Abstract
How do we acquire immune tolerance against food microorganisms and commensal bacteria that constitute the intestinal microbiota? We investigated this by stimulating the immune system of adults with commensal Lactobacillus plantarum bacteria. We studied the in vivo human responses to L. plantarum in a randomized double-blind placebo-controlled cross-over study. Healthy adults ingested preparations of living and heat-killed L. plantarum bacteria. Biopsies were taken from the intestinal duodenal mucosa and altered expression profiles were analyzed using whole-genome microarrays and by biological pathway reconstructions. Expression profiles of human mucosa displayed striking differences in modulation of NF-κB-dependent pathways, notably after consumption of living L. plantarum bacteria in different growth phases. Our in vivo study identified mucosal gene expression patterns and cellular pathways that correlated with the establishment of immune tolerance in healthy adults.
Keywords: commensal bacteria, expression profiling, host–microbe interactions, pathway analysis
Human intestinal epithelia are continuously exposed to the luminal microbiota. In healthy humans, this exposure to non-self stimuli activates balanced and interconnected innate and adaptive immune responses (1–5), leading to tolerance toward nonpathogenic bacteria. Tolerance is a general term describing the state by which the immune system is rendered nonreactive toward self- or non-self antigens. However disproportionate responses associate with diseases such as Crohn's (6, 7).
Intestinal epithelial cells play a central role in mediating balanced immune responses, among others by activating immune cells that reside in the lamina propria (3, 5, 8). Whereas pathogens induce mainly persistent inflammatory responses (9–11), commensal bacteria induce transient, noninflammatory responses. Commensal Lactobacillus species may stimulate the polarization of immune T cells toward regulatory T (Treg) cells (12). Such noninflammatory responses can be comediated by secreted proteins (13) or via wall fragments as found for L. plantarum (14, 15).
Lactobacillus plantarum WCFS1 is a common, sequenced food bacterium (16). Several studies have addressed its adaptive responses to the mammalian gastrointestinal tract (17–19). To current knowledge, L. plantarum is perceived via Toll-like receptor (TLR)2–4, CD14 antigen, and nucleotide-binding oligomerization domain-containing 2 (NOD2) (20–22). L. plantarum is a nonspecific stimulator of the immune response, initially identified as the major determinant of the adjuvanticity of a mistletoe preparation (23, 24), which in animal and human in vitro models promotes the secretion of the cytokines tumor necrosis factor-alpha (TNF-α) and interleukin(IL)-12 (23, 25). These cytokines are also produced by human monocytes upon stimulation by L. plantarum (21). Intriguingly, L. plantarum may induce innate or adaptive mouse immune responses, dependent on the viability of the bacteria (24).
Here, we describe the in vivo (immune) responses in humans after consumption of L. plantarum at the level of mucosal gene transcription, which comprehensively describes human responses to bacteria (26, 27). Mucosal transcriptional responses were determined in vivo in the duodenum, the proximal part of the small intestine, after 6 h consumption of L. plantarum, according to a randomized double-blind placebo-controlled cross-over design. Duodenal mucosa are the first small intestinal areas coming in contact with L. plantarum, minimizing the adaptive changes the bacteria might go through during passage of the intestinal tract. In addition, the duodenum is relatively accessible and does not require severely invasive sampling techniques. Finally, this intestinal region contains the lowest endogenous microbiota colonization level, ensuring that the measured responses are as specific as can be achieved. Considering the possible differential adjuvanticity response to different L. plantarum growth phases (24, 28) 3 preparations of bacteria were tested: (i) the logarithmic-phase of growth (exponentially growing; “midlog”), (ii) the stationary-phase of growth (“stationary”), and (iii) heat-killed “stationary” bacteria (10 min 85 °C; “dead”). By comprehensive pathway analysis, detailed differences between in vivo mucosal expression profiles could be related to modulation of nuclear factor (NF-) κB cascades and its antagonists. The midlog and stationary bacterial preparations provide molecular leads to the bacterial factors mediating these NF-κB-dependent mucosal responses because the molecular composition of L. plantarum cells at different growth stages is reported in refs. 29 and 30 and is studied in several transcriptome (31) and proteome (32) studies (18). This study provides a model for the in vivo establishment of immune tolerance in the small intestines of healthy adults after consumption of substantial amounts of bacteria with prominent adjuvanticity.
Results
Profiling Mucosal Gene Expression, Using Microarrays, Quantitative PCR and Laser Capture Microdissection.
