Abstract
Anxa4 belongs to the multigenic annexin family of proteins which are characterized by their ability to interact with membranes in a calcium-dependent manner. Defined as a marker for polarized epithelial cells, Anxa4 is believed to be involved in many cellular processes but its functions in vivo are still poorly understood. Previously, we cloned Xanx4 in Xenopus laevis (now referred to as anxa4a) and demonstrated its role during organogenesis of the pronephros, providing the first evidence of a specific function for this protein during the development of a vertebrate. Here, we describe the strict conservation of protein sequence and functional domains of anxa4 during vertebrate evolution. We also identify the paralog of anxa4a, anxa4b and show its specific temporal and spatial expression pattern is different from anxa4a. We show that anxa4 orthologs in X. laevis and tropicalis display expression domains in different organ systems. Whilst the anxa4a gene is mainly expressed in the kidney, Xt anxa4 is expressed in the liver. Finally, we demonstrate Xt anxa4 and anxa4a can display conserved function during kidney organogenesis, despite the fact that Xt anxa4 transcripts are not expressed in this domain. This study highlights the divergence of expression of homologous genes during Xenopus evolution and raises the potential problems of using X. tropicalis promoters in X. laevis.
Key Words: annexin 4, kidney, liver, evolution, paralog and ortholog, Xenopus laevis, Xenopus tropicalis, morpholino, organogenesis
Introduction
Annexin proteins are members of an evolutionarily conserved, multigenic family, identified in eukaryotes ranging from protists and fungi, to plants and higher vertebrates. The family consists of more than 160 members.1–4 Each annexin has a specific expression pattern and sub-cellular localization.5 Annexin proteins consist of two domains; the divergent NH2-terminal ”head,” thought to specify the individual annexin properties in vivo, and the conserved 34kDa COOH-terminal protein core, which possesses the calcium and membrane binding sites. The annexin protein core structure, consisting of the common tetrad of internal repeats, has been strictly conserved for over 1200 million years, implying a basic functional role for this domain.6 Characteristic protein specific differences in the four imperfect repeats have also remained evolutionarily conserved, enabling each annexin to bind to cell membranes containing a mixture of specific lipids and carbohydrates with high affinity.7,8 Annexins provide novel, calcium-dependent cellular mechanisms to reversibly alter membrane characteristics such as fluidity and permeability, the ability of membranes to aggregate and fuse, the regulation of ion conductance activity and the organisation of the extracellular matrix.9,10
Annexin 4 (anxa4), first identified in human placental homogenates,11 has been identified in a number of vertebrates,4,12,13 but its precise function is yet to be determined. The anxa4 protein is expressed in a wide range of tissues, associated with polarized epithelial cells and is proposed to be involved in the regulation of chloride ions across the epithelial membrane.2,14,15 Temporal and spatial expression studies in X. laevis show that anxa4a (previously referred to as Xanxa4) is expressed on the luminal surface of the pronephric tubules.3 The anxa4 gene expression in mouse and in zebrafish is also localized to the kidney in addition to the floor plate whilst, in humans it is expressed within the tubulus epithelial cells.4,16,17 These patterns of expression are consistent with a role in chloride conductance.
Cloning of the X. laevis annexin 4 (anxa4a) gene has led to the first direct functional analysis of this gene during development. Depletion of anxa4a protein, using morpholino antisense oligonucleotides, led to altered kidney organogenesis typified by a shortened, enlarged tubule. This defect could be rescued by the co-injection of anxa4a mRNA.3 This suggests that anxa4a plays a role in normal pronephrogenesis although its precise role is unclear.
In this paper we investigate the functional and evolutionarily conservation of this developmentally important anxa4a gene by protein homologies, temporal and spatial expression patterns and functional approaches. We first identify the common similarities possessed by the anxa4 protein by phylogenetic analysis and demonstrate the strict conservation of functional protein domains. We then identify an additional anxa4 transcript in Xenopus laevis which we call anxa4b. This transcript has a temporal and spatial expression pattern which is different from anxa4a. We then show that X. tropicalis anxa4 (Xt anxa4) is never expressed in the pronephric kidney but is highly expressed in the embryonic liver and so differs in its expression domain from either of the anxa4a or anxa4b transcripts. In addition we demonstrate that Xt anxa4 can functionally rescue the anxa4a MO phenotype which disrupts pronephric development indicating conservation of function, despite the lack of conservation of spatial distribution, in these closely related species. This work is the first demonstration of the divergence of patterns of orthologous gene expression between X. laevis and tropicalis.
Materials and Methods
Bioinformatics.
Anxa4 sequences (see Table 1 for Accession numbers) were identified by BLAST performed on the NCBI website. To examine the pattern of evolution of the anxa4 gene across vertebrate species, the available nucleotide and protein sequences were aligned using the GeneBee algorithm and a phylogenetic tree produced using the PHYLIP program.18 Pairwise alignments were performed using the Needle program based on Needleman-Wunsch global alignment algorithm.19 Analysis on protein sequences were performed using the ScanProsite program.20
Table 1.
Accession numbers of the anxa4 sequences used in this study
| Species | cdnA | Protein |
| X. laevis anxa4a | AY039235 | AAK83461 |
| X. laevis anxa4b | BC073582 | AAH73582 |
| X. tropicalis | CR762357 | CAJ83505 |
| H. sapiens | NM_001153 | NP_001144 |
| M. musculus | U72941 | NP_038499 |
| R. rattus | NP_077069 | |
| B. taurus | BAA11243 | |
| S. scrofa | P08132 | |
| C. familiaris | P50994 | |
| D. rerio | AY178798 | AAO20272 |
| O. latipes | CAA72122 |
Embryo culture and microinjection.
Embryos were obtained and cultured as described previously (ref. 21). All fine dissections of X. laevis embryos were performed in Barth X using watchmakers forceps and an eyebrow hair mounted in a hypodermic needle.
Microinjections were performed into a V2 blastomere of 8-cell stage X. laevis embryos. β-galactosidase (LacZ) (2 ng) (A. Philpott, Cambridge) was co-injected as lineage tracer. X-Gal staining was carried out as described in Bourguignon et al.22 The embryos were staged according to Nieuwkoop and Faber.23
Morpholinos
The anxa4a morpholino 2 (MO2) (5′-tccgagtgctgccatgatgtccacc-3′) was designed and supplied by GeneTools, LLC. This MO is designed against more 5′upstream sequences than MO1 described in Seville et al.3 (Fig. 6A). The MO (5 ng/nl) was injected (20 ng) alone or in combination with mRNA (5 ng). The random control MO (cMO) designed by GeneTools was used as a control.
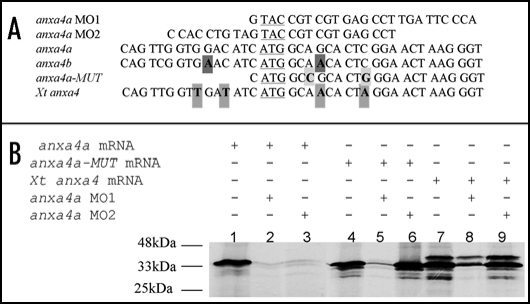
Figure 6.
