Abstract
There are clinical parallels between the nature and course of depressive symptoms in major depressive disorder (MDD) and those of inflammatory disorders. However, the characterization of a possible immune system dysregulation in MDD has been challenging. Emerging data support the role of T-cell dysfunction. Here we report the association of MDD and antidepressant response to genes important in the modulation of the hypothalamic-pituitary-adrenal axis and immune functions in Mexican Americans with major depression. Specifically, single nucleotide polymorphisms (SNPs) in two genes critical for T-cell function are associated with susceptibility to MDD: PSMB4 (proteasome β4 subunit), important for antigen processing, and TBX21 (T bet), critical for differentiation. Our analyses revealed a significant combined allele dose-effect: individuals who had one, two and three risk alleles were 2.3, 3.2 and 9.8 times more likely to have the diagnosis of MDD, respectively. We found associations of several SNPs and antidepressant response; those genes support the role of T cell (CD3E, PRKCH, PSMD9 and STAT3) and hypothalamic-pituitary-adrenal axis (UCN3) functions in treatment response. We also describe in MDD increased levels of CXCL10/IP-10, which decreased in response to antidepressants. This further suggests predominance of type 1 T-cell activity in MDD. T-cell function variations that we describe here may account for 47.8% of the attributable risk in Mexican Americans with moderate MDD. Immune function genes are highly variable; therefore, different genes might be implicated in distinct population groups.
Keywords: TBX21, PSMB4, major depression, genetic, SNP, cytokine
Introduction
Major depressive disorder (MDD) is a common and complex disorder of unknown etiology that affects about 15% of the population.1 Despite recent scientific advances and its enormous social costs,2 MDD is still currently thought to be a gene-environment disorder of polygenic nature with a descriptive diagnosis and no known biomarkers.1 Although the contributions of immune mediators to the pathophysiology and treatment of psychiatric disorders may be traced back to over 80 years with the work of Nobel laureate Julius Wagner-Jauregg,3 evidence from clinical and basic research have recently supported a role for dysregulation of the immune system in MDD.4 Both acute stress and MDD are states of hyperarousal, in which a sustained focus on the threatening stimulus, fear-related behaviors and stereotyped states of cognition and affect are matched with indices of physiological hyperarousal, such as activation of the hypothalamic-pituitary-adrenal (HPA) axis and sympathetic activation, and inhibition of counterproductive neurovegetative functions during life-threatening situations.5,6 Depression-like symptoms have been associated with activation of the HPA axis, sympathetic nervous system and inflammatory response characterized by hypercortisolaemia,7 increased central corticotropin-releasing hormone (CRH)8-10 and norepineprhine11,12 functions, increased numbers of peripheral leukocytes, positive acute phase proteins and proinflammatory cytokines.13
We and others have proposed a role for proinflammatory cytokines in the pathophysiology of MDD, with activation of the immune system and of the cellular immune response.14,15 Even though still underexplored in MDD, the T-cell arm of the immune system has been emerging as the centerpiece of the continued debate over the role of the immunomediators in depression.15,16 Data supporting either a predominance of cytokine-producing helper T-cells, type 1 (Th1) or type 2 (Th2) have accumulated. The overactivity of the hallmarks of Th1 immunity, such as interferon-γ (IFNγ), tumor necrosis factor-α or interleukin-1 (IL-1),17 or predominant Th2 patterns of production supported by increased levels of IL-6, IL-10 or IL-1316 have continued to fuel this discussion. At least two fundamental processes may contribute to the role of the T-cell arm of adaptive immunity in MDD: T-cell programmed differentiation and antigen processing.
Naive CD4+ T lymphocytes proliferate and differentiate into two main lineages defined by distinct cytokine profiles18 after encountering antigen-carrying dendritic cells in secondary lymphoid organs. The balance of two main subtypes of cytokine-producing Th1/Th2 determines the immune response to pathogens. Clinically, Th1 patterns of cytokine production are associated with inflammation and autoimmune disease, whereas Th2 patterns are related to allergic responses and asthma.19 Th1 cell-lineage commitment is controlled by the key transcriptional factor TBX21 (Tbet),20 which is rapidly produced early in Th1 differentiation and gradually decreases at later stages.21 TBX21 has the ability to simultaneously drive Th1 genetic programs and repress the development of the opposing Th2 subset; it may also redirect fully polarized Th2 cells into Th1 cells.
Proteasomes (prosome, macropain) are the major intracellular extralysosomal organelle for protein degradation and a central source of antigenic peptides in the endogenous pathway; they are utilized in major histocompatibility complex molecules class I (MHC1) antigen processing and protein degradation. Proteasomes are highly abundant in the cytosol and nucleus and are organized as multiunit protease complexes. Protein degradation is central to many important biological functions, including cell-cycle progression, apoptosis, synaptic reorganization, DNA repair, normal immune surveillance mechanisms and immune response networks. Disruption of the proteasomal degradation pathway has been implicated in a wide range of human disorders; immune abnormalities include the development of CD8 + T lymphocytes and MHC1 molecules, and MHC1-restricted antigen presentation have been described in mice lacking proteasome subunits.22 Independent lines of research have supported the role of protein synthesis/degradation in MDD-like neuropsychiatric symptoms in autoimmune disorders23 and in the central actions of antidepressant drugs.24
We used a combination strategy consisting of genetic analyses and functional assays to assess the association of pivotal elements of acquired immunity relevant to the HPA axis modulation and T-cell function with susceptibility to MDD and antidepressant response. We genotyped a panel of single nucleotide polymorphisms (SNPs) focused on the steroid pathway25 and on proteasome subunit genes (Table 1). We also conducted multiplex assays of 21 circulating cytokines in a subset of our patients.
Table 1.
Number of SNPs investigated
| Symbols | Steroid pathway genes | No. of SNPs |
|---|---|---|
| ABCB1 | ATP-binding cassette, subfamily B (MDR/TAP), member 1 | 9 |
| CD3E | CD3e molecule, epsilon (CD3-TCR complex) | 2 |
| CD4 | CD4 molecule | 7 |
| CD7 | CD7 molecule | 2 |
| CRH | Corticotropin-releasing hormone | 8 |
| CRHBP | Corticotropin-releasing hormone-binding protein | 4 |
| CRHR2 | Corticotropin-releasing hormone receptor 2 | 16 |
| CYP3A4 | Cytochrome P450, family 3, subfamily A, polypeptide 4 | 2 |
| GTF2F1 | General transcription factor IIF, polypeptide 1 | 3 |
| IL18BP | Interleukin 18-binding protein | 7 |
| IPO13 | Importin 13 | 7 |
| JUND | Jun D proto-oncogene | 2 |
| MFNG | MFNG O-fucosylpeptide 3-β-N-acetylglucosaminyltransferase | 6 |
| NR3C1 | Nuclear receptor subfamily 3, group C, member 1 | 9 |
| PFKFB4 | 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 | 3 |
| POMC | Proopiomelanocortin | 4 |
| PRKCSH | Protein kinase C substrate 80K-H | 4 |
| RAC2 | Ras-related C3 botulinum toxin substrate 2 | 6 |
| CDC42SE2 | CDC42 small effector 2 | 3 |
| TBX21 | T-box 21 | 9 |
| STAT3 | Signal transducer and activator of transcription 3 | 4 |
| UCN | Urocortin | 1 |
| UCN2 | Urocortin 2 | 4 |
| UCN3 | Urocortin 3 | 2 |
| Proteasome subunit genes | ||
| α | A1, A6, A7 | 6 |
| β | B2, B4, B5, B8 | 6 |
| 26S (non-ATPase) | D1, D2, D3, D5, D9, D13, D14 | 19 |
| Inhibitor | F1 | 6 |
| Total | 161 |
Abbreviation: SNPs, single nucleotide polymorphisms.
Methods
Genetic study
Study population
This study was approved by the institutional review boards of the University of California Los Angeles and the University of Miami, and it has been registered in the public database ClinicalTrials.gov (NCT00265292). The study population consisted of 284 depressed Mexican Americans enrolled in a pharmacogenetic study of antidepressant treatment response as previously described.26,27 We also studied 331 control individuals recruited from the same Mexican-American community in Los Angeles and studied by the same bilingual clinical research team. Controls for our genomic studies were in general good health but were not screened for medical or psychiatric illness. All patients were Mexican-American men and women aged 21-68 years, with a current episode of major depression as diagnosed by DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, 4th edn).28 In our study, all Mexican-American subjects had at least three grandparents born in Mexico.26 All patients had comprehensive psychiatric and medical assessments. We used diagnostic and ratings instruments that have been fully validated in English and Spanish, and conducted all assessments in the subject's primary language.