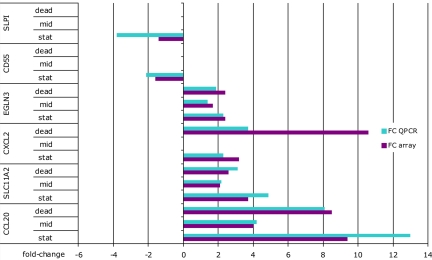
Here, we present genome-wide epithelial transcriptional profiles of the proximal duodenum of healthy adults in response to ≈2 × 1012 Lactobacillus plantarum WCFS1 cells. L. plantarum is safe for human consumption (MSDS, Public Health Agency Canada). None of the volunteers experienced any discomfort during or after the 6-h consumption period, leading to duodenal tissue sampling (biopsies) by standard flexible gastroduodenoscopy. Expression profiles were analyzed on whole-genome microarrays. Normalized expression datasets are deposited at GEO (National Center for Biotechnology Information), series number GSE11355. Array data were validated for 6 genes for each person and each intervention by quantitative reverse-transcription PCR (QPCR). QPCR confirmed the expression levels for the selected genes (Fig. 1 and Fig. S1) and did also show the subject-to-subject variation measured by the arrays (SI Results). The specificity of gene expression in epithelia was evaluated using enriched epithelial cell pools (consisting for >80% of epithelial cells; Fig. S2) obtained by laser capture microdissection (LCM; 33). Expression of genes encoding typical epithelial functions was indeed higher in epithelial cell-fractions obtained by LCM (SI Results). The quality of the microarray datasets was therefore sufficient to measure modest changes in mucosal gene transcription.
Fig. 1.
Validation of microarray results by QPCR, using total RNA from biopsies (pooled data from 8 persons). After consumption of midlog and dead bacteria, amounts of mRNA for the genes CD55 and SLPI were low. In all figures and text, down-regulated genes are indicated with negative fold-changes for ease of interpretation and comparison, although this is mathematically incorrect.
Differential Transcriptional Response in Duodenal Mucosa upon Exposure to Different Preparations of L. plantarum.
Between 400 (stationary vs. placebo) and 800 (midlog vs. placebo) genes were differentially regulated after consumption of bacteria (SI Results). The largest fold-changes were observed in the comparison dead-placebo, varying between −6.5 and 10.5; overlaps between the differentials were between 10 and 35% (SI Results). Shared genes were mainly involved in general cellular functions and metabolism.
To gain insight into the mucosal processes that were altered after consumption of bacteria, significantly overrepresented Gene Ontology (GO) classes (34) were calculated (Table S1). Dead bacteria induced especially genes involved in innate and adaptive human immune responses. Living bacteria induced immune response-related genes as well, but also genes involved in basal cellular activity and metabolism. Strikingly, there was a pronounced difference in mucosal responses to L. plantarum harvested at midlogarithmic (“midlog”) or stationary phase of growth (“stationary”). Responses to stationary bacteria were associated with induction of genes regulating immune responses and stimulation of cellular physiology, whereas responses to midlog bacteria were associated with nucleic acid metabolism, cytoplasm organization and (ribosome) biogenesis (Table S1).
We also performed a parallel gene set enrichment analysis (GSEA) to focus on coexpressed groups of genes that function in specific biochemical, metabolic, or signal transduction routes (35). Especially dead bacteria, and to a lesser extent stationary bacteria, induced expression profiles associated with immune responses and modulation of cell death. In contrast, consumption of midlog bacteria induced responses associated with cellular growth, proliferation and biogenesis (Table S2).
These expression profiles show that consumption of L. plantarum leads to regulation of hundreds of duodenal mucosal genes that modulate immune responses, cellular metabolism and biogenesis. They also underline that mucosa respond differentially to different preparations of the same bacterial strain.
Mucosal Cellular Pathways Altered After Consumption of L. plantarum Cluster Around the NF-κB Transcription Factor Complex.