The anxa4 MO2 knocks down anxa4a but not Xt anxa4 translation. (A) Alignment of the 5′ sequences of X. laevis wild type and mutant anxa4 cDNA and its X. tropicalis homolog showing the complementarity of anxa4a MO1 and MO2 in relation to these cDNAs. The ATG is underlined and mismatch residues are indicated in bold. Four nucleotides (in grey) differ between Xt anxa4 cDNA and the MO2 sequence. Two mismatch nucleotides created in anx4a-MUT cDNA are highlighted in light grey. Two nucleotides (in dark grey) are different between anxa4b and MO2 sequences. (B) anxa4a MO2 does not interfere with Xt anxa4 translation; autoradiograph of an SDS-PAGE analysis gel of in vitro translated 35S-Methionine radio-labelled anxa4 proteins. 0.5 µg of anxa4a mRNA was incubated alone (lane 1) or in combination with 10 µg of anxa4a MO1 (lane 2) or MO2 (lane 3). Both MO block the translation of anxa4a mRNA. 0.5 µg of anxa4a-MUT mRNA was incubated either alone (lane 4) or in combination with 10 µg of anxa4a MO1 (lane 5) or MO2 (lane 6). Translation of the mutant mRNA is severely affected by MO1 but unaffected by MO2. 0.5 µg of Xt anxa4 mRNA was incubated alone (lane 7) or in combination with 10 µg of anxa4a MO1 (lane 8) or MO2 (lane 9). MO2 does not block the translation of Xt anxa4 whereas the addition of MO1 affects its translation. These results demonstrate that Xt anxa4 can be used to rescue MO2.
Expression clones, mRNA synthesis.
Xt anxa4 full length cDNA clone was obtained from the X. tropicalis full length cDNA library.24 X. laevis anxa4a-MUT (anxa4a-MUT) cDNA was generated by PCR from the anxa4a cDNA and cloned into pCS2+. Two third-base modifications were introduced and the 5′UTR was removed in order that the anxa4a MO2 did not recognize and bind to this new mRNA. Each mRNA was transcribed using a mMessage mMachine kit (Sp6 RNA polymerase, Ambion) from NotI linearized template.
In vitro translation of in vitro trancribed mRNA.
mRNA (0.5 µg) was translated in vitro in the Rabbit Reticulocyte Lysate System (Promega) according to manufacturer's protocol in presence of 10 µCi of [35S] Methionine alone or with 10 µg of MO. Translation products (5 µl) were separated by SDS-PAGE and analyzed by autoradiography (Kodak) after an overnight exposure.
RT-PCR.
Total RNA extraction and cDNA synthesis from whole X. laevis embryos or from isolated kidneys and livers were performed as previously described (ref. 25). PCR's were carried out using specific primers for anxa4a (couple 5U/5D, Supplementary Table 1) and anxa4b (U-5′GCACTTATGACTCCGTACAC-3′; D-5′GACATTGCTGCTCTGATCTC-3′) in cycles of 94°C for 1 min, x for 1 min and 72°C for 1 min, where n and x are respectively: 26–27 and 60°C for anxa4a; 26–29 and 59°C for anxa4b. Direct sequencing of these PCR products confirmed the specificity of each reaction. Each PCR contained -RNA, -RT and -cDNA negative controls and a linearity range to show the PCR was in the linear range. The quantity of input cDNA was determined by equalization of the ODC signal.26
In situ hybridization.
Wholemount in situ hybridization was performed by standard methodology.27
X. laevis and tropicalis embryos were fixed in MEMFA (0.5 M MOPS pH 7.4, 100 mM EGTA, 1 mM MgSO4, and 4% formaldehyde) and hybridized with antisense and sense RNA probes. The antisense anxa4a probe was generated as described previously (ref. 3). The full length of the clone BC073582 (anxa4b) was subcloned into the plasmid pBSKS and antisense and sense probes were transcribed with T3 and T7 RNA polymerase from the template linearized with BamHI and KpnI respectively. The Xt anxa4 antisense and sense probes were transcribed with T3 and Sp6 RNA polymerase from the full length clone CR762357 previously linearized with EcoRI and NotI respectively.
Probes were synthesized and labelled using a DIG labelling kit (Roche) and the hybridization visualized using sheep anti-DIG-alkaline phosphatase antibody (Roche) and 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolylphosphate substrate (NBT/BCIP; Roche). Embryos were bleached following standard protocols.
Immunohistochemistry.
Whole mount immunohistochemistry was performed on injected embryos fixed in MEMFA as described previously (ref. 3). Embryos were immunostained with pronephric tubule-specific monoclonal antibody 3G8.28 The secondary antibody was alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma) and the color reaction was performed by using NBT/BCIP (Roche).
Nuclear staining was carried out as previously described (ref. 3). Cell counts were performed on digital images recorded using a Nikon Optiphot/ Diginet camera system or a Leica SP2 confocal microscope.
Acrylamide embedding and cryostat sectioning.
X. laevis embryos were embedded and sectioned at 18 µm thickness as described previously (ref. 3).
Results
The identification of a new X. laevis anxa4 transcript, anxa4b.
We have previously reported the cloning and functional analysis of the anxa4a gene in pronephric development in X. laevis.3 Blast analysis on the NCBI website identified a new cDNA for anxa4 in X. laevis (NCBI Accession no BC073582). This clone is 1160 bp long, contains an open reading frame of 966 nucleotides encoding for a protein of 321 amino acids. This new cDNA is 88.5% identical to our original sequence, anxa4a. The predicted translated protein is 92.8% identical to anxa4a (Table 2A), with 23 different amino acids between these two proteins located along the length of the coding region (Fig. 1).
Table 2.
anx4a and anx4b percentage identity
| A | Relatedness between anxa4b and vertebrate anxa4 proteins | |
| anxa4a | anxa4b | |
| X. laevis (anxa4a) | 100 | 92.8 |
| X. tropicalis | 93.5 | 92.2 |
| H. sapiens | 71.7 | 72.4 |
| M. musculus | 71.3 | 72 |
| R. norvegicus | 72 | 73.2 |
| B. taurus | 74.1 | 74.5 |
| S. scrofa | 73.2 | 73.5 |
| C. familiaris | 72 | 71.7 |
| O. latipes | 67.3 | 64.8 |
| D. rerio | 66.7 | 64.2 |
| B | Relatedness between anxa4b and X. laevis anxa family proteins |
| anxa4b | |
| anxa1 | 46.9 |
| anxa2 | 46.5 |
| anxa4a | 92.8 |
| anxa5 | 51.9 |
| anxa6 (1/2) | 52.9 |
| anxa7 | 32.4 |
| anxa9 | 34.2 |
| anxa13 | 45.3 |
These tables indicate percentage of identity determined by EMBOSS paired alignment using the Needleman-Wunsch global alignment algorithm (ref. 19).
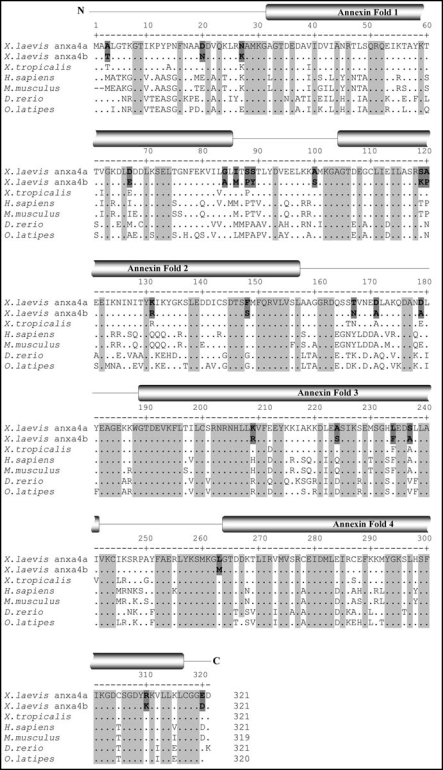
Figure 1.
Multiple alignment of anxa4 proteins. The sequence of the three Xenopus anxa4 (anxa4a, anxa4b and Xt anxa4) proteins were aligned against other vertebrate anxa4 proteins using the CLUSTALW software. The annexin fold domains are marked with reference to anxa4a sequence. Identical residues are indicated by (.). Letters highlighted in light grey are conserved between all seven anxa4 proteins. The numbering above the amino acid position refers to anxa4a sequence. Gaps are indicated by dashes. Bold residues highlighted in dark grey indicate the 23 unconserved amino acids between anxa4a and anxa4b proteins.
