Abstract
The processes promoting disease in wild animal populations are highly complex, yet identifying these processes is critically important for conservation when disease is limiting a population. By combining field studies with epidemiologic tools, we evaluated the relationship between key factors impeding southern sea otter (Enhydra lutris nereis) population growth: disease and resource limitation. This threatened population has struggled to recover despite protection, so we followed radio-tagged sea otters and evaluated infection with 2 disease-causing protozoal pathogens, Toxoplasma gondii and Sarcocystis neurona, to reveal risks that increased the likelihood of pathogen exposure. We identified patterns of pathogen infection that are linked to individual animal behavior, prey choice, and habitat use. We detected a high-risk spatial cluster of S. neurona infections in otters with home ranges in southern Monterey Bay and a coastal segment near San Simeon and Cambria where otters had high levels of infection with T. gondii. We found that otters feeding on abalone, which is the preferred prey in a resource-abundant marine ecosystem, had a very low risk of infection with either pathogen, whereas otters consuming small marine snails were more likely to be infected with T. gondii. Individual dietary specialization in sea otters is an adaptive mechanism for coping with limited food resources along central coastal California. High levels of infection with protozoal pathogens may be an adverse consequence of dietary specialization in this threatened species, with both depleted resources and disease working synergistically to limit recovery.
Keywords: Enhydra lutris nereis, Sarcocystis neurona, Toxoplasma gondii, diet specialization, wildlife disease
Disease has been increasingly identified as a limiting force in wildlife populations (1–3), but elucidating the processes promoting disease is an inherently complex problem. For pathogens that can persist in the environment outside of their host, characterizing disease dynamics is particularly problematic. Given the obvious limitations of laboratory-based experimental studies for defining pathogen–host interactions in free-ranging wildlife, observational studies of live animals may provide important clues to risky behaviors that increase the probability of pathogen infection. In this way, specific behaviors and contacts with potential sources of pathogens can be evaluated for their relationship with health outcomes to explain the distribution of disease in populations, much in the way human epidemiologic studies have advanced public health.
Southern sea otters (Enhydra lutris nereis) have struggled to recover in California, and infectious disease has been implicated as an important cause of death (4, 5). Peritonitis caused by acanthocephalan parasite infection and protozoal encephalitis caused by Sarcocystis neurona or Toxoplasma gondii infection have been specifically identified as potentially important factors (4, 6–9). Both S. neurona and T. gondii are apicomplexan protozoa that have complex multihost life cycles and an infective life stage (oocysts) that can persist in the environment outside of the host. Transmission typically occurs in carnivores by ingestion of intermediate hosts infected with the tissue cyst stage of the parasites. However, both parasites have a broad host range because any endothermic vertebrate may become infected after ingestion of viable oocysts in the environment (10, 11). Both parasites are surprisingly common in sea otters and a variety of other marine mammal species (12–14), especially because the only known definitive hosts that support sexual multiplication and shed infective oocysts into the environment are terrestrial mammals, namely opossums (Didelphis virginiana) for S. neurona and felids for T. gondii (10, 11). The life cycle propagating these pathogens in the marine environment has been a fascinating problem because ectotherms, which constitute all but the apex of the marine food web, are unlikely intermediate hosts. Laboratory studies have determined that filter-feeding bivalves can accumulate T. gondii in experimentally-contaminated water (15, 16). However, a field study screening >1,000 sea otter invertebrate prey items (mussels, clams, and sand crabs) detected type X T. gondii in just 1 outplanted sentinel California mussel (17). This low detection rate reflects the potentially low concentrations of oocysts in transport hosts and the limited sensitivity of current detection assays (18). Because direct measurement of oocysts in transport hosts or the environment is currently impractical, we instead used observational studies of live sea otters, combined with extensive health monitoring, to determine risk factors for infection, specifically determining whether individuals preying on filter-feeding bivalves or other invertebrates are more likely to be infected.
Modern advances in telemetry and biometric technology used to observe animal movements, behavior, reproduction, and survival have revolutionized our perception and understanding of the behavioral complexity of wild animals (19). Long-term telemetry-based field studies of living sea otters have identified specialized behaviors and diet preferences in individuals (20, 21) that may allow a direct view into the complex processes promoting disease in this population. The need to combine tools across disciplines to solve complex problems is broadly recognized, and here we apply this transdisciplinary approach to gain insight into the ecological and behavioral drivers promoting pathogen infection in sea otters. Specifically, by using longitudinal data on individual animal habitat use, daily movements, and diet composition to explain patterns of infection with T. gondii and S. neurona, we demonstrate how individual behavior influences the risk of disease in sea otters.
Results
Overview of Datasets.
