Abstract
The aryl hydrocarbon receptor nuclear translocator (ARNT) is a basic helix–loop–helix Period/ARNT/Single-minded (bHLH-PAS) protein that controls various biological pathways as part of dimeric transcriptional regulator complexes with other bHLH-PAS proteins. The two PAS domains within ARNT, PAS-A and PAS-B, are essential for the formation of these complexes because they mediate protein–protein interactions via residues located on their β-sheet surfaces. While investigating the importance of residues in ARNT PAS-B involved in these interactions, we uncovered a point mutation (Y456T) on the solvent-exposed β-sheet surface that allowed this domain to interconvert with a second, stable conformation. Although both conformations are present in equivalent quantities in the Y456T mutant, this can be shifted almost completely to either end point by additional mutations. A high-resolution solution structure of a mutant ARNT PAS-B domain stabilized in the new conformation revealed a 3-residue slip in register and accompanying inversion of the central Iβ-strand. We have demonstrated that the new conformation has >100-fold lower in vitro affinity for its heterodimerization partner, hypoxia-inducible factor 2α PAS-B. We speculate that the pliability in β-strand register is related to the flexibility required of ARNT to bind to several partners and, more broadly, to the abilities of some PAS domains to regulate their activities in response to small-molecule cofactors.
Keywords: alternative conformations, NMR, Per-ARNT-Sim, proline isomerization, protein folding
It is well understood that cell survival depends on the ability to detect and respond to fluctuations in the environment. Environmental factors, such as redox potential, blue light, and oxygen levels, trigger changes in signal transduction pathways that rely on protein–protein interaction domains. One family of such domains are the Per-ARNT-Sim (PAS) domains (1), named after the three proteins where they were first identified: Period (Per), aryl hydrocarbon receptor nuclear translocator (ARNT), and Single-minded (Sim) (2). Since being recognized in these three eukaryotic transcription factors, PAS domains have been found in proteins from all three kingdoms of life that perform functions as diverse as DNA binding, phosphorylation, ion conduction, and others (1).
Despite these differences, PAS domains share a common role as protein–protein interaction domains, binding PAS or non-PAS partners either in cis or in trans. To achieve this, PAS domains have chiefly conserved a common structure composed of mixed α/β-fold of ≈110–140 aa featuring several α-helices packed against one side of a five-stranded, antiparallel β-sheet (1). This β-sheet plays a particularly critical role in PAS structure and function because its solvent-exposed side is the binding site for a wide range of PAS-bound protein targets (3). Notably, some PAS domains control the binding affinity of this interface via changes in the conformation or occupancy of environmentally sensitive cofactors bound on the opposite, internal side of the sheet. This principle is exemplified by a group of PAS-based photosensors that bind flavin chromophores in their cores. These cofactors are noncovalently bound in the dark but form a protein–flavin bond when exposed to blue light (4), generating conformational changes within the domain. As shown in the Avena sativa photropin1 LOV2 domain (5) and the Neurospora crassa Vivid protein (6), this change is transmitted through the β-sheet, displacing or rearranging other protein elements bound to the exterior.
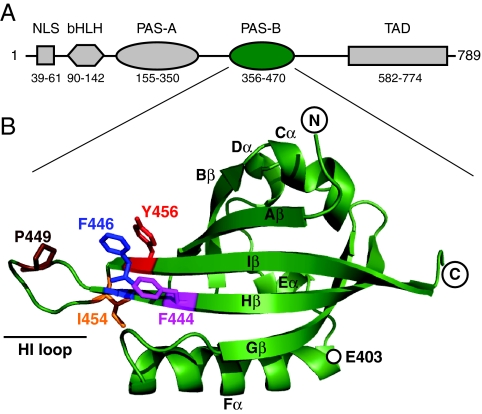
Although these data suggest that PAS domain β-sheet surfaces are inherently flexible in ligand-regulated PAS domains, it is unclear whether this holds for constitutively active PAS domains lacking internal cofactors. An example of such a domain is provided by ARNT, one of the three founding members of the PAS family (2). First identified as an essential component in the xenobiotic-sensing aryl hydrocarbon receptor (AHR) pathway (7), ARNT also plays a key role in developmental processes and response to environmental factors including hypoxia (8–11). ARNT is a member of the bHLH-PAS family of eukaryotic transcription factors, named for the presence of a N-terminal basic helix–loop–helix (bHLH) DNA-binding domain and two PAS domains (PAS-A, PAS-B) (Fig. 1A). ARNT constitutively dimerizes with other bHLH-PAS family members that themselves are regulated, including AHR and hypoxia-inducible factors (HIFs). PAS domains are essential for stable heterodimer formation, as supported by biochemical studies that show that deletion of PAS domains from ARNT (12, 13) or its partners [e.g., HIF-2α (14)] impairs the formation and activity of regulatory complexes. Biophysical studies of PAS domains from HIF-2α and ARNT have shown that the C-terminal PAS-B domains from these proteins bind each other via an antiparallel β-sheet–β-sheet packing (15, 50). Using structure-based point mutations in these β-sheets, we have demonstrated that this interaction is relevant for full-length proteins in cells (14). A broader survey of PAS-mediated protein–protein interactions further supports the role of this β-sheet surface as a general site for PAS domains to interact with a wide variety of intra- and intermolecular partners (3).
