic helix-loop-helix (bHLH) transcription factors that regulate reproductive tract development, including stigma, style, septum, and transmitting tract (Gremski et al., 2007). No significantly similar sequences (cutoff e-value of ≤10−5) were identified for HEC1 and HEC2. In contrast, the cluster TOBS006B10 encodes a bHLH family transcription factor with significant similarity (4e−06) to HEC3 (At5g09750). However, its best hit in the TBLASTX analysis at TAIR is the At1g68920 gene (3e−58), suggesting that the protein encoded by TOBS006B10 does not correspond to the HEC3 homologous sequence. The Arabidopsis NTT (At3g57670) gene encodes a C2H2/C2HC zinc finger transcription factor specifically expressed in the transmitting tract, which is required for its development (Crawford et al., 2007). There is no TOBEST sequence similar to NTT (cutoff e-value of ≤10−5). The absence of these genes in the TOBEST database may imply intrinsic differences in the development of the stigma/style specialized tissues related to pollen-pistil interaction between dry and wet stigma species. On the other hand, these genes may be expressed in the tobacco pistils but were not represented in our stigma/style cDNA library. In any case, the TOBEST represents, to our knowledge, the first large-scale investigation of gene expression in pistils of wet stigma species and provides an important resource for the study of pistil development and pollen-pistil interaction in Solanaceae as well as in other wet stigma species.
Secreted and Membrane-Bound Proteins of the Tobacco Stigmas/Styles
From the 6,177 clusters comprising the TOBEST database, 1,958 (31.7%) are full-length sequences and contain the initial Met, as revealed by the comparison of the deduced amino acid sequences of the clusters with their homologous sequences established by BLASTX (for the criterion used, see “Materials and Methods”). As the specialized tissues of the tobacco stigmas/styles are responsible for the exudate production, which in turn has been shown to contain important components to directional pollen tube growth (Goldman et al., 1994; Cheung et al., 1995), it is very interesting to identify the genes encoding putative secreted proteins. For this purpose, we have analyzed the full-length sequences using the SignalP program (Bendtsen et al., 2004; Emanuelsson et al., 2007). From 1,958 sequences, 195 (10%) contain a predicted signal peptide and 74 (4%) contain a predicted signal anchor (Supplemental Table S1). These proteins are good candidates to have a role in the cross talk between pollen and pistil during the pollination process, either as proteins of the exudate or membrane-bound proteins involved in signal transduction. It is remarkable that several proteins already depicted as important for the reproduction role performed by the pistil have been exposed by this analysis. The clusters TOBC057B05 for CELP (Wu et al., 1993), TOBC042D04 for FST (Gu et al., 1992), and TOBC054G04 for the trypsin proteinase inhibitor (Atkinson et al., 1993) are only a few examples of proteins highlighted by the presence of a signal peptide. Additionally, this analysis also revealed clusters homologous to sequences that may be important for pollen-pistil interaction in wet stigmas and that were not previously described in Nicotiana. The obtusifoliol-14-demethylase, encoded by TOBS068H04, may have a role in lipid signaling in the tobacco pistil, as has been proposed for the CYP51G1-Sc gene of S. chacoense that is induced by pollination and fertilization (O'Brien et al., 2005). The cluster TOBC088G08 encodes a protein highly similar (1e−82 and 96% overall amino acid identity) to the tomato (Solanum lycopersicum) LAT52, which is an anther-specific extracellular protein (Twell et al., 1989) that has been shown to interact with pollen-specific receptor kinases (Tang et al., 2002). To investigate the expression profile of the LAT52 tobacco homolog, we performed a real-time RT-PCR experiment in which its stamen-preferential expression was demonstrated (Supplemental Fig. S3). Despite its very low expression level in stigmas/styles, its presence was revealed in the TOBEST database, and the study of its stigma/style role is an interesting subject for further investigation.
The analysis of signal anchor proteins has revealed five clusters (TOBC092G02, TOBC109B10, TOBC109F07, TOBS055A01, and TOBS124G05) encoding the putative transport protein, SEC61 γ-subunit, and two clusters (TOBC124B09 and TOBS011B05) for the putative SEC61 β-subunit, both constituents of the general secretory pathway, a highly conserved system for intracellular trafficking, protein secretion, and membrane insertion. Additionally, one cluster (TOBS117F05) for an endoplasmic reticulum lumen-retaining receptor family-like protein and two clusters (TOBC055D02 and TOBC130D05) for the microsomal signal peptidase 12-kD subunit family protein were identified, demonstrating the possibility of finding new proteins important for the secretory pathway among the sequences listed in Supplemental Table S1.
From the 195 clusters identified as encoding proteins with signal peptides, for at least 72 sequences there are no indications of their molecular functions. Furthermore, 44 sequences from the ones containing a putative signal anchor are also for proteins of unknown function. Therefore, the TOBEST sequences identified here form a key group of candidates to participate in pollen-pistil interaction to be further studied in the future. These proteins may have functions related to pollen adhesion, hydration, and germination as well as pollen tube nutrition and directional growth, all functions provided by the secretory tissues of the stigmas/styles.
Analyses to Identify Genes Preferentially Expressed in Stigmas/Styles
Differential macroarray analysis was used as an approach to identify genes that are preferentially expressed in tobacco stigmas/styles. Plasmid DNAs of representative clones from 782 independent randomly selected clusters (for details, see “Materials and Methods”) were spotted onto nylon membranes (four repetitions of each clone and two identical membranes for each plate). cDNA probes for stigmas/styles and nonreproductive organs (a mixture of roots, stems, leaves, sepals, and petals) were prepared, and equal amounts of radioactively labeled samples were separately hybridized to the nylon membranes (for an example of hybridized membranes, see Supplemental Fig. S5). The signals were converted to relative intensity values, quantified, and statistically analyzed (for details, see “Materials and Methods”; Supplemental Table S2). After removal of the probes, the membranes were hybridized again with swapped probes prepared from independent biological replicates of each sample. In our analysis, we did not include ovary samples in the negative probe to avoid exclusion of genes expressed in the whole pollen tube path. The rationale is that stigmas/styles and ovaries belong to a single structure, the female reproductive organ, and that the pollen tube path starts in the stigma, continues through the style, and goes until the ovules in the ovary.
A total of 65 clusters were preferentially expressed in stigmas/styles, among which 46 have shown an expression ratio at least 2-fold higher in stigmas/styles than in nonreproductive organs (Table III). Eight of the genes identified in the macroarray have been previously described as stigma/style specific in Nicotiana species: PELP class III (TOBC020F02; Goldman et al., 1992), NaPRP4 (TOBC132G06; Chen et al., 1993), TTS (TOBC033H04 and TOBC091B07; Cheung et al., 1993), STIG1 (TOBC054D09 and TOBC079H03; Goldman et al., 1994), and AGPNa3 (TOBC040F11 and TOBC022C03; Du et al., 1996), demonstrating the success of our macroarray approach. The importance of some of these proteins in pollen-pistil interaction has been proposed based on accumulated evidence. PELP class III protein is located in the extracellular matrix of the stylar transmitting tissue and, after pollination, is translocated into the pollen tube callose walls, in accordance with a role in pollen tube growth (de Graaf et al., 2003). Additionally, it has been shown that the STIG1 protein from tomato binds to the pollen-specific receptor kinases (LePRK1 and LePRK2) and promotes pollen tube growth in vitro, which is in agreement with a role in pollen-pistil interaction (Tang et al., 2004). Petunia (Petunia hybrida) STIG1 knockout mutant plants and tobacco transgenic plants in which STIG1 has been silenced showed an accelerated deposition of exudate onto the stigmatic surface, suggesting that the STIG1 protein plays a role in the temporal regulation of exudate secretion (Verhoeven et al., 2005).
Table III.
Genes revealed by the macroarray analysis as preferentially expressed in tobacco pistils (stigmas/styles)
The table shows the genes that are at least 2-fold up-regulated in tobacco stigma/style in comparison with a pool of nonreproductive organs (roots, stems, leaves, sepals, and petals). A TBLASTX analysis was performed against the nonredundant NCBI database, and the results were filtered using the expectation value of 1 e−05.