Sea otter infection with T. gondii and S. neurona, as determined by detection of antibodies on a serologic assay, was common among the 118 southern sea otters captured for this study. The diagnostic assay used here detects infection with persistent tissue cyst stages (22), but not clinical disease, and all otters were apparently healthy at capture. We detected T. gondii infection in 48% of otters (56/118) and S. neurona infection in 33% of otters (39/118). Among sea otters sampled, 44 were adult males, 57 were adult females, 4 were juvenile males, and 13 were juvenile females. The duration of follow-up for otters ranged from 159 to 2,473 days (mean 2.4 years) postcapture, indicating that otters included in this study were able to survive pathogen infection. Fifty-six of the otters were captured more than once, and 22 otters were captured 3 or more times. Three S. neurona-seropositive sea otters (2 pups and 1 adult) were negative when first captured but seroconverted during the study period. One juvenile otter seroconverted to both T. gondii and S. neurona, but all other otters were either seropositive to T. gondii at first capture or remained seronegative on subsequent resampling.
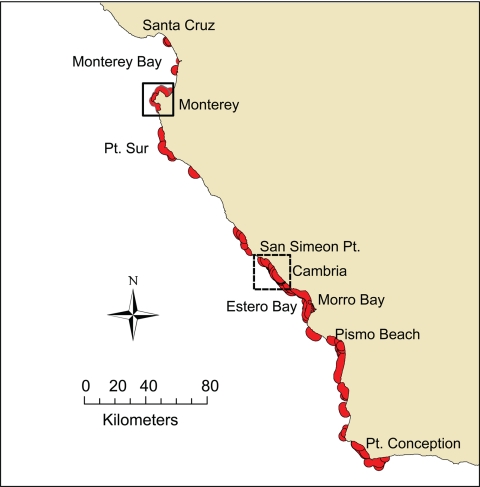
The distribution of radio-telemetered otters represented more than half of the range of the California sea otter population (Fig. 1). We used numerous parameters to characterize sea otter demographics, range use, habitat use, prey preference, foraging behavior, and movement patterns (see Materials and Methods). Males and females displayed distinctly different movement behaviors. The mean 90-day move length for males was 45 km, with 20 males maintaining >1 home-range center, and 13 of these having 3 or more centers of use. In contrast, the mean 90-day move length for females was 7 km, and only 3 of 70 females had a second home range center, which was apparently used as a secondary site for foraging.
Fig. 1.
Map of the geographic distribution of sea otters in California showing individual home ranges (red) of 118 radio-telemetered otters. High-risk areas for T. gondii (dashed square) and S. neurona (solid square) are enlarged in Figs. 2 and 3, respectively.
Because long-term daily observation of highly mobile males was more difficult than for the more sedentary females, we were able to characterize prey preference for 49 females but only 12 males. Our study population consumed a diverse diet of >50 species of macroinvertebrates, but most individual otters exhibited dietary specialization by preying predominantly on a subset of the following invertebrates: cancer crabs (Cancer spp.), abalone (Haliotis spp.), kelp crabs (Pugettia producta), sea urchins (Strongylocentrotus spp.), bivalve mollusks, small snails (Tegula spp.), sea stars (primarily Pisaster spp.), and cephalopod mollusks.
Factors Increasing Risk of T. gondii Infection.
Sea otter infection with T. gondii varied according to individual animal diet composition and range use. Risk factors associated with T. gondii infection by multivariate logistic regression included use of a coastal segment offshore from San Simeon and Cambria (Fig. 2) and a diet including ≥10% marine snails (Table 1). Only 1 of 8 otters with ≥10% abalone in their diet were infected with T. gondii, and the logistic model was improved by including a covariate describing ≥10% abalone [likelihood ratio test χ2 = 3.49, P = 0.062 with reduced Akaike information criterion (AIC) and Bayesian information criterion (BIC)]. The logistic model with covariates describing the proportion of marine snails and abalone in the diet and use of the coastal segment from San Simeon to Cambria demonstrated good overall fit (Hosmer-Lemeshow goodness-of-fit χ2 = 0.40, P = 0.940), indicating that there was little remaining unexplained variation in sea otter infection with T. gondii. Based on this model, otters consuming ≥10% marine snails were 12 times more likely to have been infected with T. gondii compared with otters consuming fewer marine snails, and otters that foraged between San Simeon to Cambria were 4 times more likely to have been infected with T. gondii than otters that foraged elsewhere. By estimating predictions using the logistic model for T. gondii, we found that among otters foraging near San Simeon and Cambria, otters preying on abalone had a 22% probability of infection with T. gondii, compared with 72% for otters with a primarily nonabalone diet and 95% for otters consuming marine snails. Outside of this high-risk area, otters preying on abalone had a 7% probability of infection, whereas otters feeding on other prey had a 45% probability of infection, except for otters consuming marine snails, which had an 85% probability of infection with T. gondii.
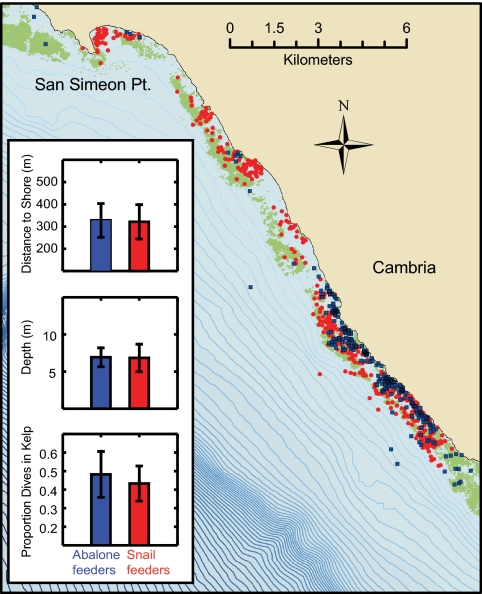
Fig. 2.
