Summary
Hydrogels based on peptide self-assembly form an important class of biomaterials that find application in tissue engineering and drug delivery. It is essential to prepare peptides with high purity to achieve batch-to-batch consistency affording hydrogels with reproducible properties. Automated solid-phase peptide synthesis coupled with optimized Fmoc (9-fluorenylmethoxycarbonyl) chemistry to obtain peptides in high yield and purity is discussed. Details of isolating a desired peptide from crude synthetic mixtures and assessment of the peptide’s final purity by high-performance liquid chromatography and mass spectrometry are provided. Beyond the practical importance of synthesis and primary characterization, techniques used to investigate the properties of hydrogels are briefly discussed.
Keywords: Biomaterial, HPLC, hydrogel, peptide self-assembly, solid-phase peptide synthesis
1. Introduction
Self-assembly forms an important process in the bottom-up approach to the design of nanostructural architectures (1,2). For example, peptide self-assembly has been extensively utilized to design intricate well-ordered structures such as nanotubes (3–12) and ribbons (13–18). Peptide self-assembly has also been employed in the development of hydrogels, heavily hydrated materials composed of dilute networks of assembled peptide. These materials are finding use in a variety of biomedical applications (19–21). In addition, peptides can be designed to undergo triggered self-assembly, leading to the formation of hydrogel material in response to physiologically relevant changes in their external environment; this allows material formation to take place with temporal resolution (22). The unique ability of peptide sequences to fold into specific secondary, tertiary, and quaternary structures has been exploited, and hydrogels based on the self-assembly of α-helices (23), β-sheets (24–29), coiled coils (30–32), and collagen mimetic peptides (33,34) have been reported in the literature. Moreover, biological function has been incorporated into hydrogels prepared from traditional polymers by ligating short peptide sequences to synthetic scaffolds (35–37).
For the reliable use of materials derived from peptides, it is essential to synthesize and purify the building blocks with high fidelity. With respect to hydrogels, batch-to-batch consistency can be achieved using peptides of high purity. In this chapter, solid-phase peptide synthesis is discussed in the context of the β-hairpin peptide hydrogels studied in our lab (25,38–42). Guidelines for the purification and primary characterization of the peptides are provided along with a brief discussion of select techniques utilized in the biophysical, structural, and mechanical characterization of the hydrogels. An example of the ramifications of purity on the material properties is also provided.
2. Materials
2.1. Reagents
2.1.1. Solid-Phase Peptide Synthesis
N-Methyl pyrrolidone (NMP) (EMD Biosciences).
Rink amide resin (PL-Rink Resin, loading = 0.64mmol/g, 75–150 μm or 100–200 mesh) (Polymer Laboratories, Amherst, MA) (see Note 1).
Appropriately side-chain-protected 9-fluorenylmethoxycarbonyl (Fmoc) amino acids (Novabiochem).
1H-Benzotriazolium1-[bis(dimethylamino)methylene]-5-chloro hexafluorophosphate (1-),3-oxide (HCTU) (Peptides International): 0.45M solution prepared in N,N-dimethylformarmide (DMF) (Fisher).
Diisopropylethylamine (DIPEA) (Acros Organics): 2M solution prepared in NMP.
20% piperidine (Sigma) in NMP or 19% piperidine in NMP containing 1% 1,8-diazabicyclo[5.4.0]-undec-7-ene (DBU) (Sigma).
Capping solution: 5% acetic anhydride (Ac2O) (Acros Organics) in NMP.
Methylene chloride or dichloromethane (DCM) (Fisher).
Methanol (MeOH) (high-performance liquid chromatographic [HPLC] grade; Fisher).
2.1.2. Peptide Resin Cleavage and Side-Chain Deprotection
Trifluoroacetic acid (TFA) (Acros Organics).
Thioanisole (Acros Organics).
Ethanedithiol (Acros Organics).
Anisole (Acros Organics).
Diethyl ether (Fisher).
Nylon filter paper (MAGNA, nylon, supported, plain, 0,45 μm, 47 mm) (GE Water and Process Technologies).
2.1.3. Reverse-Phase High-Performance Liquid Chromatography
Solvent A: 0.1% TFA in water (see Note 2).
Solvent B: 90% acetonitrile (HPLC grade; Fisher), 10% water, and 0.1% TFA.
Protein or peptide C18 column (Vydac): For analytical purposes, the use of a 250-mm length, 4.6-mm internal diameter (id) column packed with 5μm particles is used; for semipreparative scale, a column 250mm long, 22-mm id packed with 10-μm particles is used.
2.1.4 Lyophilization
For lyophilization, use liquid N2.
2.2. Instrumentation
A 433A peptide synthesizer from Applied Biosystems with SynthAssist software was used to carry out the synthesis of the peptides described here.
HP 1100 series HPLC equipment from Agilent Technologies equipped with a Vydac C18 peptide/protein column was utilized to perform analytical HPLC.
Bulk purification was carried out on Waters 600 series modular semipreparative-scale HPLC equipment with a Vydac C18 peptide/protein column.
Flexi-Dry freeze dryers from FTS, New York, were used for lyophilization.
Electrospray ionization mass spectrometry (ESI-MS) was carried out on a Thermo Finnigan LCQ mass spectrometer to characterize peptide mass.
3. Methods
3.7. Solid-Phase Peptide Synthesis
Although the following procedures are specific to the ABI 433A peptide synthesizer, they can be easily adapted for other automated synthesizers (43).
The ABI 433A is an automated batch peptide synthesizer that can perform syntheses from 0.1 to 1 mmol scale; however, the procedure described here is specific to a 0.25-mmol scale synthesis. In general, for the ABI 433A, the solid support resin is placed inside a reaction vessel. Filtered solvents or reagents are delivered to or drained from the reaction vessel by the application of N2 pressure. NMP is used as the universal solvent (44). During synthesis, vortexing of the reaction vessel or bubbling N2 through the reaction mixture facilitates mixing.