Pairwise alignment between this new protein sequence and other available annexins in X. laevis demonstrated that this new protein is more related to anxa4a than to any other annexin. The percentage of identity between this protein and other annexin family members is ≤53% (Table 2B). Moreover, this protein contains the four characteristic conserved annexin motifs but also the putative sites for posttranslational modifications in its N terminus region, as found in anxa4a protein. Based on this sequence analysis, we called this new gene anxa4b and conclude anxa4b and anxa4a are duplicated genes, since X. laevis is a pseudo-tetraploid species.
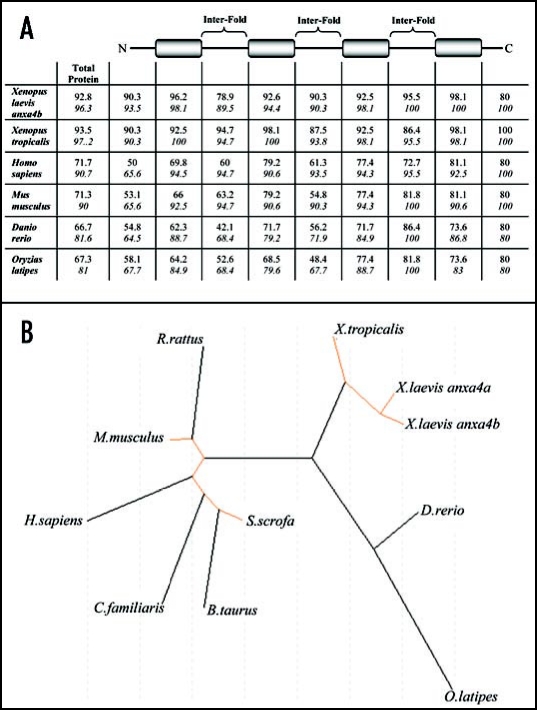
Bioinformatic analysis of Xenopus anxa4 proteins reveals they are highly conserved within Xenopus species (>90% identity) but also during vertebrate evolution (Fig. 1 and Table 2A). The anxa4b protein shows the highest degree of conservation with mammalian anxa4 whereas anxa4a is more related to the fish anxa4 protein and more importantly to X. tropicalis anxa4. The anxa4a protein is 93.5% identical to Xt anxa4 whereas anxa4b is only 92.2% identical to Xt anxa4.
The anxa4b protein has the characteristic four calcium-binding annexin fold repeats, spaced by inter-fold domains. These folds are located in identical places with respect to each other in all sequences analyzed and are typically 53 amino acids in length (Fig. 1). All of the four annexin folds retain a high level of similarity (≥79.2%) with reference to anxa4a sequence (Fig. 2A). However, the percentage of identity is less, especially for annexin fold 1 with only 62.3% of identity between zebrafish and anxa4a sequences. Amino acid substitutions occur between species, however, the replacing amino acid possesses the same characteristic as the substituted amino acid in the majority of cases e.g., basic, acid, neutral and polar, or neutral and hydrophobic enabling overall properties of the annexin folds to remain conserved (Fig. 1). In contrast, the inter-fold regions generally show a lower level of conservation with respect to amino acid identity. The inter-fold 3 shows the greatest degree of conservation, from 72.7% of identity. The most conserved region is located in the C-terminus which binds to membranes (100% similarity and ≥80% identity) whereas the N-terminus is more variable with only 50% of identity between human and Xenopus sequences.
Figure 2.
Phylogeny of vertebrate anx4 proteins. (A) Identity and similarity comparisons between structural domains of the anxa4 proteins. Percentage identities are in regular type and similarities are in italics. Annexin fold domains are represented by shaded boxes and the N- and C-terminal and inter-fold domains are represented by horizontal lines, using anx4a as the reference sequence. (B) Molecular phylogenetic tree of Xenopus orthologs and vertebrate anxa4 proteins. The unrooted tree was produced for all the available anxa4 proteins (see Table 1 for Accession numbers) using the PHYLIP program on the Genebee Internet Site.
Phylogenetic analysis was carried out on all the available anxa4 protein sequence for multiple vertebrate species (Fig. 2B). This analysis shows a clear separation in sequence divergence between the classes of mammals, amphibian and fish. The amphibian and mammalian anxa4 proteins formed closely related, distinct groups, whilst anxa4 in fish appeared to be more divergent.
Anxa4b displays a different expression pattern from anxa4a.
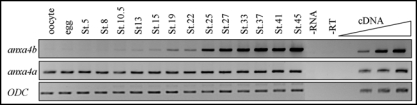
The temporal expression of two anxa4 genes was analyzed and compared by RT-PCR (Fig. 3). As the primers used in our first study amplify both Xenopus anxa4 genes,3 new specific gene primers were designed. The specificity of the primers was analyzed by sequencing the RT-PCR products which confirmed each primer pair only amplified its intended target sequence. The anxa4a gene was expressed maternally and its expression level overall remained constant during the development of X. laevis. However, no maternal expression was seen for anxa4b. Zygotic expression was first detected during gastrulation at stage 10.5 but remained at a low level until stage 19. Expression was upregulated at stage 19 and again at stage 25 and maintained at high level throughout tailbud and tadpole stages.
Figure 3.
Comparison of anxa4a and anxa4b temporal expression patterns in X. laevis embryos. RT-PCR was performed with total RNA extracted from X. laevis oocytes and embryos at different stages. Anxa4a is maternal and expressed in whole embryos at the same level throughout development until stage 45, the last stage tested. Anxa4b is expressed weakly during gastrulation. Its expression is upregulated, at both neurula and tailbud stages.
We then analyzed the spatial expression of anxa4b by whole-mount in situ hybridization in comparison to anxa4a (Fig. 4A). No specific 5′ or 3′ UTR probe could be made due to the short length of these regions in the available EST's. Therefore, the full length anxa4b cDNA was used as a template in each case, as previously used for anxa4a.3
Figure 4.
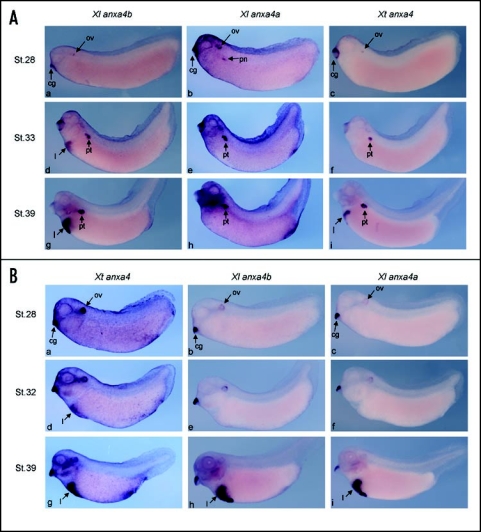
Comparison of in situ expression patterns of the three orthologs in X. laevis and tropicalis embryos. (A) anxa4 probes from each the Xenopus orthologs identify distinct in situ expression domains on X. laevis embryos. anxa4a and anxa4b can be detected from stage 28 in the cement gland (cg) and weakly in the otic vesicles (ov) (a and b). Expression of anxa4a in the pronephric anlagen (pn) can be first observed at stage 28 (b) and from stage 33 in the developing tubules (pt) (e and h). anxa4b is also in the pronephric tubules (d and g) but in a transitory manner, being lost from this region in later stage embryos (data not shown). anxa4b is expressed strongly in the developing liver (l) from stage 33 (d and g). A combination of the two expression patterns is detected with Xt anxa4 probe, which shows similar homology to both X. laevis genes. Strong staining in the pronephric tubules (pt) can be observed from stage 33 (f) and in the liver (l) at stage 39 (i). (B) Anxa4 probes from each of the Xenopus orthologs show identical in situ expression patterns on X. tropicalis embryos. Xt anxa4 is first detected in the cement gland (cg) and otic vesicle (ov) at stage 28 (a) and remains expressed in these two organs throughout their development (d and g). From stage 32, expression can be seen in the liver (l) (d). Xt anxa4 remains strongly expressed in this organ at later stages (g). The same expression pattern is detected with both anxa4a (c, f and i) and anxa4b (b, e and h) probes, although as expected the signal is weaker since probes are being used cross-species, confirming the homology of these genes.
