Abstract
Covariation between sites can arise due to a common evolutionary history. At the same time, structure and function of proteins play significant role in evolvability of different sites that are not directly connected with the common ancestry. The nature of forces which cause residues to coevolve is still not thoroughly understood, it is especially not clear how coevolutionary processes are related to functional diversification within protein families. We analyzed both functional and structural factors that might cause covariation of specificity determinants and showed that they more often participate in coevolutionary relationships with each other and other sites compared to functional sites and those sites that are not under strong functional constraints. We also found that protein sites with higher number of coevolutionary connections with other sites have a tendency to evolve slower. Our results indicate that in some cases coevolutionary connections exist between specificity sites that are located far away in space but are under similar functional constraints. Such correlated changes and compensations can be realized through the stepwise coevolutionary processes which in turn can shed light on the mechanisms of functional diversification.
Keywords: Coevolution, correlated mutation, covariation, functional diversification, specificity determinants, subfamily specificity, mutual information, protein evolution
INTRODUCTION
Coevolution between residues in proteins is a very important factor in the molecular evolution which has not been studied extensively and taken into account in the modeling of evolutionary processes. Coevolution occurs when residues in one site change depending on the residues at another site1–3. Many studies aimed at the detection of correlations between protein residues at different sites2,4–15. Despite the comparative success in prediction of protein secondary, tertiary structures4,6,16,17 and protein interaction partners18–20 coevolution has been found to be rather weak in many cases, with the strongest signal coming from the sites in alpha helical stacking2 and from charge compensating covariations8. Indeed, coevolution is difficult to detect due to the variable nature of compensatory mutations, the strong dependence of covariations on evolutionary distances, number of sequences in the alignment and residue environment. Moreover, the coevolution signal must be separated from strong background signals resulting from various correlations between the non-coevolving residues.
The apparent covariation between different sites can arise due to several factors. Associations between sites can persist over time as a natural result of a common evolutionary history (phylogenetic linkage). At the same time, structure, folding and function put certain constraints on the evolvability of the sites that are not directly connected with the common ancestry. For example, sites that are within the proximity from each other in a native structure or come into contact upon folding do not evolve independently from each other as they need to maintain amino acid interactions important for protein stability and foldability. Function is another source of covariability and functionally important sites which are not necessarily in direct contact might covary because they bind to the same ligand or participate in the allosteric regulation of functional activity. It is very difficult to separate structural and functional signals from the phylogenetic linkage21–24. It has been shown that the removal of the background phylogenetic signal improves the contact prediction in some cases although does not have much effect in the others21.
The nature of forces which cause residues to coevolve is still not thoroughly understood; especially it is unclear how coevolutionary process is related to functional changes. Some protein functional sites are under stringent evolutionary constraints to preserve their amino acid identity while other functional sites are under less evolutionary pressure. They can mutate in unison with certain sites from the same or a different (interacting) protein to maintain the overall protein functionality. It has been argued that the change in the conservation or evolutionary rate at a particular site is connected with the functional divergence after the gene duplication. After duplication of a gene, its one copy evolves under relaxed evolutionary constraints which allow it to accumulate changes and develop new functions and specificities 25,26. Two types of functional specificity have been distinguished (type I and type II)27. In the first case, a group of proteins (protein subfamily) develops recognition specificity towards certain types of ligands while maintaining an overall function of a protein family. It results in the conservation of this binding site in one subfamily and its variability in the rest of the family. The second type of specificity occurs when protein subfamilies in parallel evolve different recognition specificities. In this case purifying selection leads to similar levels of site conservation in the subfamilies but different amino acid types. It is very difficult to experimentally detect these specificity determining sites and recently new computational algorithms28–34 have been developed which predict specificity determining sites and provide the foundation for detailed analysis of evolutionary mechanisms of protein specificity.
In this paper we examined the coevolution of specificity determining sites (called “subsites” hereafter) that have been annotated in the literature or predicted by algorithms reported earlier30,32,35. These sites generally determine the protein specificity either by binding to specific substrate/inhibitor or through interaction with specific protein partner. Our ultimate goal is to understand what changes cause the evolution of new specificity which would require a redesign of the finely tuned interaction network within the protein molecule to maintain structure-function relationship. To achieve this goal we analyzed how such successive changes and compensations can be realized through the stepwise coevolutionary processes by looking at functional and structural factors that cause covariation of subsites in a protein family.
We showed that specificity determining sites covary more often with each other and other sites compared to sites that are not found under strong functional constraints. We found that individual sites that establish larger number of coevolutionary connections with other sites have a lower variability pointing to slower evolutionary rate. Our findings also suggest possible long range evolutionary coupling mechanisms in some families which might explain the coevolution between subsites that are located far away in space but are under similar functional constraints.