| TOBEST Identifier | Result of TBLASTX at NCBI | e-Value/% Identity/% Similarity |
|---|---|---|
| 10-fold up-regulated in tobacco stigma/style | ||
| C003D12 | AJ004957, N. alata mRNA for P18 protein (plant invertase/pectin methylesterase inhibitor) | 3e-12/40/56 |
| C033H04a | Z16404, N. tabacum TTS-2 mRNA | 2e-82/100/100 |
| C040F11a | NAU25628, N. alata stigma-specific arabinogalactan protein precursor (AGPNa3) mRNA | 6e-77/84/88 |
| C091B07a | Z16403, N. tabacum TTS-1 mRNA | 4e-47/98/98 |
| C092C05 | AP009261, S. lycopersicum genomic DNA, chromosome 8, clone C08HBa0005L01 (putative hydrolase involved in cell wall metabolism) | 4e-95/91/98 |
| S004A06 | NM_118062, Arabidopsis pectin acetylesterase family protein (AT4G19420) | 4e-36/60/75 |
| S008H02 | NM_128311, Arabidopsis ATFD3 (FERREDOXIN 3); electron carrier (ATFD3) | 3e-46/76/90 |
| S020F12 | AM431130, V. vinifera contig VV78X078279.6 (unknown protein) lanthionine synthetase C-like family protein | 2e-40/60/69 |
| S053D05 | NM_001056182, O. sativa (japonica group) Os03g0265300 mRNA (putative peroxisomal membrane carrier protein) | 5e-61/70/82 |
| 4-fold up-regulated in tobacco stigma/style | ||
| C020F02a | Z14019, N. tabacum mRNA for pistil extensin-like protein | 0.0/100/100 |
| C022C03a | NAU25628, N. alata stigma-specific arabinogalactan protein precursor (AGPNa3) | 6e-74/82/87 |
| C074A03 | AC148819, Medicago truncatula chromosome 2 clone mte1-11g7 (unknown protein) | 2e-19/59/80 |
| S001C11 | AF492629, Capsicum annuum auxin-induced SAUR-like protein | 6e-52/73/79 |
| S042D12 | NM_124535, Arabidopsis Leu-rich repeat transmembrane protein kinase, putative (AT5G51560) mRNA | 2e-78/71/85 |
| S046C04 | AJ306825, Populus tremula × Populus tremuloides mRNA for auxin/IAA protein (IAA2 gene) | 5e-33/77/90 |
| S073B06 | AY464053, Gossypium hirsutum clone CHX015K18 mRNA sequence (similar to ECERIFERUM5, an ABC transporter of the WBC subfamily) | 5e-29/69/80 |
| S118H02 | AM484567, V. vinifera contig VV78X017002.16 (hypothetical protein) | 4e-89/82/92 |
| 2-fold up-regulated in tobacco stigma/style | ||
| C005C12 | AF389848, N. tabacum nictaba (NT1) mRNA | 3e-70/90/92 |
| C015C07 | L46681, S. lycopersicum aspartic protease precursor mRNA | 2e-98/88/97 |
| C023B06 | M. truncatula mRNA for nodulin MtN3 gene | 7e-68/65/82 |
| C048F04 | AK070726, O. sativa (japonica group) cDNA clone J023059E17 (similar to root hair-defective 3 GTP-binding family protein) | 6e-33/80/91 |
| C054D09a | X77823, N. tabacum STIG1 gene | 2e-118/100/100 |
| C061A12 | AF321141, N. tabacumS-adenosylmethionine decarboxylase mRNA | 1e-49/86/87 |
| C061G07 | AC006259, Arabidopsis bacterial artificial chromosome F21J6 from chromosome V (nodulin-like protein) | 5e-39/66/83 |
| C065A09 | AC171731, S. lycopersicum chromosome 10 clone C10HBa0041K23 (similar to nonspecific lipid-transfer protein) | 1e-21/49/68 |
| C079H03a | X77823, N. tabacum STIG1 gene | 2e-91/90/95 |
| C110D03 | AK224692, S. lycopersicum cDNA, clone FC08AF07, unknown protein from tomato fruit | 3e-26/68/77 |
| C116D05 | NM_124077, Arabidopsis senescence-associated protein-related (At5g47060) mRNA | 2e-26/59/72 |
| C130F10 | AF173863, N. tabacum putative PLA2 (F8) mRNA (phospholipase A2) | 2e-96/95/95 |
| C132G06a | X70441, N. alata NaPRP4 mRNA | 1e-43/56/70 |
| S003A04 | BT014332, S. lycopersicum clone 133605F, mRNA sequence (unknown protein) | 2e-93/92/97 |
| S004E12 | EF417990, O. sativa (japonica group) purple acid phosphatase PAP2 mRNA | 9e-89/78/86 |
| S010A10 | NM_180553, Arabidopsis D111/G-patch domain-containing protein (At5g26610) | 6e-52/68/81 |
| S013F08 | AJ012662, N. tabacum mRNA for proliferating cell nuclear antigen | 1e-106/100/100 |
| S013H02 | NM_116135, Arabidopsis ATMRP10 (Arabidopsis multidrug resistance-associated protein 10) | 5e-56/74/86 |
| S013H03 | AY572222, Solanum virginianum auxin-repressed protein (ARP1) mRNA | 1e-59/73/83 |
| S026F05 | DQ321489, Nicotiana benthamiana AGO1-2 mRNA | 1e-49/94/96 |
| S035G04 | BT014006, S. lycopersicum clone 133058F (similar to senescence-associated protein-like Arabidopsis TETRASPANIN8) | 2e-60/80/92 |
| S049F11 | BT014514, S. lycopersicum clone 133901F, mRNA sequence from fruit (similar to calcium-binding EF hand family protein and to regulatory subunits of type 2A protein phosphatases from Arabidopsis) | 2e-76/96/99 |
| S066E06 | AB075550, N. tabacum NtPDR1 mRNA for pleiotropic drug resistance-like protein | 1e-69/75/84 |
| S068D08 | AK224748, S. lycopersicum cDNA, clone FC12BB08 from fruit mRNA | 8e-31/98/100 |
| S072H12 | AF416289, S. lycopersicum auxin-regulated protein mRNA | 8e-40/65/74 |
| S074B04 | NTU64922, N. tabacum geranylgeranylated protein NTGP1 mRNA | 3e-93/99/99 |
| S074F07 | AJ278743, S. lycopersicum partial mRNA for putative β-1,3-glucanase (ga2 gene) | 2e-60/65/76 |
| S086F04 | LEU19886, S. lycopersicum predominantly leaf-expressed protein mRNA (unknown protein) | 5e-78/90/96 |
| S132D06 | Z97064, Citrus paradisi mRNA for hypothetical protein (putative glyoxalase I) | 2e-86/84/91 |
Sequences previously described as pistil specific or preferential in Nicotiana species.
Using two or more as the cutoff value, we have identified a total of 38 new stigma/style preferentially expressed genes, from which at least six transcripts/genes encode proteins of unknown functions. It is interesting that among the genes preferentially expressed in stigmas/styles there are four genes encoding putative auxin-related proteins (TOBS001C11, TOBS046C04, TOBS013H03, and TOBS072H12, with similarity to an auxin-induced SAUR-like protein, an auxin/indole-3-acetic acid [IAA] protein, an auxin-repressed protein, and an auxin-regulated protein, respectively). Auxin has been shown to be involved in pistil development (Nemhauser et al., 2000; Cecchetti et al., 2004), and it is known that the auxin response factors ARF6 and ARF8 are expressed in style and transmitting tract (Wu et al., 2006). In the TOBEST database, there are at least 20 genes encoding auxin-related genes, including the one encoding the ARF8 homolog (TOBC075G08), and four putative auxin-response transcription factors. However, not all of these genes were included in the macroarray membranes. The overrepresentation of auxin signaling components has also been found for rice stigmas (Li et al., 2007). Our results corroborate the importance of this plant hormone in pistil development in different species, regardless of their stigma type, wet or dry.
Interestingly, three ATP-binding cassette (ABC) transporters were shown to be pistil preferential (TOBS013H02 encoding a protein from the MRP subfamily, TOBS066E06 encoding a member of the PDR subfamily, and TOBS073B06 encoding a transporter belonging to the WBC subfamily). We have previously characterized the NtWBC1 gene, which is highly expressed in the tobacco stigmatic secretory zone and is probably involved in lipid transport to the exudates (Otsu et al., 2004). It is possible that the product of TOBS073B06, which shows similarity to ECERIFERUM5, an ABC transporter of the WBC subfamily, interacts with the NtWBC1 protein to form a functional lipid transporter. Arabidopsis eceriferum mutants display pollen grains with defects in the tryphine lipid droplets of the pollen coat and specifically lack a functional pollen-stigma recognition system, resulting in male sterility (Hulskamp et al., 1995). It has already been proposed that the proteins found on the pollen coat of dry stigma species fulfill a similar role as those present in the exudate of species with wet stigmas (Wolters-Arts et al., 1998). It is also possible that similar proteins have been recruited to perform the secretion function of the pollen coat in dry stigma species and of the exudate in wet stigma species. In this case, the TOBS073B06 protein could be the stigma functional equivalent of the pollen ECERIFERUM5.
Among the pistil-preferential genes identified by macroarray there are three genes encoding proteins of cell wall metabolism (TOBC092C05 for a hydrolase, TOBS004A06 for a pectin acetylesterase, and TOBC003D12 for a pectin methylesterase inhibitor). As has been described previously, the Nicotiana secretory cells of the stigma have thin walls, mainly composed of pectin and a low amount of cellulose (Cresti et al., 1986). At maturity, the cells are loosely arranged, with large intercellular spaces, and very easily separated (Cresti et al., 1986). The possible roles of the enzymes identified could be to determine the level of cell adhesion and contribute to the controlled loosening of the cells of the stigmatic secretory zone and transmitting tissue, allowing the accumulation of exudate in the intercellular spaces and facilitating posterior pollen tube growth.
An additional pistil-preferential gene identified produces a nonspecific LTP (TOBC065A09). An LTP (SCA) from lily (Lilium longiflorum) exudates has been shown to be important for pollen tube adhesion (Park et al., 2000; Park and Lord, 2003) in conjunction with a stylar pectin (Mollet et al., 2000), and this may also be the function of the TOBC065A09-encoded protein. An alternative possibility is that this LTP is involved in loosening of the stigma/style cells, as has been shown for another LTP (Nieuwland et al., 2005). Several different LTPs have been identified as stigma specific in Arabidopsis (Swanson et al., 2005; Tung et al., 2005). Despite the large percentage of genes that may be related to lipid metabolism encountered in the rice stigma, no LTP was included (Li et al., 2007).
An interesting candidate for further studies, identified by the macroarray, is the TOBS042D12-encoded protein, a Leu-rich repeat transmembrane protein kinase. Due to its predicted plasma membrane localization, it might interact with ligands held on the pollen grain or the pollen tube surface and transduce intracellular responses into the pistil cells. The TOBS042D12-encoded protein may have an important role in cell-cell communication and signal transduction in the context of pollen-pistil interactions, like the LePRKs (Tang et al., 2004).
