Abstract
Proline utilization A (PutA) is a membrane associated multifunctional enzyme that catalyzes the oxidation of proline to glutamate in a two step process. In certain Gram-negative bacteria such as Pseudomonas putida, PutA also acts as an auto repressor in the cytoplasm when an insufficient concentration of proline is available. Here the N-terminal residues 1–45 of PutA from P. putida (PpPutA45), are shown to be responsible for DNA binding and dimerization. The solution structure of PpPutA45 was determined using NMR methods, where the protein is shown to be a symmetrical homodimer (12 kDa) consisting of two ribbon-helix-helix (RHH) structures. DNA sequence recognition by PpPutA45 was determined using DNA gel mobility shift assays and NMR chemical shift perturbations. PpPutA45 was shown to bind a 14 base-pair DNA oligomer (5′-GCGGTTGCACCTTT-3′). A model of the PpPutA45-DNA oligomer complex was generated using Haddock 2.1. The antiparallel β-sheet that results from PpPutA45 dimerization serves as the DNA recognition binding site by inserting into the DNA major groove. The dimeric core of four α-helices provides a structural scaffold for the β-sheet from which residues Thr5, Gly7, and Lys9 make sequence specific contacts with the DNA. The structural model implies flexibility of Lys9 which can either make hydrogen bond contacts with guanine or thymine. The high sequence and structure conservation of the PutA RHH domain suggest interdomain interactions play an important role in the evolution of the protein.
Keywords: Pseudomonas putida, NMR solution structure, PutA, ribbon-helix-helix (RHH) structures, PutA-DNA complex
INTRODUCTION
Proline utilization A (PutA) is a multifunctional enzyme that allows Gram-negative bacteria, such as Escherichia coli and Pseudomonas putida, to utilize proline as a carbon and nitrogen source.1,2 PutA converts proline into glutamate in a two-step process that requires a proline dehydrogenase domain (PRODH) and a Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase domain (P5CDH).3 PRODH and P5CDH are separate enzymes in Gram-positive bacteria, archea, and eukaryotes.4 Proline is first oxidized to P5C coupled with the reduction of the FAD cofactor by the PutA PRODH domain.1,5,6 The reduced FADH2 cofactor transfers the electrons to the electron transport chain system in the cytoplasmic membrane.7,8 The P5CDH domain then catalyzes the NAD+-dependant oxidation of P5C to glutamate.1,5,9
PutA also functions as an autogenous transcriptional repressor10 by binding to multiple sites in the put regulatory region.11 Because the enzymatic activity of PutA requires PutA to be peripherally membrane bound, PutA function is regulated by proline-dependent switching of its intracellular location from the cytoplasm to the membrane.3,12 PutA can bind to the put control DNA in the absence (KD ~45 nM) and presence (KD~ 100 nM) of proline suggesting changes in PutA-DNA binding affinity are not a major factor in functional switching.9 Rather, proline reduction of the FAD cofactor activates tight PutA-lipid binding (KD < 0.01nM) leading to sequestration of PutA on the membrane.7,9 The tight membrane interaction thus prevents PutA from binding DNA. Previous studies have shown redox-dependent conformational changes occur in a linker region between the DNA binding and PRODH domains. Thus, a coupled conformational change involving the PRODH and DNA binding domains may be part of the redox mechanism by which PutA transfers from the cytoplasm to the membrane.13 The PutA DNA binding domain in E. coli is localized to the N-terminal 47 amino acids and is separated from the PRODH domain (261–612) by a flexible domain of unknown function (residues 141–260).6,14 This flexible domain incurs a significant conformational change upon proline binding,9 where membrane association of PutA is primarily driven by a hydrophobic interaction.7
In PutA from E. coli, the DNA binding domain was shown to form a ribbon-helix-helix (RHH) fold, consisting of a β–stand formed by residues 3–11 (β1) followed by two α–helices formed by residues 12–25 (αI) and 29–46 (αII).6,15 The functional unit of the RHH domain is a dimer,1 in which the β-strands form an antiparallel β-sheet in the dimer. The RHH domain is predicted to be conserved among organisms where PutA has transcriptional regulatory activities.16 Sequence specific DNA binding by β-sheet residues is a defining characteristic of the RHH superfamily. The RHH β-sheet is positioned into the DNA major groove to enable specific interactions with the unique operator sequence.16,17 E. coli PutA binds to five binding sites in the put regulatory region, which all contain a GTTGCA sequence.18 This GTTGCA sequence has been identified in the put regulatory regions in all organisms where PutA has a demonstrated regulatory function.18 A site predicted to have optimal binding affinity was selected for our NMR studies.18
P. putida is a non-pathogenic organism with utility in bioremediation because of its metabolic diversity and ability to metabolize a wide-range of carbon sources.19,20 PutA from P. putida is a 1315 amino acid polypeptide and functions as a transcriptional repressor of the put regulon in a manner similar to E. coli PutA.2 Thus, the DNA binding domain of P. putida PutA is predicted to contain an RHH fold and bind to put regulatory regions similar to E. coli PutA. In order to better understand the structural basis of the DNA binding interaction of PutA and the evolutionary conservation of PutA structures among Gram-negative bacteria, we have determined the solution structure of the dimeric DNA binding domain of PutA from P. putida corresponding to residues 1 to 45 (PpPutA45) and its complex to a 14 base-pair DNA oligomer using heteronuclear triple resonance NMR methods.
MATERIALS AND METHODS
Protein Purification
The N-terminal 45 residues of PutA from Pseudomonas putida (PpPutA45) were amplified by PCR using primers 5′-GCAGCCATATGGCGACTACCACC-3′ and 5′-CCACCCTCGAGCTTCTCCAGG-3′ that incorporated the NdeI and XhoI restriction endonuclease sites, respectively. The PCR product was then cloned into the vector pET23b using NdeI and XhoI. The resulting construct, PpPutA45-pET23b, was verified by nucleic acid sequencing.
Uniformly 13C, 15N-enriched PpPutA45 was expressed as a C-terminal 6xHis tag fusion protein (6 kDa monomer) from PpPutA45-pET23b in E. coli strain BL21 DE3 pLysS in M9 minimal medium containing (15NH4)2SO4 and U-13C-glucose as sole nitrogen and carbon sources. Initial growth was carried out at 37 °C until the OD600 of the culture reached about 0.8. The incubation temperature was then decreased to 20 °C and protein expression was induced by the addition of IPTG (isopropyl-β-D-thiogalactopyranoside) at a final concentration of 0.5 mM. Following overnight incubation at 20 °C, cells were harvested by centrifugation at 7500 × g for 10 min. The resuspended cells were broken by sonication on ice using a pulse sequence of 15 sec on and 45 sec off (5 min total pulse time). The broken cell debris was removed by centrifugation at 34000 × g for 40 min at 4 °C. The cell free extract was then applied to a Ni2+ NTA affinity column and PpPutA45 was eluted by standard methods. After purification, PpPutA45 was dialyzed into a 50 mM phosphate buffer at pH 6.2 with 0.2 M NaCl. The 6xHis tag was retained for subsequent experiments. The concentration of the PpPutA45 was determined using the BCA method (Pierce) with bovine serum albumin as the standard and spectrophotometrically using a newly estimated molar extinction coefficient at 280 nm for PpPutA45 (4690 M−1 cm−1). The purity of the protein is above 99% as judged by SDS-PAGE analysis. The C-terminal 6xHis tag has been shown previously not to interfere with PutA-DNA binding.15 The dissociation constants for the EcPutA52-DNA and PpPutA45-DNA (P. putida put control DNA) complexes range from 10–30 nM, respectively, for the non-His tagged and C-terminal 6xHis tagged proteins (data not shown).
Analytical ultracentrifugation
The oligomeric state of PpPutA45 was measured at 20 °C by analytical ultracentrifugation using an Optima XL-I analytical ultracentrifuge (Beckerman Coulter, Inc., Fullerton CA) equipped with an eight-hole An50 Ti rotor. PpPutA45 was dialyzed against 50 mM sodium phosphate buffer (200 mM NaCl, pH 6.2) overnight and passed through a 0.22 μm filter to remove any large aggregates prior to loading. Three loading concentrations of 0.1 mg/ml, 0.3 mg/ml and 0.9 mg/ml in 110 μl were used and the reference cells were filled with 125 μl of the dialyzate. Radial scans at 280 nm were recorded at 22 h, 24 h, 26 h and 28 h at 25,000 RPM and represent the average of ten measurements at each radial position with a spacing of 0.001 cm. Data were fit by global analysis to a self-association model using SEDFIT.21 The partial specific volume of PpPutA45 used for best-fit analysis of the data was 0.736 calculated from SedTerp.
NMR data analysis and P. putida PutA45 structure calculation
All NMR spectra were recorded at 25 °C on a Bruker 600 MHz NMR system. Spectra were processed using the NMRPipe software package22 and analyzed with PIPP.23 The assignments of 1H, 15N, and 13C resonances were determined based on the following standard experiments: HSQC, HNCO, HNCA, CBCA(CO)NH, CBCANH, C(CO)NH, HC(CO)NH, HCCH-COSY, and 3D 13C-edited NOESY.24
NOE assignments were obtained by using 3D 15N-edited NOESY and 3D 13C-edited NOESY. NOE intensities were sorted visually into four classes: strong (1.8–2.5), medium (1.8–3.0), weak (1.8–4.0), very weak (3.0–5.0). Upper distance limits for distances involving methyl protons and nonstereospecifically assigned methylene protons were corrected appropriately for center averaging.25 Distance constraints from intramolecular NOEs were assigned to both the A and B chains in the PpPutA45 dimer structure. NOEs between residues predicted to be involved in the dimer interface from a PpPutA45 homology model were initially assigned as both intra- and intermolecular NOEs. This maintained a symmetric PpPutA45dimer structure at the early stages of refinement when a minimal number of distance constraints were identified. Constraints that exhibited high violations (> 0.5 Å) in the PpPutA45 monomer structure were then only assigned as intermolecular NOEs. Similarly, constraints that exhibited high violations (> 0.5 Å) between the monomer structures were then only assigned as intramolecular NOEs.