METHODS
Dataset of domain family alignments and subsites
We have collected reliable alignments of twelve protein families, for majority of which experimental evidence was available on the locations of subsites. The alignments were constructed by existing alignment methods and were subjected to the additional round of careful manual curation. The family alignments were grouped into subfamilies based on different criteria including sequence and structural properties, kinetic properties, substrate specificity, taxonomy and function (see SM for details). This dataset with the exception of one protein family was used in our previous study for prediction of subsites35. Five of the test families were used for validation of previously published prediction methods and seven other families were taken from the version 2.10 of the Conserved Domain Database36.
Subsites were assigned based on extensive literature search for experimental evidences (several subsites were based on consistent predictions made by several methods)29,31 (actual subsites, see SM for details on data collection) and predicted by the SPEER algorithm which was devised in our previous work35 (predicted subsites). SPEER algorithm makes predictions of subsites’ locations based on amino acids’ physico-chemical properties, evolutionary rate, and combined relative entropy. Altogether our test set contained 97 actual subsites and 180 predicted subsites (only top 15 predicted sites were taken). A complete list of the dataset families together with the number and locations of actual subsites is provided in Table SM1. The family alignments and subsite information can be can be also downloaded via ftp site ftp://ftp.ncbi.nih.gov/pub/SPEER.
All specificity determining sites were categorized into three groups, type I, type II and marginally conserved (MC) sites27,35. Type I sites were defined as those conserved for one subfamily and variable in another while type II sites were defined as those where different types of amino acids were conserved across different subfamilies. Here we considered a site to be conserved for one subfamily if any amino acid type is represented more than 75% of the time. The sites that failed to satisfy the above criteria are marked as MC (none of the subfamilies are conserved in this site). For families with more than two subfamilies, sites were categorized into different types based on the category assigned to the majority of subfamily pairs. To distinguish subsites from globally conserved sites we excluded from the subsite set those highly conserved positions within the overall alignment where any amino acid type was represented more than 80% of the time (only one highly conserved subsite was present).
Sites which are globally important for the general function of a family (called “functionally important sites” or FIS thereafter) were also extracted from six CDD families. 181 functionally important sites (Table SM2) were manually annotated based on literature and experimental data as being catalytic, metal-binding, nucleic acid or protein binding sites36. Representative 3D structures were collected for each family from the PDB database37. Spatial distances between two residues (minimum distances between the nearest atoms coordinates) were calculated by in-house scripts utilizing the 3D coordinates supplied in individual PDB files.
Identification of coevolving residue pairs
Mutual information (MI) is a measure of reduction of uncertainty38. MI of pairs of positions in the multiple sequence alignments (MSA) has been used successfully to analyze coevolved protein positions in previous studies39,40. For this study the Homolmapper program41 was applied to calculate the normalized mutual information Z-score for each pair of ungapped positions in the multiple sequence alignment. The entropy of a site “c” in the alignment was determined as:
Here, p(xi) is the observed frequency of amino acid xi. The joint entropy Hcd (c and d are two columns in the alignment) was calculated by the same method using the frequencies of occurrence of each combination of residues in positions c and d. Joint entropy scores range from the maximum of Hc or Hd to the sum, Hc + Hd. Mutual information (MI) was calculated as: MI = Hc + Hd − Hcd, where MI scores range from 0 in the case if amino acid patterns in two columns do not correlate with each other to the minimum of Hc or Hd. Since the greater entropy leads to greater MI values, the raw MI values were normalized by dividing by the joint entropy, Hcd, to reduce the influence of entropy on MI40. At the same time the MI scores were converted to the Z-scores. It has been shown that such normalizations increase the ability of MI to detect coevolved sites and has been implemented in the Homolmapper41 program. In the current study, two individual sites were considered to be coevolved if the MI-Z score between them was greater than or equal to 3. Selection of a more stringent MI-Z score threshold might result in a poor sensitivity and/or coverage as was ascertained earlier39. Additionally, our aim is to emphasize the relative differences in coevolutionary behavior between subsites and other sites and recognize even subtle tendencies toward coevolution.
To quantify the relative tendency of sites to coevolve, the fraction of coevolution (FC) for a given site i was calculated as: , where is the number of sites coevolved with site i, N is the number of sites in a protein family/alignment, (N−1) is the number of all possible site pair combinations of any given site with all other sites. Fraction of coevolution has been calculated between subsites (normalized by possible combinations between all subsites), and between subsites and all other sites.
