Abstract
Antimicrobial drug development is increasingly lagging behind the evolution of antibiotic resistance, and as a result, there is a pressing need for new antibacterial therapies that can be readily designed and implemented. In this work, we engineered bacteriophage to overexpress proteins and attack gene networks that are not directly targeted by antibiotics. We show that suppressing the SOS network in Escherichia coli with engineered bacteriophage enhances killing by quinolones by several orders of magnitude in vitro and significantly increases survival of infected mice in vivo. In addition, we demonstrate that engineered bacteriophage can enhance the killing of antibiotic-resistant bacteria, persister cells, and biofilm cells, reduce the number of antibiotic-resistant bacteria that arise from an antibiotic-treated population, and act as a strong adjuvant for other bactericidal antibiotics (e.g., aminoglycosides and β-lactams). Furthermore, we show that engineering bacteriophage to target non-SOS gene networks and to overexpress multiple factors also can produce effective antibiotic adjuvants. This work establishes a synthetic biology platform for the rapid translation and integration of identified targets into effective antibiotic adjuvants.
Keywords: antibiotic adjuvants, antibiotic resistance, bacterial persistence, bacteriophage therapy, synthetic biology
Bacterial infections are responsible for significant morbidity and mortality in clinical settings (1). Many infections that would have been cured easily by antibiotics in the past now are resistant, resulting in sicker patients and longer hospitalizations (1, 2). The economic impact of antibiotic-resistant infections is estimated to be between $5 billion and $24 billion per year in the United States (3). Antibiotic resistance can be acquired genetically (e.g., via mutations in antibiotic targets) or result from persistence, in which a small fraction of cells in a population exhibits a non-inherited, phenotypic tolerance to antimicrobials (1, 4, 5).
New classes of antibiotics and more effective antimicrobial agents are needed, but few are in pharmaceutical pipelines (1, 6). High-throughput methodologies combined with traditional molecular biology techniques have enabled the discovery of potential drug targets for new antibiotics and antibiotic potentiators (7, 8). However, translating these targets from identification to actual drug compounds requires a significant amount of additional work and investment. Moreover, antibiotic drugs typically do not take advantage of targets that need to be up-regulated to achieve antimicrobial activity. As a result, a significant gap remains between target identification and drug development.
In this work, we engineered bacteriophage to overexpress proteins to target gene networks to enhance bacterial killing by antibiotics. Phage therapy to kill bacteria has been in use since the early 20th century (9). Phage can lyse bacteria or be modified to express lethal genes to cause cell death (10–14). However, phage that are directly lethal to their bacterial hosts can select for phage-resistant bacteria in a short time (10, 11, 15). To reduce the development of phage resistance, we sought to develop engineered phage that would exert minimal evolutionary pressures. Instead of overexpressing lethal genes, our design targets nonessential genes and the networks they regulate that are not directly attacked by antibiotics. Combination therapy with different antibiotics, different bacteriophage, or antibiotics plus phage may reduce the incidence of phage resistance and/or antibiotic resistance (16–20). Therefore, by using a combination of engineered antibiotic-enhancing phage and antibiotics, we hoped to reduce the incidence of antibiotic resistance and enhance bacterial killing.
Results
Targeting the SOS DNA Repair System.
Bactericidal antibiotics (e.g., quinolones such as ofloxacin) induce hydroxyl radical formation that leads to DNA, protein, and lipid damage and ultimately to cell death (8). DNA damage induces the SOS response (21, 22), which results in DNA repair (Fig. 1A). It has been shown that bacterial killing by bactericidal antibiotics can be enhanced by knocking out recA and disabling the SOS response (8). Here we took an alternative approach and engineered M13mp18 phage to overexpress lexA3, a repressor of the SOS response (23). Overexpression of lexA to suppress the SOS system has been demonstrated to inhibit the emergence of antibiotic resistance (24). We used M13mp18, a modified version of M13 phage, as our substrate, because it is a non-lytic filamentous phage and can accommodate DNA insertions into its genome (supporting information (SI) Fig. S1) (25).
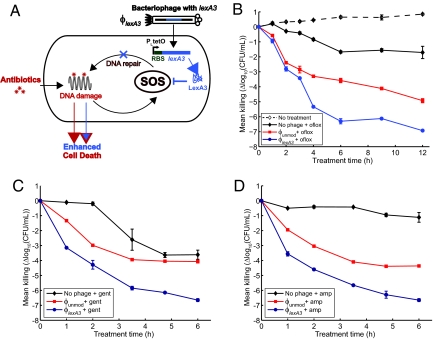
Fig. 1.