Although the instrumental software supports Fmoc-based synthesis using HBTU activation, we employ HCTU activation, which provides improved synthetic outcome. For any given synthesis, 1 mmol of dry powdered Fmoc-protected amino acid is packed in each cartridge, and the cartridges are sequentially arranged on a guideway. When the instrument couples an Fmoc amino acid to a resin-based free amine, first a pneumatic injector ruptures the cartridge septum to deliver NMP required for dissolving the amino acid. HCTU and the base (i.e., DIPEA) (stored in separate reservoirs) are also mixed with the amino acid to prepare active esters before delivery to the resin. A fourfold excess of the amino acid ensures that each coupling reaction reaches more than 99% completion. This is crucial since an accumulated decrease in the coupling efficacy can negatively affect the final yield and purity of even small peptides, such as the 20-residue sequences prepared here.
Typically, coupling is allowed to proceed for 15 min, after which the resin is washed several times with NMP. Neat piperidine from the reagent bottle is diluted with NMP in 1:4 ratio, resulting in a 20% piperidine solution, and is delivered to the reaction vessel for Fmoc deprotection. Fmoc deprotection is actively monitored by measuring the UV absorbance (301 nm) of the dibenzofulvene-piperidine adduct released from the resin-bound peptide. Based on the real-time deprotection data, successive rounds of deprotection are implemented automatically via the instrumental software. In addition, these extended rounds of deprotection automatically implement the application of a capping cycle (acetic anhydride) after the coupling of the next amino acid in the sequence is complete to minimize the formation of deletion sequences.
3.1.1. General Considerations
Scale of synthesis: Due to the time and expense of synthesizing peptides, we routinely synthesize new sequences initially in small scale (0.1 mmol) to map out difficult sequential couplings (see Subheading 3.1.2.).
Selection of the correct reaction vessel size: For efficient swelling, mixing, and washing of the resin, an appropriate size reaction vessel for the desired scale of synthesis should be used. A size that affords maximal resin swelling and mixing while minimizing the dead volume is desired.
Instrumental calibration: Routine instrumental calibration ensures optimal delivery of reagents, minimizing unsuccessful syntheses due to instrumental error.
Preparation of reagents: In our experience, reagents such as HCTU and piperidine do not store well in solution at room temperature; therefore, we recommend using fresh reagents for each new synthetic procedure (see Note 1).
3.1.2. Synthesis Optimization
Peptide sequences that have not been prepared previously in the lab are initially synthesized using a standard nonoptimized protocol. This allows problematic sequential positions to be identified where Fmoc amino acids may need to be double coupled to the growing resin-bound sequence. In this nonoptimized protocol, each residue of the sequence is single coupled to the growing chain, and based on the Fmoc deprotection profile, the instrument will conditionally cap the growing chain at problematic sequential positions. HCTU activation and 20% piperidine in NMP are used for the coupling and Fmoc deprotection steps, respectively.
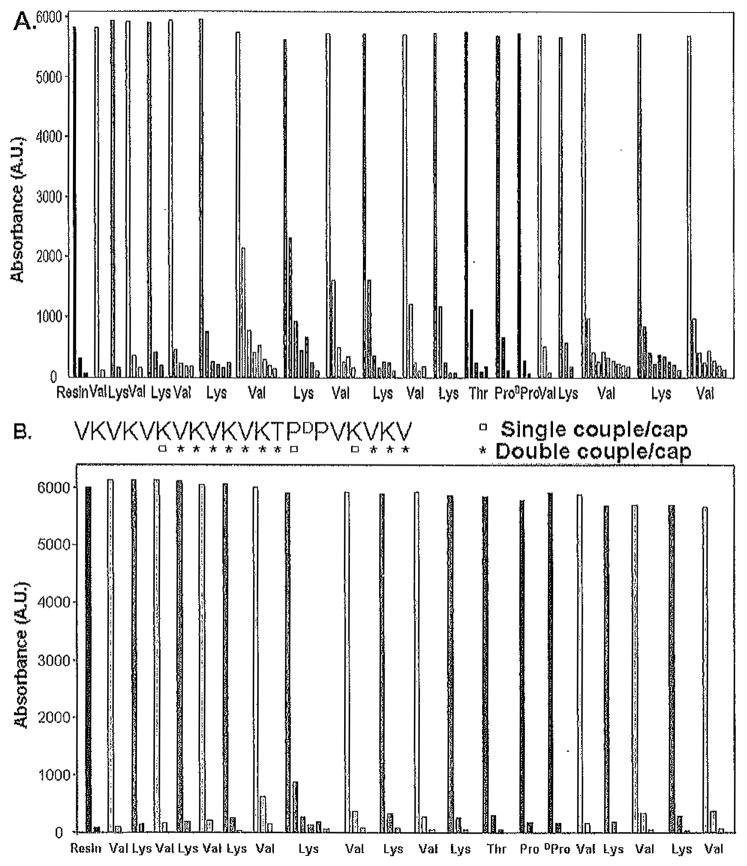
Figure 1A shows the Fmoc deprotection profile for peptide A (VKVKVDPPTKVKVKVKVKVKV-NH2), which was prepared using this nonoptimized protocol. Here, the absorbance at 301 nm monitors the release of the Fmoc group (dibenzofulvene-piperidine adduct) as a function of sequential position. As can be seen in the figure, the first valine is nearly quantitatively Fmoc deprotected after two successive rounds of treatment with 20% piperidine. The synthesis proceeds well until the deprotection of the valine at position 5 from the C-terminus, where five rounds of deprotection have been implemented. After these extended rounds of deprotection steps, the sequence is capped after the lysine at position 6 has been incorporated.