At stage 28, both genes are expressed weakly in the otic vesicle and cement gland (Fig. 4A, a and b). The anxa4a gene expression can be observed in the kidney anlagen at this stage (Fig. 4A, b) and remains expressed in the kidney and more specifically in the pronephric tubules until stage 41 (Fig. 5A, b) as shown previously (ref. 3). The expression in the cement gland and otic vesicle is not detectable from late tailbud stages. However, at stage 41, expression in the gall bladder and in the ventral lateral line can be seen (Fig. 5A, b). The latter stage expression patterns were not previously reported.3 The anxa4b gene is also expressed in the pronephric tubules at tadpole stages but only transiently (Fig. 4A, g). At stage 41, anxa4b is no longer expressed in the kidney (Fig. 5A, a). However, in contrast to anax4a, anxa4b is strongly expressed in the liver, from stage 33 throughout stage 41, the last stage tested (Fig. 4A, d and g; Fig. 5A, a). At stage 41, staining in the anterior part of the liver and in the gall bladder can be observed (Fig. 5A, a). No staining was observed with the sense probe at any stages tested (data not shown).
Figure 5.
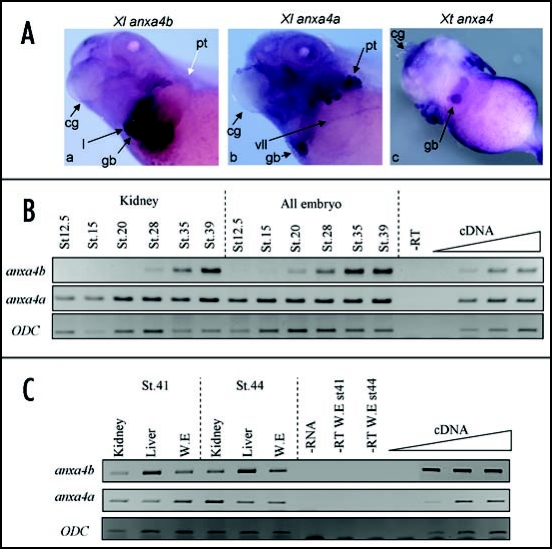
Comparison of anxa4a and anxa4b spatial expression. (A) Xenopus anxa4 genes are expressed in a common domain, the gall bladder, in stage 41 embryos. In situ hybridization was carried out on X. laevis (a and b) and X. tropicalis (c) embryos. anxa4a (b) is expressed strongly in the pronephric tubules (pt) and also in the gall bladder (gb). anxa4b (a) is detected in the anterior part of the liver (l), in the gall bladder (gb) but not expressed in the pronephros (pt) (white arrow). Xt anxa4 (c-ventral view) is only expressed in the gall bladder (gb). The cement gland (cg) is indicated to allow orientation of the embryos. (B) anxa4a and anxa4b expression is different during pronephric development. RT-PCR was performed with total RNA extracted from dissected kidneys from different stages of X. laevis embryos. anxa4a is expressed in the kidney throughout its formation whereas anxa4b is only expressed in this organ from stage 28. (C) anxa4a and anxa4b display different spatial expression at late tadpole stages. RT-PCR was performed with total RNA extracted from dissected pronephric kidneys and livers from stages 41 and 44 X. laevis embryos (W.E.). At stage 41, anxa4a is slightly more expressed in the kidney than the liver and anxa4b is more highly expressed in the liver. At stage 44, this difference of expression is more highly pronounced.
RT-PCR analysis was performed on kidney and liver dissections to confirm that the in situ hybridization analysis was specific to each transcript. Pronephric tissues were dissected at different key stages during the development of the kidney (Fig. 5B).29 The anxa4a transcript is expressed throughout the development of the pronephros, from stage 12.5 (specification of the pronephric tubules) to stage 39 (functional kidney) whereas anxa4b expression in the pronephros can be weakly detected from stage 28 and increases during pronephric differentiation until stage 39, the last stage tested in this experiment.30 At stage 41, anxa4a is expressed slightly more in the kidney than in the liver, whereas anxa4b is preferentially expressed in the liver (Fig. 5C). By stage 44, the specific spatial expression of these two paralogs clearly illustrates anxa4a as a kidney marker and anxa4b a liver marker.
X. tropicalis anxa4 (Xt anxa4) is expressed in the embryonic liver and never in the pronephros.
It has been suggested that the spatial expression pattern of homologous genes is usually conserved between X. laevis and tropicalis.31 We have identified a difference in expression patterns between the two anxa4 genes in X. laevis, and therefore investigated the expression pattern of Xt anxa4 by in situ hybridization (Fig. 4B).
At tailbud stages, Xt anxa4 expression can be detected in the otic vesicle and cement gland (Fig. 4B, a). At tadpole stages, Xt anxa4 becomes strongly expressed in the liver (Figs. 4B, d and g) and in contrast to X. laevis, no staining can be seen in the kidney at any stages studied. At stage 41, there is no detectable expression in the liver, however the gall bladder is highly and specifically stained (Fig. 5A, c). No staining with the sense probe was observed at any stages tested (data not shown).
Due to the fundamental difference of expression pattern seen between these two species, we decided to carry out in situ hybridization on X. tropicalis embryos using the two X. laevis anxa4 probes and vice and versa. It has been shown that a number of X. laevis probes cross hybridize with X. tropicalis, so no modification in our in situ hybridization protocols was introduced.31 As shown in Figure 4B, both X. laevis anxa4 probes strongly stained the liver of X. tropicalis embryos at later stages (Fig. 4B, h and i). No staining, at any stage, was detected in the pronephros. These data suggest that Xt anxa4 is the true homolog of both X. laevis anxa4 genes. The reverse experiment was performed (Fig. 4A). The Xt anxa4 probe hybridized to both kidney and liver in X. laevis embryos. Pronephric tubules are heavily stained from stage 33 (Fig. 4A, f), reflecting the expression pattern of anxa4a. The late stage expression in the liver is noticeable from stage 39 (Fig. 4A, i).
Xt anxa4 is functionally homologous to Anx4a.
The knock-down of anxa4a affects pronephric tubule development in X. laevis.3 Despite the different spatial expression pattern, we asked if Xt anxa4 can perform a similar function to its homolog in X. laevis. Therefore, we assessed if Xt anxa4 could rescue the phenotype induced by anx4a depletion in X. laevis.
A second MO (MO2) had already been designed against more 5′upstream sequences to the MO1 used in our previous published study (Fig. 6A).3 MO2 has already been shown to knock-down the expression of anxa4a and also to cause the same phenotype as MO1 on the pronephric tubules (Seville, 2001, Thesis Warwick University) (Fig. 6B, lanes 2 and 3). MO2 spans the ATG but contains fewer complimentary nucleotides in the coding region than MO1, a region highly conserved between X. laevis and tropicalis mRNAs. MO2 contains four mismatch nucleotides to Xt anxa4 mRNA as opposed as two in MO1 (Fig. 6A). As expected, MO2 does not block the translation of Xt anxa4 whereas MO1 can partially affect its translation (Fig. 6B, lanes 8 and 9). In order to rescue the MO2 phenotype, a X. laevis mutant anxa4a (anxa4a-MUT) mRNA was created by PCR (Seville, 2001, Thesis Warwick University). It lacks the 5′UTR and contains two third-base changes in the first 15 nucleotides of the coding region, no more than six nucleotides apart, preventing the binding of MO2 but not MO1 (Fig. 6A and B, lanes 5 and 6).