Inclusion criteria included DSM-IV diagnosis of current, unipolar major depressive episode, with a 21-item Hamilton Depression Rating Scale (HAM-D21)29 score of 18 or greater with item number 1 (depressed mood) rated 2 or greater. There was no anxiety threshold for inclusion. Subjects with any primary axis I other than MDD (for example, dementia, psychotic illness, bipolar disorder, adjustment disorder), electroconvulsive therapy in the past 6 months or previous lack of response to desipramine or fluoxetine were excluded. As anxiety can be a manifestation of depression, patients who met criteria for depression and also anxiety disorder were not excluded. Exclusion criteria included active medical illnesses that could be etiologically related to the ongoing depressive episode (for example, untreated hypothyroidism, cardiovascular accident within the past 6 months, uncontrolled hypertension or diabetes), current, active suicidal ideation with a plan and strong intent, pregnancy, lactation, current use of medications with significant central nervous system activity, which interfere with electroencephalography (EEG) activity (for example, benzodiazepines) or any other antidepressant treatment within the 2 weeks prior to enrollment, illicit drug use and/or alcohol abuse in the past 3 months or current enrollment in psychotherapy.
Depressed subjects were enrolled in an outpatient double-blind study of antidepressant treatment response to desipramine or fluoxetine.26 The treatment had two phases. Phase 1 was a 1-week, single-blind placebo lead-in phase to eliminate placebo responders. Subjects who continued to meet the inclusion criteria after phase 1 were randomly assigned to one of two treatment groups in a double-blind manner in phase 2; they received fluoxetine 10-40mg per day or desipramine 50-200mg per day for 8 weeks, with a dose escalation based on clinical outcomes. Depressed subjects had up to 9 weeks of structured follow-up assessments. The effect of antidepressants on HAM-D21 score was measured by the relative reduction computed as the difference in HAM-D21 score between pre- and post-treatment divided by the pretreatment HAM-D21 score. Responders were defined as the patients who had a higher than 50% reduction in HAM-D21 score on the final week (week 8).
Genotyping assays
Blood samples were collected into ethylenediaminetetraacetic acid (K2EDTA) BD Vacutainer EDTA tubes (Becton, Dickinson and Co., Franklin Lakes, NJ, USA) and genomic DNA was isolated from those samples using Gentra Puregene DNA purification kits (Gentra Systems Inc., Indianopolis, IN, USA). Genotyping of SNPs was performed using a SEQUENOM MassARRAY MALDITOF mass spectrometer (Sequenom, San Diego, CA, USA) for analysis of unlabeled single-base extension minisequencing reactions27 or using the Golden Gate assay (Illumina, San Diego, CA, USA) as part of a multiplex reaction as previously described.26 Our SEQUENOM protocol implemented the very short extension method proposed by Sun and colleagues30 whereby sequencing products are extended by only one base for three of the four nucleotides (due to the presence of dideoxynucleotides for three of the four nucleotides in the minisequencing reaction) and by several additional bases for the fourth nucleotide (specified in advance so as to represent one of the two alleles at a given SNP locus). This allowed for clearly delineated mass separation of the two allelic variants at a given locus. We addressed population stratification by stratifying our analysis by self-designated ethnic group. A set of random markers across the genome was also genotyped. Cleaning and filtering steps were performed as previously described26,27 Only data generated by SNP assays that were successfully genotyped on at least 80% of samples were included. Data quality was assessed by duplicates DNAs across all plates. Genotypes from nonmatching or missing duplicates were dropped.
Hardy-Weinberg equilibrium
We performed both standard asymptotic test and exact test for Hardy-Weinberg equilibrium (HWE) described by Wigginton et al.31 using PLINK program (http://pngu.mgh.harvard.edu/~purcell/plink/). For a locus with two alleles, the locus is in HWE in the population when the relationship between allele frequencies and genotype proportions follows the equation p2 + 2pq + q2 = 1, where p and q are the frequencies for major and minor allele, respectively. Exact test of HWE is a more appropriate approach when one allele is very rare. We detected deviation from HWE separately for control and depressed groups, and excluded those SNPs that were not in HWE in the control group.
Data analyses
SNP-based analyses of susceptibility to MDD
We performed allelic association tests using the PLINK program. Specifically, we employed χ2-test, or Fisher's exact test when the minor allele was rare, to examine the allelic association with depression by comparing allele frequencies between cases and controls. We used the following procedures to identify a list of SNPs statistically associated with a diagnosis of depression: (1) study population and controls were randomly divided into two groups: discovery and replication samples; (2) significance level was set at P≤0.05 for both discovery and replication samples; (3) the minor allele frequency in controls was ≥5% and (4) the Benjamini and Hochberg method was used to control for false discovery rate and the significance threshold was set at FDR_BH≤0.05.32 For the SNPs associated with depression in the discovery sample, we compared the odds of having depression in individuals having a risk allele with those homozygous for a nonrisk allele.
Odds ratios and population attributable fraction
We compared the odds of having depression given the homozygous for major and minor, or heterozygous genotype for the SNPs associated with diagnoses of depression. Odds ratios (ORs) and their 95% confidence intervals (CIs) were calculated using the PLINK program. We also calculated population attributable fraction (PAF) to estimate the proportional amount by which disease risk is due to the risk genotypes in the population.33
SNP-based analyses of drug response
We used χ2-test to investigate the allelic association with drug response status by comparing allele frequencies between responders and nonresponders; we used Fisher's exact tests when the minor allele was rare. We calculated the allelic OR for response status and its 95% CI using Woolf's method or fitting exact logistic regression model with SAS software when the frequency in a table cell is 0. For the SNPs with a P < 0.05 in responder vs nonresponder analysis or nonsynonymous SNPs close to any of these SNPs, we also employed a general linear regression model to examine the additive effect of minor allele on the relative reduction of HAM-D21 score by controlling age, gender and baseline HAM-D21 score using the PLINK program.
Haplotype analyses
We used Haploview version 4.0 program (http://www.broad.mit.edu/mpg/haploview/) and applied the four-gamete rule34 to conduct haplotype analysis with the depressed and control groups combined to test whether a certain haplotype was associated with the risk for depression. Blocks are formed by consecutive markers where only three gametes are observed, and htSNPs are defined in Haploview by using aggressive tagging (two- and three-marker haplotypes). All haplotypes > 0% were examined, and nontagging SNPs within haplotype blocks were omitted from the final analyses (Figure 1).
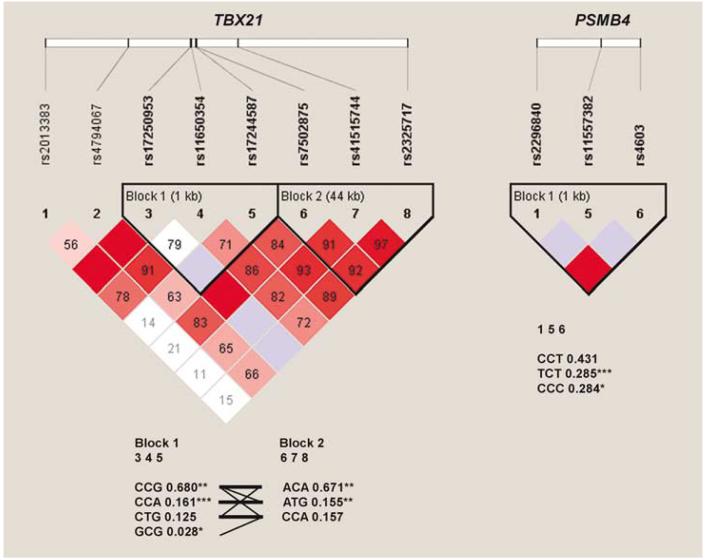
Figure 1.