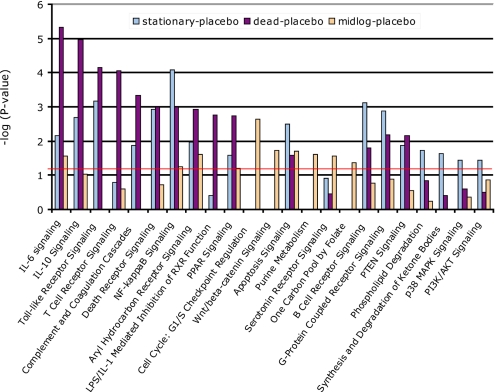
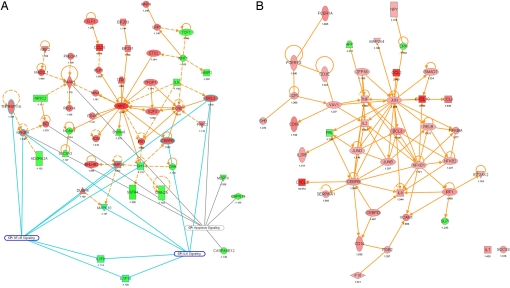
Biological pathway reconstruction provides powerful tools to analyze expression profiles (36). To this goal, all regulated genes were used as input for Ingenuity Pathway Analysis (IPA). IPA computes networks and ranks these according to a statistical likelihood approach (37). Networks with a score of at least 10 focus genes were considered to have biological relevance and to show part of the underlying biology of the mucosal responses to lactobacilli. We found 25 regulated biological networks after consumption of dead bacteria, 14 after stationary, and 24 after consumption of midlog bacteria (Table S3). In addition, 19, 8, and 16 canonical pathways were significantly modulated after consumption of dead, midlog, and stationary bacteria (P < 0.05; Fig. 2). These networks and pathways were in agreement with the GSEA and GO enrichment analysis results. We also projected transcription and pathway information on protein–protein interaction networks for specific cellular functions such as “immune response” and identified between 3 (consumption of midlog bacteria) and 5 to 8 (consumption of stationary and dead bacteria) major nodes that were central in connecting many of the genes with altered expression after consumption of bacteria. Fig. 3A shows how the major nodes v-myc oncogene homolog (MYC), poly (ADP-ribose) polymerase family, member 1 (PARP1) and cyclin D1 (CCND1) were identified for the midlog versus placebo comparison. Major nodes in the response to stationary (Fig. 3B) and dead bacteria included TNF-α and NF-κB transcription factors, and jun oncogene (JUN). Nodes emphasizing the differential role of NF-κB were also identified in an alternative analysis (SI Results).
Fig. 2.
Cellular pathways significantly modulated after bacterial consumption. Statistical significance of pathway modulation was calculated via a right-tailed Fisher's Exact test in Ingenuity Pathway Analysis and represented as −log(P value); −log values exceeding 1.30 were significant (P < 0.05).
Fig. 3.
Ingenuity protein–protein interaction networks. The interaction map was derived by plotting interacting proteins involved in specific cellular functions: immune response, immune system development and function, cell signaling, cell cycle, cell growth, cell death and proliferation. Transcriptional information was projected onto the interaction map; up-regulated genes are depicted in shades of red, down-regulated genes in shades of green. Relevant pathways that feature modulated genes were indicated as well. (A) transcriptional networks modulated after consumption of midlog bacteria. From this interaction map, it can be seen that the up-regulated PARP1, MYC, and CCND1 are important nodes associated with the mentioned pathways. (B) transcriptional networks modulated after consumption of dead bacteria. Major nodes are proteins from the CEBP, JUN, and NFKB family but also proinflammatory TNF and IFN regulatory factor, IRF1.
Pathways induced after consumption of L. plantarum correlated with immune responses, signaling, cell death modulation and lipid and fatty acid metabolism (Fig. 2). Mucosal responses to dead and stationary bacteria were more similar to each other and different from the response to midlog bacteria. Pivotal was the differential expression of NF-κB, of interest because NF-κB subunit composition determines its function (38, 39). Bacterial stimulation of TLR and NOD receptors leads to nuclear activity of a p50-p65 NF-κB complex (40, 41). Preparations of dead bacteria induced increased expression of nearly all NF-κB subunits except p65 (RelA) but also of 3 antagonists of NF-κB activity: B-cell CLL/lymphoma 3 (BCL3), TNF alpha-induced protein 3 (A20), and NF-κB inhibitor, IκB (Table S4 and Fig. S3). Preparations of stationary bacteria induced expression of p50 and p52 NF-κB subunits together with BCL3, A20 and IκB. Strikingly, midlog bacteria induced only expression of the NF-κB antagonists IκB and BCL3 (Table S4 and Fig. S3). Modulation of NF-κB activity was apparent from regulation of NF-κB target genes such as the chemokine CXCL2 (42). CXCL2 expression was not significantly increased in response to midlog bacteria, while it was among the genes with highest increased expression levels after consumption of stationary or dead bacteria (Fig. 1). Two major nodes that were identified after consumption of midlog bacteria: MYC and cyclin D1, are major regulators of the Wingless-type (Wnt)/β-catenin pathway. This proliferation and development-promoting pathway, an antagonist of the NF-κB cascade, was only modulated after consumption of midlog bacteria (Fig. 2). Differential regulation of immune response genes became very clear upon reconstruction of immune response and immune and lymphatic tissue development gene-networks (Fig. S4). Together, these findings demonstrate that NF-κB signaling and its downstream or antagonizing pathways can be differentially induced by different preparations of the same bacterial species.