We have also used alternative algorithms to predict coevolution between protein sites, namely, a continuous-time Markov process (CTMP) algorithm42 that incorporates phylogenetic information to predict coevolution between two sites. Unfortunately, the current version of the CTMP program was unable to produce results for some of the families in our dataset and the obtained coverage of coevolved sites was too low for any meaningful statistical characterization. Nevertheless, we also applied other available algorithms such as OMES43,44 that detects differences between observed versus expected frequencies of residue pairs, the McLachlan Based Substitution correlation method4,45,46 and another MI based method (MIp)47 that removes background phylogenetic signal which is caused by the phylogenetic relationships rather than by true coevolutionary relationships. Accounting for phylogenetic linkage is very important and it has been shown that MIp, correctly identifies more coevolving positions in protein families than any other methods.
Using different models of simulated protein evolution it has been observed on the whole that in order to identify the majority of true correlation events, the family should be diverse enough and contain more than 16 sequences13. It should be mentioned that except for one family (Phosphofructokinase, cd00363) which has just 11 sequence members, all these requirements are satisfied by our dataset (the average sequence identity of families in our dataset ranges from 8% to 60%). Moreover the overall family diversity is taken into account upon the MI normalization and Z-score calculation41.
Calculation of evolutionary conservation
Evolutionary conservation at each site in multiple sequence alignment was calculated by weighted sum-of-pairs measure implemented in AL2CO program48 and by maximum likelihood approach implemented in Rate4Site49 program. A maximum likelihood approach (ML) allows one to estimate evolutionary conservation taking into account the topology and branch lengths of the phylogenetic tree and the rate heterogeneity over different sites in a protein family.
RESULTS
Coevolution of specificity determining and other sites
As mentioned in the Introduction, coevolution is one of the various mechanisms which allow proteins to develop new functions and functional specificities. Since coevolved sites are clearly under selection pressure which is more pronounced for functional sites and specificity determining sites, we analyzed the coevolution between these sites 39–41. To analyse the coevolution of subsites we calculated the fraction of coevolution (FC; see Methods for definition) for each experimentally annotated and predicted subsite. Subsites were found to coevolve with up to 13% of other sites and the average fraction of coevolution for them is 1.05%. Although this is a relatively low number, the fraction of coevolution of subsites is statistically significantly higher than coevolution of non-specific sites as indicated by the two-sample t-test with p-value <0.02 (Figure 1a). A similar trend of higher fraction of coevolution for actual and predicted subsites compared to other sites is also observed when other coevolution prediction methods, like OMES43,44, McBASC4,45,46 and MIp47 were applied to our dataset (Figure SM1 in Supplemental data).
Figure 1. Fraction of coevolution (FC) for subsites and functionally important sites (FIS).
Fraction of coevolution is shown on X-axes in bins of FC (in % scale). The relative frequency (Y-axes, in % scale) for sites with a given FC is shown for different site categories: a) the fraction of coevolution of actual subsites (“Subsite”) and non-subsites (“Non_Subsites”); b) FC for top 15 predicted subsites (“Subsites (predicted)”) and FC calculated for all other sites that were not predicted as subsites (“Non_Subsites”); c) FC for FIS extracted from seven CDD families and FC for all other sites (“Non_FIS”); d) similar data as panel c after exclusion of actual subsites from the list of FIS (“FIS (w/o Subsites)”.
Subsites predicted by SPEER35 (we analyzed the top 15 high scoring sites for each family) have an average 3.0% FC (Figure 1b), which points out the ability of SPEER to identify coevolving subsites. The elevated fraction of coevolution for sites predicted by SPEER is consistent with our observation of a higher correlation (Pearson correlation coefficient: 0.54, p-value < 0.0001) between its SPEER score and FC for any given site (Figure SM2 in Supplemental data).
We analyzed the coevolution of sites that have been annotated as being functionally important (FIS, Table SM2 in Supplemental data) for the overall protein family (Figure 1c and 1d) and compared fraction coevolution of those FIS which do not overlap with the subsites with the fractions coevolution of the subsites. FIS show much lower fraction of coevolution (with an average FC of 0.52%) compared to subsites, (the difference is statistically significant with p-value <0.001). This can be explained by their invariability and low capacity to change due to the strong functional constraints, although we observed that 10% of functionally important sites actually do exhibit some coevolution of about 2% FC.