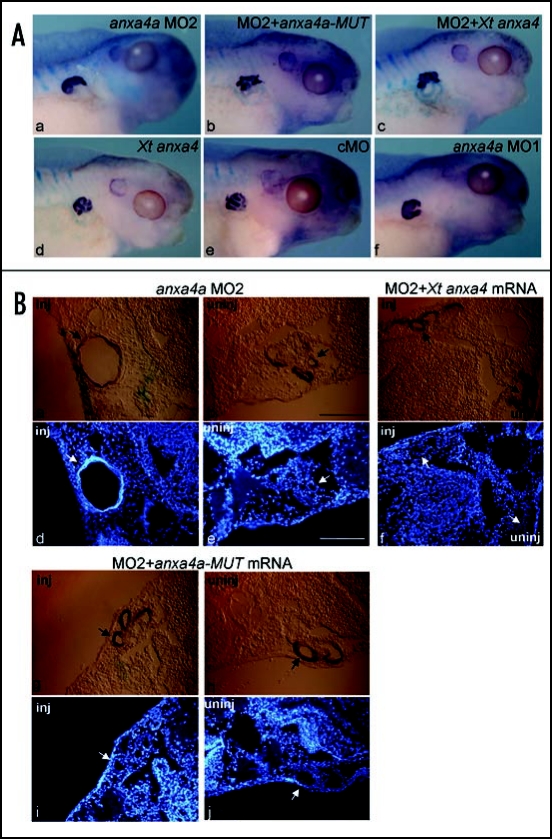
cMO, MO1, MO2 were injected into the V2 blastomere of an 8-cell stage embryo and cultured until stage 41. Tubule morphology was assessed by antibody staining with 3G8, a tubule-specific monoclonal antibody (Fig. 7A).28 As previously shown, no consistent phenotype is observed following the injection of cMO (Fig. 7A). The characteristic tubule phenotype described by Seville et al., 2002,3 was observed after injection of MO1 (Fig. 7A). Forty-one percent of the embryos (n = 20/49) display shortened and enlarged tubules on the injected side. The depletion of anxa4a by MO2 induces the same phenotype, but at a lower frequency. Enlarged tubules can be observed on the injected side of 19.3% of the embryos (n = 6/31) (Fig. 7A, a). No phenotype can be observed on the uninjected side (29/31 embryos display normal tubules).
Figure 7.
Anxa4 function is conserved between X. laevis and tropicalis. (A) Embryos were injected in the V2 blastomere of the 8-cell stage with 20 ng of anxa4a MO2 alone (a) or with 5 ng of anxa4a-MUT mRNA (b) or with Xt anxa4 mRNA (c), cultured until stage 41 and subjected to whole-mount antibody staining with the pronephric tubule specific antibody 3G8. Five nanograms of Xt anxa4 mRNA (d), 20 ng of cMO (e), 20 ng of anxa4a MO1 (f) were also injected as controls. LacZ (2 ng) was used as lineage tracer in all injections. Embryos injected with MO1 and MO2 show similar phenotype, with shortened, enlarged tubules whereas cMO injection does not affect the tubule morphology. Overexpression of Xt anxa4 induces no apparent phenotype; however, as anxa4a-MUT mRNA, it is able to rescue the MO2 phenotype. (B) Cryostat transverse sections of stage 41 Xenopus pronephroi stained with tubule-specific antibody 3G8 and counterstained with Hoechst. The slides were inspected under white light (a–c, g–h) to identify 3G8 stained tubules and the LacZ stained injected side (inj) (a, c and g) and under UV illumination to identify Hoechst nuclei staining (d–f, i–j). Embryo injected with anxa4a MO2 displays enlarged pronephric tubule phenotype on the injected side (inj) (a and d) compared to the uninjected side (uninj) (b and e). Embryo co-injected with anxa4a MO2 and anxa4a-MUT mRNA shows rescue of the phenotype [compare g and h (injected side) to i and j (uninjected side)]. No differences between the injected and the uninjected sides can be observed in embryo co-injected with anxa4a MO2 and Xt anxa4 mRNA (c and f).
We have already shown that the enlarged kidney phenotype observed as a result of anxa4a depletion is due to wider tubules consisting of a higher mean number of cells, rather than a distended tube of wild type diameter.3 Cryostat transverse sections were cut from MO2 treated embryos, counterstained with Hoechst and cell counts were performed as previously described (ref. 3, Fig. 7B). In order to count the cells, sections were analyzed by confocal microscopy. As shown in Figure 7B, pronephric tubules of anxa4a MO2 injected embryos are wider (Fig. 7B, a and d). The average number of cells contributing to the tubule was 54.75 (sd 19.26) on the injected side compared to 12 (sd 2.86) on the uninjected contralateral side (Fig. 7B, compare a and d with b and e).
Following confirmation that MO2 gives the same phenotype as that previously published for MO1, anxa4a-MUT or Xt anxa4 mRNAs was co-injected with MO2. We have previously shown that injection of anxa4a mRNA does not affect pronephric development.3 We now show that overexpression of Xt anxa4 also does not induce any apparent consistent overexpression phenotype with 77.2% of the embryos (n = 34/44) displaying normal tubule morphology on their injected side (Fig. 7A, a–d). As expected, anxa4a-MUT is able to rescue the anxa4a MO2 phenotype (Fig. 7A, a–b), 71% of the embryos (n = 27/38) display normal tubules compared to 38.7% of MO2 injected embryos. Enlarged tubules can still be observed in 5.3% of the embryos (n = 2/38). When the number of cells contributing to rescued pronephric tubules was counted, no difference can be seen between the injected and uninjected sides (Fig. 7B, compare g and i to h and j). The average number of cells forming the tubules is 10.6 (sd 2.07) on the injected side compared to 10.3 (sd 3.05) on the uninjected side. Interestingly, Xt anxa4 mRNA can almost totally rescue the MO2 phenotype. 87.5% of the embryos showed a rescued phenotype on the injected side (n = 35/40) and only 2.5% of the embryos (n = 1/40) displayed enlarged tubules (Fig. 7A, a–c). Cell counts demonstrated that the rescue is complete since the average number of cells in the pronephric tubules was 14 on both, injected and uninjected, sides (sd 4.36 and 2.65 respectively) (Fig. 7B, a–c and f).
From these data, we conclude that Xt anxa4 can rescue the phenotype induced by depletion of anxa4a in X. laevis. This indicates that Xt anxa4 has a similar function to anxa4a despite the fact that in X. tropicalis, Xt anxa4 transcripts are localized in a completely different organ system, the liver, and not in the pronephric kidney tubules.
Discussion
The anxa4 protein is highly conserved during vertebrate evolution.
We have characterized a new transcript for anxa4 in Xenopus laevis called anxa4b. Based on the sequence analysis, it seems likely that anxa4a and anxa4b are duplicated from an ancestral gene, and seem to have evolved independently at an equal rate from that ancestral gene. Twenty three amino acids are different between anxa4a and anxa4b, eight of these changes make anxa4a sequence less related to Xt anxa4 and mammal anxa4 [for example Lys27 is conserved between anx4b and 4/5 of the other anx4 studied (Fig. 1)]. The same is true for Met 86 and Ala 171. However, four amino acids which are conserved between anxa4a and the other anxa4 studied are mutated in the anxa4b sequence (Ala → Glu in position 84, Ser → Ala in position 100, Met → Lys in position 263 and Lys → Arg in position 310).