Linkage disequilibrium pattern in TBX21 and PBSM4 genes. Standard color scheme: white, D′ < 1 and logarithm of odds (LOD) < 2; blue, D′ = 1 and LOD < 2; shades of pink/red, D′ < 1 and LOD≥2; bright red, D′ = 1 and LOD≥2. D′-values represent percentages and appeared inside each diamond; values of 100% are not labeled. At the top of the figure, gene structures are illustrated schematically by a thick horizontal white rectangle. Short vertical lines indicate genotyped single nucleotide polymorphisms (SNPs), which correspond to the numbers above the triangular image for genes TBX21 and PSMB4. Haplotype blocks were defined using the four game rule and haplotype tagging SNPs (htSNPs) are shown in bold. At the bottom of the triangular figures, haplotypes are shown in blocks with frequency and connections from one block to the next; only htSNPs are displayed. Blocks are connected with thin lines if frequency is > 5% and thick lines if > 10%. Between the blocks, a value of multiallelic D′ is shown. D′ is a measure of the recombination between the two blocks. TBX21: two haplotype blocks were defined; block 1 (1 kb; SNPs 3-5: rs17250953, rs11650354 and rs17244587) and block 2 (44 kb; SNPs 6-8: rs7502875, rs41515744 and rs2325717). Haplotype CCA in block 1 is the most significantly association with major depressive disorder (MDD) diagnosis (P < 0.0001). PSMB4: one haplotype block was defined; haplotype TCT was significantly associated with MDD diagnosis (P = 0.0001). *P < 0.05, **P < 0.01 and ***P≤0.0001.
Analyses of combined effect
We used Rothman's synergy index (S) to assess the joint effect of the two polymorphisms.35 The S index is the ratio of the observed joint effect divided by the expected joint effect assuming additivity of the effects, defined as: S = (OR11 1)/(OR10+OR-01-2) in which subscript 0 denotes the absence of the risk genotype at the SNP and OR denotes the odds ratio. No interaction corresponds to S = 1, whereas S >1 (S < 1) can be interpreted as a measure of relative increase (decrease) in the effect among those exposed to risk genotypes at both SNPs. In addition, we conducted the Cochran-Armitage trend test to examine dose-effect relationship between the sum of risk alleles at both SNPs and the OR for depression. We used SAS Proc Genmod to calculate ORs and their 95% Wald CIs (SAS version 9.1.3;SAS Institute, NC, USA).
Immunoassay study
Study population
A subset of the genetic study population, consisting of 68 Mexican-American MDD patients (51 women (36.0±8.3 years old and body mass index (BMI) 28.8±5.8; mean s.d.) and 17 male (36.1±10.1 years old and BMI 28.2±5.2; mean+s.d.)) and 18 Mexican-American controls (12 women (36.1±9.2 years old and BMI 28.9±3.6; mean+s.d.) and 6 men (31.5±9.4 years old and BMI 29.1±6.1; mean+s.d.)), was assessed by 21-plex cytokine assay. Controls were free of ongoing physical illness and showed no evidence of major psychiatric illness in clinical and structured interviews. Fasting blood was collected for cytokine assays one time in controls and two times in MDD patients (pretreatment at week-1 and post-treatment at week 8). Among 68 patients, 29 were assessed for the cytokines at week 8 and 1 patient was assessed only at week 8.
Immunoassays
Plasma samples were collected before the initiation of antidepressant treatment in MDD patients. We used Human Cytokine 21 PLEX-Premixed immunoassay kits (Linco Research Inc., St Charles, MO, USA) and a multi-analyte detection system (Luminex 100 instrumentation and xMAP technology; Luminex Corp., Austin, TX, USA) to simultaneously obtain the level of several cytokines and chemokines. Assays were performed accordingly to the manufacture's instructions. Assays were run in duplicates and coefficient of variance was ≥15%.
Data analyses
Pearson's χ2-test was performed to test for the difference of gender frequency and Student's t-test was conducted to compare the mean difference in age and BMI between MDD patient and control groups. No significant difference was found in age and BMI means and gender frequency between the two groups. We excluded 10 analytes from the analyses because their frequencies of undetectable level were over 30%. Of the remaining 11 analytes, we used logarithmic transformation for chemokine CXCL10/IP10 levels because of the high kurtosis (11.14) and skewness (2.73). After transformation, the kurtosis and skewness for the log10 (CXCL10/IP10) were 1.33 and 0.27, respectively. We conducted analysis of covariance analyses based on general linear model by including age, gender and BMI as covariates to compare cytokine levels between controls and pretreatment MDD patients. We performed paired t-test to compare pre- and post-treatment cytokine levels in MDD patients with response to antidepressant treatment. All these analyses were performed using SAS.
Results
SNP associated with MDD
In our discovery sample, the MDD diagnosis was significantly associated with SNPs in the following genes (Table 2): PSMB4, POMC, CDC42SE2, NR3C1, ABCB1, TBX21 and GTF2F1. The association of four SNPs was confirmed in our replication sample; two of those untranslated regions (UTRs) SNPs: rs2296840 (T/C, 5′ UTR) in PSMB4 (proteasome β4 subunit, β7 hs, HN3, HsN3, PROS26, O(MIM) MIM 602177) and rs17244587 (G/A, 3′ UTR) in TBX21 (T-bet, O(MIM) MIM 604895) remained significant after Benjamini and Hochberg correction for multiple testing using the combined sample (Figure 1; Table 2). Mexican-American individuals with the minor allele T at rs2296840 in PSMB4 were 70% more likely to be in the MDD than in the control group (OR = 1.7; 95% CI: 1.3-2.1) and 23.2% of population risk could be attributable to the genotypes (TC or TT) at 2296840 (Table 2). Individuals who had the minor allele A at rs17244587 in TBX21 were twice more likely to be in the MDD than in the control group (OR = 2.0; 95% CI: 1.4-2.7) and 20.1% of population risk could be attributable to the genotypes (AG or AA) at rs17244587 (Table 2). Taken together, 47.8% of the population risk could be attributable to the risk genotypes at rs2296840 in PSMB4 or rs17244587 in TBX21 (Table 3). The joint effect of the combined genotypes of rs2296840 (recessive model) and rs17244587 (dominant model) was 26% greater than that predicted by assuming additivity of effects (S = 1.26) (Table 3).
Table 2.
Polymorphisms associated with risk of depression
|
Polymorphism |
Allelic association |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Combined sample (N = 559) |
|||||||||||
| Gene | SNP | Chromosome | Position | SNP type | P (discovery sample, Na = 280) | P (replication sample, Nb = 279) | P (FDR_BH) | Risk/nonrisk allele | Case risk allele frequency | Control risk allele frequency | OR (95% CI) |
| PSMB4 | rs2296840c | 1 | 149638671 | 5′ UTR | 0.002 | 0.002 | 0.0001 (0.007) | T/C | 0.34 | 0.24 | 1.65 (1.28, 2.12) |
| rs4603 | 149640649 | Missense | 0.07 | 0.07 | 0.01 (0.17) | A/G | 0.75 | 0.69 | 1.38 (1.07, 1.78) | ||
| POMC | rs2118404 | 2 | 25230833 | Flank | 0.02 | 0.30 | 0.02 (0.17) | T/C | 0.55 | 0.47 | 1.35 (1.06, 1.73) |
| CDC42SE2 | rs798412 | 5 | 130726373 | 3′ UTR | 0.0009 | 0.53 | 0.005 (0.12) | A/C | 0.42 | 0.34 | 1.43 (1.11, 1.83) |
| rs798416 | 5 | 130720999 | Intron | 0.0033 | 0.43 | 0.008 (0.13) | C/T | 0.41 | 0.33 | 1.40 (1.09, 1.79) | |
| NR3C1 | rs852977 | 5 | 142667687 | Intron | 0.02 | 0.12 | 0.007 (0.13) | A/G | 0.89 | 0.83 | 1.61 (1.13, 2.27) |
| ABCB1 | rs1002205 | 7 | 86979110 | Intron | 0.01 | 0.48 | 0.03 (0.25) | C/G | 0.19 | 0.14 | 1.45 (1.05, 2.02) |
| rs1922243 | 7 | 86981440 | Intron | 0.008 | 0.66 | 0.03 (0.25) | T/C | 0.20 | 0.15 | 1.45 (1.04, 1.99) | |
| TBX21 | rs17244587d | 17 | 43178034 | 3′ UTR | 0.004 | 0.005 | 0.00005 (0.007) | A/G | 0.21 | 0.12 | 1.97 (1.41, 2.74) |
| rs41515744 | 17 | 43186946 | Flank | 0.04 | 0.004 | 0.0004 (0.01) | T/C | 0.21 | 0.13 | 1.80 (1.30, 2.50) | |
| rs2325717 | 17 | 43222803 | Flank | 0.02 | 0.009 | 0.0004 (0.01) | C/T | 0.20 | 0.12 | 1.84 (1.31, 2.56) | |
Abbreviations: CI, confidence interval; FDR_BH, Benjamini and Hochberg false discovery rate; OR, odds ratio; SNP, single nucleotide polymorphism; UTR, untranslated region.