Fig. 1.
Architecture of human ARNT. (A) ARNT domain architecture, indicating the approximate boundaries of the nuclear localization sequence (NLS), bHLH domains, PAS domains (PAS-A, PAS-B), and transcriptional activation domains (TAD) as identified by hidden Markov model analysis (48). (B) Ribbon diagram of the solution structure of ARNT PAS-B domain (15). The side-chain positions of several residues discussed in the text are shown in stick representation. Secondary structure elements are labeled by using the nomenclature established by Gong et al. (49).
Assuming that ARNT PAS-B binds other bHLH-PAS proteins similarly to what is observed in the ARNT–HIF-2α complex, it is reasonable to suspect that the ARNT β-sheet is somehow adaptable to engage multiple binding partners with low sequence identity (≈10% among AHR, three HIFα isoforms, and Sim). To test this, we generated a series of point mutations in the exposed β-sheet surface of ARNT PAS-B and examined the structural and functional effects on heterodimer formation. Strikingly, we discovered a dramatic demonstration of the flexibility possible in this domain as we exposed a new conformation that coexisted in approximately equimolar concentrations with the wild-type conformation. The equilibrium between the wild-type and the new conformation was affected by additional mutations, providing us with a library of mutants that span a wide range of populations between the two. A high-resolution solution structure of the new conformation reveals that it was achieved by slipping the central Iβ strand by 3 residues, inverting the topology of this essential protein–protein interface. Accordingly, we found that the new conformation bound one of its heterodimerization partners, HIF-2α PAS-B, with >100-fold lower affinity than the wild-type conformation. We suggest that this conformational change reflects an inherent adaptability to the PAS domain fold that is used for a wide range of protein-binding activities and switchability in many systems.
Results
Point Mutations Reveal Second Conformation of ARNT PAS-B.
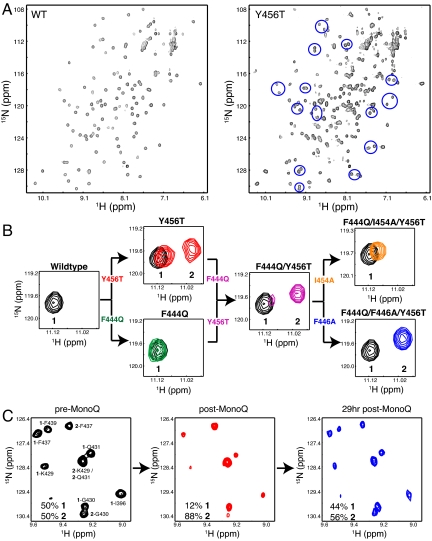
Our previous studies of ARNT PAS-B indicated that the solvent-exposed surface of the β-sheet is used both in the heterodimeric complex with HIF-2α PAS-B and a weaker homodimer (3, 15). To examine the importance of specific ARNT residues in these complexes, we designed mutations on this surface to disrupt protein–protein interactions from the β-sheet surface. Unexpectedly, a change to a solvent-exposed tyrosine on the Iβ strand (Y456T; Fig. 1B) generated a protein that exhibited cross-peak doubling in 15N-1H heteronuclear sequential quantum correlation (HSQC) spectra for ≈30 backbone amides (Fig. 2A). This doubling generated two cross-peaks of approximately equivalent intensity for each of these sites, one of which remained near the chemical shifts of the wild-type ARNT PAS-B peak. This is illustrated by the peak from the E403 backbone amide (Fig. 2B), which has one peak near the wild type (peak 1) and another shifted upfield in 1H by ≈0.1 ppm (peak 2). Notably, E403 is located 24 Å from the site of mutagenesis, consistent with a bona fide structural change. Coupled with the excellent 1H chemical shift dispersion of the new conformer 2 peaks, these data suggested that the Y456T point mutant adopted two distinct, well-folded, and equally populated conformations under these solution conditions. Sample heterogeneity, e.g., proteolysis, could have provided a trivial explanation for these results but was inconsistent with mass spectrometry data that revealed a single 13,881.7-Da species (consistent with the predicted mass of 13,880.6 Da, data not shown).
Fig. 2.
ARNT PAS-B can exist in two distinct conformations. (A) Widespread peak doubling is observed throughout 15N-1H HSQC spectra of Y456T ARNT PAS-B, as seen by comparison with spectra from the wild-type protein. Several doubled sites are highlighted with blue circles in the Y456T spectrum. (B) The E403 resonance from the Y456T ARNT mutant indicates two slowly exchanging conformations, but the relative populations of these can be perturbed by additional mutations. (C) Demonstration that conformations 1 and 2 are in equilibrium is provided by Mono Q separation of conformation 2 from a Y456T sample and monitoring the reestablishment of the equilibrium with extended incubation.