Based on our phylogenetic analysis, this study demonstrates the high degree of conservation of anxa4 during vertebrate evolution, especially after the divergence of X. laevis and X. tropicalis. As expected, the COOH -terminal protein core is the most conserved domain. All proteins show the four characteristic annexin folds and the characteristic ‘type 2’ motif [GxGt-(38 residues)-D/E] for binding calcium ion-mediated association with phospholipid membranes, suggesting a conservation of anxa4 function during evolution. However, the N-terminal domain of Xenopus anxa4 proteins is variable with respect to other vertebrate species. This N-terminal domain has already been shown to be the least conserved domain between annexin family members in the same species and was thought to confer unique functions to a given family member.2 This domain contains PKC phosphorylation and also N-myristoylation sites, suggesting possible specific posttranslational modifications for a given species; phosphorylations have already been shown to affect annexin properties.2 Moreover multiple transcripts due to alternative splicing in the 5′region have been described in mammalian species.16
In the course of these studies we have also shown that the genomic structure organisation of anxa4 genes has also been conserved during evolution (Supp. Fig. 1). The genomic structure of X. laevis anxa4 was determined by amplification of the putative intronic sequences as described in Supplementary Information (Supp. Fig. 2A). Each intron was confirmed as anxa4 sequences by Southern-Blot (Supp. Fig. 2B) and by sequencing (Supp. Fig. 3). Due to the high degree of conservation between anxa4a and anxa4b sequences, it is likely that the intronic sequences of both of these two genes were amplified during this study. However, only one clear amplification product was obtained on the agarose gel and identified positively by hybridization for each intron, except for introns 7 and 9, suggesting that the anxa4a and anxa4b genomic structures are highly similar and could not be distinguished.
The anxa4 genes in Xenopus sp., human, mouse and zebrafish exhibit the same pattern of organisation with 12 introns flanked by 13 exons, the first intron being located in the 5′UTR. The exon/intron boundaries are strictly conserved, resulting in the conservation of size of the exonic sequences between the five species studied, except for exons 1 and 2.
All anxa4 genes possess exon splice patterns and alternating intron positions, which do not reflect the annexin-fold protein domains (Supp. Fig. 1). This has led effectively to the preclusion of exon shuffling, a feature shared by the annexin family and has resulted in conservation of the C-terminus presumably due to evolutionarily pressure to conserve function.32
The three anxa4 orthologs in X. laevis and tropicalis display different expression patterns.
Despite the conservation of amino acid sequence, anxa4a and anxa4b do not display the same temporal and spatial expression pattern during X. laevis development, as demonstrated by in situ hybridization and confirmed by RT-PCR. The maternal gene anxa4a has zygotic expression restricted mainly to the pronephric tubules. However, anxa4b is only zygotically expressed, transiently in the kidney at earlier stages of development, but later found predominantly in the liver. This expression pattern in the liver is not that unexpected, since anxa4 has been shown to be expressed in epithelial cells and adult rabbit liver.1,33 At later stages of the development in Xenopus, strong expression can be detected in the gall bladder, supporting the results of our previous adult organoblot.3 Our initial published PCR data was carried out using primers which we have since determined will identify both transcripts.3 Hence the published PCR showed the combined profiles of both anxa4a and anxa4b expression patterns.
Based on the analysis of the spatial expression profile of the two transcripts rather than sequence analysis, it seems more than likely that anxa4b is closer to the ancestral anxa4 gene. Indeed, its expression pattern during development is very similar to that described for anxa4 in zebrafish.4 The zebrafish anxa4 ortholog is expressed transiently in the pronephros at 20–24 hpf and after 70–80 hpf, predominantly in the liver and gall bladder. We also demonstrated by RT-PCR that mouse anxa4 is expressed in the embryonic kidney and liver at stage 12.5 dpc (data not shown). However, surprisingly, Xt anxa4 is not expressed in the kidney at any stage, but only in the liver and gall bladder. Allotetraploidization of X. laevis has been estimated to have occurred about 30 million years ago (mya), or maybe more recently about 21 mya whereas the divergence between Silurana (including X. tropicalis) and Xenopus occurred approximately 53 million years ago.34–36 Therefore, the ancestral Xt anxa4 and X. laevis anxa4 genes could have evolved independently before the tetraploidization of X. laevis. One hypothesis to explain the difference in expression pattern could be changes in the regulatory sequences of these genes. Xt anxa4 could have lost the kidney enhancer driving the expression of this gene in the pronephros during development. Later, anxa4a and anxa4b could have diverged and anxa4a could have lost its ability to be expressed in the liver, remaining expressed in the pronephros. Interestingly, the anxa4 ortholog in the killifish medaka is strongly expressed in the developing liver during somitogenesis but not in the kidney.37
More information about the embryonic expression of anxa4 in other vertebrate species and promoter studies would be needed to explain definitively these changes in expression pattern during evolution. It is also possible that the difference in expression of anxa4a and anxa4b observed is due to differential stability of the two mRNAs, although we do not favour this interpretation.
Why are two anxa4 transcripts retained in X. laevis?
The putative allotetrapoidization that occurred in X. laevis has created a full set of paralogs, each of the same evolutionarily age as the parent species.38 It has been estimated that half of the duplicated genes have been retained in X. laevis.34 Several mechanisms have been proposed to explain the retained expression of duplicated genes.36 Neofunctionalization seems to be an unlikely reason for retention in this case since both X. laevis genes have evolved at apparently similar rates when compared to Xt anxa4. Recent studies have demonstrated that approximately 14% of the paralogous pairs show differential expression indicative of subfunctionalization.39 Therefore, we could speculate that anxa4a and anxa4b are retained in X. laevis due to temporal and spatial subfunctionalization in order to preserve the total functions of the ancestral gene. Only anxa4a is expressed maternally and therefore would fulfill the specific functions of anxa4 at these particular early stages. It has been suggested that maternal annexins may be involved in cortical granule exocytosis and we speculate anxa4a plays such a role.40
Different roles of anxa4 in polarized epithelia.
Despite many years of research, the exact functional roles for anxa4 remain unknown. The anxa4 protein can bind to membrane phospholipids in a calcium dependent manner and therefore is involved in regulating calcium ion transport activity,14,41 membrane aggregation,42 calcium-activated cellular signal transduction events,43 membrane permeability44 and recently membrane protein mobility.45 It has also been shown to promote vesicle aggregation15 and be part of a protein complex involved in exocytosis.46 Recent studies have shown anxa4 to have anti-inflammatory properties47 and anxa4 expression is upregulated in renal cancer (reviewed in ref. 48).
The anxa4 protein has been described as a marker of polarized epithelium in adult vertebrates.2 During Xenopus development, anxa4a and anxa4b is also found in tissues rich in epithelial cells; kidney, gall bladder and liver. Interestingly, the onset of expression of anxa4 follows morphological changes of these organs. The anxa4a gene expression in the kidney is detected from stage 26,3 which coincides with the time of lumen formation in the pronephric tubules and remains strongly expressed during maturation of the kidney.49 However, anxa4b is expressed in the pronephric tubules during its maturation phase. The anxa4b gene is detected in the liver around stage 33, the stage at which the bile canaliculi start to form, one of the most outstanding morphological changes which occurs in hepatocytes50 whilst anxa4a is only expressed in the gall bladder at stage 41, during maturation of the bile duct. Therefore, we speculate anxa4a and anxa4b could be involved relatively early during the organogenesis of the kidney and liver respectively but also could play a role during the late phase of maturation of polarized epithelium in the liver and kidney respectively, prior to their full functional development. Due to the expression pattern of Xt anxa4 in the liver, MO knock-down would not be expected to give a kidney phenotype in X. tropicalis, but potentially would affect liver formation. Analysis of the function of anxa4 during liver development would provide more evidence to support this hypothesis.
The function of anxa4 in the kidney is conserved.
The majority of the studies involving anxa4 function have been performed in vitro. One knock-out model for anxa4 has been generated, confirming our previous demonstrations of the role of anxa4 in the kidney formation.3 The anxa4 deficient mouse shows morphological differences in the kidney.16 However, this knock-out is a restricted loss-of-function model, since the expression of two additional anxa4 transcripts is unaffected by the knock-out strategy.