Includes 139 cases and 141 controls.
Includes 139 cases and 140 controls.
Population attributable fraction (PAF) = 23.2%.
PAF = 20.1%.
Table 3.
Combined effect of TBX21 and PSMB4 genes on the risk of depression
|
OR by total risk allele no.a |
OR by combined genotypeb |
|||||||
|---|---|---|---|---|---|---|---|---|
| No. of risk allele | Case/control | OR (95% CI) | P | rs17244587 | rs2296840 | Case/control | OR (95% CI) | P |
| 0 | 57/93 | 1.00 | - | GG | CC/TC | 133/154 | 1.00 | - |
| 1 | 123/88 | 2.28 (1.49-3.50) | 0.0002 | GG | TT | 19/8 | 2.75 (1.17-6.49) | 0.02 |
| 2 | 58/30 | 3.15 (1.82-5.47) | 0.00004 | AG/AA | CC/TC | 90/49 | 2.13 (1.40-3.23) | 0.0004 |
| 3 | 12/2 | 9.79 (2.11-45.3) | 0.004 | AG/AA | TT | 8/2 | 4.63 (0.97-22.2) | 0.05 |
Sum of risk alleles at rs17244587 (AA = 2, AG=1, GG = 0) and rs2296840 (TT = 2, TC = 1, CC = 0); Cochran-Armitage trend test: Z = -5.095, d.f. = 1, P = 1.74-E7; PAF = 47.8%.
Rothman synergy index= (4.63-1)/(2.75 + 2.13-2) = 1.26.
Trend SNPs were located in the 3′-flanking region of TBX21 in chromosome 17q21.3 (rs4151574 and rs2325717), and in the coding region of PSMB4 (rs4603) in chromosome 1q21. Figure 1 depicts that we have identified three significant risk haplotypes: CCA and ATG (TBX21, blocks 1 and 2, respectively), and TCT (PSMB4), and four protective haplotypes: CCG and GCG (TBX21, block 1), and ACA (TBX21, block 2) and CCC (PSMB4). Notably, the 5′ UTR SNP in PSMB4 is in linkage disequilibrium with the missense SNP rs4603 (C/T; Ile234Thr).
SNP associated with antidepressant response
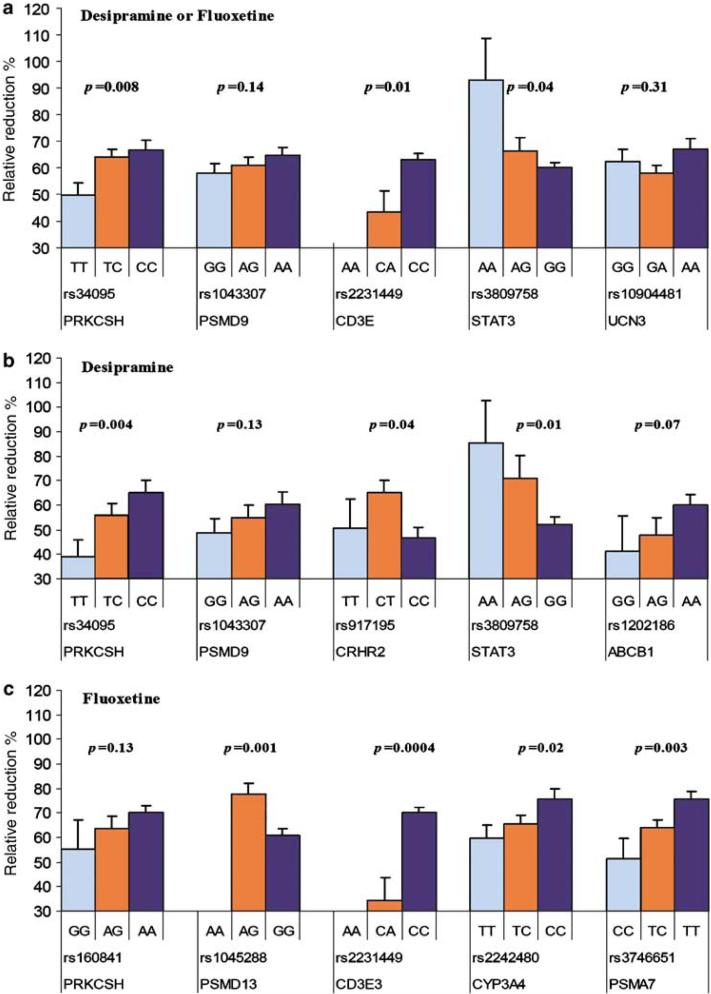
Five SNPS in the steroidal pathway and proteasome genes were significantly associated with antidepressant response within the entire depressed group treated either with desipramine or fluoxetine (Table 4). They were located in the following genes: CD3E (rs2231449, CD3 antigen-ε subunit, OMIM 186030), PRKCSH (rs34095, protein kinase C substrate 80 kD, heavy chain, OMIM 177060), PSMD9 (rs1043307, proteasome 26S non-ATPase subunit 9), STAT3 (rs3809758, signal transducer and activator of transcription 3, OMIM 102582) and UCN3 (rs10904481, urocortin III). The association of three genes remained significant in our general linear regression analyses after controlling for age, gender and baseline HAM-D score (Figure 2a). Among these polymorphisms, the two nonsynonymous, rs104330 (Glu197Gly) in PSMD9 and rs10904481 (Arg91Gly) in UCN3, and one 3′ UTR: rs2231449 (A/C) in CD3E are most likely to be functionally relevant.
Table 4.
Polymorphisms associated with response status in the treatment of depression
|
Minor allele frequency |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Medication | Gene | SNP | Chromosome | Position | SNP type | Minor/major allele | Responder | Nonresponder | or (95% CI)a | Pa |
| Desipramine or Fluoxetine | UCN3 | rs10904481 | 10 | 5405954 | Missense | G/A | 0.431 | 0.586 | 0.53 (0.30, 0.97) | 0.04 |
| CD3E | rs2231449 | 11 | 117691515 | 3′ UTR | A/C | 0.015 | 0.083 | 0.17 (0.04, 0.72) | 0.007 | |
| PSMD9 | rs1043307 | 12 | 120838179 | Missense | G/A | 0.388 | 0.591 | 0.44 (0.25, 0.77) | 0.004 | |
| STAT3 | rs3809758 | 17 | 37725506 | Intron | A/G | 0.119 | 0.031 | 4.18 (0.96, 18.2) | 0.04 | |
| PRKCSH | rs34095 | 19 | 11402685 | Intron | T/C | 0.365 | 0.625 | 0.35 (0.18, 0.64) | 0.0005 | |
| Desipramine | CRHR2 | rs917195 | 7 | 30694977 | Flank | T/C | 0.333 | 0.125 | 3.50 (1.24, 9.91) | 0.01 |
| ABCB1 | rs1202186 | 7 | 87051194 | Intron | G/A | 0.107 | 0.265 | 0.33 (0.12, 0.93) | 0.03 | |
| PSMD9 | rs1043307 | 12 | 120838179 | Missense | G/A | 0.359 | 0.591 | 0.39 (0.19, 0.81) | 0.01 | |
| STAT3 | rs3744483 | 17 | 37719964 | 3′ UTR | C/T | 0.150 | 0.000 | 9.15 (1.44, ∞) | 0.009 | |
| rs3809758 | 17 | 37725506 | Intron | A/G | 0.171 | 0.000 | 11.28 (1.81, ∞) | 0.005 | ||
| PRKCSH | rs34095 | 19 | 11402685 | Intron | T/C | 0.341 | 0.639 | 0.29 (0.13, 0.66) | 0.002 | |
| Fluoxetine | CYP3A4 | rs2242480 | 7 | 99199402 | Intron | T/C | 0.398 | 0.667 | 0.33 (0.13, 0.83) | 0.02 |
| PSMD13 | rs3817629 | 11 | 227312 | Intron | T/C | 0.170 | 0.000 | 6.17 (1.00, ∞) | 0.04 | |
| CD3E | rs2231449 | 11 | 117691515 | 3′ UTR | A/C | 0.008 | 0.125 | 0.06 (0.01, 0.60) | 0.002 | |
| PRKCSH | rs160841 | 19 | 11420158 | Intron | G/A | 0.115 | 0.292 | 0.31 (0.11, 0.89) | 0.02 | |
| PSMA7 | rs2057169 | 20 | 60145679 | Intron | C/T | 0.242 | 0.546 | 0.27 (0.11, 0.68) | 0.004 | |
| rs2057168 | 20 | 60145742 | Intron | C/T | 0.235 | 0.546 | 0.26 (0.10, 0.65) | 0.003 | ||
| rs2281740 | 20 | 60145906 | Intron | T/C | 0.231 | 0.546 | 0.25 (0.10, 0.64) | 0.002 | ||
| rs3746651 | 20 | 60151815 | 3′ UTR | C/T | 0.230 | 0.500 | 0.30 (0.11, 0.79) | 0.01 | ||
Abbreviations: SNPs, single nucleotide polymorphisms; UTR, untranslated region.