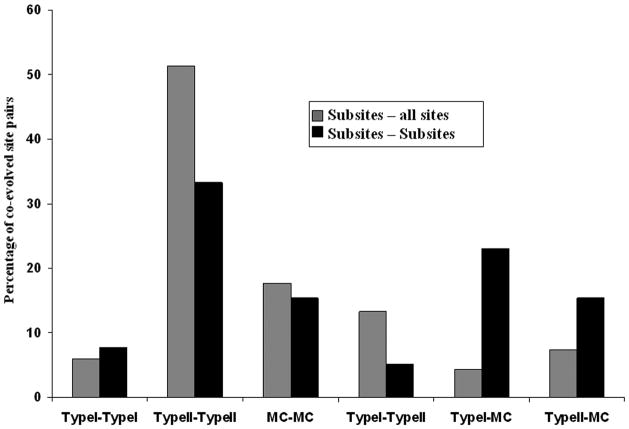
Dissecting subsites into different types shows that different types of subsites (typeI, typeII and marginally conserved (MC)) coevolve with each other to different degree (Figure 2). Indeed, different subsites can be the result of different mechanisms of functional diversification and might exhibit various degrees of conservation and other properties. For example, typeII sites coevolve the most with other typeII sites although they are also largely connected to many other non-subsites. Marginally conserved subsites show considerable coevolution with other types of subsites probably due to their frequent occurrence together with the other site types within one domain family.
Figure 2. Fraction of coevolution between different types of subsites.
All coevolved subsites were categorized into typeI, typeII and MC sites based on their sequence conservation patterns (see Methods) and percentage of each coevolved site pairs from each type category (e.g. typeI-typeI, typeI-typeII) is plotted. Percentage of coevolved pairs is calculated by dividing the number of coevolved subsite-subsite pairs of a given type by the overall number of coevolved subsite-subsite pairs. Black bars show the percent of coevolved site pairs for the actual subsites while grey bars show the percent of coevolved site pairs between the actual subsites and all other sites.
Relation between sites’covariation and evolutionary conservation
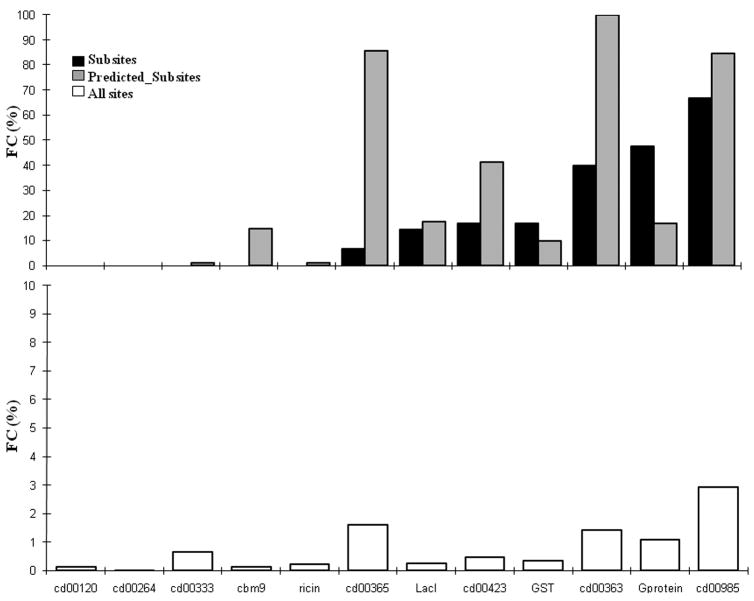
We further examined the FC for the actual and predicted subsites separately for each test family. Among families with the highest FC are the families of cd00985, cd00423, G-protein, cd00363 and GST. At the same time some families (cd00333, cd00120, cd00264 and Ricin) do not exhibit any coevolution for subsites (Figure 3). This encouraged us to investigate the correlation of FC with respect to evolutionary conservation for all coevolved sites. Evolutionary conservation of each coevolved site was calculated using weighted sum-of-pairs measures (AL2CO48 scores) and a probabilistic evolutionary model that takes into account the phylogenetic tree and the rate heterogeneity over different sites (Rate4Site49 scores). Figure 4 shows the mean values of AL2CO48 and Rate4Site49 scores together with the standard error for sites that have relatively low (<= 10 ) and high (>10 ) coevolutionary connections. We found that the sites with higher number of coevolutionary connections have a statistically significant tendency to be more conserved (t-test p-value < 10−8) compared to the sites with the smaller number of connections. These sites might act as ‘hub points’ and therefore changes in these sites would affect many other connected sites. The same trend is also observed between the number of coevolutionary links and evolutionary conservation for specificity determining sites (Figure SM3).
Figure 3. Fraction of coevolution in different families.
Average fraction of coevolution is plotted between a) actual subsites (black bar) and between predicted subsites (grey bar; top 15 predicted sites by SPEER35) and b) all coevolved sites (open bars).
Figure 4. Correlation of evolutionary conservation with the fraction of coevolution for a given site.
Fraction of coevolution for each site is plotted against evolutionary conservation (evolutionary conservation scores were calculated by the a) AL2CO48 and b) Rate4Site49 programs) where higher values indicate higher conservation. For comparison purpose, Rate4Site scores are projected on a reverse scale. Each box shows the mean value and standard error of fraction coevolution for sites with less that 10 (grey box) and more than 10 coevolutionary connections (open box).