Fig. 1.
Profiles monitoring UV absorbance of the dibenzofulvene-piperidine adduct at 301 nm for each residue in the synthesis of the peptide VKVKVDPPTKVKVKVKVKVKV-NH2. (A) Nonoptimized synthesis in which each residue is single coupled, and 20% piperidine in N-methylpyrrolidone (NMP) is used for deprotection. (B) Optimized synthesis in which amino acid double coupling and N-terminal capping with acetic anhydride are employed at the positions indicated in the sequence (written from C- to N-terminus as synthesized). In addition, 1% 1,8-diazabicyclo[5.4.0]-undec-7-ene (DBU) and 19 % piperidine in NMP is employed for 9-fluorenylmethoxycarbonyl (Fmoc) deprotection in panel B.
Peptides resulting from a nonoptimized synthesis can be purified to near homogeneity, but the purification is typically difficult and low yielding. We use the Fmoc deprotection data to generate an optimized synthetic procedure in which problematic residues are double coupled using HCTU and importantly a deprotection cocktail composed of 1 % DBU, 19% piperidine in NMP is used for Fmoc deprotection. This cocktail was reported to be superior for Fmoc deprotection as compared to 20% piperidine only, and we have found this to be true (45). In addition, the sequence may be capped with Ac2O after residues that have been double coupled in the sequence. However, for repetitive amphiphilic peptides, the sequential positions that will be capped should be carefully selected to optimize differences in hydrophobicity between the desired sequence and alternate deletion sequences, greatly simplifying the purification procedure.
Figure 1B shows the sequence of the peptide (written from the C- to N-terminus as synthesized) and the positions that have been double coupled and capped. The Fmoc deprotection profile for this optimized synthesis is shown. In comparison to the nonoptimized synthesis, fewer rounds of deprotection steps have been used for each residue, suggesting that the peptide had been prepared in a more facile manner. However, analytical HPLC is used to demonstrate the purity of the cleaved (crude) peptide as described in Subheading 3.3.1.
3.2. Peptide Resin Cleavage and Side-Chain Deprotection
After the deprotection of the final Fmoc group, the resin is washed with NMP (twice), followed by DCM (twice). The resin is then dried under vacuum for at least 1 h. It is advisable to carry out a test cleavage on a small quantity (30–40mg) of resin to ensure selection of the correct cleavage reagent mixture and reaction time. With this said, we have found that the particular cleavage cocktail discussed next is extremely versatile in effecting resin cleavage and side-chain deprotection of almost all the sequences that have been prepared in our lab.
3.2.1. Cleavage Protocol
Prepare 10 mL of the cleavage reagent by mixing TFA/thioanisole/ethandedithiol/anisole in a 90:5:3:2 volume ratio (46).
Place the dry resin in a round-bottom flask containing a magnetic stir bar and slowly add enough cleavage reagent such that it completely covers the resin. Stir the resin slowly under a N2 atmosphere for 2h. (If the sequence contains 4-methoxy-2,3,6-trimethylbenzenesulfonyl [MTR]-protected or 2,2,5,7,8-pentamethylchroman-6-sulfonyl [PMC]-protected arginine, then increase the reaction time to 4h.)
Remove the resin via filtration through a sintered glass funnel using positive N2 pressure to aid the filtration process. Wash the resin two or three times with a small amount of neat TFA to ensure that all the cleaved peptide is removed from the resin. Avoid using vacuum to aid the filtration process. This limits possible oxidation of the peptide that could result from pulling air through the apparatus.
Reduce the volume of the filtrate to 1/5 its original volume by flowing a stream of N2 across the liquid.
Precipitate the peptide by adding ice-cold diethyl ether in small portions.
Immediately collect the peptide precipitate via filtration using a nylon filter. Wash the precipitate with copious amounts of cold ether. Again, filtration may be aided by using positive N2 pressure instead of vacuum to limit possible oxidation of the peptide. The crude peptide is then dried under vacuum.
3.3. Purification and Primary Characterization
3.3.1. Initial Assessment of Peptide Purity and Establishing a Preparatory Reversed-Phase HPLC Gradient for Purification
The crude peptide obtained after resin cleavage and side chain deprotection is purified using reversed-phase HPLC (RP-HPLC). To assess the retention time of the desired peptide on a C18 peptide/protein column as well as to determine the impurity profile of the peptide synthesized, an analytical chromatogram of the crude material is collected. Typically, we analyze peptide solutions at a concentration of 1 mg/mL of solvent A (injection volume = 100μL, eluent flow rate = 1mL/min, column temperature = 20°C) on the analytical RP-HPLC. Before performing any HPLC experiment, the column is cleaned with 100% solvent B to eliminate any existing peptides adsorbed on the column. This is followed by equilibration with 100% solvent A prior to sample injection.