Torsion angle constraints were obtained by chemical shift analysis using the TALOS26 software program, and measured coupling constants from an HNHA experiment. The ranges for the ψ and ϕ dihedral angles were ± 30° and ± 50°, respectively.
Hydrogen bond constraints were determined using the (CLEANEX-PM)-FHSQC experiment.27 A total of 2048 data points were collected in the 1H dimension and 66 data points were collected in the 15N dimension. The spectrum was collected with 256 transients and a sweep width of 8012.82 Hz in the 1H dimension and 1613.424 Hz in the 15N dimension. The mixing time was set to 100 ms with a CLEANEX spinlock power of 2 KHz. The (CLEANEX-PM)-FHSQC spectrum was compared with the 2D 1H-15N HSQC spectrum, where amides with missing peaks were assigned hydrogen bond constraints. These residues were selected because the (CLEANEX-PM)-FHSQC spectrum identifies amide residues with fast water exchange rates. The hydrogen bond distance constraints were set at 2.8 Å between the carboxyl oxygen and the amide nitrogen, and 1.8 Å between the carboxyl oxygen and the amide proton. Carboxyl groups within 2.5 Å of the slowly exchanging amide groups were selected to be involved in a hydrogen bond.
A symmetric PpPutA45 dimer structure was obtained by using the non-crystallographic symmetry (NCS) function in XPLOR-NIH. NCS is an energy term that maintains a small root-mean square difference (RMSD) for the superposition of equivalent atoms from each monomer within the PpPutA45 dimer structure. All distances (including dimeric interactions) and torsion angle constraints were used as NCS constraints, with a 500 kcal/mol force constant and a RMSD limit of 0.2 Å.
A total of 100 structures were calculated using XPLOR-NIH software.28 30 of the lowest energy structures were further refined using the hybrid distance geometry dynamical-simulated annealing method29 with minor modifications30 using the program XPLOR-NIH28 adapted to incorporate pseudopotentials for 3J(HN-Hα) coupling constants,31 secondary 13Cα/13Cβ chemical shift constraints,32 radius of gyration33 and a conformational database potential.34–36 The 30 lowest energy structures were then subjected to further energy minimization with CNS using explicit water solvation that included Lennard-Jones and electrostatic potentials using a modification of the procedure and forcefield of Nilges.37 An average PpPutA45 structure was calculated from these 30 structures.
The target function that is minimized during restrained minimization and simulated annealing comprises quadratic harmonic terms for covalent geometry, 3J(HN-Hα) coupling constants, and secondary 13Cα/13Cβ chemical shift constraints, square-well quadratic potentials for the experimental distance, radius of gyration and torsion angle constraints, and a quartic van der Waals term for nonbonded contacts. The radius of gyration can be predicted with reasonable accuracy on the basis of the number of residues using a relationship determined empirically from the analysis of high resolution x-ray structures.33 The force constant for the conformational database and radius of gyration potentials were kept relatively low (0.5–1.0 kcal/mol) throughout the simulation to allow the experimental distance and torsion angle constraints to predominately influence the resulting structures. The force constant for the NOE and dihedral constraints were 30 times and 10 times stronger than the force constants used for the conformational database and radius of gyration potentials, respectively38. All peptide bonds were constrained to be planar and trans. There were no hydrogen-bonding, electrostatic, or 6–12 Lennard-Jones empirical potential energy terms in the target function.
Gel-shift assays
Gel-shift assays using fluorescently labeled put intergenic DNA were performed to test PpPutA45 binding to put control DNA from P. putida and E. coli. DNA was fluorescently labeled with IRdye-700 (LI-COR, Inc.) by PCR as previously described.6 IRdye-700 labeled put intergenic DNA (2 nM) from P. putida (PpPutC) or E. coli (EcPutC) were incubated with PpPutA45 (0–500 nM) in a total volume of 25 μl in 50 mM Tris, pH 7.5, containing 10% glycerol for 20 min at 20 °C before electrophoresis. Calf thymus competitor DNA (100 μg/ml) was added to the binding mixtures to prevent nonspecific PpPutA45-DNA interactions. The PpPutA45-DNA complexes were separated using a nondenaturing (8%) polyacrylamide gel at 4° C. The gels were visualized using a LI-COR Odyssey Imager.
Synthetic oligonucleotides of 12-bp (O2-12, 5′-GCGGTTGCACCT-3′) and 14-bp (O2-14, 5′-GCGGTTGCACCTTT-3′) were purchased from Integrated DNA Technologies. Both oligonucleotides contain the core sequence element GTTGCA and vary only in the length of the flanking sequence. Duplex DNA of each oligonucleotide was prepared by annealing the complementary oligonucleotides in 10 mM Tris buffer (pH 8.0, 50 mM NaCl, 1 mM EDTA) by first heating at 95 °C for 5 min then gradually cooling down the oligonucleotide mixture to room temperature. For gel mobility shift assays, double stranded DNA oligomers (100 nM) were incubated with PpPutA45 (0 – 400 nM) in 20 mM potassium phosphate buffer (pH 7.4, 100 mM NaCl) at 20 °C for 20 min before electrophoresis. The mixtures were then separated using a nondenaturing (8%) polyacrylamide gel at 4 °C. DNA were then stained with ethidium bromide and visualized using Bio-Rad Quantity One.
Protein-DNA Docking
Titration analysis was carried out with 85 μM PpPutA45 in a 50 mM phosphate buffer solution (pH 6.2) with 200 mM NaCl. The 14 base-pair double-stranded DNA oligomer (O2-14) was titrated into the protein solution using a 873 μM stock solution to obtain a final 1:1 molar ratio of O2-14 to PpPutA45. A 2D 1H-15N TROSY experiment was acquired for both the free PpPutA45 and the PpPutA45:O2-14 complex samples using a Bruker AVANCE 800 MHz NMR equipped with a triple-resonance Z-axis gradient cryoprobe. The magnitudes of the chemical shift perturbations (CSPs) were calculated using a common weighting approach:
| (1) |
where δN and δH represents the changes in 15N and 1H chemical shifts upon ligand binding, respectively.39 A model of the PpPutA45 molecular surface was created using VMD-XPLOR 1.8.5,40 where the surface is colored by the magnitude of the observed chemical shift changes scaled from 0 (white) to 10 (blue).
The PpPutA45:O2-14 complex was modeled using the high ambiguity driven biomolecular docking system (Haddock), which uses chemical shift perturbations to create ambiguous interaction constraints to calculate a co-structure.41 PpPutA45 residues that exhibited a missing NH peak or a > 0.05 ppm chemical shift difference in the PpPutA45:02-14 2D 1H-15N TROSY spectrum were defined as active in the Haddock rigid-body docking of PpPutA45 to the O2-14 DNA oligomer. This included PpPutA45 residues that disappeared (e.g., Lys34) or exhibited multiple peaks (e.g., His30) upon the addition of the O2-14 DNA oligomer. Other solvent accessible PpPutA45 residues, with higher than average chemical shift perturbations (> 0.03 ppm), were selected as passive in the Haddock rigid-body docking. A canonical B-DNA structure for the O2-14 sequence was constructed with the DNA model program model.it.42 All nucleotides of O2-14 sequence were selected as active residues in the Haddock docking.
A total of 2000 structures for the PpPutA45:O2-14 complex were generated by Haddock 2.1 using rigid body energy minimization. Sampling of 180° degree rotated solutions was used during the rigid body docking. The 200 lowest energy rigid body structures were further refined by semi-flexible refinement, where PpPutA45 side chains of active and passive residues were allowed to move. Similarly, all the nucleotides of the O2-14 DNA, except for the terminal bases, were allowed to move in the semi-flexibility stage. The 200 structures were then further refined using explicit solvent, an 8 Å layer of H2O. The resulting 200 structures were clustered using the backbone RMSD for both the protein and DNA structures with a cutoff of 4.5 Å. The 10 lowest energy structures from the lowest energy cluster was selected, averaged, and further analyzed.