P-values, odds ratios (OR) and 95% confidence intervals (CI) were estimated on exact logistic regression model if any cell with the frequency is 0.
Figure 2.
Genotypes and relative reduction of Hamilton Depression Rating Scale (HAM-D) score in the patients treated with desipramine and fluoxetine. Histograms represent mean and standard error of mean for relative reduction of HAM-D21 score in major depressive disorder (MDD) patients who completed 8-week antidepressant treatment with desipramine (n = 68) or fluoxetine (n = 79) by genotypes (light blue, homozygous for minor allele; orange, heterozygote; dark blue, homozygous for major allele). A general linear model was used to detect allelic additive effects on treatment response after adjustment for age, sex and baseline HAM-D21 score. The analyses were performed using all treated patients (a), desipramine-treated patients (b) and fluoxetine-treated patients (c).
Fluoxetine treatment
Eight SNPs located in five genes were associated with treatment response during fluoxetine treatment (Table 4). SNPs in CYP3A4 (rs2242480), PSMD13 (rs1045288 and rs3817629), CDE3 (rs2231449), PRKCSH (rs160841) and PSMA7 (rs2057169, rs2057168, rs2281740, rs3746651) had a difference in allele frequency with P≤0.05 for responders and nonresponders within the subjects treated with fluoxetine. The association of four genes remained significant in general linear regression analyses after controlling for age, gender and baseline HAM-D score (Figure 2c). Patients who had minor allele A at missense SNP rs1045288 (Asn13Ser) in PSMD13 had better response (β = 18.13; 95% CI: 8.29-27.96), whereas those who had minor allele C at 3′ UTR SNP rs3746651 in PSMA7 had smaller relative reduction in HAM-D21 score (β = 11.5; 95% CI: -3.34 to -18.26). Patients who had minor allele A at 3′ UTR SNP rs2231449 in CDE3 showed much worse response (β = 37.17; 95% CI: -55.69 the to -18.65), but the number in this group was very small. Two genes (CD3E and PRKCSH) associated with response in the entire depression group were also associated with response in the fluoxetine treated subjects.
Desipramine treatment
Six SNPs located in or near five genes were associated with treatment response during desipramine treatment. SNPS in ABCB1 (rs1202186), CRHR2 (rs917195), PRKCSH (rs34095), PSMD9 (rs1043307, missense) and STAT3 (rs3744483, 3′ UTR, and rs3809758) had a difference in allele frequency with P≤0.05 for responders and nonresponders within the subjects treated with desipramine (Table 4; Figure 2b). Three of these SNPs (rs1043307, rs3809758, rs34095) were associated with response in the entire depression group were also associated with response to desipramine treatment.
Immunoassays
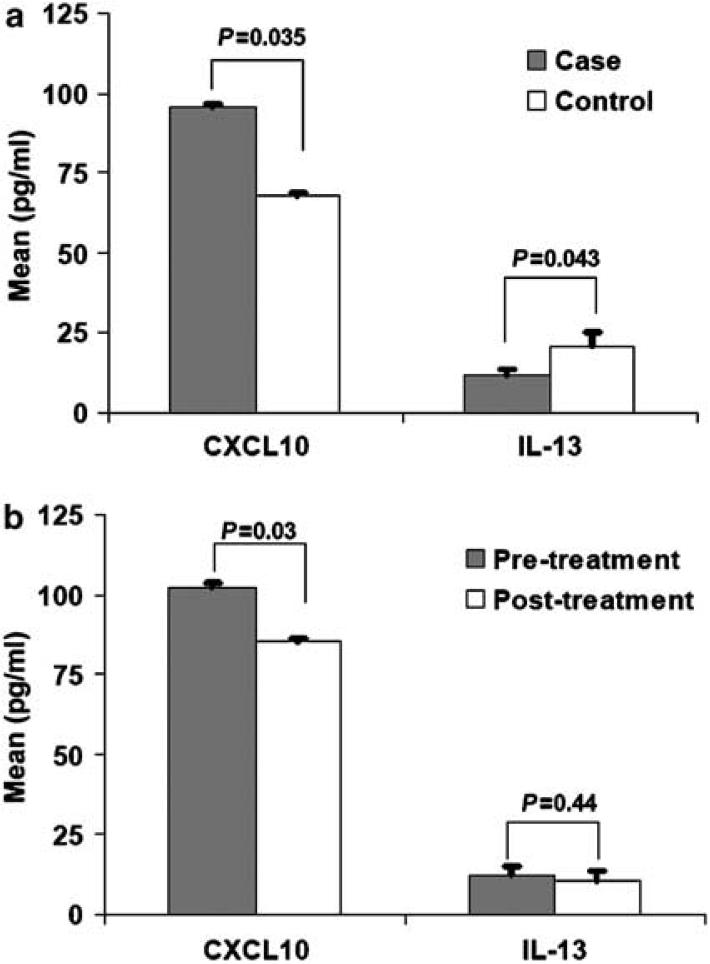
To further understand aspects of immune dysfunction relevant to MDD and/or treatment response, we examined patterns of cytokines in plasma and found increased circulating plasma levels of the IFNγ-inducible chemokine CXCL10/IP1036 in our MDD patients before initiation of antidepressant treatment when compared to controls (P = 0.035; 95.50±1.06 and 67.61±1.16 pg ml1, respectively in MDD and control groups, mean±s.d. calculated by using logarithmic transformation; Figure 3a) and significant decrements were found for IL-13 levels in MDD patients when compared to controls (P = 0.043; 11.87±11.77 and 20.82±14.77 pg ml1, respectively in MDD and control groups). Patients who responded to antidepressant treatment had a significant decrement in levels of CXCL10 (P = 0.03; 1.20±1.38 pg ml-1, paired mean difference ±s.d. calculated by using logarithmic transformation; Figure 3b).
Figure 3.
CXCL10 (IP10) and interleukin-13 (IL-13) levels in controls, major depressive disorder (MDD) patients and drug responders. (a) Histograms represent mean and standard error for CXCL10 and IL-13 levels in MDD patients before initiation of antidepressant treatment (n = 65) and controls (n = 14). Comparison levels between cases and controls were performed using general linear model after adjustment for age, sex and body mass index (BMI). Log arithmetic transformation was used for CXCL10 in our data analyses. (b) Histograms represent mean and standard error for CXCL10 and IL-13 levels before and after 8 weeks of antidepressant treatment in 19 antidepressant treatment responders (MDD patients who had higher than 50% reduction in Hamilton Depression Rating Scale (HAM-D21 score). Paired t-test was used to compare pre- and post-treatment levels in drug responders. Log arithmetic transformation was used for CXCL10 in our data analyses.
Discussion
We found that two genes that are critical for T-cell function, PSMB4 and TBX21, are associated with major depression. We found that together, 47.8% of the population risk could be attributable to the risk genotypes at rs2296840 in PSMB4 or rs17244587 in TBX21. The joint effect of the combined genotypes of rs2296840 (recessive model) and rs17244587 (dominant model) was 26% greater than that predicted by assuming additivity of effects. Our analyses revealed a significant combined allele dose-effect; therefore, individuals who had one, two and three risk alleles in PSMB4 and TBX21 were 2.3, 3.2 and 9.8 times more likely to have the diagnosis of MDD, respectively. We also found associations of several SNPs in genes relevant to HPA axis and immune function and antidepressant response and describe in MDD increased levels of CXCL10/IP-10, which decreased in response to antidepressants. These lines of evidence are indicative of a predominance of Th1 activity in MDD.