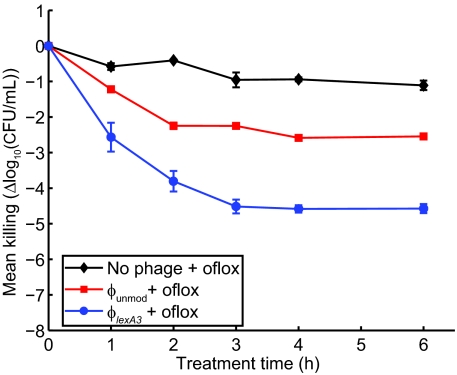
Engineered ϕlexA3 bacteriophage enhances killing of wild-type E. coli EMG2 bacteria by bactericidal antibiotics. (A) Schematic of combination therapy with engineered phage and antibiotics. Bactericidal antibiotics induce DNA damage via hydroxyl radicals, leading to induction of the SOS response. SOS induction results in DNA repair and can lead to survival (8). Engineered phage carrying the lexA3 gene (ϕlexA3) under the control of the synthetic promoter PLtetO and an RBS (27) acts as an antibiotic adjuvant by suppressing the SOS response and increasing cell death. (B) Killing curves for no phage (black diamonds), unmodified phage ϕunmod (red squares), and engineered phage ϕlexA3 (blue circles) with 60 ng/ml ofloxacin (oflox) (solid lines, closed symbols). 108 pfu/ml phage was used. A growth curve for E. coli EMG2 with no treatment (dotted line, open symbols) is shown for comparison. ϕlexA3 greatly enhanced killing by ofloxacin by 4 h of treatment. (C) Killing curves for no phage (black diamonds), ϕunmod (red squares), and ϕlexA3 (blue circles) with 5 μg/ml gentamicin (gent). 109 pfu/ml phage was used. ϕlexA3 phage greatly increases killing by gentamicin. (D) Killing curves for no phage (black diamonds), ϕunmod (red squares), and ϕlexA3 (blue circles) with 5 μg/ml ampicillin (amp). 109 pfu/ml phage was used. ϕlexA3 phage greatly increases killing by ampicillin.
To repress the SOS response, we placed the lexA3 gene under the control of the synthetic PLtetO promoter followed by a synthetic ribosome-binding sequence (RBS) (8, 23, 26, 27); we named this phage ϕlexA3 (Figs. 1A and S1B) and the unmodified M13mp18 phage ϕunmod. PLtetO, which is an inducible promoter in the presence of the TetR repressor, is constitutively on in EMG2 cells, which lack TetR. PLtetO was used for convenience for our proof-of-concept experiments described here and would not necessarily be the promoter of choice in real-world situations. We confirmed that ϕlexA3 suppressed the SOS response induced by ofloxacin treatment by monitoring GFP fluorescence in E. coli K-12 EMG2 cells carrying a plasmid with an SOS-responsive promoter driving gfp expression (Fig. S2) (8).
To test ϕlexA3's antibiotic-enhancing effect, we obtained time courses for killing of E. coli EMG2 bacteria with phage and/or ofloxacin treatment. We calculated viable cell counts by counting cfus during treatment with no phage or with 108 pfu/ml of phage and with no ofloxacin or with 60 ng/ml ofloxacin (Fig. 1B). Bacteria exposed only to ofloxacin were reduced by about 1.7 log10(cfu/ml) after 6 h of treatment, reflecting the presence of persisters not killed by the drug (Fig. 1B). By 6 h, ϕlexA3 improved the bactericidal effect of ofloxacin by 2.7 orders of magnitude compared with unmodified phage ϕunmod (≈ 99.8% additional killing) and by more than 4.5 orders of magnitude compared with no phage (≈ 99.998% additional killing) (Fig. 1B). Unmodified phage enhanced ofloxacin's bactericidal effect, a finding that is consistent with previous observations that unmodified filamentous phage augment antibiotic efficacy against Pseudomonas aeruginosa (20). Other researchers have noted that M13-infected E. coli exhibited impaired host stress responses to conditions such as acid stress (28). The mechanism by which unmodified filamentous phage can augment antibiotic efficacy is not well characterized but may involve membrane disruption or impaired stress responses. No significant bacterial regrowth was apparent with combination phage and antibiotic treatment up to 12 h (Fig. 1B) (10, 11, 15). We confirmed that both ϕunmod and ϕlexA3 replicated significantly during treatment (data not shown).
To test whether ϕlexA3 can act as an antibiotic adjuvant in different situations, we assayed for bacterial killing with varying initial phage inoculation doses (Fig. S3) and with varying doses of ofloxacin (Fig. S4) after 6 h of treatment, respectively. ϕlexA3 enhanced ofloxacin's bactericidal activity over a wide range of multiplicity of infection (MOI), from 1:1000 to 1:1 (Fig. S3). ϕlexA3's ability to increase killing by ofloxacin at a low MOI reflects rapid replication and infection by M13 phage. For ofloxacin concentrations of 30 ng/ml and higher, ϕlexA3 resulted in much greater killing compared with no phage or unmodified phage ϕunmod (Fig. S4). Thus, ϕlexA3 is a strong adjuvant for ofloxacin at doses below and above the minimum inhibitory concentration (60 ng/ml, data not shown).