Examining spectra for other ARNT PAS-B domains containing point mutations at other solvent-exposed residues near Y456, we found that these proteins adopted differing ratios of the same two distinct conformations. This is demonstrated by the F444Q mutation, which affects a position on the Hβ strand adjacent to Y456 (Fig. 1B). On its own, the F444Q change does not reveal a second conformation in the wild-type context (Fig. 2B). However, it significantly changes the relative populations of wild-type 1 and novel 2 cross-peaks when combined with Y456T (Y456T, 1:2 ratio = 50:50; F444Q/Y456T, 19:81). Further mutations at nearby sites exhibited similar behavior, generating a group of 17 proteins (containing three or fewer point mutations) that have a range of preferences for one conformation or the other spanning >99:<1 for both conformations 1 and 2 (Table 1).
Table 1.
Effect of ARNT PAS-B mutations on relative populations in conformations
| Mutation | Percentage 1 | Percentage 2 |
|---|---|---|
| Wild type | >99 | <1 |
| F444Q | >99 | <1 |
| F444Q/I454A/Y456T | >98 | <2 |
| F444Q/I454F/Y456T | >98 | <2 |
| F444Q/Y456T/I458E | >98 | <2 |
| Y456A | >98 | <2 |
| Y456S | 81 | 19 |
| Y456T | 50 | 50 |
| P449A/Y456T | 32 | 68 |
| F444Q/T445A/Y456T | 31 | 69 |
| F444Q/Y456T | 19 | 81 |
| F444Q/Y456T/T460V | 15 | 85 |
| F444Q/T445Y/Y456T | 14 | 86 |
| F444Q/N448T/Y456T | 10 | 90 |
| F444Q/N448W/Y456T | <2 | >98 |
| F444Q/Y456T/I457F | <2 | >98 |
| F444Q/F446A/Y456T | <1 | >99 |
For each species, relative populations of conformers 1 and 2 were established by 15N-1H HSQC measurements and comparisons of peak intensities at 10–18 pairs of cross-peaks. Each measurement carries ≈8% relative error on any given ratio.
Although these data indicate that these domains can adopt two distinct conformations, we set out to establish that these states interconverted in an equilibrium process. The presence of two distinct sets of peaks in our spectra indicates that such interconversion must be slow (τ >ms) (16). Although interconversion could not be directly detected in the Y456T mutant with ZZ exchange experiments (17), we found that this process was slow enough to allow chromatographic separation of the conformers at 4 °C. 15N-1H HSQC spectra collected on a Y456T sample before and after this chromatographic step demonstrated that we could significantly enrich conformation 2 to a 1:2 ratio of 12:88 starting from a Y456T sample equally populated in both states (Fig. 2C). After incubating this sample at room temperature for 29 h, peak intensities had nearly restored to their original values. These data are consistent with a slow interconversion occurring in vitro with a first-order exponential time constant of 16 h or shorter.
Solution Structure of the New Conformation 2.
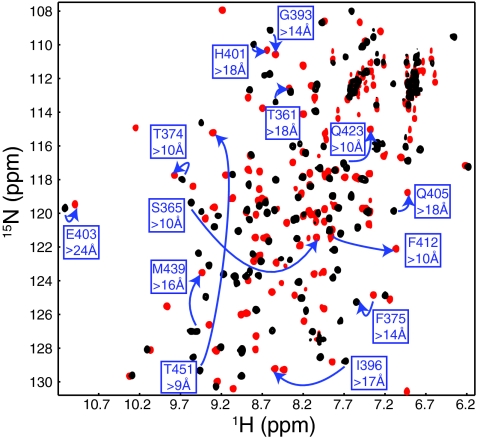
To understand the structural changes between the two conformations we observed in these domains, we determined the solution structure of a variant of human ARNT PAS-B that was stabilized in conformation 2 (>99%; Table 1) by three point mutations, F444Q/F446A/Y456T. Using standard triple-resonance NMR experiments performed on uniformly 15N, 13C-labeled protein samples, we assigned 1H, 13C, and 15N chemical shifts for >95% of NMR-observable sites in the protein. Comparing the chemical shifts of the wild-type and triple-mutant proteins, we observed numerous changes at distances over >20 Å from the locations of the mutated residues in the native structure (Fig. 3), thus reinforcing the prospect of a global change between states 1 and 2.
Fig. 3.
The two different conformations of ARNT PAS-B exhibit significant chemical shift changes at long distance. 15N-1H HSQC spectra of wild type (black) and F444Q/F446A/Y456T (red) show significant chemical shift differences. Several sites with large changes at significant distance (>10 Å from the nearest mutated residue) are highlighted.