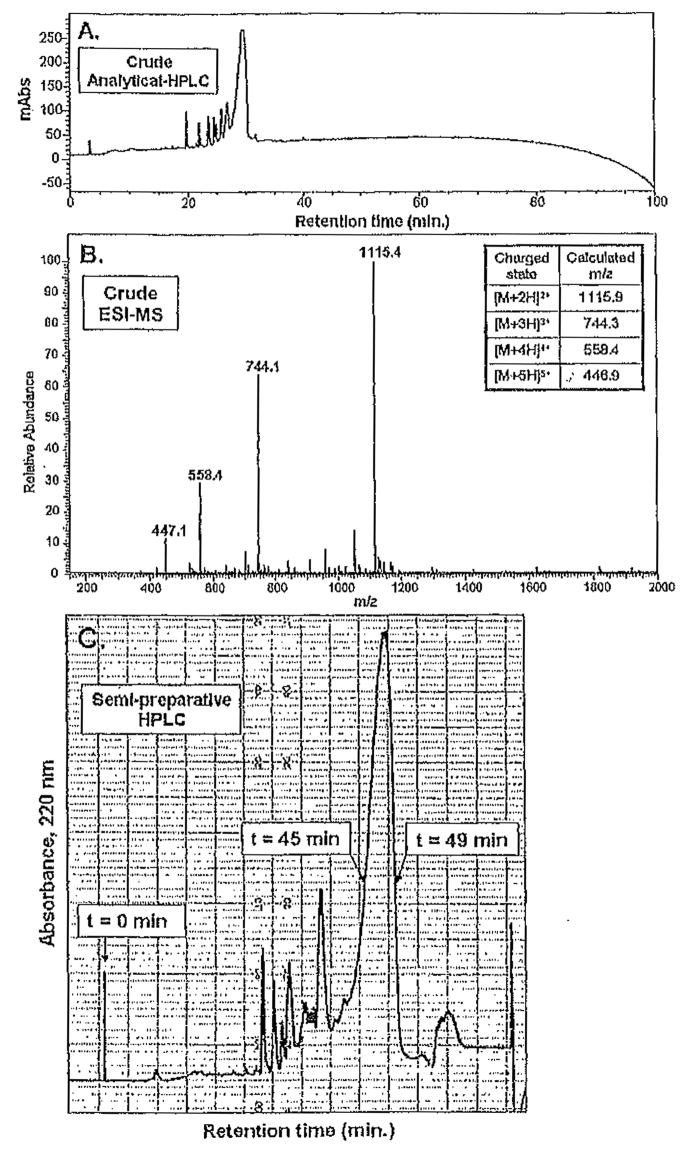
Figure 2A,B depicts an analytical HPLC trace and ESI-MS of the crude material obtained from the optimized synthetic procedure (Fig. 1B). In Fig. 2A, a linear gradient from 0% to 100% solvent B over 100min is employed. The UV absorbance at 220 nm is monitored with respect to the retention time of the eluting species from the column. We typically monitor 220, 254, and 280nm to detect peptide as well as aromatic species derived from the resin cleavage reaction. Here, we show only the data at 220 nm for clarity. Each eluted peak is manually collected as a separate fraction. Mass spectrometry of all the collected fractions indicates the retention time of the desired peptide. In the example discussed, the desired sequence elutes as the largest peak at 30 min, as shown in Fig. 2A. This indicates that an eluent mixture of 30% solvent B and 70% solvent A is necessary to elute the peptide from the C18 column since a linear gradient of 0% to 100% solvent B over 100 min was employed.
Fig. 2.
(A) Reversed-phase high-performance liquid chromatographic (RP-HPLC) trace of the crude material isolated from the resin cleavage reaction, Absorbance at 220 nm is monitored versus retention time for peptide A (VKVKVDPPTKVKVKVKVKVKV-NH2) on a C18 column employing a linear gradient of 0% to 100% solvent B in 100min. Desired peptide elutes at 30min. (B) ESI-MS (electrospray ionization mass spectrometry) of crude material isolated from resin cleavage reaction. The molecular ions of the peptide are labeled and defined (inset). Unlabeled peaks in the spectrum are indicative of the impurities in the crude material. (C) Representative sernipreparative RP-HPLC chromatogram. The desired peptide fraction is collected from 45 to 49 min.
It should be noted that despite the amphiphilic nature of the peptide, it is synthesized in high yield via the optimized synthetic protocol. The purification stage involves separation of the desired fraction from impurities on the semipreparative scale. For semipreparative-scale purification, we initially dissolve the crude material in solvent A (1–4mg/mL) and inject 5-mL portions of this solution onto the column. A distinct semipreparative gradient is calculated and used for the HPLC purification of the peptide.
Typically, we will decrease the gradient steepness from 1% solvent B per minute, which was used for analytical HPLC, to either 0.5% solvent B per minute or 0.25% solvent B per minute depending on how similar the retention times of any impurities are to that of the desired peptide. In this example, we employ a gradient of 0.25% solvent B per minute. We have found through experience that small peptides should have a retention time greater than 30 min on the Vydac C18 columns used in our lab to maximize elution resolution. Therefore, a semipreparative gradient for peptide A, which eluted at 30% solvent B, can be calculated as follows:
| (1) |
| (2) |
However, injecting crude material onto a column at high percentages of solvent B (in this example, 22% solvent B) decreases resolution; we typically introduce solutions of crude material to the column at 0% solvent B and subsequently rapidly approach the initial conditions using a steep gradient. The final gradient employed for peptide A is shown in Table 1.
Table 1.
Semipreparative High-Performance Liquid Chromatographic (HPLC) Gradient Calculated for Peptide A
| Time (min) | Solvent B (%) | Gradient |
|---|---|---|
| 0 | 0 | — |
| 22 | 22 | 1% solvent B per min |
| 312 | 100 | 0.25% solvent B per min |
The semipreparative chromatogram shown in Fig. 2C resulted from a protocol nearly identical to that discussed here. The peptide began to elute, after about 40 min (Fig. 2C). A fraction was manually collected from 45 to 49 min. After this time, the gradient was aborted and the column immediately washed with 100% solvent B followed by reequilibration with 100% solvent A; the process was repeated for the remaining crude material. The isolated fractions from HPLC purification were combined and lyophilized, affording a white powder.