We next determined whether our engineered phage could increase killing by classes of antibiotics other than quinolones. We tested ϕlexA3's antibiotic-enhancing effect for gentamicin, an aminoglycoside, and ampicillin, a β-lactam antibiotic. ϕlexA3 increased gentamicin's bactericidal action by more than 2.5 and 3 orders of magnitude compared with ϕunmod and no phage, respectively (Fig. 1C). ϕlexA3 also improved ampicillin's bactericidal effect by more than 2 and 5.5 orders of magnitude compared with ϕunmod and no phage, respectively (Fig. 1D). For both gentamicin and ampicillin, ϕlexA3's strong antibiotic-enhancing effect was noticeable after 1 h of treatment (Fig. 1 C and D). These results are consistent with previous observations that ΔrecA mutants exhibit increased susceptibility to quinolones, aminoglycosides, and β-lactams (8) and indicate that engineered phage such as ϕlexA3 can act as general adjuvants for the 3 major classes of bactericidal drugs.
We also found that engineered phage ϕlexA3 is capable of reducing the number of persister cells in populations already exposed to antibiotics as well as enhancing antibiotic efficacy against bacteria living in biofilms. For example, ϕlexA3 added to a population previously treated only with ofloxacin increased the killing of bacteria that survived the initial treatment by ≈ 1 and 1.5 orders of magnitude compared with ϕunmod and no phage, respectively (Fig. S5). In addition, simultaneous application of ϕlexA3 and ofloxacin improved killing of biofilm cells by about 1.5 and 2 orders of magnitude compared with ϕunmod plus ofloxacin and no phage plus ofloxacin, respectively (Fig. S6).
Enhancing Killing of Antibiotic-Resistant Bacteria.
In addition to killing wild-type bacteria with increased efficacy, engineered phage can enhance killing of bacteria that already have acquired antibiotic resistance. We applied ϕlexA3 with ofloxacin against E. coli RFS289, which carries a mutation (gyrA111) that renders it resistant to quinolone antibiotics (7, 29). ϕlexA3 increased the bactericidal action of ofloxacin by more than 2 and 3.5 orders of magnitude compared with ϕunmod and no phage, respectively (Fig. 2). These results demonstrate that antibiotic-enhancing phage can be used to combat antibiotic-resistant bacteria and therefore may have the potential to bring defunct antibiotics back into clinical use.
Fig. 2.
Engineered ϕlexA3 bacteriophage enhances killing of quinolone-resistant E. coli RFS289 bacteria by ofloxacin. Killing curves for no phage (black diamonds), unmodified phage ϕunmod (red squares), and engineered phage ϕlexA3 (blue circles) with 1 μg/ml ofloxacin (oflox). 108 pfu/ml phage was used. ϕlexA3 greatly enhanced killing by ofloxacin by 1 h of treatment.
Increasing Survival of Mice Infected with Bacteria.
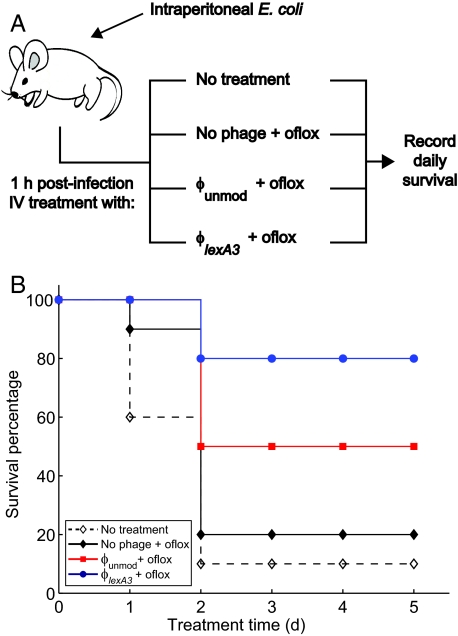
To determine the clinical relevance of antibiotic-enhancing phage in vivo, we tested the ability of our engineered phage with ofloxacin to prevent death in mice infected with bacteria. Mice were injected with E. coli EMG2 i.p. 1 h before receiving different i.v. treatments (Fig. 3A). Eighty percent of mice that received ϕlexA3 with ofloxacin survived, compared with 50% mice that received ϕunmod plus ofloxacin and 20% of mice that received ofloxacin alone (Fig. 3B). The in vivo efficacy of our antibiotic-enhancing phage in rescuing infected mice from death demonstrates the feasibility of our designs for clinical use.
Fig. 3.
Engineered ϕlexA3 bacteriophage increases survival of mice infected with bacteria. (A) Female Charles River CD-1 mice were inoculated with i.p. injection of 8.8 * 107 cfu/mouse E. coli EMG2 bacteria. After 1 h, the mice received no treatment or i.v. treatment with 0.2 mg/kg ofloxacin plus no phage, plus unmodified phage ϕunmod, or plus engineered phage ϕlexA3 (109 pfu/mouse phage was used). The mice were observed for 5 days, and deaths were recorded at the end of each day to generate survival curves. [Mouse drawing reproduced under a Creative Commons Attribution 2.5 license (53).] (B) Survival curves for infected mice treated with phage and/or ofloxacin demonstrate that engineered phage ϕlexA3 plus ofloxacin (closed blue circles with solid line) significantly increases survival of mice compared with unmodified phage ϕunmod plus ofloxacin (closed red squares with solid line), no phage plus ofloxacin (closed black diamonds with solid line), or no treatment (open black diamonds with dashed line).