Genes Essential in Mediating Appropriate Immune Responses Are Regulated in Human Mucosa After Consumption of L. plantarum.
Factors such as A20 (43), triggering receptor expressed on myeloid cells-like 1 (TREML1; 44), BCL3 (45), suppressor of cytokine signaling (SOCS) 3 (46) and secretory leukocyte peptidase inhibitor (SLPI; 3,8) protect tissues from overly aggressive immune responses. Other factors such as a proliferation inducing ligand (APRIL), thymic stromal lymphopoietin (TSLP), TREM1 (47), and B-cell activating factor (BAFF; 48) stimulate or amplify inflammatory responses, proliferation and survival of immune cells (3, 4, 48). The genes encoding A20, BCL3, IκBα, and SOCS3, negative regulators of NF-κB activity, were induced together with NF-κB after consumption of dead or stationary L. plantarum (Table S4). APRIL (48) was induced after consumption of stationary bacteria; the often coexpressed BAFF was not expressed. TREM1 was down-regulated after consumption of midlog L. plantarum; only this preparation induced expression of adrenomedullin (ADM), a peptide involved in immune tolerance (49). The costimulatory TREML1 was down-regulated after consumption of dead and midlog bacteria. SLPI and TSLP together regulate class-switching of Ig (Ig)A leading to production of broadly reactive IgA and IgG antibodies, a process that is also dependent on the enzyme AID (8). Only very low basal expression of TSLP and AID was found after consumption of bacteria (Table S4); SLPI was down-regulated after consumption of stationary and dead bacteria. In other words, no coordinated up-regulation of crucial genes driving the activation of inflammatory immune responses was apparent from the expression profiles. Based on our analyses, the mucosal response model depicted in Fig. 4 was compiled. To validate the outcome of this model, biopsy sections from 3 of the volunteers taken after consumption of the bacterial preparations and placebo control were microscopically examined. No infiltration or increased activity of immune cells (e.g., large-scale degranulation of neutrophils) was visible after consumption of bacteria (Fig. 5). Taken together, our study shows that consumption of L. plantarum by healthy adults leads to induction of a duodenal mucosal gene expression program largely determined by NF-κB-, JUN- and TNF-dependent pathways after consumption of stationary and dead bacteria, and by MYC- and cyclin D1-dependent pathways after consumption of midlog bacteria. Histological examination of biopsy sections showed that none of these preparations led to infiltration of immune cells.
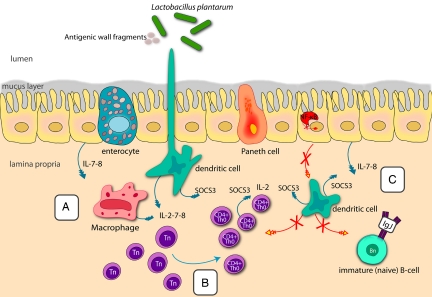
Fig. 4.
Proposed model of cellular proteins and processes associated with immune tolerance induced in the proximal small intestine of healthy adults after consumption of L. plantarum. (A and B) Because of up-regulation of factors such as A20 and IκBα and lack of p65 (RelA), no continuous NF-κB driven proinflammatory gene transcription is expected. Interleukins (blue arrows) are exclusively up-regulated during the interaction with dead bacteria (A) and are proposed to stimulate the maturation of CD4+ cells (B); overt stimulation by ILs is probably balanced by the suppressor SOCS3. (C) The crossed-out red arrows between IECs and dendritic, T and B cells indicate that no stimulatory or activating contact takes place between these cells through lack of expression of necessary genes such as BAFF, AID, IL-1, and TSLP.
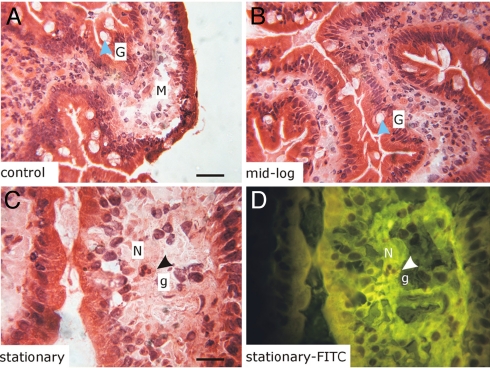
Fig. 5.
Histological examination of human biopsies does not show evidence for infiltration of immune cells after consumption of L. plantarum. The bright-field images (A–C) are from representative areas of biopsy sections; the FITC fluorescence image (D Bottom Right) is from the same area of the section labeled “stationary” (C) and shows autofluorescent granules. G, goblet cell; M, macrophage; N, neutrophil; g, granules. (Magnifications: A and B, 500×; C and D, 1,000×. Scale bars: A, 50 μm; C, 25 μm.) Images are representative for at least 12 sections from biopsies taken after consumption of the 3 bacterial preparations and placebo control by 3 individuals.