3.4. Assessment of Purity
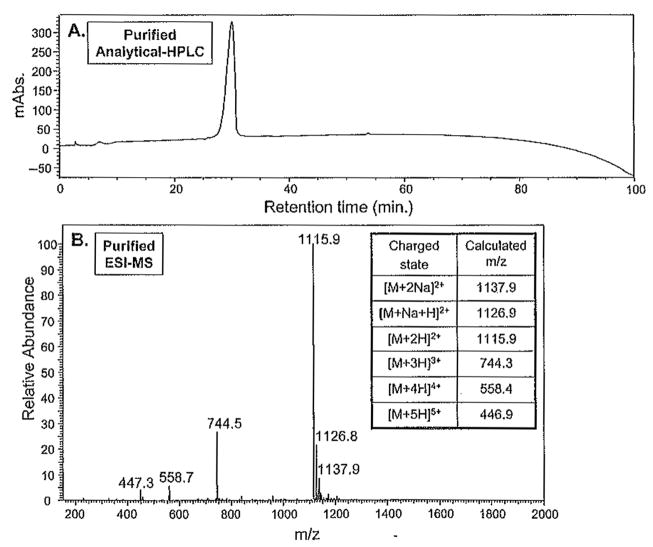
Following lyophilization, the purity of the peptide is determined by analytical RP-HPLC and ESI-MS. A 1 mg/mL solution of the peptide is prepared in solvent A. Of this solution 100 μL are injected onto the analytical C18 column, and an analytical HPLC experiment employing a linear gradient of 0–100% solvent B in 100 min is carried out. Figure 3 A depicts the analytical HPLC chromatogram of peptide A purified with the aforementioned semipreparative HPLC gradient. Peptide A was effectively separated from the prepeak and postpeak impurities present in the crude after resin cleavage and side-chain deprotection (Fig. 2A). The purity of this peptide was further assessed by mass spectrometry, as shown in Fig. 3B. The observed molecular mass ions at 1115.9,744.5, 558.7, and 447.3 correspond to the +2, +3, +4, and +5 charged states of the peptide, respectively. The peptide mass determined from ESI-MS is in agreement with the calculated masses established from the sequence.
Fig. 3.
Assessment of the peptide purity following semipreparative-scale high-performance liquid chromatographic (HPLC) separation. (A) Analytical reversed-phase HPLC of the lyophilized peptide carried out using a linear gradient of 0% to 100% solvent B in 100min on a C18 column. (B) ESI-MS (electrospray ionization mass spectrometry) of the purified peptide and appropriate calculated masses.
Both HPLC and mass spectral analysis suggest that peptide A has been purified to near homogeneity. Typically, hydrogels prepared from β-hairpin peptides of this level of purity afford consistent batch-to-batch material properties. There is always the possibility that an impurity may coelute with the purified peptide that is not observed by mass spectrometry. This uncommon scenario is usually realized after observing inconsistent properties from a given batch of peptide. This problem can usually be remedied by repurifying the peptide by RP-HPLC using a shallower gradient or isocratic conditions or warming or cooling the column. As long as the impurities have different temperature-dependent retention times, performing the purification at two different temperatures provides an excellent means of producing extremely pure samples. Of course, different column types may also be employed to maximize differences in retention times.
3.5. Importance of Purity
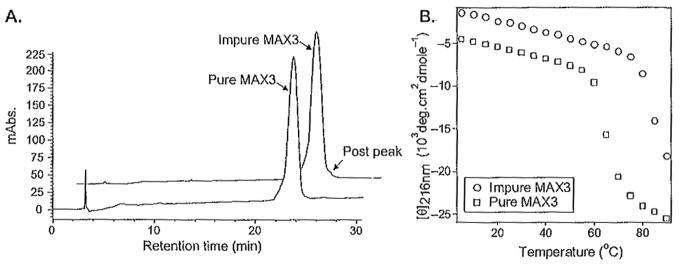
The importance of purifying peptides to the highest possible level with respect to achieving reproducible physical and biophysical properties is exemplified in Fig. 4.
Fig. 4.
(A) Analytical high-performance liquid chfomatographic (HPLC) chromatograms of distinct batches of peptide MAX3. Impure MAX3 has a small postpeak not seen in the pure trace. (B) Temperature-dependant circular dichroism spectra of 150 μM peptide at pH 9.0, 125 mM borate, 10mM NaCl with purity corresponding to the chromatograms in (A). Figure depicts the secondary structure transition from random coil to β-sheet by monitoring the mean residue ellipticity at 216nm as a function of temperature. Impure MAX3 folds and assembles at a higher temperature.
MAX3 (VKVKVKTKVDPPTKVKTKVKV-NH2) is a β-hairpin peptide that was designed to undergo a thermally triggered intramolecular folding and self-assembly event, which affords hydrogel material (38). Figure 4A shows the analytical chromatograms of two different batches of MAX3 that had been purified on separate occasions. The seemingly insignificant impurity (seen as a postpeak) in the “impure” batch grossly influences the temperature at which folding and consequent self-assembly occurs. The circular dichroism (CD) data in Fig. 4B show the mean residue ellipticity at 216nm, an indicator of β-sheet structure, as a function of temperature. At low temperatures, peptide from both batches exists in random-coil conformations. As the temperature is increased, MAX3 folds and self-assembles into a β-sheet-rich hydrogel. It is clear from the data that the temperature at which this folding/assembly transition takes place is batch dependent; a small amount of impurity increased the temperature necessary to initiate peptide folding and self-assembly. Repurifying this batch to remove the impurity restored the peptide’s normal temperature-dependent behavior. When possible, our lab routinely publishes an analytical HPLC chromatogram and the mass spectrum of each peptide discussed in a given manuscript; these data are usually contained in the supporting information. This is important in that it establishes the level of purity needed to realize the observed biophysical/material properties reported.
3.6. Beyond Primary Characterization
Although this chapter is mainly concerned with the synthesis and primary characterization of peptides used in self-assembly, a brief introduction to several techniques that are common to the study of self-assembled peptide-based hydrogels is provided next. These techniques offer insight into the secondary structure of the peptide in the self-assembled state, the nanoscale morphology of the assembled structures that constitute the hydrogel, as well as the bulk mechanical properties of the hydrogel itself. These brief introductions are meant to acquaint those new to the field; comprehensive descriptions of each technique can be found in the literature as indicated.