Reducing the Development of Antibiotic Resistance.
Exposure to subinhibitory concentrations of antibiotics can lead to initial mutations that confer low-level antibiotic resistance and eventually to more mutations that yield high-level resistance (30). We hypothesized that engineered phage, as antibiotic adjuvants, could reduce the number of antibiotic-resistant mutants that result from a bacterial population exposed to antimicrobial drugs. To test this hypothesis, we grew E. coli EMG2 in media with no ofloxacin for 24 h, with 30 ng/ml ofloxacin for 24 h, with 30 ng/ml ofloxacin for 12 h followed by ϕunmod plus ofloxacin treatment for 12 h, or with 30 ng/ml ofloxacin for 12 h followed by ϕlexA3 plus ofloxacin treatment for 12 h (Fig. S7). Then, we counted the number of mutants resistant to 100 ng/ml ofloxacin for each of the 60 samples under each growth condition. Growth in the absence of ofloxacin yielded very few resistant cells (median = 1) (Fig. S7). However, growth with subinhibitory levels of ofloxacin produced a high number of antibiotic-resistant bacteria (median = 1592) (Fig. S7). Treatment with unmodified phage ϕunmod decreased the number of resistant cells (median = 43.5); however, all samples contained > 1 resistant cfu, and more than half of the samples had > 20 resistant cfus (Fig. S7). In contrast, ϕlexA3 treatment dramatically suppressed the level of antibiotic-resistant cells (median = 2.5), resulting in a majority of samples with either no resistant cfus or < 20 resistant cfus (Fig. S7).
Flexible Targeting of Other Gene Networks.
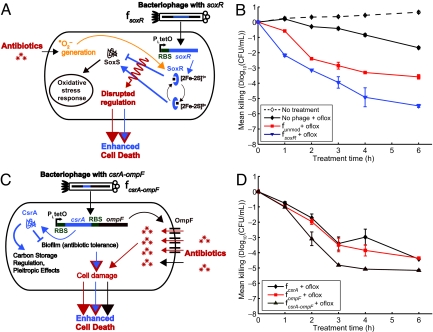
Our phage platform can be used to target many different gene networks to produce effective antibiotic adjuvants. To demonstrate this feature, we engineered phage to express proteins that regulate non-SOS gene networks (e.g., SoxR and CsrA) or modulate sensitivity to antibiotics (e.g., OmpF) (Fig. 4 and Fig. S1) (27). For example, the soxRS regulon controls a coordinated cellular response to superoxide (31). SoxR contains a [2Fe-2S] cluster that must be oxidized for it to stimulate SoxS production, which then controls the transcription of downstream genes that respond to oxidative stress (31). Because quinolones generate superoxide-based oxidative attack (7, 8), we surmised that engineering phage to overexpress wild-type SoxR (ϕsoxR) might affect this response and improve ofloxacin's bactericidal activity (Fig. 4A). As shown in Fig. 4B, ϕsoxR enhanced killing by ofloxacin compared with unmodified phage ϕunmod and no phage (Fig. 4B). However, the exact mechanism underlying the ability of SoxR overexpression in ϕsoxR to enhance antibiotic killing is not clear. Overexpression of SoxR may provide additional iron-sulfur clusters that could be destabilized to increase sensitivity to bactericidal antibiotics (7, 8). Alternatively, because SoxR usually is kept at relatively low levels in vivo that are unchanged by oxidative stress (32), overexpressing large amounts of SoxR may interfere with signal transduction in response to oxidative stress by titrating intracellular iron or oxidizing species or by competing with oxidized SoxR for binding to the soxS promoter (32–34).
Fig. 4.
Engineered bacteriophage targeting single and multiple gene networks (other than the SOS network) as adjuvants for ofloxacin treatment (oflox). (A) Ofloxacin stimulates superoxide generation, which normally is countered by the oxidative stress response, coordinated by SoxR (8). Engineered phage producing SoxR (ϕsoxR) enhances ofloxacin-based killing by disrupting regulation of the oxidative stress response. (B) Killing curves for no phage (black diamonds), unmodified phage ϕunmod (red squares), and engineered phage ϕsoxR (blue downward-pointing triangles) with 60 ng/ml ofloxacin (solid lines, closed symbols). 108 pfu/ml phage was used. The killing curve for ϕunmod and a growth curve for E. coli EMG2 with no treatment (dotted line, open symbols) are reproduced from Fig. 1B for comparison and show that ϕsoxR enhances killing by ofloxacin. (C) CsrA suppresses the biofilm state in which bacterial cells tend to be more resistant to antibiotics (35). OmpF is a porin used by quinolones to enter bacterial cells (37). Engineered phage producing both CsrA and OmpF simultaneously (ϕcsrA-ompF) enhances antibiotic penetration via OmpF and represses biofilm formation and antibiotic tolerance via CsrA to produce an improved dual-targeting adjuvant for ofloxacin. (D) Killing curves for ϕcsrA (black diamonds), ϕompF (red squares), and ϕcsrA-ompF (brown upward-pointing triangles) with 60 ng/ml ofloxacin. 108 pfu/ml phage was used. Phage expressing both csrA and ompF (ϕcsrA-ompF) is a better adjuvant for ofloxacin than phage expressing csrA (ϕcsrA) or ompF alone (ϕompF).