Electrostatic potential calculation
Electrostatic potentials for the PpPutA45 structure were calculated and visualized in GRASP.43 The surface-mapped potential is graded from −5 kT/e (red) to 5 kT/e (blue). Electrostatic potentials for the PpPutA45-DNA models were calculated using the software program Gemstone (http://gemstone.mozdev.org). The structures were visualized using VMD-XPLOR with the APBS 1.1.44
Similarity Search and Phylogenetic Mapping
The sequence alignment of the 8 RHH DNA binding domains was obtained by using PSI-BLAST (http://www.ebi.ac.uk)45 and the E. coli sequence as the template. The phylogenetic trees for the 66 known RHH DNA binding domains and PRODH domains was generated using CLUSTALW.46
RESULTS AND DISCUSSION
P. putida PutA45 is a dimer in solution
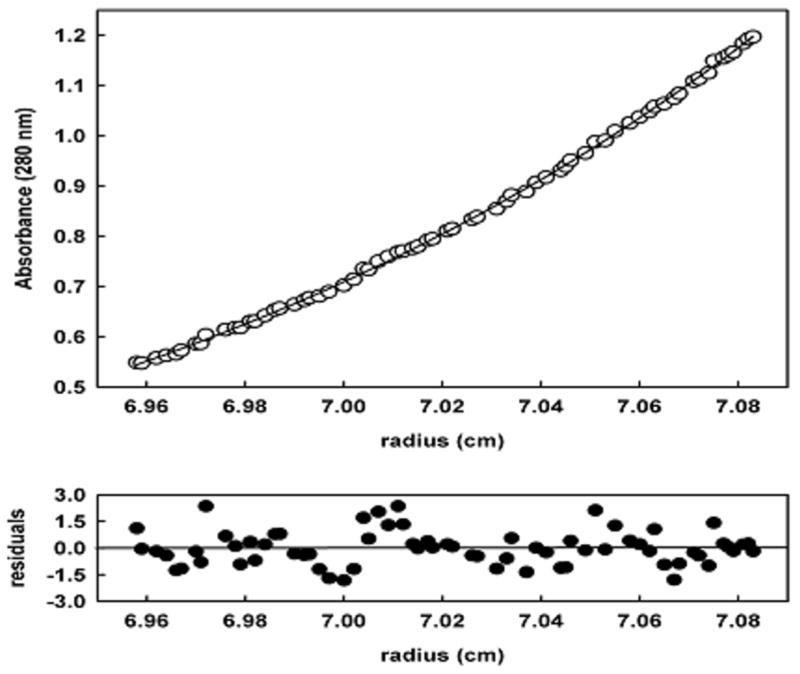
The oligomeric state of PpPutA45 was determined by analytical centrifugation using conditions similar to the NMR studies. Sedimentation velocity experiments verified that a dimeric species of PpPutA45 was present in our NMR sample. Sedimentation equilibrium was then performed with three different concentrations of PpPutA45. A molecular weight of 12,244 Da was determined for PpPutA45 by a global fitting analysis of the three datasets to a self-association model using SEDFIT.21 The calculated molecular weight from the PpPutA45 sequence is 6,007 Da, thus PpPutA45 is a dimer in solution. Fig. 1 shows the results for a sedimentation equilibrium experiment with PpPutA45 (0.9 mg/ml).
Fig. 1.
Sedimentation analysis of PpPutA45 (0.9 mg/ml). PpPutA45 was centrifuged at 25000 rpm for up to 28 h at 20 °C. The solid line through the points is the weighted nonlinear least-squares fit for a single species of 12.0 kDa.
Extent of PpPutA45 NMR assignments and secondary structure analysis
The 2D 1H-15N HSQC and other NMR spectra indicate PpPutA45 is a symmetric homodimer since a single-set of NMR assignments were observed. Essentially complete backbone and side-chain assignments were obtained for PpPutA45, where 41 out of 43 non-proline amide residues were assigned. The N-terminal residues M1 and T3 were not observed in the 2D 1H-15N HSQC spectrum. 44 out of 45 Cα, 44 out of 45 Cβ, and 40 out of 45 C′ were assigned for PpPutA45 where M1 and the C-terminal 6xHis tag were unassigned. The side-chain assignments are complete except for R27 and P29 which are located in a loop region and were only partially assigned. The PpPutA45 NMR structure consists of two α-helical regions located at residues 12–24 (αI) and 30–45 (αII). The secondary structure of PpPutA45 was determined using Cα/Cβ secondary shifts, NOE data involving HN, Hα, Hβ, and 3J(HN-Hα) coupling constants.47,48 The observed secondary structure is consistent with the conformation of the RHH family, further confirming the accuracy of the NMR assignments. All available backbone and side chain chemical shift assignments are deposited in the BioMagResBank Database (accession code 7082).49
PpPutA45 NMR structure calculation and analysis
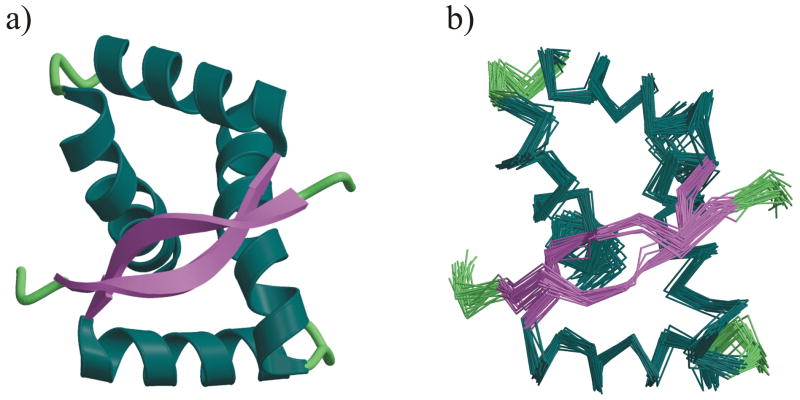
The PpPutA45 solution structure was calculated using a total of 1410 distance constraints, 167 backbone dihedral angle constraints, 50 3J(HN-Hα) coupling constant constraints, and 162 13Cα/13Cβ chemical shift constraints, and a radius of gyration of 8.84 Å estimated from the PpPutA45 sequence length.33 A total of 100 structures were calculated for PpPutA45, where the 30 lowest energy structures were selected for further analysis. The resulting PpPutA45 structures are consistent with the NMR data as evident by the low rms deviations from experimental distance, dihedral, 13Cα/13Cβ chemical shift and 3J(HN-Hα) coupling constant constraints. Also there are no distance violations > 0.5 Å or dihedral angle violations > 5°. The average root-mean square deviation (RMSD) of the 30 lowest energy structures about the mean coordinate positions is 0.62 ± 0.17 Å for all backbone atoms and 1.20 ± 0.19 Å for all heavy atoms (Fig. 2b). The final restrained minimized average structure of PpPutA45 has an RMSD about the mean coordinate positions of 0.19 Å for all backbone atoms and 0.49 Å for all heavy atoms. The structural statistics and analysis of the results are listed in Table I.
Fig. 2.
a) Ribbon diagram of the restrained minimized average NMR structure of the homodimeric P. putida PutA45 protein colored by secondary structure. b) Superposition of the backbone (N,C,C′) atoms of the 30 lowest energy NMR structures calculated for PpPutA45. The figures were generated with MOLSCRIPT 57 and rendered with Raster3D 58.
Table I.
Structural Statistics and Atomic rms Differencesa
| A. Structural Statistics | ||||
|---|---|---|---|---|
| <SA> | ( )r | |||
| rms deviations from experimental distance restraints (Å) | ||||
| all (1410) | 0.032 ± 0.035 | 0.022 | ||
| interresidue sequential (|i−j| = 1) (524) | 0.029 ± 0.005 | 0.024 | ||
| interresidue short range (1 <|i−j| # 5) (662) | 0.023 ± 0.003 | 0.023 | ||
| interresidue long-range (|i−j| > 5) (124) | 0.011 ± 0.008 | 0.0 | ||
| H-bonds (100)b | 0.031 ± 0.006 | 0.029 | ||
| rms deviation from exptl dihedral restraints (deg) (167)c,d | 0.184 ± 0.160 | 0.301 | ||
| rms deviation from exptl Cα restraints (ppm) (82) | 0.89 ± 0.01 | 0.90 | ||
| rms deviation from exptl Cβ restraints (ppm) (80) | 0.76 ± 0.03 | 0.70 | ||
| rms deviation from 3JNHα restraints (Hz) (50) | 0.87 ± 0.07 | 0.62 | ||
| FNOE (kcal mol−1)d | 50 ± 11 | 39 | ||
| Ftor (kcal mol−1)d | 1.10 ± 0.61 | 0.93 | ||
| Frepel (kcal mol−1)e | −135 ± 22 | −125 | ||
| FL-J (kcal mol−1)f | −3102 ± 111 | −3194 | ||
| deviations from idealized covalent geometry | ||||
| bonds (Å) (1500) | 0.004 ± 0 | 0.005 | ||
| angles (deg) (2730) | 0.666 ± 0.029 | 0.792 | ||
| impropers (deg) (756)g | 0.639 ± 0.034 | 0.666 | ||
| PROCHECKh | ||||
| Overall G-Factor | 0.28 ± 0.04 | 0.24 | ||
| % Residues in most favorable region of Ramachandran plot | 95.1 ± 2.0 | 97.5 | ||
| H-bond energy | 0.95 ± 0.08 | 0.50 | ||
| Number of bad contacts/100 residues | 0.2 ± 0.6 | 0.0 | ||
| B. Atomic rms Differences (Å) | ||||
| Residues 1–45 | secondary structurei | |||
| backbone atoms | all atoms | backbone atoms | all atoms | |
|
| ||||
| <SA> vs SA | 0.62 ± 0.17 | 1.20 ± 0.19 | 0.65 ± 0.18 | 1.20 ± 0.20 |
| <SA> vs (SA)r | 0.65 ± 0.17 | 1.29 ± 0.21 | 0.68 ± 0.19 | 1.29 ± 0.22 |
| (SA)r vs | 0.19 | 0.49 | 0.22 | 0.48 |
The notation of the structures is as follows: <SA> are the final 30 simulated annealing structures; is the mean structure obtained by averaging the coordinates of the individual SA structures best fit to each other; and ( )r is the restrained minimized mean structure obtained by restrained minimization of the mean structure (57). The number of terms for the various restraints is given in parentheses.