Circular dichroism (CD) can be used to determine the secondary structure of peptides in the self-assembled state of optically clear hydrogels (47). Characteristic dichroic signatures for α-helical, β-sheet, and β-turn secondary structures as well as random-coil conformations are easily detected. Importantly, CD spectroscopy provides an excellent means of monitoring changes in the secondary structure of peptides in response to changes in solution conditions (e.g., pH, temperature, ionic strength, chaotropes, etc.). However, obtaining spectra of hydrogel samples can sometimes be challenging due to the small path length cells that must be employed if the concentration of the peptide constituting the gel is high. If using small path length cells proves to be problematic, dilute preparations of assembled peptide can be studied employing larger path length cells as long as light scattering is minimized; this is the case in data that are shown in Fig. 4B.
Fourier transform infrared (FTIR) spectroscopy is another convenient technique to study the secondary structure of peptides in the self-assembled state (48). One advantage of FTIR is that, unlike CD, it is less sensitive to light scattering; as a result, greater concentrations of peptides can be studied. Well-characterized absorptions are known for helical and β-sheet structures as well as random-coil conformations. Possible limitations of this technique are that TFA salts of peptides cannot be used since TFA absorbs strongly in the amide I′ region. In addition, H2O cannot be used as a solvent for the same reason. Therefore, the TFA counterions of peptides are typically exchanged by dissolving the peptide in 0.1M HCl followed by lyophilization. The resulting HCl-peptide salt is subsequently dissolved in D2O and lyophilized several times to exchange the water. Hydrogels can then be prepared using D2O and studied.
Oscillatory rheology can be used to study the mechanical properties of peptide-based hydrogels (49). Commonly, the mechanical rigidity of the hydrogel is assessed by measuring the storage and loss modulus of the gel as a function of time, frequency, or strain. In addition, detailed insight into the physical nature (crosslink type and density, response to shear strain, etc.) of the gels can be gleaned by performing rheological measurements.
Transmission electron microscopy (TEM) can be used to characterize the local nanostructure of self-assembled peptides (50). Typically, dilute suspensions of assembled peptide are placed on grids and allowed to dry. Contrast-enhancing agents are often used to study the fine details of the nanostructure. Importantly, drawing appropriate conclusions from TEM necessitates that enough observations are recorded to provide meaningful statistics. For hydrogels, one possible limitation in employing conventional TEM is that samples are dehydrated; thus, inferences must be made to relate the self-assembled structure observed on the grid to that which actually exists in the hydrated state. In the limiting case, the observed structure may be different from that in the hydrated state. To overcome this possible limitation, the in situ structure of gels can be studied by cryogenic TEM. Here, the water in the hydrogel is vitrified to preserve the in situ nanostructure. However, this technique is difficult and lies in the hands of experts.
Complementary to TEM, atomic force microscopy (AFM) can be an important tool in studying the local nanostructure of self-assembled materials (51). AFM is particularly well suited to define the height of assemblies deposited on a surface, a dimension not amenable to TEM analysis. Conversely, AFM is limited in its capacity to accurately measure in the XY dimension, which defines the width of a given assembly. For soft materials, taking measurements in the tapping mode provides a means of minimally invasive interrogation.
Small-angle neutron scattering (SANS) is an extremely powerful tool for globally defining the self-assembled structure present in a given hydrogel at both the local and network scales (52). Scattering intensity is measured as a function of the reciprocal space, and the resulting data can be fit using different form factors that reveal information about the morphology and network properties of the self-assembly. In addition, SANS can be potentially used to track, in real time, the developing assembled structure; this affords information regarding the assembly mechanism. An important point worth mentioning is the use of ultra-small-angle neutron scattering (USANS) to characterize hydrogel morphology on the microscale. Such information can be critical for characterizing hydrogels used in biological applications.
These techniques, when used together, offer a powerful suite of analysis that allows relationships to be drawn among peptide sequence, secondary structure, and self-assembly mechanisms that ultimately dictate the assembly morphology and bulk material properties. These techniques can help establish the rules that govern the assembly of appropriately purified peptides so that custom materials can be fabricated for targeted applications.
4. Notes
Handling of synthesizer reagents: The resins, Fmoc amino acids, as well as HCTU should be stored under refrigeration to prevent degradation over time. All reagents are weighed and handled in the hood to minimize personal exposure.
Ultrapure water having a resistivity of 18.2MΩ-cm obtained from a MilliQ (Millipore) purification system should be used to prepare all solutions and is referred to as “water” in this chapter.
Acknowledgments
We acknowledge the National Institutes of Health grant R01 DE016386-01. We also thank Lisa A. Haines-Butterick for optimization of the synthesizer chemistry and her helpful discussions for this chapter as well as Karthikan Rajagopal for performing the MAX3 studies.