CsrA is a global regulator of glycogen synthesis and catabolism, gluconeogenesis, and glycolysis, and it also represses biofilm formation (35). Because biofilm formation has been linked to antibiotic resistance, we hypothesized that csrA-expressing phage (ϕcsrA) would increase susceptibility to antibiotic treatment (Fig. 4C) (36). In addition, because OmpF is a porin used by quinolones to enter bacteria (37), we hypothesized that ompF-expressing phage (ϕompF) would increase killing by ofloxacin (Fig. 4C). After 6 h, both ϕcsrA and ϕompF increased ofloxacin's bactericidal effect by ≈ 1 and 3 orders of magnitude compared with ϕunmod and no phage, respectively (Fig. 4D).
Systems biology analysis often results in the identification of multiple antibacterial targets that are not easily addressed by traditional drug compounds. In contrast, engineered phage are well suited for incorporating multiple targets into a single antibiotic adjuvant. To demonstrate this capability, we designed an M13mp18 phage to express csrA and ompF simultaneously (ϕcsrA-ompF) to target csrA-controlled gene networks and increase drug penetration (Fig. 4C) The multitarget phage was constructed by placing an RBS and ompF immediately downstream of csrA in ϕcsrA (Fig. S1F) (27). ϕcsrA-ompF was more effective in enhancing ofloxacin's bactericidal effect than were its single-target relatives, ϕcsrA and ϕompF, in planktonic (Fig. 4D) and biofilm (Fig. S8) settings. Together, these results demonstrate that engineering phage to target non-SOS genetic networks and/or overexpress multiple factors can produce effective antibiotic adjuvants.
Discussion
Our work demonstrates that combination therapy coupling antibiotics with antibiotic-enhancing phage has the potential to be a promising antimicrobial strategy. Moreover, we have shown that antibiotic-enhancing phage should have clinical relevance because of their in vivo effectiveness in rescuing infected mice. Thus, phage can be engineered to act as effective antibiotic adjuvants in vitro and in vivo and may help close the gap between antimicrobial target identification and implementation. By targeting nonessential gene networks, a diverse set of engineered bacteriophage can be developed to supplement other antimicrobial strategies.
Despite the potential benefits described earlier in the text, phage have yet to be accepted into clinical practice because of a number of issues, such as phage immunogenicity, efficacy, target bacteria identification and phage selection, host specificity, and toxin release (9–11, 38, 39). To reduce the risk of leaving lysogenic particles in patients after treatment, our adjuvant phage could be modified to be nonreplicative, as has been described previously (11). A potential concern with the use of engineered M13mp18 prototype phage described here is the development of phage resistance resulting from the loss of the F-plasmid required for infection (10). We have developed our prototype phage as a proof of concept for antibiotic adjuvants and recognize that real-world usage may necessitate the use of phage cocktails to ensure efficacy and the ability to treat non-F-plasmid−containing bacteria. Phage cocktails that target different, multiple bacterial receptors may reduce the development of phage resistance by invading bacteria through different means. Using phage cocktails with multiple antibiotics also could enhance bacterial killing and reduce resistance to both phage and antibiotics.
Our phage platform for the development of effective antibiotic adjuvants is a practical example of the application of synthetic biology to important real-world biomedical issues. Synthetic biology is focused on the rational and modular engineering of organisms to create novel behaviors. The field has produced many reports of synthetic gene circuits and systems with interesting characteristics (40–45). More recently, synthetic biologists have begun to address important industrial and medical problems (16, 46–48). To extend our work beyond proof-of-concept experiments, libraries of natural phage could be modified to target gene networks and pathways, such as the SOS response, in different bacterial species (49, 50). This process would require the isolation and genetic modification of natural phage with the ability to infect the bacterial species being targeted. With current DNA sequencing and synthesis technology, an entire engineered bacteriophage genome carrying multiple constructs to target different gene networks could be synthesized for less than $10,000, a price that is sure to decrease in the future (51). These technologies should enable large-scale modifications of phage libraries to produce antibiotic-enhancing phage that can be applied with different antibiotic drugs against a wide range of bacterial infections. Targeting clinical bacterial strains with libraries of engineered phage will be a crucial step in applying this strategy against real-world infections.
Engineered phage may be adopted more readily in industrial, agricultural, and food processing settings where bacterial biofilms and other difficult-to-clear bacteria are present (16). Applying engineered phage as antibiotic adjuvants in nonmedical settings could be economically advantageous, reduce community-acquired antibiotic resistance, and be a prudent first step toward gaining acceptance for clinical use (52).