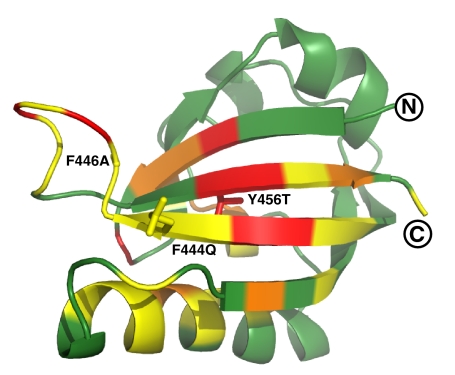
For structure determination, we used >3,000 interproton distance, dihedral angle, and hydrogen bond constraints [supporting information (SI) Table S1]. Using ARIA/CNS (18, 19), we obtained a high-precision ensemble of the 20 lowest-energy structures that were all consistent with these data (Fig. S1). From this group, it is clear that the mutant ARNT PAS-B domain retains the standard PAS topology while exhibiting extensive backbone amide chemical shift differences at long range (>10 Å) in the β-sheet and adjacent Fα-helix (Fig. 4).
Fig. 4.
Chemical shift changes between conformations of ARNT PAS-B. Ribbon diagram of structure closest to the mean in the ensemble is shown. Changes in chemical shift between the two conformations are color-coded onto the ribbon diagram (green, <0.3 ppm; yellow, 0.3–0.6 ppm; orange, 0.6–0.9 ppm; red, >0.9 ppm). Molecular graphics were generated by using MOLMOL and PyMOL (http://pymol.sourceforge.net/).
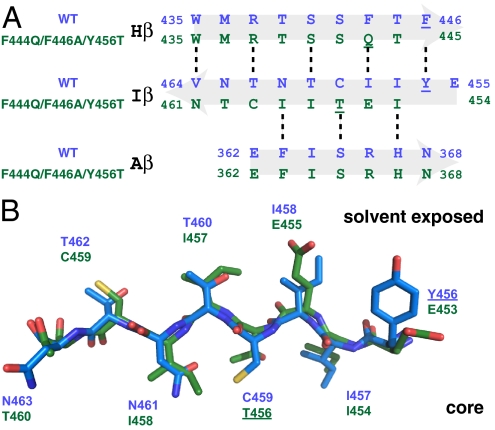
On closer inspection, we found that the source of our long-range changes was a 3-residue register shift of the Iβ strand, corresponding to a movement of ≈10 Å toward the C terminus (Fig. 5A). As expected by the odd-numbered change in register, this shift inverts the topology of the Iβ strand, burying several exposed side chains (and vice versa; Fig. 5B). Notably, only one position has the same residue type in both conformations (conformer 1 I457 = 2 I454), necessitating the packing of different side chains into similar conformations (Fig. 5B; Figs. S2 and S3). These changes include the mutated Y456T, whose side chain adopts an interior position held by C459, placing it in an environment with a mix of polar and hydrophobic neighbors. This process also significantly remaps residues presented by ARNT at the HIF-α-binding surface, suggesting changes in binding affinity (see below). Aside from this, the remainder of the protein retains an overall structure very similar to that of the wild-type domain (15), with a 1.08 Å pairwise backbone rmsd between the wild-type and triple-mutant ensembles for residues 360–443 (omitting the HI loop and Iβ strand).
Fig. 5.
Residues adopt a similar conformation in both structures. (A and B) Sequence (A) and structure (B) alignment of both mutant and wild-type Iβ strands. Shifted side chains adopt conformations of wild-type residues despite changes in identity or sequence. This is clearly seen by residue C459, where in the wild-type conformation it is buried in the core but in the mutant conformation, it slips 3 residues, becoming solvent-exposed and filling the space occupied by T462.
To accommodate the Iβ strand register shift, the preceding HI loop shortens from 8 to 5 residues. This is associated with a trans to cis isomerization of the peptide bond between N448 and P449 within the HI loop. This peptide bond, which is in a trans configuration for the wild-type domain, switches to cis in the triple mutant as established by strong N448 Hα-P449 Hα NOEs (Fig. S4) and large (>10 ppm) differences between P449 13Cβ and 13Cγ chemical shifts (20) in the mutant domain. Two lines of evidence suggest that this is a secondary effect to the Iβ strand slip and not causative. First, a P449A point mutation in a Y456T background leads to only minor changes in the 1:2 equilibrium (P449A/Y456, 32:68; Y456T, 50:50 corresponding to ΔΔG ≈1.5 kcal mol−1; Table 1). Second, when the same P449A mutation is made in a F444Q/F446A/Y456T background, we observed that the N448–A449 peptide bond is in a trans configuration (Fig. S4) while remaining predominantly in the +3 slipped Iβ strand conformation (1:2 ratio, 7:93). Taken together, these data suggest that the ability to slip the Iβ strand is primarily established by residues in the β-sheet itself rather than in the neighboring loops.
Effect of Slipped Conformation on Domain Stability.