For backbone NH-CO hydrogen bond there are two restraints: rNH-O = 1.5–2.3 Å and rN-O = 2.5 – 3.3 Å. All hydrogen bonds involve slowly exchanging NH protons.
The torsion angle restraints comprise 84 φ and 83ψ.
The values of the square-well NOE (FNOE) and torsion angle (Ftor) potentials (cf. eqs 2 and 3 in (58)) are calculated with force constants of 50 kcal mol −1 Å−2 and 200 kcal mol−1 rad−2, respectively.
The value of the quartic van der Waals repulsion term (Frep) (cf. eq 5 in (59)) is calculated with a force constant of 4 kcal mol −1 Å−4 with the hard-sphere van der Waals radius set to 0.9 times the standard values used in the CHARMM (60) empirical energy function (57,61,62).
EL-J is the Lennard-Jones-van der Waals energy calculated with the CHARMM empirical energy function.
The improper torsion restraints serve to maintain planarity and chirality.
These were calculated using the PROCHECK program.
The residues in the regular secondary structure are: 12–24 (α1), 30–45 (α2), 3–10 (β1).
The quality of the PpPutA45 NMR structure was analyzed using PROCHECK.50 The results show that PpPutA45 has an overall G-Factor of 0.24, hydrogen bond energy of 0.50 and no bad contacts, which are all consistent with a good quality structure. Also, all non-glycine dihedral angles lie within the expected region of the Ramachandran plot, where 97.5% of the backbone dihedral residues lie within the most favorable region. The consistency of the dihedral angles between each monomer further illustrates the quality of the structure since, as expected; a symmetric PpPutA45 dimer was obtained. The 30 lowest energy structures and the restrained-minimized average structure were deposited into the PDB (2JXG, 2JXH).
Description of the PpPutA45 NMR structure
The NMR structure of PpPutA45 (Fig. 2a) consists of two well defined α-helices corresponding to residues 12–24 (αI) and residues 29–45 (αII) separated by a βI -turn at residues 25–28, which is consistent with the expected RHH fold. The two monomers are intertwined where the hydrophobic face for each set of α-helices forms a stable hydrophobic core. The N-terminal β-strands (residues 3–10) from each monomer interact to form an antiparallel β-sheet.
Examination of the 30 superimposed PpPutA45 structures (Fig. 2b) illustrates well defined α-helices and β-sheet secondary structures. The β-sheet contains 8 hydrogen bonds between residues 3–10 of each strand, and has a slightly twisted orientation. The two helical regions exhibit amphipathic characteristics, which is important for stability of the PpPutA45 core structure. Helix I has a negatively charged N-erminus and terminates at Ser24 before entering the βI -turn that connects helix I to helix II. Pro29 caps the N-terminus of helix II, which contributes to the 90° orientation of helix II with respect to helix I. The N-terminus of helix II presents a positively charged face with residues His30 and Lys34. These N-terminal residues of helix II are proposed to be part of the PpPutA45 DNA interaction site that also includes residues Thr5, Gly7 and Lys9 from the β-sheet and Arg15 from helix I. The PpPutA45 hydrophobic core for each monomer is comprised of residues Leu18 and Ala22 from the C-terminal end of helix I and Leu32 from the N-terminal end of helix II. The backbone carbonyl group of Ala22 forms hydrogen bonds with the backbone amides of residues Ile25 and Asp26, which are in the βI -turn that connects helix I and II. Pro29, at the N-terminus of helix II, is also in a unique position to provide favorable hydrophobic interactions with residues Ala22, Leu18 and Thr28 that further stabilize the helical core of PpPutA45.
The PpPutA45 dimer interface forms a large hydrophobic core composed of residues Leu10, Leu18, Ala21, Ala22, Leu32, Ile33, Ala36, Ile37, Tyr40, and Leu41 from each monomer. Helix II residues Ile33, Ala36, Ile37, and Tyr40 from both chains form a tight interaction in the dimer interface. Leu41 from chain A interacts with Leu18 and Ala21 from chain B. Since PpPutA45 is a symmetric dimer, Leu41 from chain B also interacts with Leu18 and Ala21 from chain A. A third hydrophobic interaction site involves the β-sheet, where the side chains of Leu6 and Val8 from both β-strands extend deeply into the hydrophobic pocket between the intertwined helix II from both chains.
Identification of the PpPutA45 DNA interactions
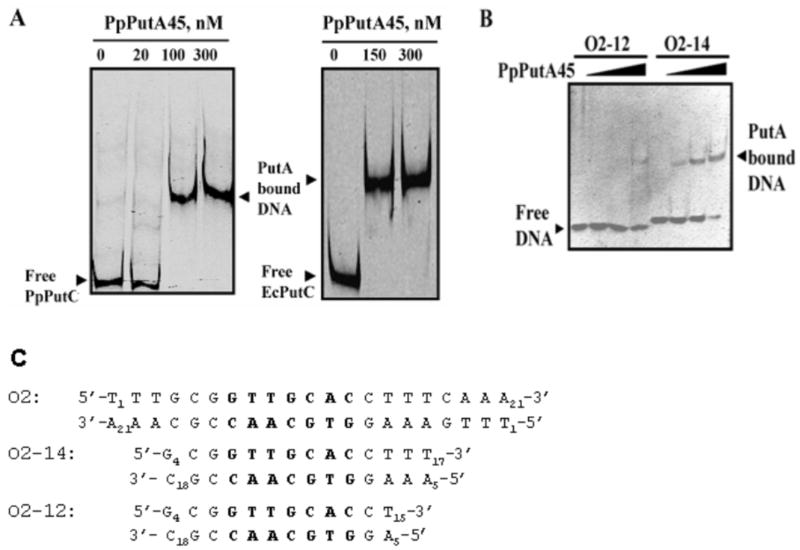
Gel-shift experiments in Fig. 3a show that PpPut45 binds to put intergenic DNA from both P. putida (361 base pairs) and E. coli (419 base pairs). Sequence analysis of the put intergenic DNA region from P. putida revealed it contains five repeats of the 5′-GTTGCA-3′ sequence motif similar to that found in the put control DNA region of E. coli. EcPutA displays high affinity to a site designated as operator 2 (O2).18 A crystal structure of the EcPutA RHH domain complexed to a 21-bp oligomer (5′-T1TTGCGGTTGCACCTTTCAAA21-3′) containing site O2 and flanking sequences was recently solved.18 Using this PutA-DNA binding site, we sought to determine the minimal oligonucleotide length required for PpPutA45 binding to DNA for NMR studies of PpPutA45-DNA interactions. PpPutA45 binding to oligomers of 12-bp (O2-12bp) and 14-bp (O2-14) were tested by gel-shift assays. Fig. 3b shows that no significant binding of PpPutA45 to O2-12 was observed while binding to O2-14 was clearly evident at 400 nM of PpPutA45 in the binding mixture. A dissociation constant of ~ 340 nM for the PpPutA45-O2-14 complex was determined by a reciprocal plot of the fraction of DNA-bound versus PpPutA45 concentration. These results as well as visualizing the EcPutA-DNA model shows T16 and T17 may play a role in PpPutA45 binding to the DNA. The two nucleotides are part of the minor groove where there is a possible interaction with Trp31. Removing the two nucleotides may decrease binding activity, due to the lack of interactions between the minor groove and Trp31.
Fig. 3.
Gel-mobility shift assays of PpPutA45. A, Gel-shift assays in which increasing concentrations of PpPutA45 were added to binding mixtures containing IRdye-700 labeled put intergenic DNA (2 nM) from P. putida (PpPutC) or E. coli (EcPutC) and 100 μg/ml of nonspecific calf thymus DNA at 23 °C. B, Gel-shift assays of PpPutA45 with O2-12 and O2-14. Double stranded O2-12 and O2-14 (100 nM) were incubated with increasing concentrations of PpPutA45 (0 – 400 nM) in 20 mM potassium phosphate buffer (pH 7.4, 100 mM NaCl). The protein-DNA complexes were then separated using a nondenaturing polyacrylamide gel (8%) native gel at 4 °C. C, Nucleotide sequence of the O2 site from E. coli put intergenic DNA and the 14-bp (O2-14) and 12-bp (O2-12) oligomers used in panel B for the gel-shift analysis. Oligomer O2-14 was used for the NMR studies of the PpPutA45-DNA complex. Highlighted in bold is the base pair region that makes contacts with PutA residues in the PpPutA45-DNA structure.