Materials and Methods
Bacterial Strains, Phage, and Chemicals.
E. coli K-12 EMG2 cells, which lack O antigens, were obtained from the Yale Coli Genetic Stock Center (CGSC #4401). E. coli RFS289 cells, which contain a gyrA111 mutation rendering them resistant to quinolones, were obtained from the Yale Coli Genetic Stock Center (CGSC #5742). M13mp18 phage was purchased from New England Biolabs. E. coli XL-10 cells used for cloning, amplifying phage, and plating phage were obtained from Stratagene. Chemicals were obtained from sources described in SI Materials and Methods.
Engineering M13mp18 Phage to Target Genetic Networks.
To construct engineered phage, lexA3, soxR, csrA, and ompF genes were first placed under the control of the PLtetO promoter in the pZE11G vector (23, 27). Details are described in SI Text. All PLtetO-gene constructs were followed by terminator T1 of the rrnB operon and preceded by a stop codon; they were PCR amplified from the respective pZE11 plasmids with primers 5′ aataca GAGCTC cTAA tccctatcagtgatagagattg 3′ and 5′ taatct CGATCG tctagggcggcggat 3′ and cloned into the SacI and PvuI sites of M13mp18 (Fig. S1) (25, 27). Resulting phage genomes were transformed into XL-10 cells, mixed with 200 μl overnight XL-10 cells in 3 ml top agar, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and 40 μl of 20 mg/ml X-Gal, and poured onto LB agar + chloramphenicol (30 μg/ml) plates for plaque formation and blue-white screening. After overnight incubation of plates at 37 °C, white plaques were scraped and placed into 1:10 dilutions of overnight XL-10 cells and grown for 5 h. Replicative form (RF) M13mp18 DNA was collected by DNA minipreps of the bacterial cultures. All insertions into M13mp18 were verified by PCR and restriction digests of RF DNA. Infective phage solutions were obtained by centrifuging infected cultures for 5 min at 16,100 × g and collecting supernatants followed by filtration through Nalge #190–2520 0.2 μm filters (Nalge Nunc International).
Determination of Plaque-Forming Units.
To obtain pfus, we added serial dilutions of phage performed in 1X PBS to 200 μl of overnight XL-10 cells in 3 ml top agar, 1 mM IPTG, and 40 μl of 20 mg/ml X-Gal, and poured the mixture onto LB agar + chloramphenicol (30 μg/ml) plates. After overnight incubation at 37 °C, plaques were counted.
Determination of Colony-Forming Units.
To obtain cfu counts, 150 μl of relevant cultures were collected, washed with 1X PBS, recollected, and resuspended in 150 μl of 1X PBS. Serial dilutions were performed with 1X PBS and sampled on LB agar plates. LB agar plates were incubated at 37 °C overnight before counting.
Flow Cytometer Assay of SOS Induction.
To monitor ϕlexA3's suppression of the SOS response (Fig. S2), we used a plasmid containing an SOS-response promoter driving gfp expression in EMG2 cells (PLlexO-gfp) (7) with a basic protocol described in the SI Text.
Ofloxacin Killing Assay.
To determine the antibiotic-enhancing effect of engineered phage for ofloxacin (Figs. 1B, 4 B and D), we grew 1:500 dilutions of EMG2 cells overnight for 3 h and 30 min at 37 °C and 300 rpm (model G25 incubator shaker; New Brunswick Scientific) to late-exponential phase and determined initial cfus, which were in the range of ≈ 109 cfu/ml. Then, we added 60 ng/ml ofloxacin alone or in combination with 108 pfu/ml phage (unmodified phage ϕunmod or engineered ϕlexA3, ϕsoxR, ϕcsrA, ϕompF, or ϕcsrA-ompF phage), and treated at 37 °C and 300 rpm. At indicated time points, we determined cfus as described earlier. Mean killing (Δlog10(cfu/ml)) was determined by subtracting mean initial log10(cfu/ml) from mean log10(cfu/ml) after treatment to compare data from different experiments. This protocol was replicated with E. coli RFS289 to determine the ofloxacin-enhancing effect of engineered ϕlexA3 phage against antibiotic-resistant bacteria (Fig. 2).
Dose–Response Assays.
The initial phage inoculation dose–response experiments (Fig. S3) were conducted using the same protocol as the ofloxacin killing assay, except that 60 ng/ml ofloxacin was added with varying concentrations of phage. Cultures were treated for 6 h before obtaining viable cell counts. The ofloxacin dose–response experiments (Fig. S4) also were obtained using the same protocol as in the ofloxacin killing assay, except that 108 pfu/ml phage was added with varying concentrations of ofloxacin, and viable cell counts were obtained after 6 h of treatment.
Gentamicin and Ampicillin Killing Assays.
To determine the antibiotic-enhancing effect of engineered phage for gentamicin and ampicillin, we used the same protocol as in the ofloxacin killing assay, except we used 109 pfu/ml initial phage inoculations. Five μg/ml gentamicin and 5 μg/ml ampicillin were used in Fig. 1 C and D, respectively.