Map of the high-risk San Simeon to Cambria coastal segment (35.756 N, 121.325 W–35.437 N, 120.901 W) defined for T. gondii infection, showing the distribution of observed foraging events for individual otters classified by prey choice and T. gondii infection status. Individual otters that specialized on snails in this area (n = 5; red) were all positive for T. gondii, whereas individuals that consumed primarily abalone (n = 3; blue) were negative. The difference in prevalence between these 2 groups was especially striking given that there was no obvious spatial segregation, all but 1 of the animals were adult females, and foraging microhabitat was generally similar for snail and abalone feeders as measured by distance to shore, dive depth, and proportion of feeding dives within kelp canopy cover. Depth contours are shown at 5-m intervals and surface kelp canopy is shown as green polygons. To improve resolution, only 25% randomly selected foraging events are shown for each individual. (Inset) Bar graphs show average ±1 SD for proportion dives, depth, and distance to shore for abalone vs. snail feeders.
Table 1.
Risk factors influencing sea otter infection with protozoal pathogens, T. gondii and S. neurona, as identified by logistic regression analyses
| Predictors | Odds ratio (95% C.I.) | P |
|---|---|---|
| For T. gondii | ||
| Diet >10% abalone | 0.2 (0.02–1.5) | 0.107 |
| Diet >10% snails | 11.7 (1.3–109.9) | 0.031 |
| Observed San Simeon to Cambria | 3.8 (1.1–12.5) | 0.031 |
| For S. neurona | ||
| Diet >10% abalone | None (n = 8) exposed | |
| Home range in Monterey cluster | 40.5 (3.9–416.9) | 0.002 |
| Foraging in Rocky Cliff habitat | 5.9 (0.7–47.7) | 0.099 |
| Males | 11.7 (2.0–70.1) | 0.007 |
| Mean 90-day move >1 km | 24.0 (1.7–328.8) | 0.018 |
Factors Increasing Risk of S. neurona Infection.
Sea otter infection with S. neurona was similarly influenced by otter diet composition, range use, habitat use, and mobility. We detected a significant geographic cluster of sea otter infection with S. neurona in a 16.4-km diameter area of the Monterey Peninsula (Fig. 3). Of 24 otters with their home range in this area, 18 (75%) were exposed to S. neurona, whereas only 9 such exposures were expected if this pathogen was distributed randomly along the coastline (spatial scan statistic, P = 0.001). By evaluating foraging habits of otters both inside and outside of this spatial cluster, we found that sea otters foraging in this area used soft sediment habitat along developed shoreline more commonly than otters foraging elsewhere (28% of foraging events within this cluster occurred in soft sediment/developed shoreline habitat compared with 1% in the rest of the range). Sea otters within the cluster also preyed more commonly on clams, fat innkeeper worms (Urechis caupo), and cephalopods than otters outside of this area.
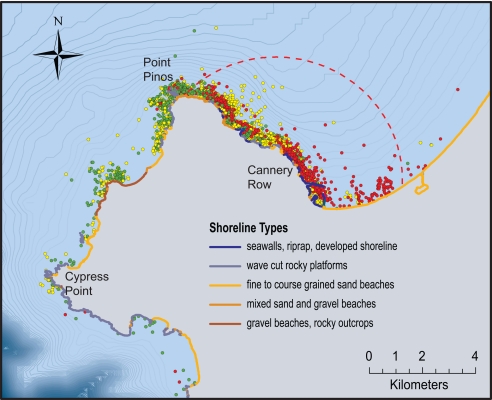
Fig. 3.
Map of the significant spatial cluster for S. neurona in Monterey, California (center, 36.658 N, 121.827 W), showing the boundaries of the spatial cluster (dashed red line) and the distribution of observed foraging events for S. neurona-positive otters (red), S. neurona-negative otters that specialized on abalone (green), and S. neurona-negative otters consuming all other diets (yellow). Shoreline habitat types based on Environmental Sensitivity Index maps are shown. Developed shoreline predominates in the area of highest risk. All sea otters that ate ≥10% abalone were not infected with S. neurona, including those animals foraging within the high-risk cluster. Depth contours are shown at 5-m intervals. Only 25% randomly selected foraging events are shown for each individual.