Covariation of subsites with respect to spatial and sequence distance
Intuitively, one would expect that coevolving residues should interact with each other in a protein molecule. A few studies addressed this question and showed a certain tendency for residues exhibiting higher covariation to be structurally coupled39,42. We have attempted to go further and evaluate the degree of coevolution of functionally important sites (FIS) and specificity determining sites with respect to their spatial distance between each other. There is a very broad distance distribution for all coevolved pairs with the average spatial distance between coevolved sites in proteins from our dataset being 19.6 Å with an average distance along the sequence of 96 residues (see Figures SM3 and SM4 in Supplemental data for details). It should be noted that structural variability between different structures of the same family is very limited with the average root mean square deviation (RMSD) of less than 2.5Å and loop (dis)similarity metric50 of less than 5Å (with one exception), the average values of structural similarity measures are listed in Table SM1. The distance distributions shown in Figure 5a for coevolved subsites and overall subsite-subsite distances are statistically different (p-value < 0.003). Coevolved subsites show two preferable distance regions: at small distances (less than 10 Å) there are 44% of coevolved subsite pairs compared to 35% of all subsite pairs, whereas at larger distances (more than 20 Å) there are 41% of coevolved subsite pairs compared to only 20% of all subsite pairs. The majority of these distantly located coevolved subsites belong to three families, Gprotein, Phosphofructokinase and LacI, each of which interacts with multiple ligands and/or proteins at different binding sites. Figure 5b also demonstrates a great variability in distance distributions of coevolved subsites for different families.
Figure 5. Spatial distance distribution for all subsites and coevolved subsites.
Panel a) shows spatial distances between all actual subsites (grey bars) and between coevolved subsites (open bars). Panel b) shows average spatial distances (along with standard errors) between all actual subsites (grey bars) and between coevolved actual subsites (open bars) for each protein family in our dataset.
Three dimensional graphical representations of the spatial locations of actual and predicted subsites of three families are provided in Figure 6. It shows a varying degree of coevolutionary connections within actual and top predicted subsites: forming a very dense network of coevolutionary connections (Maf_HamI) in one case and a sparse network in another (LacI/PurR). However, this graphical representation of coevolutionary connections for actual and predicted subsites emphasizes the observation that some sites can covary despite their large spatial distances if they are selected under similar functional constraints. In the LacI/PurR family the specificity of the transcription factors is regulated by binding to small molecules, such as nucleotides. For example, sites such as 221Phe and/or 160Asp in PurR protein (PDB code: 1WET), involved in nucleotide binding seem to coevolve with the DNA binding sites (15Thr and/or 16Thr) that are located on the opposite sides of the protein molecule.
Figure 6. Examples of families with different number of coevolutionary connections among the actual and predicted subfamily specific sites.
Coevolutionary connections between two sites are shown as line while the actual and predicted subsites are outlined as black circles and blue boxes, respectively. Panels A and B show top 15 predicted subsites (including actual subsites), panel C shows top 25 predicted subsites (including actual subsites) which are projected on the representative structures of A) Maf_Ham1 family (cd00985; 2MJP, chain A), B) Pterin binding enzyme family (cd00423; 1AJ0) and C) LacI/PurR family (1WET). Color coding on 3D structures (residues shown as space fill models) indicates the range of SPEER score. Ligands such as ATP analog (panel A), p-aminobenzoic acid (pABA) analog (panel B) and guanine and DNA (panel C) are shown in purple (stick representation).
DISCUSSION
As a result of our analysis we found that subsites more often participate in coevolutionary connections with each other and other sites compared to functionally important sites or protein sites that are not under strong functional constraints. It was also reported earlier that in about half of the studied proteins (13 out of 25) functionally important sites were near coevolving positions42. Unlike functionally important sites which are mostly highly conserved and important for an entire family, specificity determining sites are not globally conserved and subsequent correlated changes at these sites can lead to functional diversification. Interestingly, we found that the networks of coevolutionary connections between the subsites can be quite dense (87 coevolutionary connections between 15 sites for the Maf_Ham1 family; Figure 6A) and subsites with many coevolutionary connections have a tendency to change slower in evolution compared to subsites with a few connections (this trend is also observed for all other sites). These sites might act as ‘hub points’ and therefore, could be responsible for establishing new coevolutionary connections or rewiring the existing finely tuned coevolutionary network.