Mouse Survival Assay.
Female Charles River CD-1 mice (weighing 18–20 g) received i.p. injections with 8.8 * 107 cfu/mouse E. coli EMG2 cells in a volume of 0.5 ml with 8% mucin (Fig. 3). After 1 h, the mice received either no treatment or i.v. infusions of ofloxacin alone (0.2 mg/kg), 109 pfu/mouse unmodified phage ϕunmod with ofloxacin (0.2 mg/kg), or 109 pfu/mouse engineered ϕlexA3phage with ofloxacin (0.2 mg/kg). Ten mice were used per treatment group. The mice were observed over 5 days, and deaths were recorded at the end of each day. All mouse materials were provided by ViviSource Laboratories, a facility approved by the United States Department of Agriculture and by the Office of Laboratory Animal Welfare, where all in vivo experimental work was performed.
Persister Killing Assay.
Persister killing (Fig. S5) was assayed using a basic protocol described in SI Text.
Biofilm Killing Assay.
Biofilm killing (Fig. S6 and Fig. S8) was assayed using a previously reported protocol described in SI Text (16).
Antibiotic Resistance Assay.
To analyze the effect of subinhibitory concentrations of ofloxacin on the development of antibiotic-resistant mutants, we grew 1:108 dilutions of EMG2 cells overnight in LB media containing either no ofloxacin or 30 ng/ml ofloxacin (Fig. S7). After 12 h of growth at 37 °C and 300 rpm (model G25 incubator shaker, New Brunswick Scientific), we split the cells grown in no ofloxacin into 100-μl aliquots with no ofloxacin into 60 wells in 96-well plate format (Costar 3370; Fisher Scientific). We also split the cells grown in 30 ng/ml ofloxacin into 100-μl aliquots in 60 wells with no phage and 30 ng/ml ofloxacin, with ϕunmod and 30 ng/ml ofloxacin, or with ϕlexA3 and 30 ng/ml ofloxacin in 96-well plates. We placed the 96-well plates in 37 °C and 300 rpm with plastic bags to minimize evaporation. After 12 h of treatment, we plated cultures from each well on LB agar + 100 ng/ml ofloxacin to select for mutants that developed resistance against ofloxacin. To compare results, we constructed box-and-whisker plots using the 60 individual observations for each treatment condition (Fig. S7).
Statistical Analysis.
All cfu data were log10-transformed before analysis. For all data points in all experiments, 3 samples were collected except where noted. Error bars in figures indicate standard error of the mean.
Supplementary Material
Acknowledgments.
We thank Andrew Slee and Tim Murphy from ViviSource Laboratories for their assistance with our in vivo mouse studies. We greatly appreciate the reviewers' critique of our research. This work was supported by the National Institutes of Health Director's Pioneer Award Program (Grant DP1 OD00344), the National Science Foundation Frontiers in Integrative Biological Research Program, and the Howard Hughes Medical Institute. T.K.L. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship and a Harvard-MIT Health Sciences and Technology Medical Engineering/Medical Physics Fellowship.
Footnotes
Conflict of interest statement: We have submitted a patent disclosure regarding the work described in this paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800442106/DCSupplemental.
References
- 1.Wise R. The relentless rise of resistance? J Antimicrob Chemother. 2004;54(2):306–310. doi: 10.1093/jac/dkh340. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nature Reviews Microbiology. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 3.Hall BG. Predicting the evolution of antibiotic resistance genes. Nature Reviews Microbiology. 2004;2(5):430–435. doi: 10.1038/nrmicro888. [DOI] [PubMed] [Google Scholar]
- 4.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. Persister cells, dormancy and infectious disease. Nature Reviews Microbiology. 2007;5(1):48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 6.Walsh C. Where will new antibiotics come from? Nature Reviews Microbiology. 2003;1(1):65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 7.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Molecular Systems Biology. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130(5):797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 9.Merril CR, Scholl D, Adhya SL. The prospect for bacteriophage therapy in Western medicine. Nature Reviews Drug Discovery. 2003;2(6):489–497. doi: 10.1038/nrd1111. [DOI] [PubMed] [Google Scholar]
- 10.Hagens S, Blasi U. Genetically modified filamentous phage as bactericidal agents: A pilot study. Letters in Applied Microbiology. 2003;37(4):318–323. doi: 10.1046/j.1472-765x.2003.01400.x. [DOI] [PubMed] [Google Scholar]
- 11.Hagens S, Habel AvAU, von Gabain A, Blasi U. Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob Agents Chemother. 2004;48(10):3817–3822. doi: 10.1128/AAC.48.10.3817-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westwater C, et al. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: An alternative therapy for treatment of bacterial infections. Antimicrob Agents Chemother. 2003;47(4):1301–1307. doi: 10.1128/AAC.47.4.1301-1307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heitman J, Fulford W, Model P. Phage Trojan horses: A conditional expression system for lethal genes. Gene. 1989;85(1):193–197. doi: 10.1016/0378-1119(89)90480-0. [DOI] [PubMed] [Google Scholar]
- 14.Brüssow H. Phage therapy: The Escherichia coli experience. Microbiology. 2005;151(Pt 7):2133–2140. doi: 10.1099/mic.0.27849-0. [DOI] [PubMed] [Google Scholar]