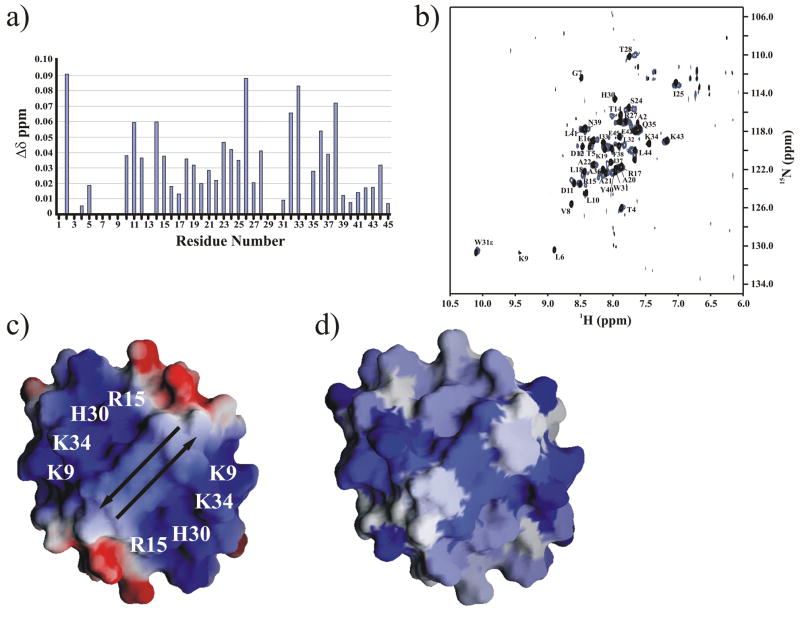
NMR chemical shift perturbation analysis was used to identify the DNA binding site on the molecular surface of PpPutA45 and generate a PpPutA45-DNA model using the O2-14 oligomer. A free PpPutA45 sample and a PpPutA45-O2-14 mixture with DNA in slightly molar excess were used to collect 2D 1H-15N TROSY experiments on a 800 MHz NMR spectrometer. An initial 2D 1H-15N HSQC spectrum at 600 MHz of the PpPutA45:O2-14 complex yielded surprisingly broad NMR peaks, presumably due to slow exchange of the complex because of the relatively high salt concentration (200 mM NaCl) required to stabilize PpPutA45. The 2D 15N-TROSY experiments at 800 MHz resolved this issue and a binding interaction was clearly evident by the presence of numerous chemical shift changes from comparing the 2D 1H-15N TROSY spectra of the free PpPutA45 with the PpPutA45:O2-14 complex. Fig. 4a shows a chemical shift difference plot for the PpPutA45 residues perturbed by the presence of O2-14. Nearly all the PpPutA45 amide peaks in the 2D 15N-TROSY spectra incurred a chemical shift change upon the addition of O2-14 (Fig. 4b). This includes residues involved in the hydrophobic dimer interface, suggesting a potential change in the orientation of the monomers. Residues Ala2, Asp26, and Ile33 displayed the greatest chemical shift changes in the PpPutA45:O2-14 complex. Thr5, Gly7 and Lys9 were expected to bind to the major groove of O2-14. The amide NMR peaks in the 2D 1H-15N TROSY spectrum for Gly7 and Lys9 disappeared in the presence of O2-14, but Thr5 only exhibited a small chemical shift change. His30 and Lys34 also disappeared when bound to O2-14. Therefore, the disappearance of peaks due to interactions with the DNA is not displayed in the plot of chemical shift changes in Fig. 4a. The unique changes in the 2D 15N-TROSY spectra displayed by Gly7, Lys9 and His30 suggest a direct interaction with O2-14.
Fig. 4.
a) NH chemical shift differences between the PpPutA45 (85 μM) and PpPutA45:O2-14 DNA (82 μM) complex are plotted against the PpPutA45 sequence. b) Comparison of the 2D 1H-15N TROSY HSQC spectra of the free PpPutA45 (black) and PpPutA45 bound to O2-14 (blue). The backbone amide resonances are assigned. c) Electrostatic surface of PpPutA45 was calculated using GRASP 43. The arrows show the direction of the β-sheet. The labeled positively charged residues bind to the DNA major groove. d) GRASP molecular surface of PpPutA45 colored by the NMR chemical shift changes plotted in (a). The intensity of the blue color corresponds to the magnitude of the chemical shift change.
Electrostatic interactions play an important role in protein-DNA binding, in which positively charged protein residues bind to the negatively charged phosphate DNA backbone. Comparison of the electrostatic potential with the observed chemical shift changes on the GRASP molecular surface of PpPutA45 (Fig. 4c–d) indicates this expected correlation. A strong positive electrostatic potential resulting from residues Lys9, Arg15, His30, and Lys34 surrounds the PpPutA45 β-sheet. This positive electrostatic potential surface also corresponds to large chemical shift changes observed in the PpPutA45:O2-14 complex. Therefore, the PpPutA45 molecular surface where a strong positive electrostatic potential surrounds a β-sheet is the proposed O2-14 binding site.
Structural model of the PpPutA45-DNA complex
Five clusters were obtained from the 200 calculated structures after water refinement. The average intermolecular energies for the clusters are −508, −507, −477, −470, −479 kcal/mol−1, respectively. The lowest energy cluster contained a total of 25 structures, where the 10 lowest energy structures were averaged and minimized for further analysis. The Haddock docking of the dimeric PpPutA45 structure to the canonical B-DNA structure of O2-14 yielded a robust model with no steric clashes (Fig. 5). Low van der Waal and electrostatic energies of −638 ± 11 kcal/mol−1 and −4,194 ± 53 kcal/mol−1, respectively, were observed for the ensemble of structures. Similarly, the PpPutA45:O2-14 structures exhibited a backbone RMSD from the average structure of 0.88 ± 0.36 Å. The PpPutA45 backbone RMSD in the complex is 0.66 ± 0.20 Å, which is comparable to the RMSD observed for the NMR refinement of the PpPutA45 structure (Table I). An apparently high number of AIR violations of 34.9 ± 2.9 were observed for the complex. This is due to the unbiased selection of all the residues displaying a chemical shift greater than 0.03 ppm that was combined with all but the terminal nucleotides as possible interactions in the complex. Clearly a sub-set of PpPutA45 residues experience a chemical shift change in the PpPutA45:O2-14 complex that was not due to a direct interaction with O2-14. All of these residues would contribute to the AIR violation number. Table II summarize the results for the ensemble of the 10 lowest energy structures. The PpPutA45: O2-14 model was deposited into the PDB (2JXI).
Fig. 5.
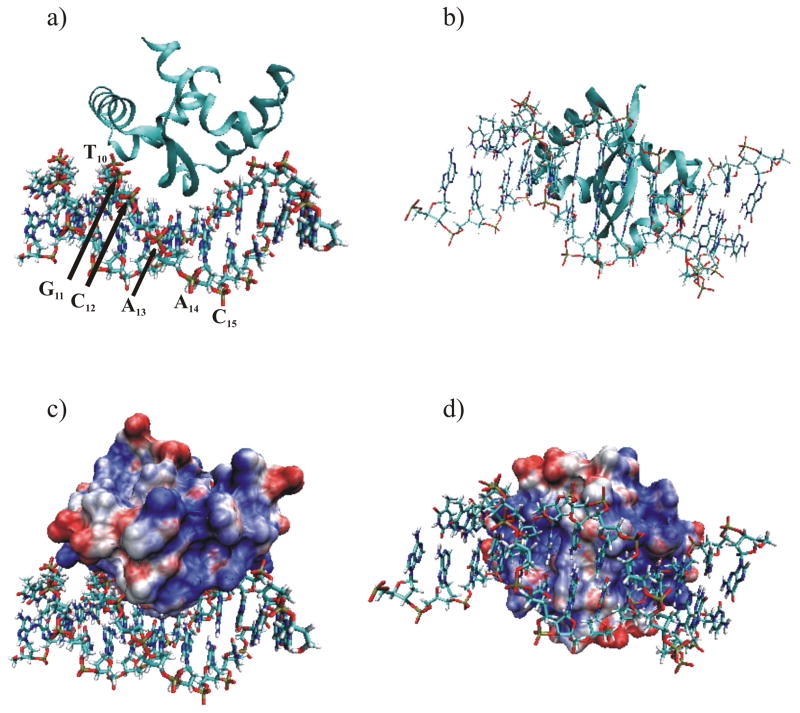
Ribbon diagram of the (a) side view and (b) bottom view of the PpPutA45:O2-14 DNA complex. The PpPutA45 β-strand is inserted into the DNA major groove where it interacts with the conserved GTTGCA DNA sequence. The DNA is shown as solid bonds colored by atom-type. (c and d) Electrostatic molecular surface of PpPutA45 calculated using Gemstone and VMD-XPLOR with APBS plugin 44. Basic residues are shown in blue, acidic residues are shown in red, and neutral residues are shown in white. The DNA is shown as solid bonds colored by atom-type. Same view is shown as in (a) and (b), respectively.
Table II.
Structural Statistics of PpPutA45-O2-14 Complexa
| Docking Statistics | |
|---|---|
| Evdwb (kcal/mol) | −638 ± 11 |
| Eelecb (kcal/mol) | −4194 ± 53 |
| Cluster populationc | 25 |
| AIR-energy (kcal/mol) | 103 ± 8 |
| AIR-violations > 0.5 Å | 34.9 ± 2.9 |
| AIR RMS (Å) | 1.61 ± 0.07 |
|
| |
| Structural Statistics
| |
| RMSD backboned | |
| Interface all | 0.87 ± 0.37 |
| All | 0.88 ± 0.36 |
| PutA45 | 0.66 ± 0.20 |
| O2-14 | 1.29 ± 0.25 |
| BSAe (Å) | 1779 ± 134 |
|
| |
| Ramachandran Analysis
| |
| Most favored (%) | 91.5 ± 1.3 |
| Additional allowed (%) | 8.3 ± 1.3 |
| Generously allowed (%) | 0.12 ± 0.37 |
| Disallowed (%) | 0 |
The reported values for the solution structure are averages and standard deviations over the 10 final structures.