Similar to previous studies performed on all protein sites39,42, we observed that almost half of all coevolved subsites are located at distances less than 10Å which might be the result of coevolution through compensatory mutations, an important mechanism which shapes the finer details of the specificity in evolution. In addition, we found that forty percent of coevolved subsites are located at distances of more than 20 Å, the effect which is most pronounced for three families of Gprotein, Phosphofructokinase and LacI/PurR. This observation points to the possibility of allosteric mechanisms other than preservation of the stereochemistry of residue contacts. Indeed, several other studies also showed that the residues which are important for the specificity of binding are not necessarily located within a distance of direct interaction with the ligand or other protein51–63, although statistical coupling inferred from multiple sequence alignments is not necessarily a true reporter of allosteric changes64. Another explanation of long-range coevolutionary coupling between subsites comes from the consideration that many proteins function as homooligomers and sites, which are located far from each other in a monomer, can form contacts between the monomers at non-symmetrical homodimer interface.
Supplementary Material
Acknowledgments
We thank one of the anonymous reviewers for the interesting comment about the possible effect of homodimerization on long-range coupling. This work was supported by the Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS.
References
- 1.Pollock DD, Taylor WR. Effectiveness of correlation analysis in identifying protein residues undergoing correlated evolution. Protein Eng. 1997;10:647–657. doi: 10.1093/protein/10.6.647. [DOI] [PubMed] [Google Scholar]
- 2.Pollock DD, Taylor WR, Goldman N. Coevolving protein residues: maximum likelihood identification and relationship to structure. J Mol Biol. 1999;287:187–198. doi: 10.1006/jmbi.1998.2601. [DOI] [PubMed] [Google Scholar]
- 3.Yanofsky C, Horn V, Thorpe D. Protein Structure Relationships Revealed by Mutational Analysis. Science. 1964;146:1593–1594. doi: 10.1126/science.146.3651.1593. [DOI] [PubMed] [Google Scholar]
- 4.Gobel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 1994;18:309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 5.Neher E. How frequent are correlated changes in families of protein sequences? Proc Natl Acad Sci U S A. 1994;91:98–102. doi: 10.1073/pnas.91.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shindyalov IN, Kolchanov NA, Sander C. Can three-dimensional contacts in protein structures be predicted by analysis of correlated mutations? Protein Eng. 1994;7:349–358. doi: 10.1093/protein/7.3.349. [DOI] [PubMed] [Google Scholar]
- 7.Taylor WR, Hatrick K. Compensating changes in protein multiple sequence alignments. Protein Eng. 1994;7:341–348. doi: 10.1093/protein/7.3.341. [DOI] [PubMed] [Google Scholar]
- 8.Chelvanayagam G, Eggenschwiler A, Knecht L, Gonnet GH, Benner SA. An analysis of simultaneous variation in protein structures. Protein Eng. 1997;10:307–316. doi: 10.1093/protein/10.4.307. [DOI] [PubMed] [Google Scholar]
- 9.Pazos F, Helmer-Citterich M, Ausiello G, Valencia A. Correlated mutations contain information about protein-protein interaction. J Mol Biol. 1997;271:511–523. doi: 10.1006/jmbi.1997.1198. [DOI] [PubMed] [Google Scholar]
- 10.Fukami-Kobayashi K, Schreiber DR, Benner SA. Detecting compensatory covariation signals in protein evolution using reconstructed ancestral sequences. J Mol Biol. 2002;319:729–743. doi: 10.1016/S0022-2836(02)00239-5. [DOI] [PubMed] [Google Scholar]
- 11.Govindarajan S, Ness JE, Kim S, Mundorff EC, Minshull J, Gustafsson C. Systematic variation of amino acid substitutions for stringent assessment of pairwise covariation. J Mol Biol. 2003;328:1061–1069. doi: 10.1016/s0022-2836(03)00357-7. [DOI] [PubMed] [Google Scholar]
- 12.Korber BT, Farber RM, Wolpert DH, Lapedes AS. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc Natl Acad Sci U S A. 1993;90:7176–7180. doi: 10.1073/pnas.90.15.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard L, Bladon P, Mitchell JMO, Dufton MJ. Evaluation of a novel method for the identification of coevolving protein residues. Protein Eng. 2001;14:549–555. doi: 10.1093/protein/14.8.549. [DOI] [PubMed] [Google Scholar]
- 14.Tuff P, Darlu P. Exploring a phylogenetic approach for the detection of correlated substitutions in proteins. Mol Biol Evol. 2000;17:1753–1759. doi: 10.1093/oxfordjournals.molbev.a026273. [DOI] [PubMed] [Google Scholar]
- 15.Dutheil J, Pupko T, Jean-Marie A, Galtier N. A model-based approach for detecting coevolving positions in a molecule. Mol Biol Evol. 2005;22:1919–1928. doi: 10.1093/molbev/msi183. [DOI] [PubMed] [Google Scholar]