Complementing these studies of the structural changes in ARNT PAS-B introduced by this β-strand slip, we characterized the effect on protein stability by using equilibrium denaturation and 2H exchange measurements. Using the intrinsic fluorescence properties of two tryptophan residues (W436, W438) in the Hβ strand, we characterized the equilibrium denaturation behavior of the wild-type, Y456T, and F444Q/F446A/Y456T ARNT PAS-B domains by using guanidine hydrochloride (Gdn·HCl). As expected for denaturant-induced unfolding, increasing Gdn·HCl concentrations were accompanied by shifts of the maximum intensity of fluorescence emission to longer wavelengths (344–354 nm) (Figs. S5 and S6). All three proteins exhibited similar cooperative and reversible unfolding behaviors with transitions centered near 2.3 M Gdn·HCl. Although these titrations potentially involve three states (folded states 1 and 2 plus a denatured state), the similarities of the emission spectra of proteins that populate 1 or 2 reduce these data to being sensitive to the relative populations of only two states, a combined native state (1 + 2), and an unfolded state. Combined with the similarities of the denaturation profiles (Fig. S6) of all three proteins, we do not anticipate significant differences in the global stabilities of conformations 1 and 2, allowing us to interpret these data in a pseudo two-state model involving a folded and an unfolded state. Such analyses indicated that the wild-type, Y456T, and F444Q/F446A/Y456T domains all had very similar global stabilities, ΔGu = 9.01, 9.37, and 9.20 kcal mol−1, respectively. In parallel, 2H exchange measurements conducted under EX2 conditions demonstrated comparable protection factors for the wild-type, Y456T, and F444Q/F446A/Y456T, with the only significant changes being the expected changes in residues in the Iβ strand caused by the +3 register shift (Fig. S6). These data indicate that proteins in either conformation 1 or 2 (or interconverting between both) are equivalently stable, despite the significant change in β-sheet register.
Functional Effect of Iβ Slip.
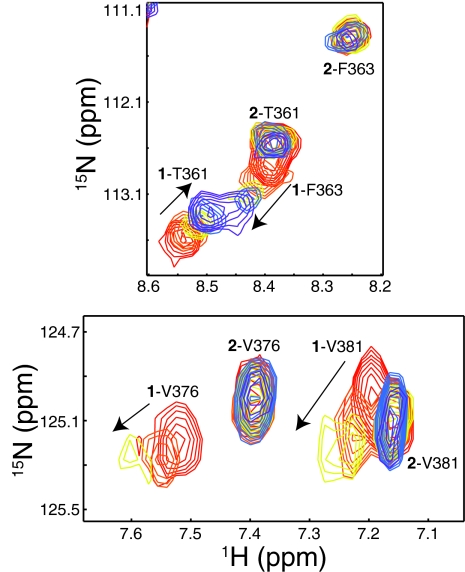
To examine functional differences between conformations 1 and 2, we took advantage of the favorable interconversion kinetics and equivalent populations of Y456T ARNT PAS-B to monitor binding of both conformations to HIF-2α PAS-B simultaneously. Complex formation can be monitored by chemical shift and peak intensity changes in the 15N-1H HSQC spectra of one domain as the other partner is added (3, 15). Titrating HIF-2α PAS-B into a 15N-labeled sample of Y456T, we observed dimerization-associated spectral changes only with the wild-type conformation 1 (Fig. 6). Examining the concentration dependence of these changes, we estimate that conformation 1 binds HIF-2α PAS-B with a Kd = 90 μM affinity. In contrast, conformation 2 shows no binding when HIF-2α PAS-B was added at concentrations up to 950 μM. Given these data and the minimum dimer concentration that would give a detectable spectral change, we suggest a minimum Kd ≈10 mM for this interaction, showing that the inversion of the Iβ strand topology effectively abolishes the HIF-2α–ARNT PAS-B interaction. Notably, we did not observe a significant change in the 1:2 equilibrium during these titrations, perhaps related to the slow interconversion between conformations (Fig. 2).
Fig. 6.
HIF-2α PAS-B selectively binds to conformation 1 of the ARNT PAS-B Y456T point mutant. Sections of 15N-1H HSQC spectra of a titration of unlabeled HIF-2α PAS-B (red, 0 μM; orange, 40 μM; gold, 100 μM; blue, 200 μM; violet, 950 μM HIF) into 75 μM 15N-labeled ARNT-PAS B Y456T are shown. Peaks are labeled with conformation (1 or 2) and residue name/number. The wild-type conformation 1 of Y456T still binds HIF-2α as shown by HIF-dependent chemical shift and intensity changes, whereas signals from the new conformation 2 are unaffected.