The non-bonded energies Evdw and Eelec were calculated with an 8.5 Å cutoff using the OPLS59 nonbonded parameters from the parallhdg5.3.pro parameter file.60
Number of structures in the lowest energy cluster out of 200 structures
Average RMSD from the average structure
Buried Surface area (calculated with NACCESS61)
The PpPutA45:O2-14 Haddock model indicates that the PpPutA45 β-sheet fits into the major groove of O2-14, where it interacts with the conserved GTTGCA sequence element. Helices I from both PpPutA45 chains are positioned approximately parallel to the phosphate backbone, where the N-terminal residues of helices II are perpendicularly directed towards the phosphate backbone. This positions residues Arg15, Thr28, and His30 to provide a positive electrostatic interaction between PpPutA45 and the phosphate backbone of O2-14. These salt-bridge interactions may be stabilized by hydrophobic shielding from the solvent by Trp31.51 An electrostatic potential surface of PpPutA45, within the model complex (Fig. 5c–d), illustrates that the highly positive charged surface of PpPutA45 perfectly matches the O2-14 phosphate DNA backbone. Conversely, the PpPutA45 surface opposite to the DNA-binding face consists mostly of polar regions and a few charged areas, which presumably contribute to the solubility of the complex and are not involved in DNA binding.
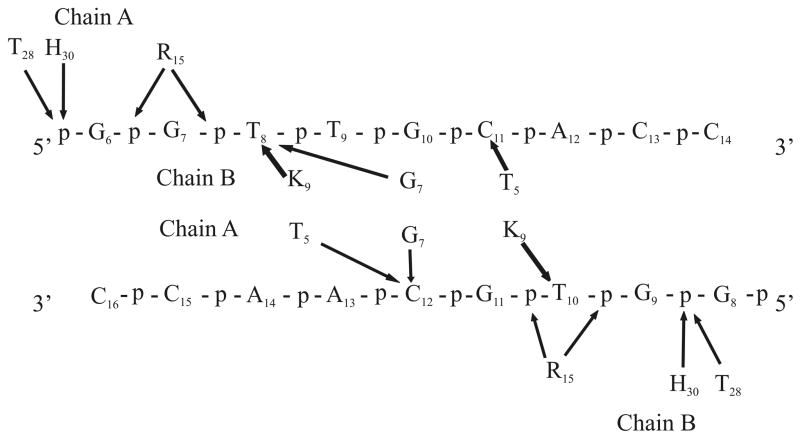
Direct contacts between PpPutA45 and O2-14 in the model of the PpPutA45-DNA complex are diagrammed in Fig. 6. PpPutA45 β-sheet residues Thr5, Gly7 and Lys9 interact with the major groove base pairs or phosphate groups of the nucleotides G7TTGCAC13. Lys9 of PpPutA45 chain B interacts with the thymine (T8) base on the 5′ strand while Lys9 from chain A interacts with the thymine (T10) on the 3′ strand. Thr5 from chains A and B interacts symmetrically with cytosine bases C11 and C12 on the 5′ and 3′ strands, respectively. Gly7 from chain A interacts with cytosine (C12) on the 3′ strand, and Gly7 from chain B interacts with thymine (T8) on the 5′ strand.
Fig. 6.
Schematic representation of the PpPutA45 and O2-14 residues involved in specific interactions in the PpPutA45:O2-14 DNA complex. Arrows indicate interaction with either phosphate group (p) or the base group (base-pair number).
Comparison of the E. coli and P. putida PutA and PutA-DNA structures
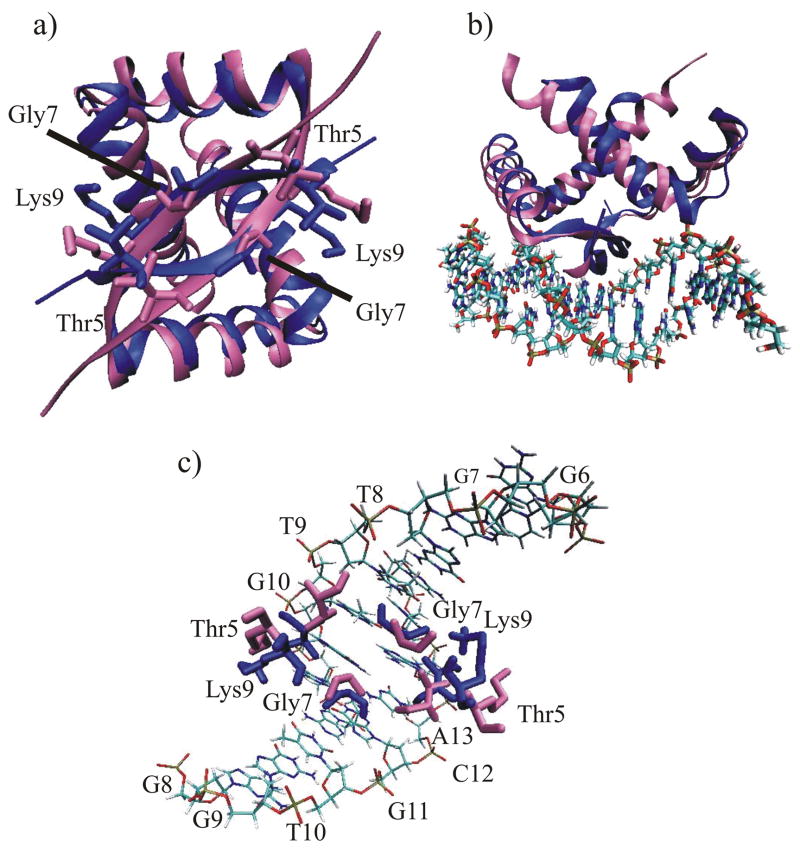
The PpPutA45 NMR structure was compared to the x-ray structure of EcPutA52 from E. coli (Fig. 7a).15 The sequence identity between the two RHH domains of PpPutA and EcPutA is nearly 77%. The superposition of the backbone atoms from both structures yielded a RMSD of 1.72 Å. Some differences between the structures, however, are observed due to variations in specific residues such as the substitution of Ala13 in EcPutA with Pro in PpPutA. The relatively large difference in the φ (16°) between Pro13 in PpPutA and Ala13 in EcPutA results in the displacement of helix I in the PpPutA45 NMR structure relative to the EcPutA52 structure. The displacement of helix I is propagated in PpPutA45 causing the orientation of helix II to be slightly bent and out of register with helix II of EcPutA52. The bend of helix II in PpPutA45 allows for proper positioning of helix II residues (Leu32, Ile33, Ala36, Ile37, Tyr40, and Leu41) that form the hydrophobic core of the RHH domain.
Fig. 7.
a) P. putida PutA45 (blue) superimposed on the x-ray structure of E. coli PutA52 (pink). The side chains for residue T5, G7, and K9, which bind to the GTTGCA DNA sequence, are shown as licorice bonds. b) PpPutA45:O2-14 NMR based complex (blue) superimposed on the E. coli PutA52-DNA complex (pink). c) Expanded view of the superimposed sidechains for P. putida PutA45 (blue) and E. coli PutA52 (pink) involved in a direct interaction with DNA.
The overall structural model of the P. putida PutA45:O2-14 complex is also similar to the E. coli PutA52-DNA x-ray crystal structure supporting the validity of the Haddock approach. Superposition of the backbone atoms from the PpPutA45 and EcPutA52 DNA complexes yielded and an RMSD of 2.14Å (Fig. 7b). The footprint of PpPutA-DNA and EcPutA-DNA complex shows similar binding in a 9-bp region. The side chains of Thr5 and Lys9, which are involved in DNA recognition, extend toward the same general location in both structures but some differences are observed (Fig. 7c). In EcPutA, Lys9 of both chains makes symmetrical hydrogen bond contacts with G6 and G7 of the 5′-3′ strand and G8 and G9 of the 3′-5′ strand. In the PpPutA45-DNA structure, Lys9 of chain A makes contacts with T10 of the 3′-5′ strand while Lys9 of chain B interacts with T8. Thus, the PpPutA45-DNA complex model suggests that Lys9 can sample various confirmations allowing for diverse RHH-DNA interactions. A peculiar feature noted in the EcPutA-DNA structure was the unusual involvement of Gly7 in DNA recognition. In EcPutA, Gly7 of chain A interacts with G11 of the 3′-5′ strand and Gly7 of chain B interacts with T9 of the 5′-3′ strand. In PpPutA45, Gly7 of chain A interacts with C12 of the 3′-5′ strand and Gly7 of chain B interacts with T8 of 5′-3′ strand. A non-varying interaction appears to be Thr5 which contacts C11 and C12 of the 5′-3′ and 3′-5′strands, respectively, in the PpPutA45-DNA complex model similar to that found in the EcPutA52-DNA complex structure. Protein interactions with the phosphate backbone of the DNA are mediated by His30 and Thr28 similar to the EcPutA52-DNA complex but additional phosphate backbone contacts in the PpPutA45-DNA model are evident for Arg15.
The differences observed between the PpPutA45-DNA and EcPutA-DNA complexes may be functionally meaningful or may just represent conformational sampling in PutA-DNA recognition patterns. In put control DNA from various organisms with the same genetic organization as that found in P. putida and E. coli, the base pairs flanking the conserved sequence element GTTGCA vary at the different binding sites. From the x-ray crystal structure of the EcPutA-DNA complex, operators with the GGTTGCACC sequence were proposed to be the optimal binding sites and exhibit higher PutA-binding affinity than other operators that do not have the flanking guanine and cytosine base pairs.18 In P. putida, two operator sites have the sequence AGTTGCACC while the other three operator sites have the proposed optimal binding sequence.18 From the PpPutA45 NMR structure, it appears that Lys9 has inherent flexibility and can form hydrogen bonds with the neighboring thymine base pair presumably with the C4 carbonyl. Thus, in operators that lack the optimal sequence Lys9 may help maintain sequence specific recognition by forming alternative hydrogen bond interactions with nearby bases such as thymine.