- 16.Valencia A, Pazos F. Computational methods for the prediction of protein interactions. Curr Opin Struct Biol. 2002;12:368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 17.Kundrotas PJ, Alexov EG. Predicting residue contacts using pragmatic correlated mutations method: reducing the false positives. BMC Bioinformatics. 2006;7:503. doi: 10.1186/1471-2105-7-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh CS, Bogan AA, Joachimiak M, Walther D, Cohen FE. Co-evolution of proteins with their interaction partners. J Mol Biol. 2000;299:283–293. doi: 10.1006/jmbi.2000.3732. [DOI] [PubMed] [Google Scholar]
- 19.Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- 20.Jothi R, Cherukuri PH, Tasneem A, Przytycka TM. Co-evolutionary analysis of domains in interacting proteins reveals insights into domain-domain interactions mediating protein-protein interactions. J Mol Biol. 2006;362:861–875. doi: 10.1016/j.jmb.2006.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atchley WR, Wollenberg KR, Fitch WM, Terhalle W, Dress AW. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol Biol Evol. 2000;17:164–178. doi: 10.1093/oxfordjournals.molbev.a026229. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira L, Paiva AC, Vriend G. Correlated mutation analyses on very large sequence families. Chembiochem. 2002;3:1010–1017. doi: 10.1002/1439-7633(20021004)3:10<1010::AID-CBIC1010>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 23.Tillier ER, Lui TW. Using multiple interdependency to separate functional from phylogenetic correlations in protein alignments. Bioinformatics. 2003;19:750–755. doi: 10.1093/bioinformatics/btg072. [DOI] [PubMed] [Google Scholar]
- 24.Wollenberg KR, Atchley WR. Separation of phylogenetic and functional associations in biological sequences by using the parametric bootstrap. Proc Natl Acad Sci U S A. 2000;97:3288–3291. doi: 10.1073/pnas.070154797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohno S. Evolution by gene duplications. Berlin: Springer-Verlag; 1970. [Google Scholar]
- 26.Doolittle RF. Similar amino acid sequences: chance or common ancestry? Science. 1981;214:149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- 27.Gu X. Statistical Methods for Testing Functional Divergence after Gene Duplication. Mol Biol Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- 28.Hannenhalli SS, Russell RB. Analysis and prediction of functional sub-types from protein sequence alignments. J Mol Biol. 2000;303:61–76. doi: 10.1006/jmbi.2000.4036. [DOI] [PubMed] [Google Scholar]
- 29.Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J Mol Biol. 2002;321:7–20. doi: 10.1016/s0022-2836(02)00587-9. [DOI] [PubMed] [Google Scholar]
- 30.Kalinina OV, Novichkov PS, Mironov AA, Gelfand MS, Rakhmaninova AB. SDPpred: a tool for prediction of amino acid residues that determine differences in functional specificity of homologous proteins. Nucleic Acids Res. 2004;32:W424–428. doi: 10.1093/nar/gkh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirovano W, Feenstra KA, Heringa J. Sequence comparison by sequence harmony identifies subtype-specific functional sites. Nucleic Acids Res. 2006;34:6540–6548. doi: 10.1093/nar/gkl901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei J, Cai W, Kinch LN, Grishin NV. Prediction of functional specificity determinants from protein sequences using log-likelihood ratios. Bioinformatics. 2006;22:164–171. doi: 10.1093/bioinformatics/bti766. [DOI] [PubMed] [Google Scholar]
- 33.Capra J, Singh M. Characterization and Prediction of Residues Determining Protein Functional Specificity. Bioinformatics. 2008 May 1; doi: 10.1093/bioinformatics/btn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8:R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakrabarti S, Bryant SH, Panchenko AR. Functional specificity lies within the properties and evolutionary changes of amino acids. J Mol Biol. 2007;373:801–810. doi: 10.1016/j.jmb.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchler-Bauer A, Anderson JB, Derbyshire MK, Deweese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, Flippen-Anderson JL, Ionides J, Kamada C, Krissinel E, et al. Remediation of the protein data bank archive. Nucleic Acids Res. 2008;36:D426–433. doi: 10.1093/nar/gkm937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cover TA, Thomas JA. Elements of Information Theory. John Wiley and Sons; New York: 1991. [Google Scholar]
- 39.Gloor GB, Martin LC, Wahl LM, Dunn SD. Mutual information in protein multiple sequence alignments reveals two classes of coevolving positions. Biochemistry. 2005;44:7156–7165. doi: 10.1021/bi050293e. [DOI] [PubMed] [Google Scholar]