Genetic variations in PSMB4 and TBX21 may also be relevant to two immune disorders, psoriasis37 and asthma,38 that are known to be comorbid with MDD. These two disorders are polygenic and reactive to psychosocial stressors. Susceptibility to psoriasis has been associated to the area of chromosome 1q21 (PSORS4) that encodes PSMB4,30 and susceptibility to asthma and nasal polyps (O(MIM) MIM 208550)14 has been associated with functional promoter SNPs in TBX21 (1993T/C), in chromosome 17q21.3.
The UTR variations in TBX21 and PSMB4 that we found to be significantly associated with MDD are in UTRs but they may nevertheless impact on immune response in our patients. Several roles in gene expression have been attributed to UTRs, including mRNA stability, localization and translational efficiency. The 5′ UTR, also known as the leader sequence, is a particular section of the mRNA that usually contains a ribosome-binding site; it is a major site of translational regulation and may affect the stability or translation of mRNA and gene expression. Evidence implicating the 3′ UTR of mRNA in the regulation of gene expression has accumulated recently. The 3′ UTR may influence transcript cleavage, polyadenylation and nuclear export, which determine transcript stability, level of translation and mRNA targeting.39 It is therefore plausible that SNPs associated with treatment response may have contributed to the increased plasma levels of the IFNγ-inducible chemokine CXCL10 found in our patients.
CXCL10 is a potent angiostatic factor with anti-fibrotic properties40 and its elevation is congruent with elevated leukocyte counts in peripheral blood that have been shown to be dependent on severity and treatment outcome in MDD.41 Inflammatory immune mediators and specifically CXCL10 have also been implicated in arteriosclerosis, and they may be a link between the presence of depressive symptoms and stress, and increased risk of, morbidity and mortality in myocardial infarction.42 The increase of an IFNγ-inducible chemokine supports the presumption of a predominance of Th1 type activity during the symptomatic phase of MDD, as well as its role in the pathophysiology, therapeutic outcome of this disorder and immunoregulatory effects of antidepressants.43
We found that genetic variations affecting T-cell function and HPA axis regulation were associated with antidepressant treatment response. The following T-cell functions may be implicated in treatment response: T-cell development (CD3E, T-cell antigen receptor-ε subunit of T3),44 antigen processing/degradation (PSMD9: proteasome 26S subunit, non-ATPase,9,45 and intracellular signaling (STAT3: signal transducer and activator of transcription 3).46 The association of a variation in the urocortin III or stresscopin gene (UCN3)47 suggests a possible role for the adaptive stress response that mediates endocrine, autonomic, cardiovascular and immune systems in treatment outcome. The association of a SNP in the CRHR2 in the treatment response to desipramine indicates that HPA axis modulation may be particularly important for tricyclic antidepressants. Notably, some of the SNPs associated with treatment response could lead to differences in immune response such as nonsynonymous variations in the PSMD9 and UCN3 genes, and 3′ UTR SNPs in CD3E, STAT3 and PSMA7 genes. Somatic variations in some of those genes have been implicated in immunodeficiencies (CD3E),48,49 polycystic liver disease (PRKCSH, protein kinase C substrate, 80 kD, heavy chain,50-52 type 2 diabetes53 or autosomal dominant hyper-immunoglobulin E (IgE) syndrome, also called `Job Syndrome'.54-56
We found no clear Th1 or Th2 cytokine patterns in our patients. Our results of decreased IL-13 levels in MDD contrast with a recent report of increased levels of Th2 cytokines IL-13 and IL-4 and decreased levels of Th1 cytokines.16 Several factors could account for this discrepancy, from differences in gender and age composition to differences in environment/pathogens or differences in the phases of neuroendocrine, counterregulatory systems or severity and stage of the disorder. Moreover, cytokine profiling in Th1 and Th2 cytokine expression seem to be relative, not absolute as inconsistencies between cytokine profiles, antibody and total serum IgE have been reported.57 Therefore, chemokines (such as CXCL10), which are low molecular weight chemotactic molecules, are emerging as a major communication system in the brain58 as their serum and CSF levels may be correlated.59 Chemokines are key mediators of inflammation that have major effects on migration of cells to inflammation sites as well as activation of recruited and resident central nervous system (CNS) cells, which have been implicated in a number of human pathophysiological systemic and CNS conditions60 and their level or expression has been linked to the activity of CNS disease.
Figure 4 summarizes our results of genetic variations associated with the diagnosis of MDD. These implicate that specific UTR variations in TBX21 or PSMB4 increase the risks for and characterize a T-cell dysfunction in MDD in Mexican Americans. These genetic variations may be involved in the immune system dysregulation described in this disorder and in known comorbidity disorders such as psoriases38 and asthma.37 Our patients had increased peripheral levels of the chemokine CXCL10, which decreased with response to antidepressant treatment.
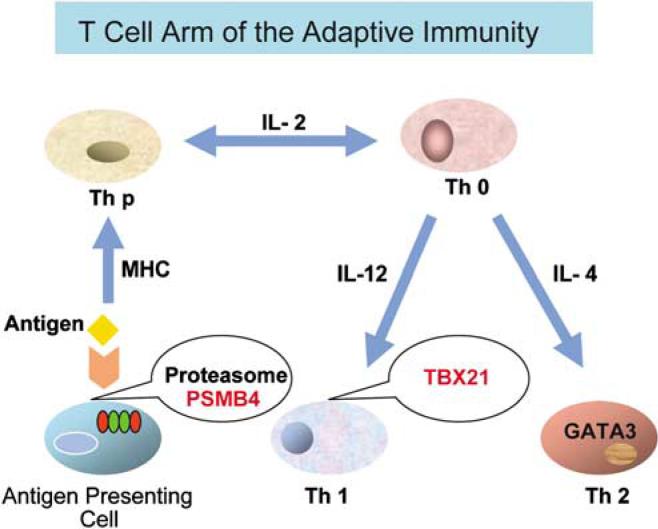
Figure 4.
Schematic of sites where variations in TBX21 or PSMB4 could influence the T-cell arm of the adaptive immunity and contribute to susceptibility to major depressive disorder (MDD): Two crucial functions, specifically antigen processing and T cell-programmed differentiation are involved in Mexican Americans with MDD and are highlighted in red. A naive helper T-cell precursor (Th p) can become either a Th1 or Th2 cell under the instructive influence of interleukin-12 (IL-12) or IL-4, respectively; Th1 cell expresses TBX21 and Th2 expresses GATA3.
These results lead to the presumption that an imbalance of Th1/Th2 activity toward a predominance of Th1 response is present in the symptomatic phase of mild to moderate forms of MDD. Replication of our findings in other ethnic groups is needed to validate the role of TBX21 and PSMB4 in major depression here reported in Mexican Americans. Because genes involved in immune function are highly polymorphic in human populations,61 allele frequency may vary considerably in different ethnic populations, and variations of T-cell function may result from common variations in other genes/gene regions, which may cause a predominance of net Th1 activity. The allele frequency for rs17244587 (TBX21) in our subjects was similar to European populations; however, rs2296840 and rs4603 (PSMB4) were significantly less frequent (respectively 0 and 10%) in Europeans than in the Mexican Americans we studied (24% in Mexican-American controls and 34% in MDD). It is therefore unlikely that the PSMB4 variations described here are significant in the susceptibility to MDD in individuals of predominant European descendant. Consequently, characterization of neuroimmune profiles may vary depending on specific genes and SNPs involved in T-cell function variations in different populations. Moreover, given our n and limited numbers of patients in the desipramine and fluoxetine treatment groups, these results need to be taken with caution, pending replication by other independent studies.
Because chemokine networks already represent potentials targets for new therapies in several CNS and systemic conditions,58 further studies are needed to fully clarify the extent of CNS immunedysregulation in the pathophysiology of MDD.