Discussion
This in vivo study used a double-blind placebo-controlled randomized cross-over design to investigate human responses to a common food bacterium. L. plantarum has been the subject of biochemical and immunological studies since the early 1960s and potently stimulates human immune cells. Here, we present biological networks and pathways with special attention for mucosal immune responses, and histological examination of human duodenal mucosa after consumption of L. plantarum. The expression profiles correspond to biopsies obtained from healthy human adults after consumption of 3 different preparations containing ≈2 × 1012 stationary, midlog or dead L. plantarum. L. plantarum reaches relatively high cell densities, exceeding 5 × 109 colony forming units per ml under standard culture conditions. The dosage used in this study could be expected to be present in relatively normal sized food-portions of somewhat >200 mL. Although consumed in a live state, some proportion of the L. plantarum culture will probably not survive stomach passage. Nevertheless, several studies show that L. plantarum displays relatively favorable survival in the human intestinal tract (50, 51). The results imply that consumption of nonpathogenic L. plantarum modulates in vivo transcriptional profiles of the proximal small intestinal mucosa involved in innate and adaptive immune responses. Biological context was provided for expression datasets by transcriptome-based pathway reconstruction that clusters modulated genes in cellular pathways that when compiled into a cell- or tissue-“snapshot,” can be compared between treatments (36). After consumption of L. plantarum, genes that prevent overt adaptive immune responses were induced, whereas genes involved in establishing and amplifying inflammatory immune responses were not expressed or not modulated. Analogously, histological tissue-examination revealed no infiltration of immune cells after consumption of bacteria. The transcriptional profiles presented here therefore seem to represent an expression “blueprint” for establishing immune tolerance in healthy humans.
The cell-wall of L. plantarum contains peptidoglycan and teichoid acids (TAs) that may bear glycosyl and d-alanine decorations (30, 52). In vitro, TA-coupled d-alanine of L. plantarum strongly stimulates cultured lymphocytes and monocytes (15). Antigenicity of the L. plantarum wall TA depends on its association with the protein fraction (14). Heat treatment of L. plantarum may release substantial amounts of immunogenic wall components (53) including immunogenic proteins and wall TAs (14) and can be expected to increase TA-accessibility for mucosal cells (15, 29, 30). This might in part explain the prominent comodulation of immune response-related processes after consumption of preparations containing heat-killed L. plantarum.
Differential immune responses toward living and dead L. plantarum are described in ref. 24. Bloksma and colleagues showed that s.c. mouse injections with preparations of viable L. plantarum induced mild inflammatory responses, whereas dead bacteria induced antibody formation by plasma cells and infiltration of polymorphonuclear cells (24). Although our study differs drastically from these mouse model studies, the human expression profiles show correlation; only consumption of dead L. plantarum leads to increased expression of TNF-α and of genes involved in T cell activation, antigen processing and presentation. In addition, only consumption of dead L. plantarum induced expression of genes involved in attraction and maturation of T cells such as the genes encoding the interleukins 2, 7, and 8. However, possibly through action of immune-modulatory factors such as SOCS3, no infiltration of immune cells was observed in human biopsy sections.
One major finding of this study was the differential regulation of NF-κB subunits after consumption of different preparations of L. plantarum. Dead and stationary bacteria induce mucosal expression of genes encoding NF-κB subunits. In the absence of inhibitory factors such as BCL3 or A20 and in presence of the cytokine TNF-α, nuclear activity of heterodimerised p50-p65 subunits is a major cause of increased transcription of genes involved in inflammatory responses (38, 39, 45). In our study, the p65 subunit (RelA) was not induced and only showed basal expression. The genes encoding p50-p52 and RelB-p52 complexes (the latter after ingestion of dead bacteria) showed elevated mucosal expression. The RelB-p52 dimer drives transcription of genes involved in lymphoid organogenesis (38) that belongs to the category of “Immune and lymphatic system development and function,” which is among the strongest regulated cellular functions induced after consumption of dead L. plantarum (Table S5). RelB may associate with the aryl hydrocarbon receptor (AHR) and together regulate responses to nonself molecules (54). The AHR signaling pathway was regulated after consumption of all 3 bacterial preparations suggesting that in the proximal duodenum of healthy humans, RelB-dependent NF-κB signaling and AHR signaling may participate in regulating responses to food bacteria, potentially with roles for the NF-κB inhibitors A20, IκB, BCL3, and SOCS3.