Taking advantage of the tobacco EST projects that are in progress at the European Sequencing of Tobacco (ESTobacco; http://www.estobacco.info/) and at the Tobacco Genome Initiative (TGI; http://tgi.ncsu.edu/), we have undertaken a complementary approach to analyze the TOBEST database and identify the stigma/style transcripts/genes that are not expressed in other tobacco organs. We performed an in silico comparison of the stigma/style-expressed genes with the ESTs from vegetative organs (leaves, roots, and seedlings), flowers, and seeds using the BLAST program (cutoff e-value of ≤10−5). The results are shown in Supplemental Figure S4. Despite the different number of ESTs found in the databases of these projects, the overlap between the stigma/style and vegetative organs is around 60% to 70% (66.6% for ESTobacco and 61.8% for TGI). In contrast, the TOBEST database contains from 27.6% (ESTobacco) to 36.1% (TGI) transcripts/genes that were not found in any of the other tobacco organs examined in these projects, and at least some of them may represent stigma/style-specific sequences.
Real-Time RT-PCR Experiments and Validation of the Macroarray Results
A key requirement for qRT-PCR analysis of gene expression is the identification of a reference gene with transcript levels that are relatively stable across the samples in study and, therefore, suitable for use as a good internal control. As there is no internal control previously established as adequate for studies with the different tobacco organs/tissues, we first performed qRT-PCR experiments with three genes commonly considered as constitutive and used in the literature as internal controls: β-actin, GAPDH, and polyubiquitin. The results were analyzed by the NormFinder algorithm (Andersen et al., 2004), and the stability value was calculated. According to this analysis, the lower the stability value, the lower the estimated expression variation and, therefore, the higher the constancy of the expression among the samples tested, in our case tobacco roots, stems, leaves, sepals, petals, stamens, stigmas/styles, and ovaries. As can be seen in Table IV, both the β-actin and the GAPDH genes have the same stability value, indicating that they are equally suitable as internal controls for the study of gene expression in the different tobacco vegetative and floral organs. Therefore, all of our qRT-PCR data were normalized to β-actin gene expression.
Table IV.
Stability values for three candidate reference genes
Real-time RT-PCR experiments were performed for the genes listed in RNA samples extracted from tobacco vegetative and floral organs (roots, stems, leaves, sepals, petals, stamens, stigmas/styles, and ovaries), and their stability values were determined by the NormFinder algorithm (Andersen et al., 2004). Primer sequences of the three candidate reference genes are listed in Table VI.
| Gene Name | TOBEST Identifier | Stability Value |
|---|---|---|
| β-Actin | C110E12 | 0.99 |
| GAPDH | C010F09 | 0.99 |
| Polyubiquitin | C018H09 | 1.82 |
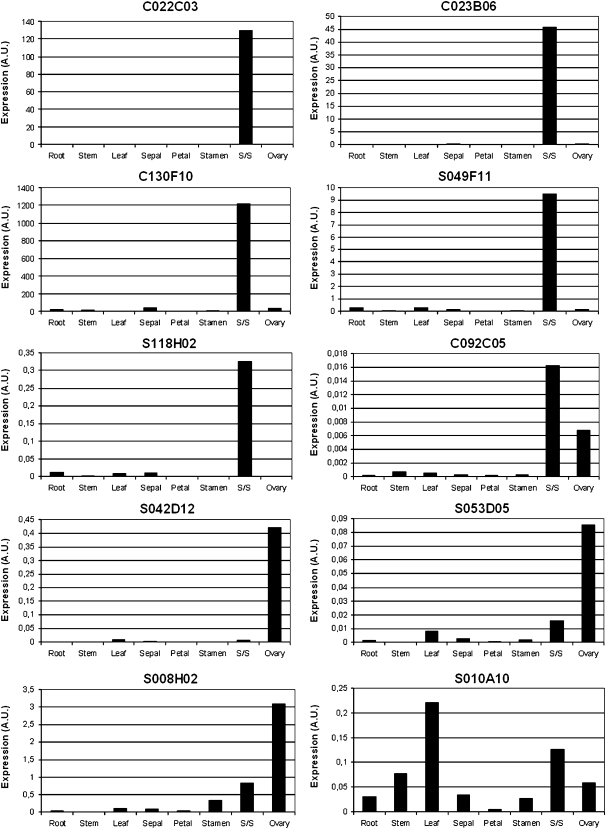
Direct validation of our macroarray results was obtained by qRT-PCR analysis for nine uncharacterized genes and one gene (TOBC022C03) homologous to a sequence previously identified as stigma specific in the self-incompatible species N. alata (AGPNa3). These 10 genes were representative of the three categories of genes with distinct levels of fold up-regulation (Table III). The genes TOBC092C05, TOBS008H02, and TOBS053D05 belong to the 10-fold up-regulation category; the TOBC022C03, TOBS042D12, and TOBS118H02 genes have shown a 4-fold up-regulation in the macroarray analysis; and the TOBC023B06, TOBC130F10, TOBS010A10, and TOBS049F11 genes are from the 2-fold up-regulation category. As shown in Figure 2, nine of the 10 genes (90%) examined have demonstrated pistil-preferential expression. The genes corresponding to the clusters TOBC022C03 (to an arabinogalactan protein similar to AGPNa3), TOBC023B06 (similar to nodulin MtN3), TOBC130F10 (phospholipase A2), TOBS049F11 (similar to calcium-binding EF hand family protein and to regulatory subunits of type 2A protein phosphatases from Arabidopsis), and TOBS118H02 (hypothetical protein) are expressed exclusively at stigmas/styles. TOBC092C05 (putative hydrolase involved in cell wall metabolism) is preferentially expressed in stigmas/styles, but it is also expressed at intermediary levels in ovaries.
Figure 2.
Expression patterns of genes identified as preferentially expressed in the pistil by macroarray analysis. qRT-PCR experiments were performed with cDNA templates prepared from roots, stems, leaves, sepals, petals, stamens, stigmas/styles (S/S), and ovaries. The relative expression levels are represented in arbitrary units (A.U.) normalized to the expression level of the β-actin gene, used as a reference, in each RNA organ sample.
The clones TOBS042D12 (for a Leu-rich repeat transmembrane protein kinase), TOBS008H02 (encoding ferredoxin 3), and TOBS053D05 (putative peroxisomal membrane carrier protein) are expressed at higher levels in ovaries than in stigmas/styles (Fig. 2). Ovaries, which are the lower part of the pistils, also contain specialized tissues for conducting the pollen tube, as a continuation of the stigmatic secretory zone and stylar transmitting tissue. In addition to their role in producing the female gametophyte, ovaries also have to provide appropriate conditions for nutrition of the pollen tube and directional growth, as the stigmas/styles. Therefore, it was expected that some of the stigma/style-preferential genes would be expressed at the ovary as well. The fact that the macroarray analysis has revealed pistil-specific genes with higher expression levels in ovaries than in stigmas/styles can be explained based on the methodology used, in which the negative probe was a mixture of the vegetative organs (roots, stems, leaves, sepals, and petals) and the comparisons of the hybridization signals were performed solely with stigmas/styles (positive probe). Supplemental Figure S6 shows the expression profiles of these genes evaluated by real-time RT-PCR on RNA samples prepared like those used for the positive and negative probes of the macroarray experiment. The gene corresponding to TOBS008H02 is also expressed at a lower level in stamens and is expressed exclusively at the reproductive organs.
The single gene (TOBS010A10, for a D111/G-patch domain-containing protein) that did not confirm the macroarray result had shown an expression ratio of stigma/style to nonreproductive organs very close to the cutoff value established as meaningful in our experiments. The fact that this gene has a high expression level in leaves and low expression levels in other nonreproductive organs like petals resulted in the dilution of its transcripts in our negative probe and might explain the 2-fold up-regulation ratio we encountered in the macroarray analysis (Supplemental Fig. S6).
The gene TOBC022C03 encoding the tobacco homolog of AGPNa3 is stigma/style specific and developmentally regulated, displaying its higher expression level at stage 11 of tobacco flowers (data not shown), which precedes anthesis (Koltunow et al., 1990). This expression pattern is the same as that shown for AGPNa3 in N. alata, a self-incompatible species (Du et al., 1996). The prevalence of AGPs in reproductive tissues has led to speculation that they play significant functional roles ranging from serving as nutrient resources to cell-cell recognition in plant reproduction (Cheung and Wu, 1999). Our results corroborate the idea that this AGP has an important role in plant reproduction and is not restricted to self-incompatible species.
Multiple secretory phospholipase A2 genes have been identified in plants and encode isoforms with distinct regulatory and catalytic properties (Lee et al., 2005). Plant phospholipases A2 have been suggested as being involved in several important plant processes, including lipid metabolism and plant signaling. Some of the Arabidopsis isoforms were found to be predominantly or exclusively expressed in the flower tissues, suggesting that they might play a role in the process of reproduction (Lee et al., 2005). Therefore, the identification of a stigma/style-specific phospholipase A2 (TOBC130F10) is consistent with a role in some reproduction process taking place in the pistil.