In spite of the limitations of this study, our data support the hypothesis that key T-cell functions leading to Th1 net activity are features of immune dysfunction in MDD and may also have a role in antidepressant treatment response. Different genes and polymorphisms might characterize MDD immune dysfunctions in distinct populations, as genes that influence immune functions are highly polymorphic and their allele frequency varies across human populations. We suggest that interferon-γ-inducible chemokines, such as CXCL-10, may provide viable biomarkers and might also be useful in predicting/following antidepressant response. Our findings provide a basis for conceptually innovative pharmacological approaches to MDD with a focus on T-cell function dysregulation and variations in T-cell programmed differentiation, antigen processing and cellular proteasome organelle function.
Acknowledgments
This study was supported by NIH grants GM61394, RR017365, MH062777, RR000865, RR16996, HG002500 and DK063240, and institutional funds from the University of Miami, Department of Psychiatry & Behavioral Sciences. We thank the Mexican American individuals who have participated in this study. We are grateful for the contributions to the care of our patients from Dr Israel Alvarado, Dr Deborah Flores and Dr Anil Sharma; our nursing staff Rita Jepson and Lorraine Garcia-Teague; our social workers Patricia Reyes and Gabriela Marquez at the Semel Institute for Neuroscience and Human Behavior, University of California, Los Angeles (UCLA) and staff of the UCLA GCRC. We thank Dr Kristopher Irizarry (UCLA), Dr Luciana Ribeiro (University of Miami) and Dr Joao Busnello (University of Miami) who have helped us with bioinformatics and database aspects of the work. We are also grateful for the contributions of Fiona O'Kirwan and Sarika Thakur (Semel Institute), and Dr Rita Cantor, Department of Genetics, UCLA, in preliminary statistics analyses. We also thank Dr Scott Weiss for facilitating our interactions with the Channing Laboratory, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, and Dr Panos Deloukas for facilitating genotyping work at the Wellcome Trust Sanger Institute, UK.
References
- 1.Wong ML, Licinio J. Research and treatment approaches to depression. Nat Rev Neurosci. 2001;2:343–351. doi: 10.1038/35072566. [DOI] [PubMed] [Google Scholar]
- 2.Wong ML, Licinio J. From monoamines to genomic targets: a paradigm shift for drug discovery in depression. Nat Rev Drug Discov. 2004;3:136–151. doi: 10.1038/nrd1303. [DOI] [PubMed] [Google Scholar]
- 3.Raju TN. The Nobel chronicles. 1927: Julius Wagner-Jauregg (1857-1940) Lancet. 1998;352:1714. doi: 10.1016/s0140-6736(05)61500-0. [DOI] [PubMed] [Google Scholar]
- 4.Marques-Deak AH, Neto FL, Dominguez WV, Solis AC, Kurcgant D, Sato F, et al. Cytokine profiles in women with different subtypes of major depressive disorder. J Psychiatr Res. 2007;41:152–159. doi: 10.1016/j.jpsychires.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Gold PW, Goodwin FK, Chrousos GP. Clinical and biochemical manifestations of depression. Relation to the neurobiology of stress (2) N Engl J Med. 1988;319:413–420. doi: 10.1056/NEJM198808183190706. [DOI] [PubMed] [Google Scholar]
- 6.Gold PW, Chrousos GP. The endocrinology of melancholic and atypical depression: relation to neurocircuitry and somatic consequences. Proc Assoc Am Phys. 1999;111:22–34. doi: 10.1046/j.1525-1381.1999.09423.x. [DOI] [PubMed] [Google Scholar]
- 7.Sachar EJ, Hellman L, Fukushima DK, Gallagher TF. Cortisol production in depressive illness. A clinical and biochemical clarification. Arch Gen Psychiatry. 1970;23:289–298. doi: 10.1001/archpsyc.1970.01750040001001. [DOI] [PubMed] [Google Scholar]
- 8.Gold PW, Loriaux DL, Roy A, Kling MA, Calabrese JR, Kellner CH, et al. Responses to corticotropin-releasing hormone in the hypercortisolism of depression and Cushing's disease. Pathophysiologic and diagnostic implications. N Engl J Med. 1986;314:1329–1335. doi: 10.1056/NEJM198605223142101. [DOI] [PubMed] [Google Scholar]
- 9.Nemeroff CB, Widerlov E, Bissette G, Walleus H, Karlsson I, Eklund K, et al. Elevated concentrations of CSF corticotropin-releasing factor-like immunoreactivity in depressed patients. Science (New York, NY) 1984;226:1342–1344. doi: 10.1126/science.6334362. [DOI] [PubMed] [Google Scholar]
- 10.Holsboer F, Von Bardeleben U, Gerken A, Stalla GK, Muller OA. Blunted corticotropin and normal cortisol response to human corticotropin-releasing factor in depression. N Engl J Med. 1984;311:1127. doi: 10.1056/NEJM198410253111718. [DOI] [PubMed] [Google Scholar]
- 11.Wong ML, Kling MA, Munson PJ, Listwak S, Licinio J, Prolo P, et al. Pronounced and sustained central hypernoradrenergic function in major depression with melancholic features: relation to hypercortisolism and corticotropin-releasing hormone. Proc Natl Acad Sci USA. 2000;97:325–330. doi: 10.1073/pnas.97.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veith RC, Lewis N, Linares OA, Barnes RF, Raskind MA, Villacres EC, et al. Sympathetic nervous system activity in major depression. Basal and desipramine-induced alterations in plasma norepinephrine kinetics. Arch Gen Psychiatry. 1994;51:411–422. doi: 10.1001/archpsyc.1994.03950050071008. [DOI] [PubMed] [Google Scholar]
- 13.Musselman DL, Lawson DH, Gumnick JF, Manatunga AK, Penna S, Goodkin RS, et al. Paroxetine for the prevention of depression induced by high-dose interferon alfa. N Engl J Med. 2001;344:961–966. doi: 10.1056/NEJM200103293441303. [DOI] [PubMed] [Google Scholar]
- 14.Licinio J, Wong ML. The role of inflammatory mediators in the biology of major depression: central nervous system cytokines modulate the biological substrate of depressive symptoms, regulate stress-responsive systems, and contribute to neurotoxicity and neuroprotection. Mol Psychiatry. 1999;4:317–327. doi: 10.1038/sj.mp.4000586. [DOI] [PubMed] [Google Scholar]
- 15.Kronfol Z, Remick DG. Cytokines and the brain: implications for clinical psychiatry. Am J Psychiatry. 2000;157:683–694. doi: 10.1176/appi.ajp.157.5.683. [DOI] [PubMed] [Google Scholar]
- 16.Pavon L, Sandoval-Lopez G, Eugenia Hernandez M, Loria F, Estrada I, Perez M, et al. Th2 cytokine response in major depressive disorder patients before treatment. J Neuroimmunol. 2006;172:156–165. doi: 10.1016/j.jneuroim.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Sluzewska A, Rybakowski J, Bosmans E, Sobieska M, Berghmans R, Maes M, et al. Indicators of immune activation in major depression. Psychiatry Res. 1996;64:161–167. doi: 10.1016/s0165-1781(96)02783-7. [DOI] [PubMed] [Google Scholar]
- 18.Moser M, Murphy KM. Dendritic cell regulation of TH1-TH2 development. Nat Immunol. 2000;1:199–205. doi: 10.1038/79734. [DOI] [PubMed] [Google Scholar]
- 19.Akahoshi M, Obara K, Hirota T, Matsuda A, Hasegawa K, Takahashi N, et al. Functional promoter polymorphism in the TBX21 gene associated with aspirin-induced asthma. Hum Genet. 2005;117:16–26. doi: 10.1007/s00439-005-1285-0. [DOI] [PubMed] [Google Scholar]
- 20.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8T cells. Science (New York, NY) 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 21.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science (New York, NY) 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 22.Monaco JJ, Nandi D. The genetics of proteasomes and antigen processing. Annu Rev Genet. 1995;29:729–754. doi: 10.1146/annurev.ge.29.120195.003501. [DOI] [PubMed] [Google Scholar]