Evaluating the expression of factors that amplify or costimulate inflammatory immune responses after consumption of L. plantarum was considered highly relevant because proper expression of genes encoding such factors appears to be compromized in persons suffering from intestinal diseases (1, 55). The epithelial cytokine TSLP stimulates, together with AID, APRIL and BAFF, dendritic cells to induce class switching (8). Expression of TSLP was not altered after consumption of L. plantarum and only expressed at very low levels in all persons. Increased expression of APRIL was induced only after consumption of stationary bacteria. Overall, analysis of mucosal expression profiles after consumption of L. plantarum suggests that these profiles are not consistent with a systematic promotion of proinflammatory immune responses but are more consistent with balanced duodenal mucosal responses to commensal bacteria (7, 10).
One striking finding after consumption of midlog L. plantarum was the lack of induction of immune response-associated genes but instead, induction of genes associated with anti-inflammatory activities, such as BCL3, IκB and ADM. The proximal duodenal mucosal response to midlog L. plantarum bacteria is also unique in the up-regulation of MYC, PARP1 and cyclin D1, potent positive regulators of cell proliferation. These findings, together with up-regulation of pathways modulating metabolic functions and growth (via Wnt/β-catenin signaling; Fig. 2), suggest that midlog bacteria stimulate proliferation in the proximal region of the small intestine. Remarkably, L. plantarum has been shown to stimulate cell growth and proliferation of the mouse spleen and liver (24).
We measured an acute 6-h response to L. plantarum. In view of the short time between reconstitution of bacteria and passage through the gut, the L. plantarum cells are not expected to go through major changes of their cell wall composition during these experiments. Our preliminary results using L. plantarum suggests that one of the most prominent changes upon transition from midlogarithmic to stationary phase of growth relates to cell envelope and exopolysaccharide- (EPS-) associated functions. Such cell wall differences between the bacterial preparations might at least partially explain the different responses induced in vivo in human intestinal mucosa. Current transcriptomic, proteomic and glycomic analyses are aimed to unravel the molecular characteristics of L. plantarum cell walls in more detail. Thereby, this study shows the potential for obtaining molecular leads toward differential modulation of the NF-κB network. Experiments testing the immunomodulatory effects of food bacteria that present different wall antigens appear to be within reach.
Materials and Methods
Bacterial Preparations.
Lactobacillus plantarum was cultured at 37 °C in MRS medium (Merck). To obtain stationary phase or midlog cultures, bacteria were cultured overnight or until an optical density at 600 nm of 1.0 was reached. Dead bacteria were heat-treated (10 min 85 °C) stationary bacteria. Maltodextrin and glucose were added to a final concentration of 20% and 2% (wt/vol) respectively to obtain bacterial preparations; placebo controls only contained the two sugars. The freeze-dried midlog and stationary-phase bacterial preparations contained ≈0.8 and 0.9 × 1011 CFUs per ml; this means that >85% of the bacteria present before freeze-drying did remain viable. Detailed protocols for culturing, handling and storage of bacteria and determination of viable counts can be found in SI Methods.
Volunteers.
This study was approved by the University Hospital Maastricht Ethical Committee, and conducted in full accordance with the principles of the “Declaration of Helsinki.”* All subjects gave their written informed consent before their inclusion into the study. Eight healthy nonsmoking volunteers (24 ± 4y) without a history of gastrointestinal symptoms and free of medication were investigated on 4 separate occasions (three bacterial interventions and one placebo control, randomly chosen) in a randomized placebo-controlled cross-over study. Interventions were separated by a 2-week wash-out period; this 2-week period did allow for complete healing of the biopsy sampling region. Volunteers fasted overnight (without breakfast) and were administered 1 × 150 mL at the start of the intervention, after which they were each 30 min provided with a preparation containing reconstituted freeze-dried bacteria resuspended in maltodextrin solution just before consumption, or only containing the maltodextrin solution (the placebo control), for a period of 6 h. Both the volunteers and the researchers did not know whether a bacterial preparation or a placebo control was provided (“double-blind” study). After this 6-h period, 4–5 tissue samples were obtained from the horizontal part of the duodenum by standard flexible gastroduodenoscopy, at ≈15 cm distal to the pylorus.
Transcriptome Analysis.
Total RNA, isolated from biopsies, was labeled and hybridized to human Genome U133 Plus 2.0 arrays (Affymetrix) using standard methods (see SI Methods). Transcriptome datasets were processed using statistical and functional analyses (see SI Methods).
Tissue Section Analyses.
Tissue sectioning and laser capture microdissection were used to obtain epithelial cell preparations, which were used for RNA extraction and reverse transcription quantitative PCR to assess the expression levels of selection genes (see SI Methods). Standard methodology was employed for histological and microscopic evaluation of tissue sections (for details see SI Methods).