The cluster TOBC023B06 corresponds to a stigma/style-specific gene encoding a protein similar to the nodulin MtN3. In Arabidopsis, there are at least eight members of the nodulin MtN3 family that are candidates for transmembrane putative receptors but for which the biological processes are unknown. To better characterize the gene corresponding to the cluster TOBC023B06, we have analyzed its expression in stigmas/styles at different developmental stages (data not shown). This gene is developmentally regulated, and its higher expression level occurs at stage 11 (preceding anthesis). TOBS049F11 singlet, which is almost exclusively expressed in stigmas/styles (Fig. 2), encodes a protein highly similar to calcium-binding EF hand family proteins and to regulatory subunits of type 2A protein phosphatases. Dephosphorylation and phosphorylation events are involved in most of the signaling pathways in plants and have to be in balance for optimal regulation of cellular activities (Luan, 2003). The organ specificity of TOBC023B06 and TOBS049F11 raises questions about their roles in the stigmas/styles. Future experiments will be necessary to verify if they are involved or not in some signal transduction pathway. The singlet TOBS118H02 encodes a protein highly conserved in Vitis vinifera (AM484567; 82% amino acid identity) and Arabidopsis (At1g62870), for which no function has been established so far. Therefore, it represents a novel stigma/style-specific gene.
TOBS053D05 is mainly expressed in ovaries and encodes a putative peroxisomal membrane carrier protein. In plants, the roles of the peroxisomes depend on the organ or tissue in which they occur and may be as diverse as lipid mobilization, photorespiration, and conversion of fixed N2 (Buchanan et al., 2000), so it is still very premature to propose a function for this pistil protein. The gene corresponding to clone TOBS008H02 encodes a ferredoxin 3 homolog and is specifically expressed at the reproductive organs, with a high expression level in ovaries. Interestingly, Yoshida et al. (2005) also identified a rice pistil-specific gene encoding a ferredoxin III (Os3736; AC092557). Ferredoxins act as electron carriers in photosynthesis and play an important role in electron transfer processes. The pistil is a photosynthetic organ, and the elevation of ferredoxin transcript levels in pistils might be related to active electron transport in this organ (Yoshida et al., 2005) or involved in providing reductive agents to some specific metabolic process taking place in the reproductive organs.
Our work is, to our knowledge, the first large-scale study of genes expressed in stigmas/styles of a wet stigma species, provides a basis for the comparison of genes expressed in the female reproductive organs of species with different strategies of interaction with pollen grains/tubes, and represents an important contribution to a better comprehension of the plant reproductive process.
Wet and Dry Stigmas/Styles Express Common as Well as Distinct Sets of Genes
To establish a wide and meaningful comparison between wet and dry stigmas/styles at the molecular level, we have undertaken two complementary approaches: to compare the tobacco pistil-preferential transcripts/genes, revealed by our macroarray experiment, with the Arabidopsis pistil data sets and to compare the Arabidopsis pistil data sets with the whole TOBEST database. For this purpose, we constructed two local Arabidopsis pistil databases, each containing all of the protein sequences from the genes identified by Tung et al. (2005) and Swanson et al. (2005), including supplemental data. First, we used the tobacco pistil-preferential genes and searched for similarities in the TAIR database using the TBLASTX algorithm. The Arabidopsis best hit for each tobacco gene and its gene description, as it appears in TAIR, are shown in Table V. The tobacco pistil-preferential genes were then compared by BLASTX with the databases formed from the Tung et al. (2005) and Swanson et al. (2005) sequences. This analysis revealed an overlap of only three (6.5%) “true homologous” sequences and, for this reason, probably participating in the same biological process, between the tobacco and Arabidopsis pistil data sets: TOBC092C05 and At1g64670, TOBC054D09 and At1g53130, and TOBS132D06 and At1G11840, encoding a hydrolase, STIG1 (Goldman et al., 1994), and a glyoxalase I, respectively (Table V). Additionally, eight (17.4%) sequences with equivalent protein activity (functional relationship at the molecular level) are common between Arabidopsis and tobacco: TOBS042D12 encoding for a Leu-rich transmembrane protein kinase; TOBS046C04 for an auxin/IAA protein; TOBC023B06 for a nodulin MtN3 (transmembrane putative receptor); TOBC079H03 for STIG1; TOBC116D05 for a senescence-associated protein; TOBS013H02 for an ABC transporter of the MRP subfamily; TOBS049F11 for a phosphatase type 2A; and TOBS074F07 for a 1,3-β-glucosidase. Taken together, these results indicate an overlap of 23.9% between the tobacco pistil-preferentially expressed genes and the Arabidopsis pistil data sets. These proteins probably belong to more general processes occurring in plant reproduction and are common between wet and dry stigma species. Auxin-related proteins and proteins involved in the cell wall metabolism were also found in the dry stigmas of rice (Li et al., 2007). However, for some sequences previously implicated in pollen-pistil interaction in tobacco, such as TTS (Cheung et al., 1995) and PELP class III (de Graaf et al., 2003), no similar sequences were expressed in the Arabidopsis pistils. Additionally, it is interesting to verify that some of the tobacco pistil-preferential genes, like the one for a putative pectin methylesterase inhibitor (TOBC003D12), two for the stigma-specific arabinogalactan protein AGPNa3 (TOBC040F11 and TOBC022C03), and one for phospholipase A2 (TOBC130F10), are not even present in the Arabidopsis genome, based on a TBLASTX analysis of the TAIR database (cutoff e-value of ≤10−5). It has to be mentioned that a pectin methylesterase inhibitor-related gene (At5g50040) was found to be up-regulated in Arabidopsis stigmas (Swanson et al., 2005); however, there is no sequence similarity between At5g50040 and TOBC003D12.
Table V.
Tobacco pistil-preferential genes identified in the macroarray analysis and their corresponding sequences in the Arabidopsis genome
The homologous sequences were determined by the TBLASTX program in the TAIR database (cutoff e-value of ≤10−5) and by BLASTX comparisons in our local databases created with the Arabidopsis pistil protein sequences identified by Tung et al. (2005) and Swanson et al. (2005).
| TOBEST Identifier | Arabidopsis Genome Initiative No. | e-Value | Gene Description in TAIR | Present in the Arabidopsis Pistil Gene Sets
|
|
|---|---|---|---|---|---|
| Tung et al. (2005) | Swanson et al. (2005) | ||||
| C003D12 | No hit | ||||
| C033H04a | At2g34700 | 3e-07 | Pollen Ole e 1 allergen and extensin | ||
| C040F11a | No hit | ||||
| C091B07a | At2g34700 | 3e-20 | Pollen Ole e 1 allergen and extensin | ||
| C092C05 | At1g64670 | 6e-85 | BDG1 (BODYGUARD1); hydrolase | At1g64670 | |
| S004A06 | At4g19420 | 9e-39 | Pectin acetylesterase family protein | ||
| S008H02 | At2g27510 | 7e-49 | ATFD3 (FERREDOXIN3); electron carrier | ||
| S020F12 | At2g20770 | 9e-36 | Lanthionine synthetase C-like family protein | ||
| S053D05 | At2g39970 | 1e-49 | Peroxisomal membrane protein (PMP36) | ||
| C020F02a | At1g28290 | 5e-09 | Pollen Ole e 1 allergen and extensin | ||
| C022C03a | No hit | ||||
| C074A03 | At5g57123 | 1e-20 | Similar to unknown protein [Arabidopsis] (TAIR At4g29905.1) | ||
| S001C11 | At1g29450 | 6e-26 | Auxin-responsive protein, putative | ||
| S042D12 | At5g51560 | 2e-95 | Leu-rich repeat transmembrane protein | At1g66880 | At3g14840 |
| S046C04 | At3g04730 | 6e-31 | IAA16 (indole acetic acid-induced protein 16); transcription factor | At1g04250 | |
| S073B06 | At1g51460 | 1e-26 | ABC transporter family protein | ||
| At1g51500 | 4e-25 | CER5 (ECERIFERUM5); ATPase, coupled to transmembrane movement of substances | |||
| S118H02 | At1g62870 | 2e-83 | Similar to unknown protein | ||
| C005C12 | At2g02230 | 5e-06 | ATPP2-B1 (phloem protein 2-B1) | ||
| C015C07 | At1g11910 | 2e-61 | Aspartyl protease family protein | ||
| C023B06 | At5g23660 | 6e-69 | Arabidopsis homolog of M. truncatula nodulin MTN3 | At5g53190 | At1g21460 |
| C048F04 | At3g13870 | 7e-43 | RHD3 (ROOT HAIR-DEFECTIVE3) | ||
| C054D09a | At1g53130 | 5e-18 | Stigma-specific STIG1 family protein | At1g53130 | |
| C061A12 | At3g25570 | e-155 | SAMDC (S-adenosylmethionine decarboxylase) | ||
| C061G07 | At5g25260 | 8e-69 | Similar to unknown protein [Arabidopsis] (TAIR At5g25250.1) | ||
| C065A09 | At2g18370 | 1e-06 | Protease inhibitor/seed storage/lipid-transfer protein (LTP) family protein | ||
| C079H03a | At1g11925 | 6e-18 | Encodes a stigma-specific STIG1 family protein | At1g53130 | |
| C110D03 | At5g48480 | 5e-14 | Identical to protein At5g48480 [Arabidopsis]; similar to early tobacco anther 1 [M. truncatula] | ||
| C116D05 | At5g47060 | 2e-28 | Senescence-associated protein-related | At4g17670 | |
| C130F10 | No hit | ||||
| C132G06a | At2g34700 | 2e-30 | Pollen Ole e 1 allergen and extensin | ||
| S003A04 | At3g02220 | 5e-63 | Similar to hypothetical protein [Cleome spinosa] (GB ABD96929.1) | ||
| S004E12 | At5g50400 | 6e-88 | ATPAP27, PAP27 (purple acid phosphatase 27) | ||
| S010A10 | At5g26610 | 2e-54 | D111/G-patch domain-containing protein | ||
| S013F08 | At2g29570 | 9e-87 | PCNA2 (proliferating cell nuclear 2); DNA-binding/DNA polymerase processivity factor | ||
| S013H02 | At3g62700 | 2e-58 | ATMRP10 (Arabidopsis multidrug resistance-associated protein 10) | At1g02520 | |
| S013H03 | At1g28330 | 2e-33 | DRM1 (dormancy-associated protein 1)/auxin-associated family protein | ||
| S026F05 | At1g48410 | 6e-17 | AGO1 (ARGONAUTE1) | ||
| S035G04 | At2g23810 | 6e-51 | TET8 (TETRASPANIN8) | ||
| S049F11 | At5g18580 | 5e-65 | TON2, EMB40, FS1, GDO, FASS (FASS1) | At5g44090 | |
| S066E06 | At1g15520 | 4e-61 | ATPDR12/PDR12 (pleiotropic drug resistance 12) | ||
| S068D08 | At2g20490 | 2e-30 | NOP10, EDA27 (embryo sac development arrest 27); RNA binding | ||
| S072H12 | At5g59790 | 2e-31 | Similar to signal transducer [Arabidopsis] (TAIR At3g46110.1); similar to auxin-regulated protein [S. lycopersicum] | ||
| S074B04 | At5g58060 | 1e-79 | ATGP1/YKT61 (similar to yeast SNARE YKT6 1) | ||
| S074F07 | At3g57260 | 6e-38 | PR-2, BGL2 (pathogenesis-related protein 2); glucan 1,3-β-glucosidase | At3g04010 | |
| S086F04 | At4g27450 | 5e-94 | Similar to unknown protein [Arabidopsis] (TAIR At3g15450.1) | ||
| S132D06 | At1g11840 | e-115 | ATGLX1 (glyoxalase I homolog); lactoylglutathione lyase | At1g11840 | |
Sequences previously described as pistil specific or preferential in Nicotiana species.