- 23.Schneebaum AB, Singleton JD, West SG, Blodgett JK, Allen LG, Cheronis JC, et al. Association of psychiatric manifestations with antibodies to ribosomal P proteins in systemic lupus erythematosus. Am J Med. 1991;90:54–62. doi: 10.1016/0002-9343(91)90506-s. [DOI] [PubMed] [Google Scholar]
- 24.Wong ML, O'Kirwan F, Hannestad JP, Irizarry KJ, Elashoff D, Licinio J. St John's wort and imipramine-induced gene expression profiles identify cellular functions relevant to antidepressant action and novel pharmacogenetic candidates for the phenotype of antidepressant treatment response. Mol Psychiatry. 2004;9:237–251. doi: 10.1038/sj.mp.4001470. [DOI] [PubMed] [Google Scholar]
- 25.Weiss ST, Lake SL, Silverman ES, Silverman EK, Richter B, Drazen JM, et al. Asthma steroid pharmacogenetics: a study strategy to identify replicated treatment responses. Proc Am Thorac Soc. 2004;1:364–367. doi: 10.1513/pats.200409-043MS. [DOI] [PubMed] [Google Scholar]
- 26.Wong ML, Whelan F, Deloukas P, Whittaker P, Delgado M, Cantor RM, et al. Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc Natl Acad Sci USA. 2006;103:15124–15129. doi: 10.1073/pnas.0602795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Licinio J, O'Kirwan F, Irizarry K, Merriman B, Thakur S, Jepson R, et al. Association of a corticotropin-releasing hormone receptor 1 haplotype and antidepressant treatment response in Mexican Americans. Mol Psychiatry. 2004;9:1075–1082. doi: 10.1038/sj.mp.4001587. [DOI] [PubMed] [Google Scholar]
- 28.Rabe-Jablonska J, Bienkiewicz W. [Anxiety disorders in the fourth edition of the classification of mental disorders prepared by the American Psychiatric Association: diagnostic and statistical manual of mental disorders (DMS-IV—options book] Psychiatr Pol. 1994;28:255–268. [PubMed] [Google Scholar]
- 29.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giardina E, Capon F, De Rosa MC, Mango R, Zambruno G, Orecchia A, et al. Characterization of the loricrin (LOR) gene as a positional candidate for the PSORS4 psoriasis susceptibility locus. Ann Hum Genet. 2004;68(Part 6):639–645. doi: 10.1046/j.1529-8817.2004.00118.x. [DOI] [PubMed] [Google Scholar]
- 31.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 33.Rockhill B, Newman B, Weinberg C. Use and misuse of population attributable fractions. Am J Public Health. 1998;88:15–19. doi: 10.2105/ajph.88.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothman KJ. Modern Epidemiology. 2nd edn Lippincott-Raven Publishers; Philadelphia, PA: 1998. [Google Scholar]
- 36.Luster AD, Ravetch JV. Biochemical characterization of a gamma interferon-inducible cytokine (IP-10) J Exp Med. 1987;166:1084–1097. doi: 10.1084/jem.166.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimball AB, Jacobson C, Weiss S, Vreeland MG, Wu Y. The psychosocial burden of psoriasis. Am J Clin Dermatol. 2005;6:383–392. doi: 10.2165/00128071-200506060-00005. [DOI] [PubMed] [Google Scholar]
- 38.Scott KM, Von Korff M, Ormel J, Zhang MY, Bruffaerts R, Alonso J, et al. Mental disorders among adults with asthma: results from the World Mental Health Survey. Gen Hosp Psychiatry. 2007;29:123–133. doi: 10.1016/j.genhosppsych.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular ′hotspot′ for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 40.Le Moine C, Fauchey V, Jaber M. Opioid receptor gene expression in dopamine transporter knock-out mice in adult and during development. Neuroscience. 2002;112:131–139. doi: 10.1016/s0306-4522(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 41.Seidel A, Arolt V, Hunstiger M, Rink L, Behnisch A, Kirchner H. Major depressive disorder is associated with elevated monocyte counts. Acta Psychiatr Scand. 1996;94:198–204. doi: 10.1111/j.1600-0447.1996.tb09849.x. [DOI] [PubMed] [Google Scholar]
- 42.Glassman AH, Shapiro PA. Depression and the course of coronary artery disease. Am J Psychiatry. 1998;155:4–11. doi: 10.1176/ajp.155.1.4. [DOI] [PubMed] [Google Scholar]
- 43.Maes M. The immunoregulatory effects of antidepressants. Hum Psychopharmacol. 2001;16:95–103. doi: 10.1002/hup.191. [DOI] [PubMed] [Google Scholar]
- 44.DeJarnette JB, Sommers CL, Huang K, Woodside KJ, Emmons R, Katz K, et al. Specific requirement for CD3epsilon in T cell development. Proc Natl Acad Sci USA. 1998;95:14909–14914. doi: 10.1073/pnas.95.25.14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe TK, Saito A, Suzuki M, Fujiwara T, Takahashi E, Slaughter CA, et al. cDNA cloning and characterization of a human proteasomal modulator subunit, p27 (PSMD9) Genomics. 1998;50:241–250. doi: 10.1006/geno.1998.5301. [DOI] [PubMed] [Google Scholar]
- 46.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu SY, Hsueh AJ. Human stresscopin and stresscopin-related peptide are selective ligands for the type 2 corticotropin-releasing hormone receptor. Nat Med. 2001;7:605–611. doi: 10.1038/87936. [DOI] [PubMed] [Google Scholar]
- 48.Le Deist F, Thoenes G, Corado J, Lisowska-Grospierre B, Fischer A. Immunodeficiency with low expression of the T cell receptor/CD3 complex. Effect on T lymphocyte activation. Eur J Immunol. 1991;21:1641–1647. doi: 10.1002/eji.1830210709. [DOI] [PubMed] [Google Scholar]
- 49.Soudais C, de Villartay JP, Le Deist F, Fischer A, Lisowska-Grospierre B. Independent mutations of the human CD3-epsilon gene resulting in a T cell receptor/CD3 complex immunodeficiency. Nat Genet. 1993;3:77–81. doi: 10.1038/ng0193-77. [DOI] [PubMed] [Google Scholar]
- 50.Drenth JP, te Morsche RH, Smink R, Bonifacino JS, Jansen JB. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat Genet. 2003;33:345–347. doi: 10.1038/ng1104. [DOI] [PubMed] [Google Scholar]
- 51.Reynolds AJ, Bartlett SE, Hendry IA. Molecular mechanisms regulating the retrograde axonal transport of neurotrophins. Brain Res. 2000;33:169–178. doi: 10.1016/s0165-0173(00)00028-x. [DOI] [PubMed] [Google Scholar]
- 52.Li XS, Reddy MS, Baev D, Edgerton M. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J Biol Chem. 2003;278:28553–28561. doi: 10.1074/jbc.M300680200. [DOI] [PubMed] [Google Scholar]
- 53.Gragnoli C, Cronsell J. PSMD9 gene variants within NIDDM2 may rarely contribute to type 2 diabetes. J Cell Physiol. 2007;212:568–571. doi: 10.1002/jcp.21127. [DOI] [PubMed] [Google Scholar]
- 54.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 55.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 56.Renner ED, Torgerson TR, Rylaarsdam S, Anover-Sombke S, Golob K, LaFlam T, et al. STAT3 mutation in the original patient with Job's syndrome. N Engl J Med. 2007;357:1667–1668. doi: 10.1056/NEJMc076367. [DOI] [PubMed] [Google Scholar]
- 57.Selgrade M, Boykin EH, Haykal-Coates N, Woolhiser MR, Wiescinski C, Andrews DL, et al. Inconsistencies between cytokine profiles, antibody responses, and respiratory hyperresponsiveness following dermal exposure to isocyanates. Toxicol Sci. 2006;94:108–117. doi: 10.1093/toxsci/kfl094. [DOI] [PubMed] [Google Scholar]
- 58.Adler MW, Geller EB, Chen X, Rogers TJ. Viewing chemokines as a third major system of communication in the brain. AAPS J. 2005;7:E865–E870. doi: 10.1208/aapsj070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scarpini E, Galimberti D, Baron P, Clerici R, Ronzoni M, Conti G, et al. IP-10 and MCP-1 levels in CSF and serum from multiple sclerosis patients with different clinical subtypes of the disease. J Neurol Sci. 2002;195:41–46. doi: 10.1016/s0022-510x(01)00680-3. [DOI] [PubMed] [Google Scholar]
- 60.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 61.Chanock S, Taylor JG. Using genetic variation to study immunomodulation. Curr Opin Pharmacol. 2002;2:463–469. doi: 10.1016/s1471-4892(02)00186-8. [DOI] [PubMed] [Google Scholar]