References
- 1.Rajagopal K, Schneider JP. Self-assembling peptides and proteins for nanotechnological applications. Curr Opin Struct Biol. 2004;14(4):480–486. doi: 10.1016/j.sbi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Whitesides GM, Mathias JP, Seto CT. Molecular self-assembly and nanochemistry — a chemical strategy for the synthesis of nanostructures. Science. 1991;254(5036):1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 3.Carny O, Shalev DE, Gazit E. Fabrication of coaxial metal nanocables using a self-assembled peptide nanotube scaffold. Nano Lett. 2006;6(8):1594–1597. doi: 10.1021/nl060468l. [DOI] [PubMed] [Google Scholar]
- 4.Ray S, Drew MGB, Das AK, Banerjee A. The role of terminal tyrosine residues in the formation of tripeptide nanotubes: a crystallographic insight. Tetrahedron. 2006;62(31):7274–7283. [Google Scholar]
- 5.Crisma M, Toniolo C, Royo S, Jimenez AI, Cativiela C. A helical, aromatic, peptide nanotube. Org Lett. 2006;8(26):6091–6094. doi: 10.1021/ol062600u. [DOI] [PubMed] [Google Scholar]
- 6.Leclair S, Baillargeon P, Skouta R, Gauthier D, Zhao Y, Dory YL. Micrometer-sized hexagonal tubes self-assembled by a cyclic peptide in a liquid crystal. Angew Chem Int Ed. 2004;43(3):349–353. doi: 10.1002/anie.200352259. [DOI] [PubMed] [Google Scholar]
- 7.Horne WS, Stout CD, Ghadiri MR. A heterocyclic peptide nanotube. J Am Chem Soc. 2003;125(31):9372–9376. doi: 10.1021/ja034358h. [DOI] [PubMed] [Google Scholar]
- 8.Amorin M, Castedo L, Granja JR. Self-assembled peptide tubelets with 7 angstrom pores. Chemistry. 2005;11(22):6543–6551. doi: 10.1002/chem.200500478. [DOI] [PubMed] [Google Scholar]
- 9.Block MAB, Hecht S. Wrapping peptide tubes: merging biological self-assembly and polymer synthesis. Angew Chem Int Ed. 2005;44(43):6986–6989. doi: 10.1002/anie.200502436. [DOI] [PubMed] [Google Scholar]
- 10.Lu K, Jacob J, Thiyagarajan P, Conticello VP, Lynn DG. Exploiting amyloid fibril lamination for nanotube self-assembly. J Am Chem Soc. 2003;125(21):6391–6393. doi: 10.1021/ja0341642. [DOI] [PubMed] [Google Scholar]
- 11.Gao XY, Matsui H. Peptide-based nanotubes and their applications in bionanotechnology. Adv Mater. 2005;17(17):2037–2050. doi: 10.1002/adma.200401849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolfson DN, Ryadnov MG. Peptide-based fibrous biomaterials: some things old, new and borrowed. Curr Opin Chem Biol. 2006;10(6):559–567. doi: 10.1016/j.cbpa.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 13.Aggeli A, Nyrkova IA, Bell M, et al. Hierarchical self-assembly of chiral rod-like molecules as a model for peptide beta-sheet tapes, ribbons, fibrils, and fibers. Proc Natl Acad Sci U S A. 2001;98(21):11857–11862. doi: 10.1073/pnas.191250198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bitton R, Schmidt J, Biesalski M, Tu R, Tirrell M, Bianco-Peled H. Self-assembly of model DNA-binding peptide amphiphiles. Langmuir. 2005;21(25):11888–11895. doi: 10.1021/la051811p. [DOI] [PubMed] [Google Scholar]
- 15.Deechongkit S, Powers ET, You SL, Kelly JW. Controlling the morphology of cross beta-sheet assemblies by rational design. J Am Chem Soc. 2005;127(23):8562–8570. doi: 10.1021/ja050558c. [DOI] [PubMed] [Google Scholar]
- 16.Elgersma RC, Meijneke T, Posthuma G, Rijkers DTS, Liskamp RMJ. Self-assembly of amylin(20–29) amide-bond derivatives into helical ribbons and peptide nanotubes rather than fibrils. Chemistry. 2006;12(14):3714–3725. doi: 10.1002/chem.200501374. [DOI] [PubMed] [Google Scholar]
- 17.Lowik D, Garcia-Hartjes J, Meijer JT, van Hest JCM. Tuning secondary structure and self-assembly of amphiphilic peptides. Langmuir. 2005;21(2):524–526. doi: 10.1021/la047578x. [DOI] [PubMed] [Google Scholar]
- 18.Matsumura S, Uemura S, Mihara H. Fabrication of nanofibers with uniform morphology by self-assembly of designed peptides. Chemistry. 2004;10(11):2789–2794. doi: 10.1002/chem.200305735. [DOI] [PubMed] [Google Scholar]
- 19.Zhang SG. Fabrication of novel biomaterials through molecular self-assembly. Nat Biotechnol. 2003;21(10):1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 20.Fairman R, Akerfeldt KS. Peptides as novel smart materials. Curr Opin Struct Biol. 2005;15(4):453–463. doi: 10.1016/j.sbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Bonzani IC, George JH, Stevens MM. Novel materials for bone and cartilage regeneration. Curr Opin Chem Biol. 2006;10(6):568–575. doi: 10.1016/j.cbpa.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Mart RJ, Osborne RD, Stevens MM, Ulijn RV. Peptide-based stimuli-responsive biomaterials. Soft Matter. 2006;2(10):822–835. doi: 10.1039/b607706d. [DOI] [PubMed] [Google Scholar]
- 23.Nowak AP, Breedveld V, Pakstis L, et al. Rapidly recovering hydrogel scaffolds from self-assembling diblock copolypeptide amphiphiles. Nature. 2002;417(6887):424–428. doi: 10.1038/417424a. [DOI] [PubMed] [Google Scholar]
- 24.Stendahl JC, Rao MS, Guler MO, Stupp SI. Intermolecular forces in the self-assembly of peptide amphiphile nanofibers. Adv Funct Mater. 2006;16(4):499–508. [Google Scholar]