As a second approach, all 149 protein sequences from Tung et al. (2005) were searched by TBLASTN in the TOBEST database (cutoff e-value of ≤10−5), from which 30 had no hits (20.1%) and 119 showed similar sequences in the tobacco stigmas/styles. These last sequences were then compared with the Arabidopsis genome, using the TBLASTX algorithm in the TAIR database. This analysis demonstrated that only 28 sequences were the true homologs between Arabidopsis and tobacco (Supplemental Table S3), representing 18.8% of precise overlap in gene expression between these dry and wet stigma species. The remaining 91 sequences (61.1%) were considered as having similar molecular functions (functional relationship). Taken together, these protein sequences (79.9%) probably represent functions necessary for broad processes in plant reproduction and are not restricted to a certain type of pollen-pistil interaction strategy, while 20.1% may be unique to Arabidopsis pistils. It should be mentioned that at least part of these protein sequences may just be missing from the TOBEST database, because we cannot guarantee that it contains all of the stigma/style-expressed sequences.
As shown in Supplemental Table S3, from the 41 papillar cell-specific genes predicted to encode secreted proteins in Arabidopsis (Table I of Tung et al., 2005), only eight true homologous sequences (19.5%) were found in the tobacco stigma/style gene set, one for each of the following: a plantacyanin, a protein disulfide isomerase, a calnexin-like protein, a putative cytochrome P450, a Gly-rich protein, a pyrimidine nucleotide sugar transporter, Pro-rich protein ATPRP4, and an unknown protein. Additionally, from the 10 transmitting tract-specific genes predicted to encode secreted proteins in Arabidopsis (Table II of Tung et al., 2005), only one true homolog was found in the TOBEST database: a putative chitinase gene. When we searched for the 10 genes previously implicated in the pollen-pistil interactions of Brassica, Arabidopsis, and lily (Table III of Tung et al., 2005), only Arabidopsis At2g02850 had a true homolog (TOBC017H10) among the tobacco pistil-expressed genes. The Arabidopsis At2g02850 gene encodes a plantacyanin, similar to the chemocyanin that acts as a chemotropic factor for pollen tubes in lily (Kim et al., 2003), a species of wet stigmas. It is noteworthy that none of the Brassica and Arabidopsis genes involved in pollen-pistil interactions was found in the TOBEST. However, we have to consider that despite the good representation of the TOBEST database, it contains a limited number of cDNA clones and may not include all of the tobacco stigma/style-expressed genes.
The same procedure has been applied for the 679 protein sequences described by Swanson et al. (2005), from which 219 were no hits (32.3%) and 460 showed similar sequences in the tobacco stigmas/styles. These tobacco sequences were used to search the TAIR database, revealing 124 (18.3%) true homologs in the Arabidopsis genome (Supplemental Table S4) and 336 (49.5%) sequences having similar molecular functions. It is remarkable that the percentage of true homologous sequences between Arabidopsis and tobacco is very similar for Tung's (18.8%; Tung et al., 2005) and Swanson's (18.3%; Swanson et al., 2005) databases. As the complete tobacco genome sequence is not available, it is not possible to perform the reverse analysis and determine if the no-hit sequences exist in the tobacco genome and were just not represented in the TOBEST. However, as our cDNA library is one of the most extensive sequencing efforts done in a single plant organ with a restricted function, and it was not normalized, subtracted, or subjected to PCR bias, it is expected that if the Arabidopsis genes were expressed at the tobacco stigma/style, most of them would be present in the TOBEST database. In agreement with the wide representation of stigma/style genes in the TOBEST is the fact that a comparison of the 46 tobacco pistil-preferential genes with the Arabidopsis pistil data sets revealed that 76.1% of the tobacco sequences were not represented in Tung's or in Swanson's database. On the other hand, only 20.1% of Tung's sequences and 32.3% of Swanson's sequences were not present in the TOBEST database.
We decided to establish a full comparison only with the Arabidopsis pistil data sets (Swanson et al., 2005; Tung et al., 2005), due to the fact that this is a dicotyledonous species with complete flowers, like tobacco. Therefore, it is more likely that the differences encountered in gene expression in the female reproductive organs are related to the type of interaction between pollen and pistil in species with dry and wet stigmas. We have not done a detailed comparison with the rice pistil data sets (Yoshida et al., 2005; Li et al., 2007), since this is a monocotyledonous species and additional differences are expected in gene expression on top of the ones related to the strategy of pollen-pistil interaction.
Tobacco is considered an allotetraploid species, derived from the interspecific hybridization of Nicotiana sylvestris and Nicotiana tomentosiformis. For some genes previously described in tobacco, two copies were identified and proposed to be originated from its progenitors, like TTS1 and TTS2 (Cheung et al., 2000). As shown in Tables III and V, in the TOBEST we have also identified two highly homologous sequences for TTS (TOBC033H04 and TOBC091B07), STIG1 (TOBC054D09 and TOBC079H03), and AGPNa3 (TOBC022C03 and TOBC040F11). The most parsimonious explanation for these duplicated cDNA sequences is that they are the expression products of the homologous genes of the ancestral genomes, as has already been shown for the tobacco nitrate reductase (Vaucheret et al., 1989). Thus, if we reconsider the number of independent genes revealed by the tobacco macroarray analysis, there are 43 pistil-preferential genes, of which 33 (76.7%) had no similar sequences in the Arabidopsis pistil data sets (Table V), including three tobacco genes (7.0%) for which there are no homologous sequences in the Arabidopsis genome. Taken together, our results show that there are a considerable number of genes that seem to be unique to each strategy of pollen-pistil interaction.
CONCLUSIONS AND PERSPECTIVES
In this study, we generated a database composed of 6,177 nonredundant ESTs from the upper pistil of a wet stigma species. This large collection of sequences from a single organ was obtained from a wild-type plant and, therefore, is not subject to gene expression bias or artifacts resulting from the use of mutants or transgenic plants. Overall, the results presented here show, to our knowledge, the first report in which a high-throughput EST analysis was used to examine gene expression in the pistil of a wet stigma species. This large-scale approach, coupled with transcriptional profiling by macroarray, represents an efficient method to identify genes involved in plant reproduction. Despite the simplicity of the macroarray experiments performed here, the identification of genes preferentially expressed in pistils was very successful. qRT-PCR analyses of 10 selected genes confirmed the pistil-preferential expression for nine of them. Most of the genes disclosed by the macroarray analysis were not previously studied or reported to be expressed in stigmas/styles, and their potential involvement in the pollination/progamic phase opens new avenues for understanding important processes in plant reproduction. The TOBEST cDNA library, the accompanying database, and the genes highlighted in this article are valuable resources for understanding pistil development and pollen-pistil interaction in general and, more specifically, in wet stigma species.
Our results indicate that the stigma/style seems to have some common conserved molecular functions between wet and dry stigma species, such as proteins of cell wall metabolism and proteins related to auxin signaling and regulation as well as proteins involved in stress/defense responses. On the other hand, wet stigmas produce proteins secreted to the exudate and for pollen-pistil interactions (like PELP class III, TTS, and AGPNa3) that are not expressed in dry stigmas. In addition, several proteins of as yet unknown functions are expressed solely on wet stigmas or dry stigmas. Therefore, the comparisons presented here show that, at the molecular level, pollen-pistil interactions in wet and dry stigma species differ considerably.
It has been suggested that some exudate proteins of species with wet stigmas fulfill similar roles as the proteins found on the pollen coat of dry stigma species (Wolters-Arts et al., 1998). It is interesting that a protein similar to CER1 (involved in lipid biosynthesis), previously described in pollen of dry stigma species (Aarts et al., 1995), was found in our analysis of the tobacco stigma/style. This observation points to a strict coevolution of the pollination strategies in which proteins important for pollen-pistil interactions and the success of the reproduction process should be provided by the male or the female partner. Our results illustrate gene expression differences between the wet stigmas of tobacco and the dry stigmas of Arabidopsis that should be explored in future experiments. The large-scale surveys of pistil gene expression previously performed in Arabidopsis and rice, and now carried out in Nicotiana, allow a broader study of pistil development and the reproductive process and will provide a basis for elucidating the evolution of pollination strategies.