Sequence and structural comparison of the PutA RHH and PRODH domains
The solution structure of PpPutA45 verifies its membership in the RHH superfamily. Other members of this superfamily include CopG, MetJ, Arc, Mnt, Omega, FitA, NikR, CcdA, ParG, and ParD families.16 The sequence identity between each family can be as low as 15.4%, despite a high similarity in both structure and function.16 The β-sheet and the N-terminal region of helix II of the RHH structure support the recognition of specific DNA sequences.17 Most members of the RHH superfamily are transcription repressors with one exception being AlgZ, which is both an activator and a repressor in Pseudomonas aeruginosa.52
The sequence alignment of the RHH domains for the PutA family is shown in Fig. 8a. It is readily apparent that a high sequence identity exists between all members of the PutA RHH family with highly conserved residues having functionally or structurally important roles. These residues are involved in DNA binding or hydrophobic interactions within the protein core or dimer interface. Thr5, Gly7, and Lys9 of the RHH β-sheet are important for sequence specific interactions with the conserved GTTGCA DNA sequence while R15 in Helix I and Thr28 and His30 in Helix II contact the phosphate backbone of the DNA. Leu18, Ala22, Leu32 and Pro29 form part of the hydrophobic core and Ala36, Ile37 and Tyr40 are part of helix II that is involved in the dimer interface. The significant percentage of residues that are structurally or functionally critical suggests that the high conservation of the PutA RHH domain may be explained by an optimized protein architecture.
Fig. 8.
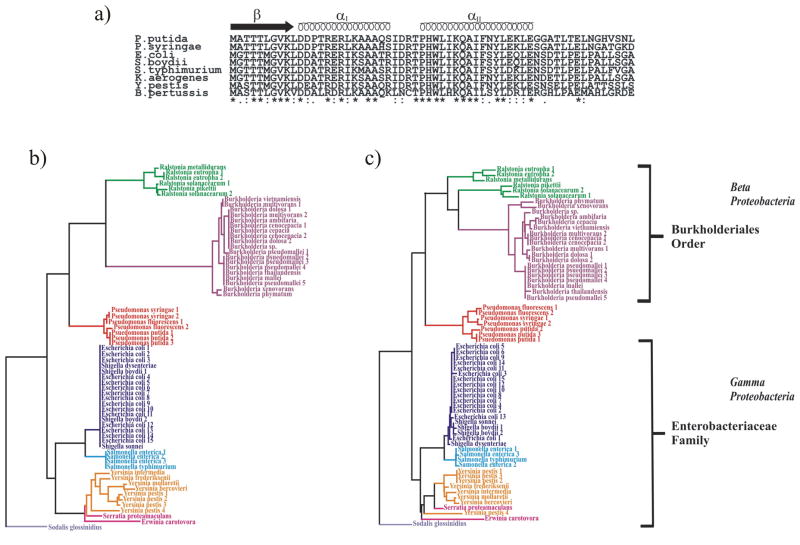
a) Amino acid sequence alignments of 8 PutA RHH domains. b) Phylogenetic tree of the RHH domain from 66 PutA sequences. c) Phylogentic tree of the PRODH domain associated with the 66 PutA domain sequences.
The majority of prokaryotes use proline as a viable energy source; however, the only organism known to inherit the RHH domain of PutA is the enteric bacteria.53,54 Although residues in the RHH domain are highly conserved among PutA proteins slight differences do exist, such as the Pro13 substitution in PutA from P. putida. The sequence variations appear to correlate with the evolutionary divergence of bacteria that contain a PutA RHH domain. A phylogenetic tree based on 66 known PutA RHH sequences is illustrated in Fig. 8b. The two major branches correspond to the beta and gamma proteobacteria classes. All the members in the gamma branch are members of the Enterobacteriacaea family illustrating that the RHH domain is highly conserved among enteric bacteria, for example, E. coli and S. boydii have identical sequences. Pseudomonas, which is a gamma proteobacteria, but not a member of the Enterobacteriacaea family, is in the beta branch. The Ralstonia and Burkholderia genus are part of a common node in the beta branch, since both belong to the Burkholderiales order. The Ralstonia genus was previously included in the Pseudomonas genus, which also explains the grouping of Pseudomonas in the beta branch. A nearly identical phylogenetic tree is obtained using the PutA PRODH domain (Fig. 8c). These results are consistent with a bacterial phylogenetic tree based on 16S rRNA gene sequences, despite the high sequence and structure conservation of the PutA protein.
Protein evolutionary rates are correlated with the dispensability of a protein instead of the essentiality of the protein.55 Additionally, protein-protein interactions have been proposed to constrain protein evolution.56 A protein with a high number of binding partners or protein interaction number (PIN) experiences a low evolutionary rate. Also, protein binding partners have correlated evolutionary rates. These factors appear to play a role in the high conservation of the PutA RHH domain. Proline utilization is not an essential biochemical function for bacteria, but presumably it provides a competitive advantage when nutrients are limited. Therefore bacteria will be preferentially selected that maintain PutA activity. Since PutA is a multidomain protein and proline binding to the PRODH domain converts the protein from a transcriptional repressor to a membrane bound metabolic enzyme, domain interactions are expected to be important in PutA functionality. The high conservation of solvent exposed residues Ile25, Asp26, Arg27, Glu35 and Phe38, which form a separate and distinct surface from the DNA binding site, is suggestive of a potential PutA interdomain interaction site. This is also consistent with the correlated high conservation of the PRODH sequence, since interacting proteins are expected to evolve at similar rates. Essentially, a high percentage of the PutA RHH residues are critical for maintaining the protein’s function either through a direct DNA interaction, by stabilizing the RHH fold or by responding to proline binding through interdomain interactions. Conversely, the functionally dispensable RHH residues experience a normal substitution rate, which account for the bacterial divergence observed in the PutA phylogenic tree. These results imply either a direct interaction between the PRODH and RHH domains or an indirect interaction modulated by the intervening flexible domain of unknown function (residues 141–262) that undergoes a large conformational change upon proline binding.9
CONCLUSION
The NMR solution structure of PpPutA45 and an experimentally derived PpPutA45-DNA model confirms the protein adopts an RHH fold and binds a DNA oligomer containing the consensus GTTGCAC sequence. The PpPutA45 and PpPutA45-DNA complex are similar to the corresponding x-ray structures of EcPutA52 and display comparable DNA binding characteristics. The NMR structure verifies that PpPutA45 is a member of the CopG/MetJ superfamily, where the two helices and the β-strand of each chain form a stable hydrophobic core. The PpPutA45 β-sheet fits into the DNA major groove and binds to the GTTGCAC region which is conserved in all five operator sites of the P. putida control DNA.18 The positive electrostatic surface that surrounds the β-sheet also plays a role in stabilizing the DNA complex by binding to the phosphate backbone. A phylogenic tree based on either the RHH or PRODH sequence predicts similar bacterial divergences as observed previously with 16S rRNA gene sequences. Nevertheless, multiple factors, including interdomain interactions, appear to explain the high sequence and structure conservation for PutA RHH, while little sequence similarity is maintained across the RHH superfamily.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB-0340912), National Institutes of Health (GM061068) and Nebraska Tobacco Settlement Biomedical Research Development Funds. This work is a contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. This publication was also made possible by NIH Grant Number P20 RR-017675-02 from the National Center for Research Resources. Research was performed in facilities renovated with support from NIH (RR015468-01) and the XL-I analytical centrifuge was funded by NSF grant DBI-0619764. We thank Prof. Gaetano Montelione (Rutgers University, Piscataway, NJ) for generously providing the 800 MHz 2D 1H-15N TROSY NMR data of PpPutA45 and its DNA complex. We also thank Dr. John Tanner (University of Missouri-Columbia) for sharing the PDB files of the E. coli PutA-DNA complex prior to publication.
The abbreviations used are
- Proline utilization A
PutA
- proline dehydrogenase domain
PRODH
- Δ1-pyrroline-5-carboxylate
P5C
- dehydrogenase domain
P5CDH
- Nuclear Overhauser effect
NOE
- ribbon-helix-helix
RHH
- root-mean square difference
RMSD
- 14 base-pair double-stranded DNA oligomer
O2-14
- 12 base-pair double-stranded DNA oligomer
O2-12
- E. coli PutA
EcPutA
- DNA binding domain of PutA from P. putida corresponding to residues 1 to 45
PpPutA45
- put intergenic DNA from P. putida
PpPutC
- put intergenic DNA from E. coli
EcPutC
Footnotes
The atomic coordinates and structure factors (code 2JXG, 2JXH, 2JXI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/). The NMR chemical shift assignments (code7082) have been deposited in the Biological Magnetic Resonance Data Bank, Department of Biochemistry, University of Wisconsin-Madison (http://www.bmrb.wisc.edu/).