Discussion
The β-sheet surfaces of many PAS domains bind a variety of inter- (15, 21, 22, 50) or intramolecular (5, 6, 23) partners, critical to their function as protein–protein interaction domains (3). Notably, environmental stimuli modulate the affinity of these binding events, harnessing changes in the conformations of cofactors inside of the domain itself (5, 6, 24, 25). These proteins suggest that intramolecular signaling can be achieved by ligand control of the β-sheet structure or stability, which in turn alters PAS/protein-binding equilibria (26). This is supported by site-directed mutagenesis studies highlighting the critical role played by Iβ strand residues in signal transduction in a subset of photosensory PAS domains via internally bound flavin chromophores (6, 27, 28).
In this context, it is perhaps surprising that the structure and function of this critical β-sheet are fragile to the point that a single point mutation (Y456T) enables the domain to slip into a different, nonfunctional Iβ strand register. Notably, this change allows ARNT PAS-B to interconvert slowly between both of these two possible Iβ registers, contrary to the usual behavior linking sequence and a single structure as encapsulated in Anfinsen's hypothesis (29). Subsequent mutations (Table 1) adjust the relative populations of the two states, shifting some forms almost completely into one conformation whereas others continue to sample both of the two structures. It should be emphasized that these changes occur without compromising the integrity of the domain itself, which remains remarkably unchanged in its overall structure and stability. As such, our data are consistent with mutations shifting the relative stabilities of conformations 1 or 2 within the context of a structurally and energetically stable fold, where the final Iβ strand effectively completes the domain topology in a manner reminiscent of receptor–ligand interactions. These thermodynamic effects on the stabilities may also be complemented by kinetic changes via alterations in the energetics of the transition state between 1 and 2, but characterization of such effects must await more precise measurements of interconversion rates between these species.
What structural features of ARNT PAS-B enable such a transition to occur? An obvious candidate is the sequence of the Iβ strand, which is well conserved among all homologs and consists of a mix of similarly sized hydrophobic and polar residues (Fig. S7). The key exception to this is Y456, suggesting that the Y456T point mutation removes a local impediment to the wild-type protein undergoing the +3 Iβ strand slip. We suggest that the bulk of the Y456 side chain itself plays a significant role in preventing the slip because we expect significant steric occlusion to hinder the burial of this side chain in the wild-type sequence (Fig. 5B). Notably, the removal of steric hindrance at this position is not the sole determinant of slip propensity, as indicated by the differences in 1:2 equilibrium among the Y456T > Y456S > Y456A mutants (Table 1). This suggests that interactions between this site (occupied by a polar residue, C459, in conformation 1) and neighboring residues also influence register choice (Fig. S3). As expected, other changes in the newly slipped Iβ or neighboring Hβ strands can also modulate the relative populations of the two conformations we observed, as demonstrated by point mutations at F444, I454, or I458 in addition to Y456 (Table 1). As expected, changes that result in the burial of charged or large groups diminish the population of the slipped conformation 2 (e.g., I458E), whereas mutations that remove unfavorable interactions in the new form promote its formation (e.g., F444Q, which is adjacent to a newly exposed E455). These data indicate that side chain–side chain tertiary interactions play a critical role in establishing β-strand register, as they do with β-strand propensity itself (30, 31).
As noted above, slippage of the Iβ strand shortens the preceding HI loop from 8 to 5 residues and leads to a trans- to cis-isomerization of the N448–P449 peptide bond in this loop. Given the minor effects this has on the equilibrium, and our confirmation that a P449A point mutant retains a trans configuration at the preceding peptide bond while in a slipped register, it is clear that the Iβ strand sequence and interactions are the primary determinants of the strand register at equilibrium. We note that this leaves open the potential for this bond isomerization to have significant kinetic effects as seen in several biologically relevant systems (32), but this remains to be fully characterized.
Looking more broadly in the context of PAS domain signaling, it is clear that some degree of flexibility in the central β-sheet is essential. This has been most vividly demonstrated among a class of light-sensitive PAS domains, known as LOV (light–oxygen–voltage) domains (33), which regulate the conformations of various regulatory elements on the external side of the sheet in response to light (5, 6). Photoactivation depends on a highly conserved glutamine (Q513 in A. sativa phototropin 1), which is located in the Iβ strand. Upon the formation of the photochemical adduct, this residue changes its conformation and hydrogen-bonding patterns with the internally bound flavin chromophore. These changes are a key element of the photodetection process as established by conformational (6, 27) and functional changes (28) caused by mutations to this residue in several LOV proteins. Although a signaling mechanism has yet to be fully detailed, the most direct path is via distortions in the central β-sheet. Additional data supporting the malleability of PAS β-sheets come from a combination of experiments and simulations with single-molecule pulling studies of photoactive yellow protein (PYP) (34, 35). Under mechanical stress, PYP slips its Aβ strand while the remainder of the protein remains intact, entirely analogous to our +3 shift in β-strand register that occurs spontaneously in Y456T ARNT PAS-B. Although the biological role of such motions remains to be established, it is tempting to speculate that this flexibility may somehow reflect inherent flaws or properties in the PAS domain fold that are equally useful for ARNT to bind different bHLH-PAS partners as they are for LOV and PYP to distort the sheet in response to photoactivation.