- 15.Summers WC. Bacteriophage therapy. Annu Rev Microbiol. 2001;55:437–451. doi: 10.1146/annurev.micro.55.1.437. [DOI] [PubMed] [Google Scholar]
- 16.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci USA. 2007;104(27):11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonhoeffer S, Lipsitch M, Levin BR. Evaluating treatment protocols to prevent antibiotic resistance. Proc Natl Acad Sci USA. 1997;94(22):12106–12111. doi: 10.1073/pnas.94.22.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chait R, Craney A, Kishony R. Antibiotic interactions that select against resistance. Nature. 2007;446(7136):668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- 19.Levy SB, Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat Med. 2004;10(12) Suppl:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 20.Hagens S, Habel A, Bläsi U. Augmentation of the antimicrobial efficacy of antibiotics by filamentous phage. Microbial Drug Resistance (Larchmont, NY) 2006;12(3):164–168. doi: 10.1089/mdr.2006.12.164. [DOI] [PubMed] [Google Scholar]
- 21.Miller C, et al. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305(5690):1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 22.Lewin CS, Howard BM, Ratcliffe NT, Smith JT. 4-Quinolones and the SOS response. Journal of Medical Microbiology. 1989;29(2):139–144. doi: 10.1099/00222615-29-2-139. [DOI] [PubMed] [Google Scholar]
- 23.Little JW, Harper JE. Identification of the lexA gene product of Escherichia coli K-12. Proc Natl Acad Sci USA. 1979;76(12):6147–6151. doi: 10.1073/pnas.76.12.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cirz RT, et al. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3(6):e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 26.Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements. Nucleic Acids Res. 1997;25(6):1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson F, Malmborg-Hager AC, Albrekt AS, Borrebaeck CA. Genome-wide comparison of phage M13-infected vs. uninfected Escherichia coli. Can J Microbiol. 2005;51(1):29–35. doi: 10.1139/w04-113. [DOI] [PubMed] [Google Scholar]
- 29.Schleif R. Fine-structure deletion map of the Escherichia coli L-arabinose operon. Proc Natl Acad Sci USA. 1972;69(11):3479–3484. doi: 10.1073/pnas.69.11.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez JL, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44(7):1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hidalgo E, Ding H, Demple B. Redox signal transduction: Mutations shifting [2Fe-2S] centers of the SoxR sensor-regulator to the oxidized form. Cell. 1997;88(1):121–129. doi: 10.1016/s0092-8674(00)81864-4. [DOI] [PubMed] [Google Scholar]
- 32.Hidalgo E, Leautaud V, Demple B. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 1998;17(9):2629–2636. doi: 10.1093/emboj/17.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181(15):4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93(19):10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson DW, et al. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol. 2002;184(1):290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 37.Hirai K, Aoyama H, Irikura T, Iyobe S, Mitsuhashi S. Differences in susceptibility to quinolones of outer membrane mutants of Salmonella typhimurium and Escherichia coli. Antimicrob Agents Chemother. 1986;29(3):535–538. doi: 10.1128/aac.29.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boratynski J, et al. Preparation of endotoxin-free bacteriophages. Cellular and Molecular Biology Letters. 2004;9(2):253–259. [PubMed] [Google Scholar]
- 39.Merril CR, et al. Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci USA. 1996;93(8):3188–3192. doi: 10.1073/pnas.93.8.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: New engineering rules for an emerging discipline. Molecular Systems Biology. 2006;2 doi: 10.1038/msb4100073. 2006.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 42.McDaniel R, Weiss R. Advances in synthetic biology: On the path from prototypes to applications. Curr Opin Biotechnol. 2005;16(4):476–483. doi: 10.1016/j.copbio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Molecular Systems Biology. 2005;1 doi: 10.1038/msb4100025. 2005.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guido NJ, et al. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 45.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130(2):363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 46.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355(4):619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 47.Loose C, Jensen K, Rigoutsos I, Stephanopoulos G. A linguistic model for the rational design of antimicrobial peptides. Nature. 2006;443(7113):867–869. doi: 10.1038/nature05233. [DOI] [PubMed] [Google Scholar]
- 48.Ro D-K, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 49.Hickman-Brenner FW, Stubbs AD, Farmer JJ. Phage typing of Salmonella enteritidis in the United States. J Clin Microbiol. 1991;29(12):2817–2823. doi: 10.1128/jcm.29.12.2817-2823.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135(4):679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker D, et al. Engineering life: building a fab for biology. Sci Am. 2006;294(6):44–51. doi: 10.1038/scientificamerican0606-44. [DOI] [PubMed] [Google Scholar]
- 52.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430(6996):242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart JB, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6(1):e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.