References
- 1.Brown ED, Wood JM. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J Biol Chem. 1992;267(18):13086–13092. [PubMed] [Google Scholar]
- 2.Vilchez S, Molina L, Ramos C, Ramos JL. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. Journal of Bacteriology. 2000;182(1):91–99. doi: 10.1128/jb.182.1.91-99.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnan N, Becker DF. Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress. J Bacteriol. 2006;188(4):1227–1235. doi: 10.1128/JB.188.4.1227-1235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phang JM. The regulatory functions of proline and pyrroline-5-carboxylic acid. Current Topics in Cellular Regulation. 1985;25:91–132. doi: 10.1016/b978-0-12-152825-6.50008-4. [DOI] [PubMed] [Google Scholar]
- 5.Menzel R, Roth J. Enzymatic properties of the purified putA protein from Salmonella typhimurium. J Biol Chem. 1981;256(18):9762–9766. [PubMed] [Google Scholar]
- 6.Gu D, Zhou Y, Kallhoff V, Baban B, Tanner JJ, Becker DF. Identification and characterization of the DNA-binding domain of the multifunctional PutA flavoenzyme. J Biol Chem. 2004;279(30):31171–31176. doi: 10.1074/jbc.M403701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Zhou Y, Becker DF. Regulation of PutA-membrane associations by flavin adenine dinucleotide reduction. Biochemistry. 2004;43(41):13165–13174. doi: 10.1021/bi048596g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scarpulla RC, Soffer RL. Membrane-bound proline dehydrogenase from Escherichia coli. Solubilization, purification, and characterization. J Biol Chem. 1978;253(17):5997–6001. [PubMed] [Google Scholar]
- 9.Becker DF, Thomas EA. Redox properties of the PutA protein from Escherichia coli and the influence of the flavin redox state on PutA-DNA interactions. Biochemistry. 2001;40(15):4714–4721. doi: 10.1021/bi0019491. [DOI] [PubMed] [Google Scholar]
- 10.Bohm A, Boos W. Transport-dependent gene regulation by sequestration of transcriptional regulators. Topics in Current Genetics. 2004;9(Molecular Mechanisms Controlling Transmembrane Transport):47–66. [Google Scholar]
- 11.Ostrovsky de Spicer P, Maloy S. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc Natl Acad Sci U S A. 1993;90(9):4295–4298. doi: 10.1073/pnas.90.9.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrahamson JLA, Baker LG, Stephenson JT, Wood JM. Proline dehydrogenase from Escherichia coli K12. Properties of the membrane-associated enzyme. European Journal of Biochemistry. 1983;134(1):77–82. doi: 10.1111/j.1432-1033.1983.tb07533.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Zhang M, Zhu W, Zhou Y, Wanduragala S, Rewinkel D, Tanner JJ, Becker DF. Redox-Induced Changes in Flavin Structure and Roles of Flavin N(5) and the Ribityl 2′-OH Group in Regulating PutA-Membrane Binding. Biochemistry. 2007;46(2):483–491. doi: 10.1021/bi061935g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, White TA, Schuermann JP, Baban BA, Becker DF, Tanner JJ. Structures of the Escherichia coli PutA Proline Dehydrogenase Domain in Complex with Competitive Inhibitors. Biochemistry. 2004;43(39):12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson JD, Jenkins JL, Schuermann JP, Zhou Y, Becker DF, Tanner JJ. Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition. Protein Sci. 2006;15(11):2630–2641. doi: 10.1110/ps.062425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme. Nat Rev Microbiol. 2007;5(9):710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- 17.Raumann BE, Rould MA, Pabo CO, Sauer RT. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994;367(6465):754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Larson J, Bottoms C, Arturo EC, Henzl MT, Jenkins J, Nix J, Becker DF, Tanner JJ. Structural Basis of the Transcriptional Regulation of the Proline Utilization Regulon by Multifunctional PutA. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.05.084. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding RC, Hill GA, Lin Y-H. Bioremediation of toluene-contaminated air using an external loop airlift bioreactor. Journal of Chemical Technology and Biotechnology. 2003;78(4):406–411. [Google Scholar]
- 20.Nwachukwu SU. Bioremediation of sterile agricultural soils polluted with crude petroleum by application of the soil bacterium, Pseudomonas putida, with inorganic nutrient Supplementations. Current Microbiology. 2001;42(4):231–236. doi: 10.1007/s002840110209. [DOI] [PubMed] [Google Scholar]
- 21.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J Magn Reson. 1991;95(1):214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Quarterly Reviews of Biophysics. 2000;33(1):29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 25.Wuthrich K, Billeter M, Braun W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J Mol Biol. 1983;169(4):949–961. doi: 10.1016/s0022-2836(83)80144-2. [DOI] [PubMed] [Google Scholar]
- 26.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 27.Hwang TL, Mori S, Shaka AJ, van Zijl P. Application of Phase-Modulated CLEAN Chemical EXchange Spectroscopy (CLEANEX-PM to Detect Water-Protein Proton Exchange and Intermolecular NOEs. J Am Chem Soc. 1997;119:6203–6204. [Google Scholar]
- 28.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 29.Nilges M, Clore GM, Gronenborn AM. Determination of three-dimensional structures of proteins from interproton distance data by hybrid distance geometry-dynamical simulated annealing calculations. FEBS Lett. 1988;229(2):317–324. doi: 10.1016/0014-5793(88)81148-7. [DOI] [PubMed] [Google Scholar]
- 30.Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Three-dimensional structure of interleukin 8 in solution. Biochemistry. 1990;29(7):1689–1696. doi: 10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- 31.Garrett DS, Kuszewski J, Hancock TJ, Lodi PJ, Vuister GW, Gronenborn AM, Clore GM. The impact of direct refinement against three-bond HN-C alpha H coupling constants on protein structure determination by NMR. J Magn Reson B. 1994;104(1):99–103. doi: 10.1006/jmrb.1994.1061. [DOI] [PubMed] [Google Scholar]
- 32.Kuszewski J, Qin J, Gronenborn AM, Clore GM. The impact of direct refinement against 13C alpha and 13C beta chemical shifts on protein structure determination by NMR. J Magn Reson B. 1995;106(1):92–96. doi: 10.1006/jmrb.1995.1017. [DOI] [PubMed] [Google Scholar]
- 33.Kuszewski J, Gronenborn AM, Clore GM. Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J Am Chem Soc. 1999;121:2337–2338. [Google Scholar]
- 34.Kuszewski J, Gronenborn AM, Clore GM. Improving the quality of NMR and crystallographic protein structures by means of a conformational database potential derived from structure databases. Protein Sci. 1996;5(6):1067–1080. doi: 10.1002/pro.5560050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuszewski J, Gronenborn AM, Clore GM. Improvements and extensions in the conformational database potential for the refinement of NMR and X-ray structures of proteins and nucleic acids. J Magn Reson. 1997;125(1):171–177. doi: 10.1006/jmre.1997.1116. [DOI] [PubMed] [Google Scholar]
- 36.Kuszewski J, Clore GM. Sources of and solutions to problems in the refinement of protein NMR structures against torsion angle potentials of mean force. J Magn Reson. 2000;146(2):249–254. doi: 10.1006/jmre.2000.2142. [DOI] [PubMed] [Google Scholar]
- 37.Linge JP, Nilges M. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J Biomol NMR. 1999;13(1):51–59. doi: 10.1023/a:1008365802830. [DOI] [PubMed] [Google Scholar]
- 38.Powers R, Mirkovic N, Goldsmith-Fischman S, Acton TB, Chiang Y, Huang YJ, Ma L, Rajan PK, Cort JR, Kennedy MA, Liu J, Rost B, Honig B, Murray D, Montelione GT. Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes. Protein Sci. 2005;14(11):2849–2861. doi: 10.1110/ps.051666705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrett D, Seok Y-J, Peterkofsky A, Clore GM, Gronenborn AM. Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry. 1997;36(15):4393–4398. doi: 10.1021/bi970221q. [DOI] [PubMed] [Google Scholar]
- 40.Schwieters CD, Clore GM. The VMD-XPLOR visualization package for NMR structure refinement. J Magn Reson. 2001;149(2):239–244. doi: 10.1006/jmre.2001.2300. [DOI] [PubMed] [Google Scholar]
- 41.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 42.Vlahovicek K, Kajan L, Pongor S. DNA analysis servers: plot.it, bend.it, model. it and IS. Nucleic Acids Res. 2003;31(13):3686–3687. doi: 10.1093/nar/gkg559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 44.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 48.Wüthrich K. NMR of proteins and nucleic acids. New York: John Wiley & Sons, Inc; 1986. p. 292. [Google Scholar]
- 49.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 51.Schmuck C. How to Improve Guanidinium Cations for Oxoanion Binding in Aqueous Solution? The Design of Artificial Peptide Receptors Coordination Chemistry. Reviews. 2006;250(23–24):3053–3067. [Google Scholar]
- 52.Ramsey DM, Baynham PJ, Wozniak DJ. Binding of Pseudomonas aeruginosa AlgZ to sites upstream of the algZ promoter leads to repression of transcription. J Bacteriol. 2005;187(13):4430–4443. doi: 10.1128/JB.187.13.4430-4443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keuntje B, Masepohl B, Klipp W. Expression of the putA gene encoding proline dehydrogenase from Rhodobacter capsulatus is independent of NtrC regulation but requires an Lrp-like activator protein. J Bacteriol. 1995;177(22):6432–6439. doi: 10.1128/jb.177.22.6432-6439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vilchez S, Manzanera M, Ramos JL. Control of expression of divergent Pseudomonas putida put promoters for proline catabolism. Appl Environ Microbiol. 2000;66(12):5221–5225. doi: 10.1128/aem.66.12.5221-5225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirsh AE, Fraser HB. Protein dispensability and rate of evolution. Nature. 2001;411(6841):1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- 56.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296(5568):750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 57.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. Journal of Applied Crystallography. 1991;24(5):945–949. [Google Scholar]
- 58.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics, Macromolecular Crystallography, Part B. Methods in Enzymology. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]