Because none of the otters with diet composed of ≥10% abalone were exposed to S. neurona, this covariate was highly correlated with the outcome variable, and all 8 otters with diet ≥10% abalone were excluded from the multivariate logistic modeling procedure. For otters not consuming abalone, risk factors significantly related to S. neurona infection in the logistic model included sex, home range within the S. neurona geographical cluster, and mobility (Table 1). Although foraging on clams was linked to S. neurona infection in the univariate analysis, this dietary risk factor was not significant in a model that already contained the high-risk cluster variable because most otters preying on clams were foraging within this cluster. Foraging in exposed rocky cliff habitat (n = 6) was marginally related to S. neurona infection in the univariate analysis (P = 0.099), and including this variable in the multivariate model improved the overall fit (likelihood ratio test χ2 = 2.85, P = 0.091 with reduced AIC and BIC), so the exposed rocky cliff habitat covariate was retained in the S. neurona logistic model. This model also demonstrated excellent overall fit with the data (Hosmer-Lemeshow goodness-of-fit χ2 = 0.80, P = 0.98).
Estimates from the S. neurona logistic model indicate that male sea otters were 12 times more likely to be exposed to S. neurona than females; otters with their home range located within the Monterey spatial cluster were 41 times more likely to have been exposed to S. neurona than otters with their home range outside of this cluster; and otters with mean 90-day move length ≥1 km were 24 times more likely to have been infected with S. neurona than those with mean move length <1 km. For sea otters with home ranges located within the high-risk S. neurona spatial cluster in Monterey, predicted probability of infection with S. neurona was 95% for males and 72% for females. Female otters with home ranges outside of this cluster had only 15% estimated probability of S. neurona exposure, compared with 25% for very site fidelic males and 84% for males with mean 90-day move >1 km.
Discussion
Previous studies based on necropsies of stranded sea otter carcasses have identified T. gondii and S. neurona as pathogens of concern for the southern sea otter population (4, 8). Although sea otters preying on abalone, a historically preferred prey species, were not likely to be infected with either pathogen, sea otters infected with S. neurona exhibited very different prey and habitat use characteristics than sea otters infected with T. gondii. Both pathogens are thought to have terrestrial origins, but these discrete spatial and behavioral patterns of infection illuminate complex processes that challenge a simple model of land to sea pathogen pollution in which sea otters are most at risk directly offshore from high freshwater runoff. We show here that most variation in pathogen prevalence can be explained by the spatial ecology and prey preference of individual otters, illustrating the power of integrating behavioral and health-related studies for understanding the factors promoting pathogen infection in wildlife populations.
Detailed spatial resolution, made possible by intense tracking of individual animal movements, led to our discovery of high T. gondii prevalence near the coastal towns of San Simeon and Cambria (Fig. 2). This finding sheds light on an area of central California noted for high T. gondii prevalence in previous studies that documented clusters of T. gondii-infected carcasses in Estero Bay in the late 1990s (4, 23). The high-risk coastal segment discovered here is adjacent but slightly to the north of Estero Bay (Fig. 1) and otters with home ranges near San Simeon and Cambria may have contributed to these previously detected clusters of beachcast carcasses. Otters that die at sea in this area may tend to drift southward with the prevailing winds and currents, accumulate within sandy embayments, and be detected in areas that are densely populated with humans (4). Although many of the otters using habitat off of San Simeon and Cambria were relatively mobile, our longitudinal data indicate that only 31% (11/36) of T. gondii-seropositive study animals were ever observed traveling as far south as Morro Bay, suggesting that most otters are being exposed to T. gondii within this coastal segment slightly to the north of Estero Bay. Unlike the area noted for high S. neurona prevalence, the San Simeon and Cambria coast is relatively unpopulated and without substantial freshwater inputs from land runoff. Potential sources of T. gondii to this coastal system are unclear at this time, and complex near shore processes, such as currents, sedimentation, and movement of detritus may also influence the distribution of oocysts in seawater and sea otter prey.
The spatial cluster of highly prevalent S. neurona infections in southern Monterey Bay is another important finding emerging from our study of live animals. This high-risk location, immediately adjacent to Fisherman's Wharf and Cannery Row in Monterey, represents a greatly altered and densely populated stretch of coast (Fig. 3). There is a preponderance of soft sediment habitat in this area, and sea otters here foraged more commonly on soft-bottom infauna, such as clams and fat innkeeper worms, than elsewhere in our study. Three of the 4 otters that seroconverted to S. neurona during the follow-up period were juveniles at first capture, and all were observed in the high-risk S. neurona geographical cluster foraging mainly on clams, sand crabs, and sand dollars. Diet preference and foraging habitat were strongly correlated among infected otters in this location, so high S. neurona prevalence here could be related to (i) high-risk diet preferences of most otters in this area, (ii) use of highly developed (urbanized) areas with impacted soft sediment habitat and altered water quality, or (iii) a point source input of S. neurona in this particular part of Monterey Bay. Infection risk drops off sharply just southwest of this cluster near Point Pinos, concurrent with a loss of developed shoreline habitat and a predominance of abalone in the diets of otters foraging there (Fig. 3). Infection with S. neurona in sea otters outside of this Monterey cluster was higher in males and more mobile individuals, who likely encountered secondary high-risk areas, especially in exposed rocky cliff habitat located along the San Simeon coast as identified in the logistic model.