Supplementary Material
Acknowledgments.
We thank J. Jansen and M. Grootte Bromhaar (Human Nutrition, Wageningen University) for excellent microarray hybridisations, M. Boekschoten and S. Keshtkar (Human Nutrition, Wageningen University) for assistance with Bibliosphere analyses and histology, J. van Lent (Laboratory of Virology, Wageningen University) for fluorescence microscopy facilities, and E. Nieuwenhuis and J. Samsom (Erasmus University Medical Centre, Rotterdam, The Netherlands) for helpful insights and critical reading of the manuscript. This work was supported by the BioRange program of the Netherlands Bioinformatics Centre, which is supported by a Besluit Subsidies Investeringen Kennisinfrastructuur grant through the Netherlands Genomics Initiative.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE11355).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809919106/DCSupplemental.
Please see www.wma.net/e/policy/b3.htm.
References
- 1.Clavel T, Haller D. Molecular interactions between bacteria, the epithelium, and the mucosal immune system in the intestinal tract: Implications for chronic inflammation. Curr Issues Intest Microbiol. 2007;8:25–43. [PubMed] [Google Scholar]
- 2.Kabelitz D, Medzhitov R. Innate immunity - cross-talk with adaptive immunity through pattern recognition receptors and cytokines. Curr Opin Immunol. 2007;19:1–3. doi: 10.1016/j.coi.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 3.McHeyzer-Williams M. Local sentries for class switching. Nat Immunol. 2007;8:230–232. doi: 10.1038/ni0307-230. [DOI] [PubMed] [Google Scholar]
- 4.MacPherson AJ, McCoy K. APRIL in the intestine: A good destination for immunoglobulin A2. Immunity. 2007;26:755–757. doi: 10.1016/j.immuni.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Fritz JH, Le Bourhis L, Magalhaes JG, Philpott DJ. Innate immune recognition at the epithelial barrier drives adaptive immunity: APCs take the back seat. Trends Immunol. 2008;29:41–49. doi: 10.1016/j.it.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Sansonetti PJ. The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006;12:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- 7.Baumgart DC, Carding SR. Inflammatory bowel disease: Cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 8.Xu W, et al. Epithelial cells trigger frontline immunoglobulin class switching through a pathway regulated by the inhibitor SLPI. Nat Immunol. 2007;8:294–303. doi: 10.1038/ni1434. [DOI] [PubMed] [Google Scholar]
- 9.Foster TJ. Immune evasion by Staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 10.Sansonetti PJ, Di Santo JP. Debugging how bacteria manipulate the immune response. Immunity. 2007;26:149–161. doi: 10.1016/j.immuni.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Schnaith A, et al. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem. 2007;282:2695–2706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- 12.Mohamadzadeh M, et al. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc Natl Acad Sci USA. 2005;102:2880–2885. doi: 10.1073/pnas.0500098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan F, et al. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterol. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knox KW, Wicken AJ. Serological studies on the teichoic acids of Lactobacillus plantarum. Infect Immun. 1972;6:43–49. doi: 10.1128/iai.6.1.43-49.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grangette C, et al. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleerebezem M, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bron PA, Grangette C, Mercenier A, de Vos WM, Kleerebezem M. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J Bacteriol. 2004;186:5721–5729. doi: 10.1128/JB.186.17.5721-5729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marco ML, Pavan S, Kleerebezem M. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol. 2006;17:204–210. doi: 10.1016/j.copbio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Marco ML, Bongers RS, de Vos WM, Kleerebezem M. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl Environ Microbiol. 2007;73:124–132. doi: 10.1128/AEM.01475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlsson H, Hessle C, Rudin A. Innate immune responses of human neonatal cells to bacteria from the normal gastrointestinal flora. Infect Immun. 2002;70:6688–6696. doi: 10.1128/IAI.70.12.6688-6696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson H, Larsson P, Wold AE, Rudin A. Pattern of cytokine responses to gram-positive and gram-negative commensal bacteria is profoundly changed when monocytes differentiate into dendritic cells. Infect Immun. 2004;72:2671–2678. doi: 10.1128/IAI.72.5.2671-2678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasegawa M, et al. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem. 2006;281:29054–29063. doi: 10.1074/jbc.M602638200. [DOI] [PubMed] [Google Scholar]
- 23.Bloksma N, van Dijk H, Korst P, Willers JM. Cellular and humoral adjuvant activity of mistletoe extract. Immunobiol. 1979a;156:309–318. [PubMed] [Google Scholar]