MATERIALS AND METHODS
RNA Extraction and cDNA Library Construction
Tobacco plants (Nicotiana tabacum ‘Petit Havana SR1’) were grown under standard greenhouse conditions in Ribeirão Preto, São Paulo, Brazil (latitude, 21°10′24″ S; longitude, 47°48′24″ W; with average temperature of 22°C in winter and 27°C in summer; the difference in daylength between summer and winter is less than 2 h). Stigmas/styles of unpollinated flowers at developmental stages 1 to 11 (Koltunow et al., 1990) were collected, frozen in liquid nitrogen, and stored at −70°C. Total RNA was extracted from frozen material essentially as described by Dean et al. (1985). The RNA was quantified by measuring optical density at 260 and 280 nm. To evaluate the RNA integrity, 10 μg of RNA was fractionated on a 2.2 m formaldehyde-1.2% agarose gel, stained with ethidium bromide, and visualized with UV light. The presence of intact 28S and 18S rRNA bands was used as the criterion for RNA integrity. Poly(A+) RNA was purified from total RNA using the kit PolyATract mRNA isolation system (Promega) according to the manufacturer's instructions. cDNA was synthesized from 2 μg of poly(A+) RNA using the Superscript Plasmid System for cDNA synthesis and the Plasmid Cloning kit (Life Technologies), directionally cloned into the pSPORT1 vector, and introduced in DH10B Escherichia coli cells by electroporation. The transformed cells were selected in the presence of ampicillin (100 μg mL−1), isopropylthio-β-galactoside, and X-gal. Individual white colonies were randomly picked and transferred to 96-well plates containing liquid Circle Grow medium (BIO101) supplemented with 100 μg mL−1 ampicillin. After growth for 22 h at 37°C, glycerol was added to a final concentration of 25%. The 96-well plates containing the glycerol stocks were stored at −70°C for further use.
Plasmid DNA Extraction and cDNA Sequencing
Bacterial clones were inoculated into Circle Grow (BIO101) on a 96-well plate from the glycerol stocks using a Boekel 96 pins replicator and incubated in a rotatory shaker (300 rpm) at 37°C for 22 h. Plasmid DNA was extracted using a 96-well alkaline lysis method and purified through a 96-well filter plate (PVDF membrane 0.2 μm; Corning), essentially as described at http://sucest.lad.ic.unicamp.br/public. Single-run sequencing was done for each cDNA clone using 2 μL of DNA (200–500 ng), 2 μL of ABI Prism BigDye Terminator sequencing kit (Applied Biosystems), 2 μL of 5 pmol/μL T7 primer (5′-TAATACGACTCACTATAGGG-3′), and 2 μL of dilution buffer (200 mm Tris-HCl, pH 9.0, 5 mm MgCl2) in a final volume of 10 μL. The reaction products were precipitated with 75% isopropanol, and the pellets were washed with 70% ethanol and dried at room temperature. The sequencing reaction products were analyzed on an ABI 3100 fluorescence automated sequencer (Applied Biosystems), generating an electropherogram for each cDNA sequence (read).
Data Handling and Sequence Analysis
A pipeline was built to analyze and assemble the tobacco EST sequences. The cDNA reads were automatically analyzed using the base-calling program Phred (Ewing et al., 1998; Ewing and Green, 1998), and the ones containing at least 300 bases with Phred quality of ≥20 were accepted as valid ESTs. These sequences were scanned by the program Crossmatch from Phrap (www.phrap.org), and the pSPORT1 vector (minmatch 12, minscore 20) and adaptors were masked. Only ESTs with at least 180 bases of cDNA sequence (Phred quality value of ≥20) were further processed. These sequences were assembled using CAP3 software (Huang and Madan, 1999), resulting in contigs (at least two ESTs) and singlets (only one EST). The consensus sequences of each contig were named after the longest EST at their 5′ ends. For the singlets, there is no consensus sequence as there is only one read. Each cluster (contig or singlet) represents a putative transcript/gene. The TOBEST database is available at http://gbi.fmrp.usp.br/mhelena.
Automated annotation of all putative transcripts (TOBEST Fasta sequences) was performed using the BLASTX algorithm and the nonredundant protein database (nr) available from the NCBI (ftp://ftp.ncbi.nih.gov/blast/db/, February 2008). The highest significant similarity score (best hit) was used for automated annotation and correlation of a putative molecular function to each cluster sequence. A cutoff e-value of ≤10−5 was defined, and sequences with greater e-values were considered without a statistically significant similarity (no significant match or no hits). Additionally, all of the clusters (TOBEST Fasta sequences) were analyzed by TBLASTX (Altschul et al., 1997) against the protein sequences of the Arabidopsis (Arabidopsis thaliana) database (TAIR; obtained at http://www.arabidopsis.org). The results of the analyses mentioned above are available on the TOBEST Web server. Additional BLAST analyses were carried out at the NCBI site with some sequences of interest and are mentioned elsewhere in this article. A categorization into functional groups was done automatically through the KOGnitor tool (http://www.ncbi.nlm.nih.gov/COG/grace/kognitor.html). The sequences categorized by KOGnitor as “No related KOG,” for which a putative molecular function was associated by the BLAST analysis, were manually revised by a curator (for details, see Supplemental Materials and Methods S1).
TOBEST clusters were considered as putative full length when their sequences had an in-frame start codon encoding a Met that aligned to a Met identified as the start codon in the homologous sequences revealed by the BLASTX analysis. These TOBEST sequences were analyzed by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) in order to verify the presence of signal peptide and signal anchor (Bendtsen et al., 2004; Emanuelsson et al., 2007).
The EST sequence information contained in the ESTobacco (http://www.estobacco.info/) and the TGI (http://tgi.ncsu.edu/) databases, as available in July 2008, was downloaded in the TOBEST Web Server and used to search for similarity to TOBEST sequences using the BLASTN program (cutoff e-value of ≤10−5). The availability of this information allowed the comparison of the tobacco stigma/style transcriptome with the transcriptome of other tobacco organs. The EST information from all organs not related to plant reproduction (leaf lamina, leaf midrib, leaf stem, seedling, and root) was considered as a single group, vegetative organs. For details about the numbers of individual ESTs from each cDNA library, see Supplemental Figure S4.
For the comparison of the Arabidopsis pistil data sets and the TOBEST sequences, two local databases were constructed, each containing all of the protein sequences listed by Tung et al. (2005) and Swanson et al. (2005), including supplemental data. The Arabidopsis sequences were obtained from the TAIR database and were used as query in TBLASTN analysis or as database in BLASTX analysis against TOBEST sequences. The results are available at http://gbi.fmrp.usp.br/mhelena. The 11,216 EST sequences were deposited in GenBank with accession numbers FG621296 to FG632511.
Preparation and Analysis of cDNA Macroarrays
A total of 792 randomly selected clones, which later were revealed to represent 782 independent clusters, were rearranged on nine 96-well plates. Plasmid DNA was extracted from each plate and spotted (400 ng spot−1) four times (replicates) onto Hybond N+ (Amersham Biosciences) membranes, using a VP 409 Multi-Blot replicator 96 pins (V&P Scientific). Two identical membranes with 384 spots each were produced per plate. Negative and positive controls were included in the last column, and the positions on each plate were as follows: 12A, pSPORT1 empty vector; 12C, ubiquitin (TOB023F02); 12D, β-tubulin (TOB023H08); 12F, Glc-6-P dehydrogenase (TOB043E01); 12G, β-actin (TOB119E07); and 12H, 400 ng of tobacco genomic DNA. Total RNAs from roots, stems, leaves, sepals, and petals were isolated separately and used in equal amounts for the synthesis of the nonreproductive organ cDNA probe labeled with [α-32P]dCTP. The same amount of total RNAs from stigmas/styles was used for the synthesis of the positive cDNA probe. The cDNA probes were prepared as described previously (Malavazi et al., 2006). Nonincorporated radioactive nucleotides were removed by spin column purification, and equal amounts of labeled probes were used in the membrane hybridizations. After hybridization for 18 h, the membranes were washed and exposed to imaging plates for appropriate periods of time and then scanned in a Fla3000-G phosphor imager (Fujifilm). The radioactive signals were completely removed from the membranes (Sambrook and Russell, 2001), and those that had been hybridized with radioactive stigma/style cDNA were probed with radioactive nonreproductive organ cDNA, and vice versa. The probes used for the experiment with swapped membranes were prepared from biological replicates of the RNA samples. The signals were evaluated again and compared with those observed in the previous hybridization experiment. Signals were quantified using Array Vision software (Imaging Research). For each membrane, the grids were predefined and manually adjusted to obtain maximum spot recognition, and the spots were then individually quantified. The local background was automatically subtracted from each spot by taking the average intensity of the area surrounding each spot and subtracting it from the intensity value of each spot (sARVol value). The values obtained for each membrane were pooled by averaging the four signal intensity values of the four replicated DNA spots in each filter and used to calculate the expression ratios between stigma/style and vegetative organs as described previously (Malavazi et al., 2006). To reduce the fluctuations among replica filters due to differences in the experimental conditions, the signal from every spot was normalized using the median of all signals on that membrane (i.e. the median was set to 1, generating the nARVol values). Genes with expression levels close to the background value were discarded. Differentially expressed genes were identified by Student's t test using the log2 of normalized values (nARVol) from the two replicated experiments (P < 0.05). The data sets generated were then imported into Microsoft Excel spreadsheets for further calculations. Means (M) of the normalized signals for the genes that showed up-regulation or down-regulation in the stigma/style were then calculated. The fold increase and log ratios for each clone were calculated as M(nARVol)Stigma/Style/M(nARVol)Vegetative. The complete data set containing the raw and normalized data for all 792 clones used in the macroarray hybridizations is available in Supplemental Table S2. The column “Indicator” in this table represents the genes up-regulated in stigmas/styles (1), the down-regulated genes (−1) or, in other words, the genes with higher expression levels in vegetative organs than in stigmas/styles, and the nonmodulated genes or genes excluded from the analysis (0).