Dietary individuality was an important determinant of infection with protozoal pathogens in sea otters. Trophic mechanisms driving protozoal pathogen exposure are supported by our discovery of both protective and high-risk prey types, whose impacts were independent of sex, home range location, habitat use, or mobility. The very low rate of seroconversion detected by subsequent recaptures of postweanling sea otters suggests that certain behaviors that influence risk of infection are sustained into adulthood. This finding is consistent with observations that individual prey preferences are often life-long and passed down matrilineally from mother to young (24). Sea otters that preyed on abalone were at substantially reduced risk of infection with both protozoal pathogens and remained uninfected throughout the observation period. In contrast, most sea otters that consumed marine snails were infected with T. gondii, regardless of location and habitat use. In fact, T. gondii-seropositive snail feeders were frequently observed foraging in the very same kelp-dominated habitat as T. gondii-seronegative abalone feeders (Fig. 2). Similarly, sea otters preying on abalone were protected from S. neurona infection even if foraging within the high-risk S. neurona cluster near Monterey (Fig. 3).
Although filter-feeding bivalves have been shown to remove T. gondii oocysts from contaminated water in laboratory experiments (15, 16), our findings do not support the hypothesis that bivalves are the primary source of T. gondii infections in wild sea otters. However, our findings are consistent with a role for filter-feeding bivalves, specifically clams and other soft-sediment infaunal invertebrates, in sea otter infections with S. neurona. In contrast, T. gondii infections in otters appear to be related to a diet rich in marine snails. It is possible that T. gondii-infected otters may have altered behavior and preferentially prey on snails, but, more likely, our findings implicate marine snails as a potential transport host for T. gondii, whereby otters acquire infections from the snails themselves or some closely associated process (see SI Text for discussion of potential dietary mechanisms of exposure).
Individual dietary specializations observed in southern sea otters represent a facultative behavioral response to food resource limitation (20, 21). The depletion of preferred prey species at high sea otter densities leads to greater dietary diversity at the population level, as individuals become diet specialists as opposed to diet generalists (21). Selection pressure for individuals to rely on more abundant but lower-quality prey (i.e., prey with low nutritional value or energy content, such as kelp-dwelling marine snails, small infuanal clams, or sand crabs), likely occurred before the nonnative definitive hosts for T. gondii and S. neurona became abundant along the California coast. Our current findings suggest that specialization on marine snails is associated with increased risk of T. gondii exposure. Similarly, trophic transmission of acanthocephalan parasites, another important disease-causing pathogen in sea otters (4, 9, 25), occurs when otters consume larval acanthocephalans within the tissues of sand crabs and spiny mole crabs (Blepharipoda occidentalis). In contrast, abalone was identified by our study as protective for both T. gondii and S. neurona. Abalone has been characterized as preferred prey for sea otters (26, 27), having high nutritional value and energy content, and is often one of the first prey types depleted when otters colonize new areas (28). Together, these patterns are suggestive of an association between pathogen exposure and consumption of lower-quality prey types. If so, elevated pathogen exposure represents a negative consequence of behavioral adaptations that have evolved to cope with limited resources. Under this scenario, disease and increasingly scarce food resources will act synergistically to decrease population-level fitness and limit recovery.
By combining detailed behavioral studies with epidemiologic tools, we have been able to identify biological and behavioral explanations behind the individual and spatial variation in sea otter infection with protozoal pathogens. Sea otters are especially well suited for observational studies, but our approach is broadly applicable to understanding the consequences of prey limitation and habitat use change in other wildlife species. Recent advances in tracking technology have greatly improved our ability to describe movements, foraging behavior, and other potentially important correlates of disease exposure in wild animals. When combined with health and disease indices, these longitudinal studies can elucidate the disease outcomes related to individual behavioral strategies, an approach well established in human epidemiology. Given the growing recognition of individual behavioral specialization strategies in many taxa (29), the methods used here have broad potential utility for better understanding the causes and consequences of disease in wildlife populations.
Materials and Methods
Data Collection.
Sea otters were captured, anesthetized and surgically implanted with VHF transmitters (ATS Inc.) and archival time depth recorders (Mk-9 model; Wildlife Computers) between 1998 and 2004 by using previously established methods (21). Sea otters were captured in 3 primary locations, Monterey, San Simeon, and Point Conception (Fig. 1). All otters underwent basic health screening at the time of capture. Serum was evaluated for antibodies to T. gondii and S. neurona by using an indirect fluorescent antibody test (IFAT) optimized for use in this species (22). For both serum tests, a positive cutoff titer of ≥1:320 was selected to identify otters with confirmed parasite brain infections. Unlike the IFAT test for T. gondii (22), the cutoff titer for S. neurona has not been validated, so seropositive test results may reflect infection with S. neurona or closely related Sarcocystis parasites with serological cross-reactivity. Sex was recorded at capture, and age class was estimated based on animal size and tooth condition. Otters were categorized as juvenile if ≤4 years old or adult if >4 years old at the time of capture.