- 24.Bloksma N, de Heer E, van Dijk H, Willers JM. Adjuvanticity of lactobacilli. I. Differential effects of viable and killed bacteria. Clin Exp Immunol. 1979b;37:367–375. [PMC free article] [PubMed] [Google Scholar]
- 25.Heinzerling L, von Baehr V, Liebenthal C, von Baehr R, Volk HD. Immunologic effector mechanisms of a standardized mistletoe extract on the function of human monocytes and lymphocytes in vitro, ex vivo, and in vivo. J Clin Immunol. 2006;26:347–359. doi: 10.1007/s10875-006-9023-5. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi SD, et al. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci USA. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaffrey RL, et al. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci USA. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bloksma N, et al. Effects of lactobacilli on parameters of non-specific resistance of mice. Med Microbiol Immunol. 1981;170:45–53. doi: 10.1007/BF02123796. [DOI] [PubMed] [Google Scholar]
- 29.Wicken AJ, Broady KW, Ayres A, Knox KW. Production of lipoteichoic acid by lactobacilli and streptococci grown in different environments. Infect Immun. 1982a;36:864–869. doi: 10.1128/iai.36.3.864-869.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wicken AJ, Evans JD, Campbell LK, Knox KW. Teichoic acids from chemostat-grown cultures of Streptococcus mutans and Lactobacillus plantarum. Infect Immun. 1982b;38:1–7. doi: 10.1128/iai.38.1.1-7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serrano LM, et al. Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb Cell Fact. 2007;6:29. doi: 10.1186/1475-2859-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen DP, et al. Proteomic analysis of log to stationary growth phase Lactobacillus plantarum cells and a 2-DE database. Proteomics. 2006;6:6485–6493. doi: 10.1002/pmic.200600361. [DOI] [PubMed] [Google Scholar]
- 33.Espina V, et al. Laser-capture microdissection. Nat Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 34.Lee HK, Braynen W, Keshav K, Pavlidis P. ErmineJ: Tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6:269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Baarlen P, van Esse HP, Siezen RJ, Thomma BP. Challenges in plant cellular pathway reconstruction based on gene expression profiling. Trends Plants Sci. 2008;13:44–50. doi: 10.1016/j.tplants.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 38.Wietek C, O'Neill LA. Diversity and regulation in the NF-κB system. Trends Biochem Sci. 2007;32:301–309. doi: 10.1016/j.tibs.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 40.Chen LF, Greene WC. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 41.Hayden MS, West AP, Ghosh S. NF-κB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 42.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Negative regulation of Toll-like receptor signaling by NF-κB p50 ubiquitination blockade. Science. 2007;317:675–678. doi: 10.1126/science.1142953. [DOI] [PubMed] [Google Scholar]
- 43.Hitotsumatsu O, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Washington AV, Quigley L, McVicar DW. Initial characterization of TREM-like transcript (TLT)-1: A putative inhibitory receptor within the TREM cluster. Blood. 2002;100:3822–3824. doi: 10.1182/blood-2002-02-0523. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Wang H, Claudio E, Brown K, Siebenlist U. A role for the IκB family member BCL3 in the control of central immunologic tolerance. Immunity. 2007;27:438–452. doi: 10.1016/j.immuni.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 47.Schenk M, Bouchon A, Seibold F, Mueller C. TREM-1-expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J Clin Invest. 2007;117:3097–3106. doi: 10.1172/JCI30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacLennan I, Vinuesa C. Dendritic cells, BAFF, and APRIL: Innate players in adaptive antibody responses. Immunity. 2002;17:235–238. doi: 10.1016/s1074-7613(02)00398-9. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Rey E, Chorny A, Delgado M. Regulation of immune tolerance by anti-inflammatory neuropeptides. Nat Rev Immunol. 2007;7:52–63. doi: 10.1038/nri1984. [DOI] [PubMed] [Google Scholar]
- 50.Vesa T, Pochart P, Marteau P. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment Pharmacol Ther. 2000;14:823–828. doi: 10.1046/j.1365-2036.2000.00763.x. [DOI] [PubMed] [Google Scholar]
- 51.Ahrné S, et al. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J Appl Microbiol. 1998;85:88–94. doi: 10.1046/j.1365-2672.1998.00480.x. [DOI] [PubMed] [Google Scholar]
- 52.Ikawa M. The partial chemical degradation of the cell walls of Lactobacillus plantarum, Streptococcus faecalis, and Lactobacillus casei. J Biol Chem. 1961;236:1087–1092. [PubMed] [Google Scholar]
- 53.Campbell LK, Knox KW, Wicken AJ. Extractability of cell wall polysaccharide from lactobacilli and streptococci by autoclaving and by dilute acid. Infect Immun. 1978;22:842–851. doi: 10.1128/iai.22.3.842-851.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogel CF, et al. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol. 2007;21:2941–2955. doi: 10.1210/me.2007-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.