More globally, β-strand slips have been observed in a variety of biological systems, where they can be triggered by a variety of stimuli, including ligand binding [e.g., ARF (36)], changes in solvent conditions [e.g., lymphotactin (37)] or proteolysis [e.g., clotting factor VIIa (38)]. Their functional role has been detailed in enzymatic regulation (38, 39) and in the conversion of β2-microglobulin and transthyretin from their soluble forms into disease-associated amyloid fibrils (40, 41). These cases exploit the long-range conformational changes that can be generated by β-strand slips, which can be particularly significant for odd-numbered shifts (as observed for all of the systems above except ARF). We suggest that the barriers that establish normal Anfinsen sequence:structure relationships (29) can be fragile for a wider variety of natural proteins, susceptible to modifications in sequence (see above) or solution conditions (37), enabling the same sequence to explore multiple stable folded conformations. In addition to providing valuable models to explore the sequence dependence of determining the equilibrium positions of strand register and interconversion kinetics, these may also reveal a greater role for such slips as part of the evolutionary record of protein structure and function (42).
Materials and Methods
Mutagenesis.
Point mutants of ARNT PAS-B were created from wild-type DNA, and primers including the desired mutation(s), with the QuikChange site-directed mutagenesis kit (Stratagene). ARNT PAS-B mutants were obtained by PCR amplification of the corresponding sequence and digestion of the parental template with DpnI, as directed by the manufacturer. The modified plasmid was transformed into the pHis-parallel bacterial expression vector (43) and sequenced to confirm the presence of the desired mutations.
Protein Expression and Purification.
DNA-encoding fragments of ARNT PAS-B (residues 356–470) were subcloned into the pHis-parallel expression vector and transformed into Escherichia coli BL21(DE3) cells. Cultures were grown in M9 minimal medium in the presence of 1 g/L 15NH4Cl for U-15N samples (and 3 g/L [13C6]glucose for U-15N,13C-labeled samples) at 37 °C to an A600 of 0.6–0.8, at which the cells were induced with 0.5 mM isopropyl β-d-thiogalactoside for 15 h at 20 °C. Proteins were purified by using a protocol described in ref. 15, as detailed in SI Materials and Methods.
Biochemical Purification of Mutant Conformation in Y456T.
ARNT PAS-B Y456T protein suspended in 50 mM Tris (pH 6.6), 17 mM NaCl buffer was injected over a 1-mL Mono Q column. The chromatogram resulted in one of three peaks that was enriched ≈88% for the mutant conformation, as established by 15N-1H HSQC spectra.
Structure Determination of ARNT PAS-B F444Q/F446A/Y456T.
NMR experiments for structure determination were carried out at 25 °C on a Varian Inova 600- and 800-MHz spectrometers with 0.8 mM samples of uniformly 15N- or 15N, 13C-labeled ARNT PAS-B F444Q/F446A/Y456T mutant in 50 mM Tris (pH 7.5), 17 mM NaCl, and 5 mM DTT solutions. Chemical shift assignments were obtained from 3D HNCACB, CBCA(CO)NH, HNCO, and HCCH-TOCSY data (16). Interproton distance restraints were obtained from 3D 15N-edited NOESY (120 ms), 3D 15N, 13C simultaneous-edited NOESY (120 ms), and 2D NOESY (120 ms) spectra. Hydrogen bond constraints (1.3 Å < dNH-O < 2.5 Å, 2.3 Å < dN-O < 3.5 Å) were defined for backbone amide protons protected from 2H exchange for >5 h (25 °C, pH 7.5) (44). φ and ψ dihedral angle constraints were obtained from analysis of backbone chemical shifts by using TALOS, with restraints set at twice the standard deviation yielded by TALOS (45) (15° minimum). Initial structures were calculated with automatic assignments of NOESY spectra by using ARIA 2.1 (18) and later redefined by using both automated and manual assignments. A total of 1,000 structures were obtained, and the 20 structures with the lowest NOE energy were evaluated with MOLMOL (46) and PROCHECK-NMR (47).
Global Stability Measurements.
Global stability measurements were made by using standard equilibrium denaturation measurements (monitored by tryptophan fluorescence) and 2H exchange measurements (monitored by NMR). Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Daniel Buehler, Leanna Steier, Amy Zhou, and Brian Zoltowski for contributions to this work and members of the Gardner laboratory for constructive comments and suggestions. This work was supported by National Institutes of Health Grant R01 GM081875 (to K.H.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors of the F444Q/F446A/Y456T ARNT mutant have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2K7S). The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 15928).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808270106/DCSupplemental.