- 25.Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. J Am Chem Soc. 2002;124(50):15030–15037. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 26.Caplan MR, Schwartzfarb EM, Zhang SG, Kamm RD, Lauffenburger DA. Control of self-assembling oligopeptide matrix formation through systematic variation of amino acid sequence. Biomaterials. 2002;23(1):219–227. doi: 10.1016/s0142-9612(01)00099-0. [DOI] [PubMed] [Google Scholar]
- 27.Collier JH, Messersmith PB. Self-assembling polymer-peptide conjugates: nanostructural tailoring. Adv Mater. 2004;16(11):907–910. [Google Scholar]
- 28.Ramachandran S, Trewhella J, Tseng Y, Yu YB. Coassembling peptide-based biomaterials: effects of pairing equal and unequal chain length oligopeptides. Chem Mater. 2006;18(26):6157–6162. [Google Scholar]
- 29.Zhang SG. Emerging biological materials through molecular self-assembly. Biotechnol Adv. 2002;20(5–6):321–339. doi: 10.1016/s0734-9750(02)00026-5. [DOI] [PubMed] [Google Scholar]
- 30.Yang JY, Xu CY, Wang C, Kopecek J. Refolding hydrogels self-assembled from N-(2-hydroxypropyl)methacrylamide graft copolymers by antiparallel coiled-coil formation. Biomacromolecules. 2006;7(4):1187–1195. doi: 10.1021/bm051002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen W, Zhang KC, Kornfield JA, Tirrell DA. Tuning the erosion rate of artificial protein hydrogels through control of network topology. Nat Mater. 2006;5(2):153–158. doi: 10.1038/nmat1573. [DOI] [PubMed] [Google Scholar]
- 32.Ciani B, Hutchinson EG, Sessions RB, Woolfson DN. A designed system for assessing how sequence affects alpha to beta conformational transitions in proteins. J Biol Chem. 2002;277(12):10150–10155. doi: 10.1074/jbc.M107663200. [DOI] [PubMed] [Google Scholar]
- 33.Lee HJ, Lee J-S, Chansakul T, Yu C, Elisseeff JH, Yu SM. Collagen mimetic peptide-conjugated photopolymerizable PEG hydrogel. Biomaterials. 2006;27(30):5268–5276. doi: 10.1016/j.biomaterials.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Kotch FW, Raines RT. Self-assembly of synthetic collagen triple helices. Proc Natl Acad Sci U S A. 2006;103(9):3028–3033. doi: 10.1073/pnas.0508783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang ZM, Liang GL, Wang L, Bing X. Using a kinase/phosphatase switch to regulate a supramolecular hydrogel and forming the supramoleclar hydrogel in vivo. J Am Chem Soc. 2006;128(9):3038–3043. doi: 10.1021/ja057412y. [DOI] [PubMed] [Google Scholar]
- 36.Jun HW, Yuwono V, Paramonov SE, Hartgerink JD. Enzyme-mediated degradation of peptide-amphiphile nanofiber networks. Adv Mater. 2005;17(21):2612–2617. [Google Scholar]
- 37.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 38.Pochan DJ, Schneider JP, Kretsinger J, Ozbas B, Rajagopal K, Haines L. Thermally reversible hydrogels via intramolecular folding and consequent self-assembly of a de Novo designed peptide. J Am Chem Soc. 2003;125(39):11802–11803. doi: 10.1021/ja0353154. [DOI] [PubMed] [Google Scholar]
- 39.Ozbas B, Kretsinger J, Rajagopal K, Schneider JP, Pochan DJ. Salt-triggered peptide folding and consequent self-assembly into hydrogels with tunable modulus. Macromolecules. 2004;37(19):7331–7337. [Google Scholar]
- 40.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled ss-hairpin peptide hydrogel surfaces. Biomaterials. 2005;26(25):5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 41.Haines LA, Rajagopal K, Ozbas B, Salick DA, Pochan DJ, Schneider JP. Light-activated hydrogel formation via the triggered folding and self-assembly of a designed peptide. J Am Chem Soc. 2005;127(48):17025–17029. doi: 10.1021/ja054719o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajagopal K, Ozbas B, Pochan DJ, Schneider JP. Probing the importance of lateral hydrophobic association in self-assembling peptide hydrogelators. Eur Biophys J Biophys Lett. 2006;35(2):162–169. doi: 10.1007/s00249-005-0017-7. [DOI] [PubMed] [Google Scholar]
- 43.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Oxford University Press; New York: 2000. [Google Scholar]
- 44.Fields GB, Fields CG. Solvation effects in solid-phase peptide-synthesis. J Am Chem Soc. 1991;113(11):4202–4207. [Google Scholar]
- 45.Kates SA, Sole NA, Beyermann M, Barany G, Albericio F. Optimized preparation of deca(L-alanyl)-L-valinamide by 9-fluorenylmethyloxycarbonyl (Fmoc) solid-phase synthesis on polyethylene glycol-polystyrene (PEG-PS) graft supports, with 1,8-diazobicyclo[5.4.0]-undec-7-ene (DBU) deprotection. Peptide Res. 1996;9(3):106–113. [PubMed] [Google Scholar]
- 46.Angell YM, Alsina J, Albericio F, Barany G. Practical protocols for step-wise solid-phase synthesis of cysteine-containing peptides. J Peptide Res. 2002;60(5):292–299. doi: 10.1034/j.1399-3011.2002.02838.x. [DOI] [PubMed] [Google Scholar]
- 47.Fasman GD. Circular Dichroism and the Conformational Analysis of Biomolecules. Plenum Press; New York: 1996. [Google Scholar]
- 48.Cantor CR, Schimniel PR. Biophysical Chemistry. Freeman; New York: 1980. [Google Scholar]
- 49.Larson RG. The Structure and Rheology of Complex Fluids. Oxford University Press; New York: 1999. [Google Scholar]
- 50.Williams DB, Carter CB. Transmission Electron Microscopy: A Textbook for Materials Science. Springer; New York: 1996. [Google Scholar]
- 51.Cohen SH, Lightbody ML. Atomic Force Microscopy/Scanning Tunneling Microscopy 2. Springer; New York: 1997. [Google Scholar]
- 52.Higgins JS, Benoît HC. Polymers and Neutron Scattering. Oxford University Press; New York: 1997. [Google Scholar]