qRT-PCR Experiments
Roots, stems, leaves, sepals, petals, stamens, stigmas/styles, and ovaries were collected and frozen for RNA extraction. For the developmental expression studies, stigmas/styles were excised from flowers at stages 1 to 12 of tobacco flower development as described previously (Koltunow et al., 1990). Total RNA was extracted and analyzed for purity and integrity as described above. The RNA samples used for these experiments are independent from those of the macroarray analysis and represent biological replicates that increase the confidence of our results. RNase-free DNase treatment was done for each sample in a final volume of 50 μL containing 1× buffer (rq1 DNase buffer; Promega), 0.5 μL of RNAse OUT (40 units μL−1; Invitrogen), 5 μL of RNase-free DNase (1 unit μL−1; Promega), 2.5 μL of 100 mM dithiothreitol, and 20 μg of total RNA. The reaction mixture was incubated at 37°C for 60 min. An aliquot was removed and checked for genomic DNA contamination in standard real-time PCR using the β-actin primers (Table VI). The DNA-free RNA was cleaned by phenol/chloroform, precipitated, and resuspended in 12 μL of diethyl pyrocarbonate-water. Each RNA sample received 1.5 μL of oligo(dT)V (100 pmol μL−1) and was incubated at 75°C for 10 min and immediately chilled on ice. For cDNA conversion, to each sample was added 5 μL of first-strand buffer, 2.5 μL of dithiothreitol (100 mm), 0.5 μL of RNAse OUT (40 units μL−1; Invitrogen), 2.5 μL of dNTP (10 mm), and 1 μL of SuperScript II (200 units μL−1; Invitrogen), and an incubation at 42°C for 60 min was performed. After the reaction, water was added to complete a volume of 70 μL. The PCRs were performed on 96-well plates using the ABI Prism 7700 Sequence Detection System (Applied Biosystems). Each reaction contained 1 μL of cDNA, 5 μL of Taq-Man Universal PCR master mix (Applied Biosystems), and 2 μL of primer 1 and 2 μL of primer 2 labeled with 6-carboxy-fluorescein. Both primers were used in a concentration of 1 pmol μL−1, and their sequences are shown in Tables VI and VII. The PCR thermal cycling conditions were as follows: an initial step at 50°C for 2 min; 10 min at 95°C; and 40 cycles, with each cycle consisting of 15 s at 95°C and 1 min at 60°C.
Table VI.
Primers and fluorescent probes for the candidate reference genes
The genes listed were tested as candidates to reference internal control of the gene expression experiments by real-time RT-PCR.
| Gene Name | Primer 1 (5′–3′) | Primer 2a (5′–3′) |
|---|---|---|
| β-Actin | CATGCCAACCATCACACCAGT | cacagcGATGATGCTCCAAGGGCTGtG |
| GAPDH | GGCAGATCAAATCAATCACACG | cacaaaTGCCTTGAGCAAGAACTTTGtG |
| Polyubiquitin | TCCGACACAATCGACAACGTAA | gacgtacTGTTGTAATCGGCTAGGGTACGtC |
Primers labeled with 6-carboxy-fluorescein, incorporated at the boldface, lowercase, underlined nucleotides. Lowercase letters (at the 5′ end) represent the nucleotides added to the sequence according to the LUX system for real-time PCR. The primer sequences were determined based on the sequence of the following TOBEST clusters: β-actin (TOBC110E12), GAPDH (TOBC010F09), and polyubiquitin (TOBC018H09).
Table VII.
Primers and fluorescent probes for the putative stigma/style-specific genes
| TOBEST Identifier | Primer 1 (5′–3′) | Primer 2a (5′–3′) |
|---|---|---|
| C022C03 | CATTTGCACAAGCACCACCA | caacagtTTTCCAGCACGTTCTGACTGtTG |
| C023B06 | AATTCTGCTGCTGGCTTTACCTT | catctgaTCTTCCTGGGAATGATTCAGAtG |
| C092C05 | TTGCGTTGCTTGTTCTCCCTTT | cggtgTCCCTCACTTTAATTGCACcG |
| C130F10 | GCGAGTCGCAACAACTTGG | gacttgaATGGCGAATGAGGAGATCAAGtC |
| S008H02 | TGGGAGACAGCATACCAATTTCA | cgacaGGGGTACGAATCTTGTGTcG |
| S010A10 | TCCATTTCTTGCTGGCGTTT | cacgatTTTCTTCCATCAGCATTATCGtG |
| S042D12 | GGAAACAAGGGAAGCAAACCTG | cgctaTGAGAGCAATGGGTTGTAGcG |
| S049F11 | GCATCGAGGATGTAAGGGATG | cactggGCATGACAATAAATCAGCCAGtG |
| S053D05 | CAGGATGCAATATCTGCTGTGG | cggtgCCCAAACTCCAGCTTCACcG |
| S118H02 | GAGGAAGCTCCCATTGCACTAA | cactgtatCTTGAACAGCCTGAGCATACAGtG |
Primers labeled with 6-carboxy-fluorescein, incorporated at the boldface, lowercase, underlined nucleotides. Lowercase letters (at the 5′ end) represent the nucleotides added to the sequence according to the LUX system for real-time PCR.
In all experiments, appropriate negative controls containing no template DNA or RNA were subjected to the same procedure to exclude or detect any possible contamination or carryover. Each sample was repeated at least twice. The results were normalized using the threshold cycle obtained for the β-actin amplifications from each RNA/cDNA sample run on the same plate. The reactions and calculations were performed according to Semighini et al. (2002).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FG621296 to FG632511.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of sequence length (bp) of the individual reads that constitute the TOBEST database.
Supplemental Figure S2. Representative ethidium bromide-stained agarose gel (1.5%) of semiquantitative RT-PCR analysis performed from serial dilutions of stigma/style cDNA and primers for the indicated TOBEST sequences.
Supplemental Figure S3. Expression of the tobacco LAT52 homolog in the different vegetative and floral organs evaluated by real-time RT-PCR.
Supplemental Figure S4. Venn diagrams showing the overlap between the transcriptome data sets contained at the ESTobacco (A) and TGI-North Carolina State University (B) sites (available in July 2008) with the TOBEST.
Supplemental Figure S5. Images obtained with the Fla3000-G phosphor imager from a pair of hybridized membranes of the macroarray analysis.
Supplemental Figure S6. qRT-PCR experiments performed with samples from vegetative organs (a mixture of equal amounts of RNA from roots, stems, leaves, sepals, and petals) and stigmas/styles (S/S).
Supplemental Table S1. TOBEST sequences containing a putative signal peptide or containing a putative signal anchor, as established by the SignalP analysis of the cDNAs complete at the 5′ end.
Supplemental Table S2. Final statistical analysis of the macroarray results obtained for the 792 clones (representing 782 transcripts/genes) in stigma/style and vegetative organ samples.
Supplemental Table S3. List of the Arabidopsis sequences from the pistil data set described by Tung et al. (2005) for which there are true homologous sequences in the TOBEST database.
Supplemental Table S4. List of the Arabidopsis sequences from the pistil data set described by Swanson et al. (2005) for which there are true homologous sequences in the TOBEST database.
Supplemental Materials and Methods S1. Details about the categorization process of the TOBEST clusters by a curator, in complementation to the automatic KOGnitor categorization.
Supplementary Material
Acknowledgments
We are grateful to Dr. Claudia T. Otsu, Dr. Everaldo R. Marques, and Dr. Marcela Savoldi for stimulating discussions and suggestions and to Davidson C.D. Ribeiro, Diogénes C.D. Ribeiro, and Danillo C.A. Silva for their contributions to the bioinformatics of the project. We thank Gerard M. van der Weerden and the Botanical Garden of the University of Nijmegen (The Netherlands) for providing us with Nicotiana seeds, Prof. Dr. Francisco Gorgonio da Nóbrega (Universidade do Vale do Paraíba, Brazil) for lending us the VP 409 Multi-Blot replicator 96 pins, and Prof. Dr. Maria Cristina Roque-Barreira (Faculdade de Medicina de Ribeirão Preto/University of São Paulo) for allowing us to use the Fla3000-G phosphor imager. Additionally, we thank the anonymous reviewers who with their comments and suggestions contributed to improving this article.