Once fitted with flipper tags and intraabdominal implants, sea otters were immediately released at the location of capture. Otters were directly observed 4–7 times per week for up to 5 years for survival, reproductive status, location, habitat association, feeding behavior, and prey selection (19–21). To obtain representative behavioral data on each individual, only otters that could be observed for >2 months postcapture were included in the study (n = 118). Otters were relocated by radio signal with VHF receivers, and precise locations were recorded by shore-based observers using compass, laser rangefinder, and global positioning satellite technology. Animals that moved outside of the intensive study areas were relocated twice monthly by a fixed-wing plane using aerial telemetry, which precluded direct observation but still provided accurate data on location and survival. We used fixed kernel techniques (30 and www.spatialecology.com/htools) to estimate annual home range size for each otter, defined as the 90% utilization probability contour (HR90; least-squares cross-validation was used for bandwidth selection). We identified the geographic center and the northernmost and southernmost boundaries of the HR90 polygon for each otter, and in the case of animals with multiple centers of use, we identified the number of geographically distinct polygons and their center point. The relative frequency of sightings of each otter within each of 17 contiguous coastal segments (spanning the entire sea otter range between Half Moon Bay in the north and Santa Barbara in the south) was calculated to describe range-use patterns at a regional scale. We also estimated each otter's mean 90-day move-length, calculated as the net linear displacement distance along the 10-m bathymetric contour between the starting and ending location of each 90-day period of observation (a 90-day cut-off was used because resampling of the dataset suggested that linear displacement distance reached an asymptote after 90 days of sampling). Each otter was then assigned a relative mobility score of 1–6, based on whether their mean 90-day move length was <1, 1–2, 3–4, 5–15, 16–50, or 51–190 km. We further collapsed this variable to just 2 levels, <1 and ≥1 km for the multivariate analyses.
Individual diet preferences have been characterized in this population by using a combination of direct observation and archival data from time-depth recorders (20, 24). Prey type was identified while otters handled prey at the surface after each observed foraging dive. For otters with ≥300 foraging dives spanning at least 1 year of observation (n = 61), we calculated the relative frequency of 14 different prey types (cancer crabs, sea stars, cephalopods, abalone, urchins, kelp crabs, snails, clams, mussels, sand crabs, worms, sand dollars, unidentified crab species, and small kelp-dwelling mollusks, including limpets and chitons) in an individual's diet. To quantify differences in feeding habitat, we calculated the relative frequency of recorded foraging dives for each animal that occurred in each of 9 habitat categories shown on the Environmental Sensitivity Index maps for central California (National Oceanic and Atmospheric Administration, Office of Response and Restoration, Seattle). Foraging habitat types included developed shore with hard substrate, developed shore with soft substrate, exposed rocky cliff, rocky bench, mixed beach and rocky bench, gravel beach, sandy beach, tidal flat, and “offshore” (if the feeding dive was >300 m from shore). The average foraging dive depth, distance to shore, and the proportion of foraging dives in kelp were calculated for each individual by joining geographic information system (GIS) coverage data with foraging dive locations. All activities were covered by an institutional animal care permit issued by University of California Santa Cruz and Federal Permit MA672624 issued by the U.S. Fish and Wildlife Service.
Statistical Analysis.
Analyses were conducted independently for T. gondii and S. neurona, because we did not assume the same risk factors influence sea otter infection with both pathogens. The purpose of our statistical analyses was to evaluate associations of diet composition, home range location, and habitat use patterns with pathogen exposure. Because there were many variables characterizing sea otter habitat use and foraging behavior, we first conducted univariate analyses for each risk factor to identify which independent variables might be related to sea otter protozoal pathogen exposure by using 2 sample t tests and χ2 tests of independence.
We also used the spatial scan statistic Bernoulli model (31) to evaluate significant geographical clustering of infection with T. gondii and S. neurona. We restricted the geographic cluster analysis to otters with narrow range use (≤25 km mean 90-day move length), because pathogen exposure in these more sedentary animals would be more likely to reflect spatially explicit patterns in the risk of exposure. Significant spatial clusters were identified where the number of cases observed significantly exceeded the number of cases expected (P ≤ 0.05) after adjusting for the nonuniform distribution of radio-tagged sea otters along the coastline. All otters were then categorized as to whether any portion of their home range was located within the boundaries of a significant spatial cluster so that this variable could be included in the multivariate analyses.