- 40.Martin LC, Gloor GB, Dunn SD, Wahl LM. Using information theory to search for co-evolving residues in proteins. Bioinformatics. 2005;21:4116–4124. doi: 10.1093/bioinformatics/bti671. [DOI] [PubMed] [Google Scholar]
- 41.Rockwell NC, Lagarias JC. Flexible mapping of homology onto structure with Homolmapper. BMC Bioinformatics. 2007;8:123–137. doi: 10.1186/1471-2105-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeang CH, Haussler D. Detecting coevolution in and among protein domains. PLoS Comput Biol. 2007;3:e211. doi: 10.1371/journal.pcbi.0030211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larson SM, Di Nardo AA, Davidson AR. Analysis of covariation in an SH3 domain sequence alignment: applications in tertiary contact prediction and the design of compensating hydrophobic core substitutions. J Mol Biol. 2000;303:433–446. doi: 10.1006/jmbi.2000.4146. [DOI] [PubMed] [Google Scholar]
- 44.Kass I, Horovitz A. Mapping pathways of allosteric communication in GroEL by analysis of correlated mutations. Proteins. 2002;48:611–617. doi: 10.1002/prot.10180. [DOI] [PubMed] [Google Scholar]
- 45.Olmea O, Rost B, Valencia A. Effective use of sequence correlation and conservation in fold recognition. J Mol Biol. 1999;293:1221–1239. doi: 10.1006/jmbi.1999.3208. [DOI] [PubMed] [Google Scholar]
- 46.Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- 47.Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- 48.Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;8:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 49.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl 1):S71–77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 50.Panchenko AR, Madej T. Analysis of protein homology by assessing the (dis)similarity in protein loop regions. Proteins. 2004;57:539–547. doi: 10.1002/prot.20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatley ME, Lockless SW, Gibson SK, Gilman AG, Ranganathan R. Allosteric determinants in guanine nucleotide-binding proteins. Proc Natl Acad Sci U S A. 2003;100:14445–14450. doi: 10.1073/pnas.1835919100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yano T, Oue S, Kagamiyama H. Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc Natl Acad Sci U S A. 1998;95:5511–5515. doi: 10.1073/pnas.95.10.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nettles KW, Sun J, Radek JT, Sheng S, Rodriguez AL, Katzenellenbogen JA, Katzenellenbogen BS, Greene GL. Allosteric control of ligand selectivity between estrogen receptors alpha and beta: implications for other nuclear receptors. Mol Cell. 2004;13:317–327. doi: 10.1016/s1097-2765(04)00054-1. [DOI] [PubMed] [Google Scholar]
- 54.Chen Z, Zhao H. Rapid creation of a novel protein functions by in vitro coevolution. J Mol Biol. 2005;348:1273–1282. doi: 10.1016/j.jmb.2005.02.070. [DOI] [PubMed] [Google Scholar]
- 55.Halperin I, Wolfson H, Nussinov R. Correlated mutations: advances and limitations. A study on fusion proteins and on the Cohesin-Dockerin families. Proteins. 2006;63:832–845. doi: 10.1002/prot.20933. [DOI] [PubMed] [Google Scholar]
- 56.Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 57.Gouldson PR, Dean MK, Snell CR, Bywater RP, Gkoutos G, Reynolds CA. Lipid-facing correlated mutations and dimerization in G-protein coupled receptors. Protein Eng. 2001;14:759–767. doi: 10.1093/protein/14.10.759. [DOI] [PubMed] [Google Scholar]
- 58.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 59.Buck MJ, Atchley WR. Networks of coevolving sites in structural and functional domains of serpin proteins. Mol Biol Evol. 2005;22:1627–1634. doi: 10.1093/molbev/msi157. [DOI] [PubMed] [Google Scholar]
- 60.Daily MD, Upadhyaya TJ, Gray JJ. Contact rearrangements form coupled networks from local motions in allosteric proteins. Proteins. 2008;71:455–466. doi: 10.1002/prot.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flynn TC, Swint-Kruse L, Kong Y, Booth C, Matthews KS, Ma J. Allosteric transition pathways in the lactose repressor protein core domains: asymmetric motions in a homodimer. Protein Sci. 2003;12:2523–2541. doi: 10.1110/ps.03188303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taraban M, Zhan H, Whitten AE, Langley DB, Matthews KS, Swint-Kruse L, Trewhella J. Ligand-induced conformational changes and conformational dynamics in the solution structure of the lactose repressor protein. J Mol Biol. 2008;376:466–481. doi: 10.1016/j.jmb.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 64.Chi CN, Elfström L, Shi Y, Snäll T, Engström A, Jemth P. Reassessing a sparse energetic network within a single protein domain. Proc Natl Acad Sci U S A. 2008;105:4679–4684. doi: 10.1073/pnas.0711732105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.