This work was supported by grants from the Fundação de Amparo à Pesquisa no Estado de São Paulo-Brazil (FAPESP; grant no. 06/54431–9) and from the Conselho Nacional de Desenvolvimento Científico e Tecnológico-Brazil (CNPq), as well as by fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (to A.C.Q., M.S.B., L.A.S.B.), FAPESP (to I.d.S., I.M., J.B.M.-M.), and CNPq (to H.C.D., G.H.G., M.H.S.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Maria Helena S. Goldman (mgoldman@ffclrp.usp.br).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aarts MG, Keijzer CJ, Stiekema WJ, Pereira A (1995) Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7 2115–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64 5245–5250 [DOI] [PubMed] [Google Scholar]
- Atkinson AH, Heath RL, Simpson RJ, Clarke AE, Anderson MA (1993) Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340 783–795 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL (2000) Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 32–37
- Cecchetti V, Pomponi M, Altamura MM, Pezzotti M, Marsilio S, D'Angeli S, Tornielli GB, Costantino P, Cardarelli M (2004) Expression of rolB in tobacco flowers affects the coordinated processes of anther dehiscence and style elongation. Plant J 38 512–525 [DOI] [PubMed] [Google Scholar]
- Chen CG, Mau SL, Clarke AE (1993) Nucleotide sequence and style-specific expression of a novel proline-rich protein gene from Nicotiana alata. Plant Mol Biol 21 391–395 [DOI] [PubMed] [Google Scholar]
- Chen Y, Peumans WJ, Hause B, Bras J, Kumar M, Proost P, Barre A, Rouge P, Van Damme EJ (2002) Jasmonic acid methyl ester induces the synthesis of a cytoplasmic/nuclear chito-oligosaccharide binding lectin in tobacco leaves. FASEB J 16 905–907 [DOI] [PubMed] [Google Scholar]
- Cheung AY, May B, Kawata EE, Gu Q, Wu HM (1993) Characterization of cDNAs for stylar transmitting tissue-specific proline-rich proteins in tobacco. Plant J 3 151–160 [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu HM (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82 383–393 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM (1999) Arabinogalactan proteins in plant sexual reproduction. Protoplasma 208 87–98 [Google Scholar]
- Cheung AY, Wu HM, di Stilio V, Glaven R, Chen C, Wong E, Ogdahl J, Estavillo A (2000) Pollen-pistil interactions in Nicotiana tabacum. Ann Bot (Lond) (Suppl A) 85 29–37 [Google Scholar]
- Crawford BC, Ditta G, Yanofsky MF (2007) The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr Biol 17 1101–1108 [DOI] [PubMed] [Google Scholar]
- Cresti M, Keijzer CJ, Ciampolini F, Focardi S (1986) Stigma of Nicotiana: ultrastructural and biochemical studies. Am J Bot 73 1713–1722 [Google Scholar]
- Cruz-Garcia F, Nathan Hancock C, Kim D, McClure B (2005) Stylar glycoproteins bind to S-RNase in vitro. Plant J 42 295–304 [DOI] [PubMed] [Google Scholar]
- Dean C, Van der Elzen P, Tamaki S, Dunsmuir P, Bedbrook J (1985) Differential expression of the eight genes of the petunia ribulose bisphosphate carboxylase small subunit multi-gene family. EMBO J 4 3055–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf BHJ, Knuiman BA, Derksen J, Mariani C (2003) Characterization and localization of the transmitting tissue-specific PELPIII proteins of Nicotiana tabacum. J Exp Bot 54 55–63 [DOI] [PubMed] [Google Scholar]
- Du H, Simpson RJ, Clarke AE, Bacic A (1996) Molecular characterization of a stigma-specific gene encoding an arabinogalactan-protein (AGP) from Nicotiana alata. Plant J 9 313–323 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protocols 2 953–971 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res 8 186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res 8 175–185 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Pelaz S, Yanofsky MF (1999) Control of carpel development and fruit development in Arabidopsis. Annu Rev Biochem 68 321–354 [DOI] [PubMed] [Google Scholar]
- Goldman MH, Goldberg RB, Mariani C (1994) Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO J 13 2976–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MH, Pezzotti M, Seurinck J, Mariani C (1992) Developmental expression of tobacco pistil-specific genes encoding novel extensin-like proteins. Plant Cell 4 1041–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MH, Seurinck J, Marins M, Goldman GH, Mariani C (1998) A tobacco flower-specific gene encodes a polyphenol oxidase. Plant Mol Biol 36 479–485 [DOI] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF (2007) The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134 3593–3601 [DOI] [PubMed] [Google Scholar]
- Gu Q, Kawata EE, Morse MJ, Wu HM, Cheung AY (1992) A flower-specific cDNA encoding a novel thionin in tobacco. Mol Gen Genet 234 89–96 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison Y, Shivanna KR (1977) The receptive surface of the angiosperm stigma. Ann Bot (Lond) 41 1233–1258 [Google Scholar]
- Hiscock SJ, Allen AM (2008) Diverse cell signalling pathways regulate pollen-stigma interactions: the search for consensus. New Phytol 179 286–317 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp M, Kopczak SD, Horejsi TF, Kihl BK, Pruitt RE (1995) Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J 8 703–714 [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Watanabe Y, Ito N, Nonaka H, Takeda N, Sakai T, Kanaya H, Okada K (2002) SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J 21 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Mollet JC, Dong J, Zhang K, Park SY, Lord EM (2003) Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc Natl Acad Sci USA 100 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2 1201–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Bahn SC, Shin JS, Hwang I, Back K, Doelling JH, Ryu SB (2005) Multiple forms of secretory phospholipase A2 in plants. Prog Lipid Res 44 52–67 [DOI] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ (2008) Genomic plasticity and the diversity of polyploidy plants. Science 320 481–483 [DOI] [PubMed] [Google Scholar]
- Li M, Xu W, Yang W, Kong Z, Xue Y (2007) Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice (Oryza sativa L.). Plant Physiol 144 1797–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S (2003) Protein phosphatases in plants. Annu Rev Plant Biol 54 63–92 [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126 469–481 [DOI] [PubMed] [Google Scholar]
- Malavazi I, Savoldi M, Di Mauro SM, Menck CF, Harris SD, Goldman MH, Goldman GH (2006) Transcriptome analysis of Aspergillus nidulans exposed to camptothecin-induced DNA damage. Eukaryot Cell 5 1688–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B, Mou B, Canevascini S, Bernatzky R (1999) A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc Natl Acad Sci USA 96 13548–13553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet JC, Park SY, Nothnagel EA, Lord EM (2000) A lily stylar pectin is necessary for pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12 1737–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Feldman LJ, Zambryski PC (2000) Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127 3877–3888 [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BA, Fasolino A, Hilbers CW, Derksen J, Mariani C (2005) Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17 2009–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien M, Bertrand C, Matton DP (2002) Characterization of a fertilization-induced and developmentally regulated plasma-membrane aquaporin expressed in reproductive tissues, in the wild potato Solanum chacoense Bitt. Planta 215 485–493 [DOI] [PubMed] [Google Scholar]
- O'Brien M, Chantha SC, Rahier A, Matton DP (2005) Lipid signaling in plants: cloning and expression analysis of the obtusifoliol-14α-demethylase, from Solanum chacoense Bitt., a pollination- and fertilization-induced gene with both obtusifoliol and lanosterol demethylase activity. Plant Physiol 139 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Sessa G, Lotan T, Himmelhoch S, Fluhr R (1990) A major stylar matrix polypeptide (sp41) is a member of the pathogenesis-related proteins superclass. EMBO J 9 3429–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu CT, daSilva I, Molfetta JB, Silva LR, de Almeida-Engler J, Engler G, Torraca PC, Goldman GH, Goldman MH (2004) NtWBC1, an ABC transporter gene specifically expressed in tobacco reproductive organs. J Exp Bot 55 1643–1654 [DOI] [PubMed] [Google Scholar]
- Park SY, Jauh GY, Mollet JC, Eckard KJ, Nothnagel EA, Walling LL, Lord EM (2000) A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lord EM (2003) Expression studies of SCA in lily and confirmation of its role in pollen tube adhesion. Plant Mol Biol 51 183–189 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Semighini CP, Marins M, Goldman MH, Goldman GH (2002) Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl Environ Microbiol 68 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Clark T, Preuss D (2005) Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex Plant Reprod 18 163–171 [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S (2002) A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 14 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kelley D, Ezcurra I, Cotter R, McCormick S (2004) LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. Plant J 39 343–353 [DOI] [PubMed] [Google Scholar]
- Tung CW, Dwyer KG, Nasrallah ME, Nasrallah JB (2005) Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol 138 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D, Wing R, Yamaguchi J, McCormick S (1989) Isolation and expression of an anther-specific gene from tomato. Mol Gen Genet 217 240–245 [DOI] [PubMed] [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vincentz M, Kronenberger J, Caboche M, Rouzé P (1989) Molecular cloning and characterisation of the two homologous genes coding for nitrate reductase in tobacco. Mol Gen Genet 216 10–15 [DOI] [PubMed] [Google Scholar]
- Verhoeven T, Feron R, Wolters-Arts M, Edqvist J, Gerats T, Derksen J, Mariani C (2005) STIG1 controls exudate secretion in the pistil of petunia and tobacco. Plant Physiol 138 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Grsic-Rausch S, Rausch T, Greiner S (2003) Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett 555 551–555 [DOI] [PubMed] [Google Scholar]
- Wolters-Arts M, Lush WM, Mariani C (1998) Lipids are required for directional pollen-tube growth. Nature 392 818–821 [DOI] [PubMed] [Google Scholar]
- Wu HM, Zou J, May B, Gu Q, Cheung AY (1993) A tobacco gene family for flower cell wall proteins with a proline-rich domain and a cysteine-rich domain. Proc Natl Acad Sci USA 90 6829–6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133 4211–4218 [DOI] [PubMed] [Google Scholar]
- Yoshida KT, Endo M, Nakazono M, Fukuda H, Demura T, Tsuchiya T, Watanabe M (2005) cDNA microarray analysis of gene expression changes during pollination, pollen-tube elongation, fertilization, and early embryogenesis in rice pistils. Sex Plant Reprod 17 269–275 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.