During this study, we validate the pronephric phenotype observed previously by the use of a second MO, confirming the potential role of anxa4 during kidney organogenesis.3 Based on the sequence analysis, both proteins, anxa4a and anxa4b should be knocked-down by MO1, since there is only one mismatch between MO1 and anxa4b sequences. However, anxa4b translation could be less effectively knocked-down by MO2 since we identified two nucleotide differences between the sequence of this second MO and anxa4b sequence. This explains the lower frequency of the enlarged tubule phenotype observed following MO2 injection. We demonstrate here, that despite a different expression domain, Xt anxa4 can rescue the phenotype induced by anxa4a MO2, highlighting the conservation of the function of this protein after the divergence between the siluranan species, X tropicalis, and other Xenopus species.
This unexpected result raises the main question of which gene fulfils anxa4 functions in the kidney in X. tropicalis. Despite being the annexin expressed highest in the kidney, anxa4 is not the only family member expressed in this organ (reviewed in ref. 51). anxa1, 2, 5 and 13b are also widely expressed in the kidney. The anxa1, 2 and 5 genes have been cloned in X. tropicalis (NCBI accession no CAJ81780; CAJ 83327, CAJ 83861 respectively) and despite the lack of expression pattern described for these genes, we could speculate that one is a candidate to be expressed in the pronephros.
Final conclusions.
Pseudo-alleles in X. laevis have been suggested to be expressed in identical domains and therefore been considered redundant. Our work demonstrates anxa4 paralogous genes display different expression patterns in this species and we suggest this could be an example of subfunctionalisation in X. laevis. Moreover, many developmental genes have been shown to display identical expression domains in X. laevis and tropicalis, the kidney markers Pax2 and Pax8, being of particular relevance to this study.31 A preliminary study covering a quarter of the Xenopus transcripts has demonstrated that these two Xenopus species have similar global gene expression patterns.52 To the best of our knowledge, this study is the first to describe two functionally identical orthologs with different expression domains in X. laevis and tropicalis, raising the possibility of divergence in regulatory sequences and the potential problems of using X. tropicalis promoters for the study of regulatory expression domains of X. laevis gene or simply for transgenesis.
Supplementary Material
Acknowledgements
We thank N. Ahmed for X. laevis genomic DNA, G. Rogers for assistance with confocal images and R. Taylor and P. Jarrett for maintenance of the breeding frogs. This work was supported by the BBSRC. The authors dedicate this work to the memory of Robert Taylor.
Abbreviations
- Anxa
Annexin
- Anxa4
Annexin4
- anxa4a
X. laevis Annexin4
- Xt anx4
X. tropicalis anxa4
- anxa4b
X. laevis annexin4 pseudoallele
- sd
standard deviation
Note
Supplemental material can be found at: www.landesbioscience.com/supplement/MasseORG3-2Sup.pdf
Footnotes
Previously published online as an Organogenesis E-publication: http://www.landesbioscience.com/journals/organogenesis/article/4945
References
- 1.Dreier R, Schmid KW, Gerke V, Riehemann K. Differential expression of annexins I, II and IV in human tissues: An immunohistochemical study. Histochem Cell Biol. 1998;10:137–148. doi: 10.1007/s004180050275. [DOI] [PubMed] [Google Scholar]
- 2.Gerke V, Moss SE. Annexins: From structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 3.Seville RA, Nijjar S, Barnett MW, Massé K, Jones EA. Annexin IV (Xanx-4) has a functional role in the formation of pronephric tubules. Development. 2002;129:1693–1704. doi: 10.1242/dev.129.7.1693. [DOI] [PubMed] [Google Scholar]
- 4.Farber SA, De Rose RA, Olson ES, Halpern ME. The zebrafish annexin gene family. Genome Res. 2003;13:1082–1096. doi: 10.1101/gr.479603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan RO, Fernandez MP. Annexin gene structures and molecular evolutionary genetics. Cell Mol Life Sci. 1997;53:508–515. doi: 10.1007/s000180050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luecke H, Chang BT, Mailliard WS, Schlaepfer DD, Haigler HT. Crystal structure of the annexin XII hexamer and implications for bilayer insertion. Nature. 1995;378:512–515. doi: 10.1038/378512a0. [DOI] [PubMed] [Google Scholar]
- 7.Burger A, Berendes R, Liemann S, Benz J, Hofmann A, Gottig P, Huber R, Gerke V, Thiel C, Romisch J, Weber K. The crystal structure and ion channel activity of human annexin II, a peripheral membrane protein. J Mol Biol. 1996;257:839–847. doi: 10.1006/jmbi.1996.0205. [DOI] [PubMed] [Google Scholar]
- 8.Sohma H, Creutz CE, Gasa S, Ohkawa H, Akino T, Kuroki Y. Differential lipid specificities of the repeated domains of annexin IV. Biochim Biophys Acta. 2001;1546:205–215. doi: 10.1016/s0167-4838(01)00140-6. [DOI] [PubMed] [Google Scholar]
- 9.Rescher U, Gerke V. Annexins-unique membrane binding proteins with diverse functions. J Cell Sci. 2004;117:2631–2636. doi: 10.1242/jcs.01245. [DOI] [PubMed] [Google Scholar]
- 10.Gerke V, Creutz CE, Moss SE. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 11.Tait JF, Sakata M, McMullen BA, Miao CH, Funakoshi T, Hendrickson LE, Fujikawa K. Placental anticoagulant proteins: Isolation and comparative characterization four members of the lipocortin family. Biochemistry. 1988;27:6268–6276. doi: 10.1021/bi00417a011. [DOI] [PubMed] [Google Scholar]
- 12.Sable CL, Riches DW. Cloning and functional activity of a novel truncated form of annexin IV in mouse macrophages. Biochem Biophys Res Commun. 1999;258:162–167. doi: 10.1006/bbrc.1999.0544. [DOI] [PubMed] [Google Scholar]
- 13.Fukuoka S, Kern H, Kazuki-Sugino R, Ikeda Y. Cloning and characterization of ZAP36, an annexin-like, zymogen granule membrane associated protein, in exocrine pancreas. Biochim Biophys Acta. 2002;1575:148–152. doi: 10.1016/s0167-4781(02)00299-3. [DOI] [PubMed] [Google Scholar]
- 14.Chan HC, Kaetzel MA, Gotter AL, Dedman JR, Nelson DJ. Annexin IV inhibits calmodulin-dependent protein kinase II-activated chloride conductance: A novel mechanism for ion channel regulation. J Biol Chem. 1994;269:32464–32468. [PubMed] [Google Scholar]
- 15.Kaetzel MA, Mo YD, Mealy TR, Campos B, Bergsma-Schutter W, Brisson A, Dedman JR, Seaton BA. Phosphorylation mutants elucidate the mechanism of annexin IV-mediated membrane aggregation. Biochemistry. 2001;40:4192–4199. doi: 10.1021/bi002507s. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Dedman JR, Kaetzel MA. Intron disruption of the annexin IV gene reveals novel transcripts. J Biol Chem. 2003;278:43276–43283. doi: 10.1074/jbc.M306361200. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann U, Balabanov S, Giebel J, Teller S, Junker H, Schmoll D, Protzel C, Scharf C, Kleist B, Walther R. Increased expression and altered location of annexin IV in renal clear cell carcinoma: A possible role in tumour dissemination. Cancer Lett. 2004;209:111–118. doi: 10.1016/j.canlet.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Brodsky LI, Vasiliev AV, Kalaidzidis YaL, Osipov YuS, Tatuzov RL, Feranchuk SI. GeneBee: The program package for biopolymer structure analysis. Dimacs. 1992;8:127–139. [Google Scholar]