The putative risk factors identified in univariate analyses were further evaluated in a multivariate framework, using 2 logistic regression models to assess the demographic and behavioral risk factors that increased the probability of sea otter infection with T. gondii and S. neurona. Our multivariate logistic regression analyses were limited to the 61 otters with extensive foraging behavior data. Because susceptible mammals can develop systemic T. gondii infection when exposed to a single oocyst (32), we classified an animal as exposed to a coastal section if we had 1 or more resights of the animal in that area, and we considered an animal exposed to a certain prey type if it represented 10% or more of food intake on the basis of consumed biomass (cut-off values established a priori reflected minimal, low, and moderate levels of exposure, but we collapsed levels that did not differ with respect to their relationship with pathogen infection). Individual use of geographic regions (coastal sections), habitat types, and prey types were thus classified for each study animal, creating a suite of categorical variables. Independent variables that were significantly related to pathogen exposure (P ≤ 0.10) in the univariate analyses and had >5 otters in a category were evaluated as main effects in the logistic model (see Table S1 for distribution of risk categories evaluated), along with all biologically plausible interactions. Likelihood ratios and information theory criterion (AIC and BIC) were used to select final models. Overall model fit was evaluated by the Hosmer and Lemeshow goodness-of-fit statistic (33). To identify the most parsimonious model predicting T. gondii and S. neurona exposure while accounting for low statistical power caused by small sample sizes, we used the likelihood-ratio test to determine whether each variable significantly improved model fit (P ≤ 0.10), compared with a nested model without that variable. Variables that improved model fit and resulted in the highest proportion of correct predicted classifications, while minimizing AIC and BIC, were included in the logistic model for each pathogen. Odds ratios with 95% C.I. were estimated for each main effect by using large-sample maximum-likelihood asymptotic methods, and we calculated model-based exposure probabilities for factors significantly influencing pathogen exposure. All statistical analyses were conducted by using STATA software (STATA SE 10; StataCorp).
Supplementary Material
Acknowledgments.
We thank the collaborating investigators and many volunteers who assisted in sea otter captures and field efforts, Jack Ames, Gena Bentall, Eva Berberich, James Bodkin, Dave Casper, Erin Dodd, George Esslinger, Mike Harris, Brian Hatfield, Marty Haulena, Alisha Kage, Mike Kenner, Ann Melli, Woutrina Miller, Dan Monson, Mike Murray, Teri Nicholson, Andrea Packham, Greg Sanders, Julie Stewart, and Laura Yeates; and Frances Gulland, Jonathon Moore, Tom Suchanek, and Julie Yee, who provided thoughtful reviews of the manuscript. Field work at the Kenneth S. Norris Rancho Marina Reserve was made possible by the UC Natural Reserve System. This work was supported by the U.S. Geological Survey's Western Ecological Research Center, California Department of Fish and Game's Marine Wildlife Veterinary Care and Research Center and Resource Assessment Program, Monterey Bay Aquarium, Minerals Management Agency Grant, and National Science Foundation Grant 0525765.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.E.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806449106/DCSupplemental.
References
- 1.Pedersen AB, Jones KE, Nunn C, Altizer S. Infectious diseases and extinction risk in wild mammals. Conserv Biol. 2007;21:1269–1279. doi: 10.1111/j.1523-1739.2007.00776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leendertz FH, et al. Pathogens as drivers of population declines: The importance of systematic monitoring in great apes and other threatened mammals. Biol Conserv. 2006;131:325–337. [Google Scholar]
- 3.Cleaveland S, et al. The role of pathogens in biological conservation. In: Hudson PJ, Rizzoli A, Grenfell BT, Heesterbeek H, Dobson AP, editors. The Ecology of Wildlife Diseases. Oxford: Oxford Univ Press; 2001. pp. 139–150. [Google Scholar]
- 4.Kreuder C, et al. Patterns of mortality in southern sea otters (Enhydra lutris nereis) from 1998–2001. J Wildl Dis. 2003;39:495–509. doi: 10.7589/0090-3558-39.3.495. [DOI] [PubMed] [Google Scholar]
- 5.Thomas NJ, Cole RA. The risk of disease and threats to the wild population. Endangered Species Update. 1996;13:23–27. [Google Scholar]
- 6.Miller MA, et al. Isolation and characterization of Sarcocystis from brain tissue of a free-living southern sea otter (Enhydra lutris nereis) with fatal meningoencephalitis. Parasitol Res. 2001;87:252–257. doi: 10.1007/s004360000340. [DOI] [PubMed] [Google Scholar]
- 7.Cole RA, et al. Biological and molecular characterizations of Toxoplasma gondii strains obtained from southern sea otters (Enhydra lutris nereis) J Parasitol. 2000;86:526–530. doi: 10.1645/0022-3395(2000)086[0526:BAMCOT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Thomas NJ, et al. Protozoal meningoencephalitis in sea otters (Enhydra lutris): A histopathological and immunohistochemical study of naturally occurring cases. J Comp Pathol. 2007;137:102–121. doi: 10.1016/j.jcpa.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Mayer KA, Dailey MD, Miller MA. Helminth parasites of the southern sea otter Enhydra lutris nereis in central California: Abundance, distribution and pathology. Dis Aquat Org. 2003;53:77–88. doi: 10.3354/dao053077. [DOI] [PubMed] [Google Scholar]
- 10.Dubey J. Advances in the life cycle of Toxoplasma gondii. Int J Parasitol. 1998;28:1019–1024. doi: 10.1016/s0020-7519(98)00023-x. [DOI] [PubMed] [Google Scholar]
- 11.Dubey JP, et al. A review of Sarcocystis neurona and equine protozoal myeloencephalitis (EPM) Vet Parasitol. 2001;95:89–131. doi: 10.1016/s0304-4017(00)00384-8. [DOI] [PubMed] [Google Scholar]
- 12.Dubey J, et al. Toxoplasma gondii, Neospora caninum, Sarcocystis neurona, and Sarcocystis canis-like infections in marine mammals. Vet Parasitol. 2003;116:275–296. doi: 10.1016/s0304-4017(03)00263-2. [DOI] [PubMed] [Google Scholar]
- 13.Miller MA. Tissue cyst-forming coccidia of marine mammals. In: Fowler M, editor. Zoo and Wild Animal Medicine, Current Therapy. 6th Ed. Philadelphia: Saunders; 2008. pp. 319–340. [Google Scholar]
- 14.Conrad PA, et al. Transmission of Toxoplasma: Clues from the study of sea otters as sentinels of Toxoplasma gondii flow into the marine environment. Int J Parasitol. 2005;35:1155–1168. doi: 10.1016/j.ijpara.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Lindsay DS, et al. Survival of Toxoplasma gondii oocysts in Eastern oysters (Crassostrea virginica) J Parasitol. 2004;90:1054–1057. doi: 10.1645/GE-296R. [DOI] [PubMed] [Google Scholar]
- 16.Arkush KD, et al. Molecular and bioassay-based detection of Toxoplasma gondii oocyst uptake by mussels (Mytilus galloprovincialis) Int J Parasitol. 2003;33:1087–1097. doi: 10.1016/s0020-7519(03)00181-4. [DOI] [PubMed] [Google Scholar]
- 17.Miller MA, et al. Type X Toxoplasma gondii in a wild mussel and terrestrial carnivores from coastal California: New linkages between terrestrial mammals, runoff, and toxoplasmosis of sea otters. Int J Parasitol. 2008;38:1319–1328. doi: 10.1016/j.ijpara.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Dubey JP. Toxoplasmosis: A waterborne zoonosis. Vet Parasitol. 2004;126:57–72. doi: 10.1016/j.vetpar.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Tinker MT, et al. Incorporating diverse data and realistic complexity into demographic estimation procedures for sea otters. Ecol Appl. 2006;16:2293–2312. doi: 10.1890/1051-0761(2006)016[2293:iddarc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Tinker MT, Costa DP, Estes JA, Wieringa N. Individual dietary specialization and dive behavior in the California sea otter: Using archival time-depth data to detect alternative foraging strategies. Deep Sea Res II. 2007;54:330–342. [Google Scholar]
- 21.Tinker MT, Estes JA, Bentall G. Food limitation leads to behavioral diversification and dietary specialization in sea otters. Proc Natl Acad Sci USA. 2008;105:560–565. doi: 10.1073/pnas.0709263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller M, et al. Evaluation and application of an indirect fluorescent antibody test (IFAT) for detection of Toxoplasma gondii in sea otters (Enhydra lutris) J Parasitol. 2002;88:594–599. doi: 10.1645/0022-3395(2002)088[0594:EOAIFA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 23.Miller MA, et al. Coastal freshwater runoff is a risk factor for Toxoplasma gondii infection of southern sea otters (Enhydra lutris nereis) Int J Parasitol. 2002;32:997–1006. doi: 10.1016/s0020-7519(02)00069-3. [DOI] [PubMed] [Google Scholar]
- 24.Estes JA, et al. Individual variation in prey selection by sea otters: Patterns, causes, and implications. J Anim Ecol. 2003;72:144–155. [Google Scholar]
- 25.Hennessy S, Morejohn G. Acanthocephalan parasites of the sea otter, Enhydra lutris, off coastal California. California Fish Game. 1977;63:268–272. [Google Scholar]
- 26.Fisher EM. Habits of the southern sea otter. J Mammal. 1939;20:21–36. [Google Scholar]
- 27.Ostfeld RS. Foraging strategies and prey switching in the California sea otter Enhydra lutris. Oecologia. 1982;53:170–178. doi: 10.1007/BF00545660. [DOI] [PubMed] [Google Scholar]
- 28.Wendell F. Relationship between sea otter range expansion and red abalone abundance and size distribution in central California. California Fish Game. 1994;80:45–56. [Google Scholar]
- 29.Bolnick DI, Svanback R, Araujo MS, Persson L. Comparative support for the niche variation hypothesis that more generalized populations also are more heterogeneous. Proc Natl Acad Sci USA. 2007;104:10075–10079. doi: 10.1073/pnas.0703743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worton BJ. Kernel methods for estimating the utilization distribution in home-range studies. Ecology. 1989;70:164–168. [Google Scholar]
- 31.Kulldorf M, Nagarwalla N. Spatial disease clusters: Detection and inference. Stat Med. 1995;14:799–810. doi: 10.1002/sim.4780140809. [DOI] [PubMed] [Google Scholar]
- 32.Dubey JP, et al. Infectivity of low numbers of Toxoplasma gondii oocysts to pigs. J Parasitol. 1996;82:438–443. [PubMed] [Google Scholar]
- 33.Hosmer DW, Lemeshow S. Applied Logistic Regression. New York: Wiley; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.