- 19.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 20.Gattiker A, Gasteiger E, Bairoch A. ScanProsite: A reference implementation of a PROSITE scanning tool. Applied Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- 21.Massé K, Eason R, Bhamra S, Dale N, Jones EA. Comparative genomic and expression analysis of the conserved NTPDase gene family in Xenopus. Genomics. 2006;87:366–381. doi: 10.1016/j.ygeno.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Bourguignon C, Li J, Papalopulu N. XBF-1, a winged helix transcription factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development. 1998;125:4889–4900. doi: 10.1242/dev.125.24.4889. [DOI] [PubMed] [Google Scholar]
- 23.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) New York: Garland Publishing, Inc.; 1994. [Google Scholar]
- 24.Gilchrist MJ, Zorn AM, Voigt J, Smith JC, Papalopulu N, Amaya E. Defining a large set of full-length clones from a Xenopus tropicalis EST project. Dev Biol. 2004;271:498–516. doi: 10.1016/j.ydbio.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 25.Barnett MW, Old RW, Jones EA. Neural induction and patterning by fibroblast growth factor, notochord and somite tissue in Xenopus. Dev Growth Differ. 1998;40:47–57. doi: 10.1046/j.1440-169x.1998.t01-5-00006.x. [DOI] [PubMed] [Google Scholar]
- 26.Bassez T, Paris J, Omilli F, Dorel C, Osborne HB. Post-transcriptional regulation of ornithine decarboxylase in Xenopus laevis oocytes. Development. 1990;110:955–962. doi: 10.1242/dev.110.3.955. [DOI] [PubMed] [Google Scholar]
- 27.Harland R. In situ hybridization: An improved wholemount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 28.Vize PD, Jones EA, Pfister R. Development of the Xenopus pronephric system. Dev Biol. 1995;171:531–540. doi: 10.1006/dbio.1995.1302. [DOI] [PubMed] [Google Scholar]
- 29.Chan T, Asashima M. Growing kidney in the frog. Nephron Exp Nephrol. 2006;103:e81–e85. doi: 10.1159/000092192. [DOI] [PubMed] [Google Scholar]
- 30.Brennan HC, Nijjar S, Jones EA. The specification of the pronephric tubules and duct in Xenopus laevis. Mech Dev. 1998;75:127–137. doi: 10.1016/s0925-4773(98)00094-x. [DOI] [PubMed] [Google Scholar]
- 31.Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, Papalopulu N, Smith JC, Zorn AM, Harland RM, Grammer TC. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- 32.Moss SE, Morgan RO. The annexins. Genome Biol. 2004;5:219. doi: 10.1186/gb-2004-5-4-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massey D, Traverso V, Rigal A, Maroux S. Cellular and subcellular localization of annexin IV in rabbit intestinal epithelium, pancreas and liver. Biol Cell. 1991;73:151–156. doi: 10.1016/0248-4900(91)90097-7. [DOI] [PubMed] [Google Scholar]
- 34.Bisbee CA, Baker MA, Wilson AC, Haji-Azimi I, Fischberg M. Albumin phylogeny for clawed frogs (Xenopus) Science. 1997;195:785–787. doi: 10.1126/science.65013. [DOI] [PubMed] [Google Scholar]
- 35.Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: Phylogeography and implications for polyploid evolution. Mol Phylogenet Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 36.Chain FJ, Evans BJ. Multiple mechanisms promote the retained expression of gene duplicates in the tetraploid frog Xenopus laevis. PLoS Genet. 2006;2:e56:478–490. doi: 10.1371/journal.pgen.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osterloh D, Wittbrodt J, Gerke V. Characterization and developmentally regulated expression of four annexins in the killifish medaka. DNA Cell Biol. 1998;17:835–847. doi: 10.1089/dna.1998.17.835. [DOI] [PubMed] [Google Scholar]
- 38.Graf JD, Kobel HR. Genetics of Xenopus laevis. Methods Cell Biol. 1991;36:19–34. doi: 10.1016/s0091-679x(08)60270-8. [DOI] [PubMed] [Google Scholar]
- 39.Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, Barber S, Babakaiff R, Dickson MC, Matsuo C, Wong D, Yang GS, Smailus DE, Wetherby KD, Kwong PN, Grimwood J, Brinkley CP, IIIrd, Brown-John M, Reddix-Dugue ND, Mayo M, Schmutz J, Beland J, Park M, Gibson S, Olson T, Bouffard GG, Tsai M, Featherstone R, Chand S, Siddiqui AS, Jang W, Lee E, Klein SL, Blakesley RW, Zeeberg BR, Narasimhan S, Weinstein JN, Pennacchio CP, Myers RM, Green ED, Wagner L, Gerhard DS, Marra MA, Jones SJ, Holt RA. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 2006;16:796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanenkov VV, Weber K, Gerke V. The expression of different annexins in the fish embryo is developmentally regulated. FEBS Lett. 1994;352:227–230. doi: 10.1016/0014-5793(94)00956-2. [DOI] [PubMed] [Google Scholar]
- 41.Kaetzel MA, Chan HC, Dubinsky WP, Dedman JR, Nelson DJ. A role for annexin IV in epithelial cell function: Inhibition of calcium-activated chloride conductance. J Biol Chem. 1994;269:5297–5302. [PubMed] [Google Scholar]
- 42.Creutz CE. The annexins and exocytosis. Science. 1992;258:924–931. doi: 10.1126/science.1439804. [DOI] [PubMed] [Google Scholar]
- 43.Raynal P, Kuijpers G, Rojas E, Pollard HB. A rise in nuclear calcium translocates annexins IV and V to the nuclear envelope. FEBS Lett. 1996;392:263–268. doi: 10.1016/0014-5793(96)00827-7. [DOI] [PubMed] [Google Scholar]
- 44.Hill WG, Kaetzel MA, Kishore BK, Dedman J, Zeidel ML. Annexin A4 reduces water and proton permeability of model membranes but does not alter aquaporin 2-mediated water transport in isolated endosomes. J Gen Physiol. 2003;121:413–425. doi: 10.1085/jgp.200308803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piljic A, Schultz C. Annexin A4 self-association modulates general membrane protein mobility in living cells. Mol Biol Cell. 2006;17:3318–3328. doi: 10.1091/mbc.E06-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willshaw A, Grant K, Yan J, Rockliffe N, Ambavarapu S, Burdyga G, Varro A, Fukuoka S, Gawler D. Identification of a novel protein complex containing annexin A4, rabphilin and synaptotagmin. FEBS Lett. 2004;559:13–21. doi: 10.1016/S0014-5793(03)01513-8. [DOI] [PubMed] [Google Scholar]
- 47.Gotoh M, Takamoto Y, Kurosaka K, Masuda J, Ida M, Satoh A, Takayama E, Kojima-Aikawa K, Kobayashi Y, Matsumoto I. Annexins I and IV inhibit Staphylococcus aureus attachment to human macrophages. Immunol Lett. 2005;98:297–302. doi: 10.1016/j.imlet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Hayes MJ, Moss SE. Annexins and disease. Biochem Biophys Res Commun. 2004;322:1166–1170. doi: 10.1016/j.bbrc.2004.07.124. [DOI] [PubMed] [Google Scholar]
- 49.Brandli AW. Towards a molecular anatomy of the Xenopus pronephric kidney. Int J Dev Biol. 1999;43:381–395. [PubMed] [Google Scholar]
- 50.Spornitz UM. Studies on the liver of Xenopus laevis. III. The ultrastructure and the glycogen content of the developing liver. Anat Embryol (Berl) 1978;154:1–25. doi: 10.1007/BF00317951. [DOI] [PubMed] [Google Scholar]
- 51.Markoff A, Gerke V. Expression and functions of annexins in the kidney. Am J Physiol Renal Physiol. 2005;289:F949–F956. doi: 10.1152/ajprenal.00089.2005. [DOI] [PubMed] [Google Scholar]
- 52.Sartor MA, Zorn AM, Schwanekamp JA, Halbleib D, Karyala S, Howell ML, Dean GE, Medvedovic M, Tomlinson CR. A new method to remove hybridization bias for interspecies comparison of global gene expression profiles uncovers an association between mRNA sequence divergence and differential gene expression in Xenopus. Nucleic Acids Res. 2006;34:185–200. doi: 10.1093/nar/gkj413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.