References
- 1.Taylor BL, Zhulin IB. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nambu JR, Lewis JO, Wharton KA, Jr, Crews ST. The Drosophila single-minded gene encodes a helix–loop–helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 3.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix–loop–helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci USA. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 5.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 6.Zoltowski BD, et al. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman EC, et al. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 8.Ema M, et al. Two new members of the murine Sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol Cell Biol. 1996;16:5865–5875. doi: 10.1128/mcb.16.10.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffett P, Reece M, Pelletier J. The murine Sim-2 gene product inhibits transcription by active repression and functional interference. Mol Cell Biol. 1997;17:4933–4947. doi: 10.1128/mcb.17.9.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lees MJ, Whitelaw ML. Multiple roles of ligand in transforming the dioxin receptor to an active basic helix–loop–helix-PAS transcription factor complex with the nuclear protein Arnt. Mol Cell Biol. 1999;19:5811–5822. doi: 10.1128/mcb.19.8.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic helix–loop–helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman-Smith A, Lutwyche JK, Whitelaw ML. Contribution of the Per/Arnt/Sim (PAS) domains to DNA binding by the basic helix–loop–helix PAS transcriptional regulators. J Biol Chem. 2004;279:5353–5362. doi: 10.1074/jbc.M310041200. [DOI] [PubMed] [Google Scholar]
- 13.Chapman-Smith A, Whitelaw ML. Novel DNA binding by a basic helix–loop–helix protein. The role of the dioxin receptor PAS domain. J Biol Chem. 2006;281:12535–12545. doi: 10.1074/jbc.M512145200. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, et al. Functions of the Per/ARNT/Sim domains of the hypoxia-inducible factor. J Biol Chem. 2005;280:36047–36054. doi: 10.1074/jbc.M501755200. [DOI] [PubMed] [Google Scholar]
- 15.Card PB, Erbel PJ, Gardner KH. Structural basis of ARNT PAS-B dimerization: Use of a common β-sheet interface for hetero- and homodimerization. J Mol Biol. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 16.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. San Diego: Elsevier; 2007. [Google Scholar]
- 17.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 18.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Linge JP, O'Donoghue SI, Nilges M. Automated assignment of ambiguous nuclear Overhauser effects with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 20.Schubert M, Labudde D, Oschkinat H, Schmieder P. A software tool for the prediction of Xaa–Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J Biomol NMR. 2002;24:149–154. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, et al. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry. 2008;47:4051–4064. doi: 10.1021/bi7021156. [DOI] [PubMed] [Google Scholar]
- 22.Yildiz O, et al. Crystal structure and interactions of the PAS repeat region of the Drosophila clock protein PERIOD. Mol Cell. 2005;17:69–82. doi: 10.1016/j.molcel.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Borgstahl GEO, Williams DR, Getzoff ED. 1.4-Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site and chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 24.Key J, Moffat K. Crystal structures of deoxy and CO-bound bjFixLH reveal details of ligand recognition and signaling. Biochemistry. 2005;44:4627–4635. doi: 10.1021/bi047942r. [DOI] [PubMed] [Google Scholar]
- 25.Lee BC, Pandit A, Croonquist PA, Hoff WD. Folding and signaling share the same pathway in a photoreceptor. Proc Natl Acad Sci USA. 2001;98:9062–9067. doi: 10.1073/pnas.111153598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-Jα photoswitch. Nat Chem Biol. 2008;4:449–450. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nozaki D, et al. Role of Gln1029 in the photoactivation processes of the LOV2 domain in Adiantum phytochrome 3. Biochemistry. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 28.Jones MA, Feeney KA, Kelly SM, Christie JM. Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J Biol Chem. 2007;282:6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 29.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 30.Minor DL, Jr, Kim PS. Context is a major determinant of β-sheet propensity. Nature. 1994;371:264–267. doi: 10.1038/371264a0. [DOI] [PubMed] [Google Scholar]
- 31.Smith CK, Withka JM, Regan L. A thermodynamic scale for the β-sheet-forming tendencies of the amino acids. Biochemistry. 1994;33:5510–5517. doi: 10.1021/bi00184a020. [DOI] [PubMed] [Google Scholar]
- 32.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 33.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 34.Zhao JM, et al. Single-molecule detection of structural changes during Per-Arnt-Sim (PAS) domain activation. Proc Natl Acad Sci USA. 2006;103:11561–11566. doi: 10.1073/pnas.0601567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nome RA, Zhao JM, Hoff WD, Scherer NF. Axis-dependent anisotropy in protein unfolding from integrated nonequilibrium single-molecule experiments, analysis, and simulation. Proc Natl Acad Sci USA. 2007;104:20799–20804. doi: 10.1073/pnas.0701281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg J. Structural basis for activation of ARF GTPase: Mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 37.Tuinstra RL, et al. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci USA. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eigenbrot C, et al. The factor VII zymogen structure reveals reregistration of β-strands during activation. Structure. 2001;9:627–636. doi: 10.1016/s0969-2126(01)00624-4. [DOI] [PubMed] [Google Scholar]
- 39.Maun HR, et al. Disulfide locked variants of factor VIIa with a restricted β-strand conformation have enhanced enzymatic activity. Protein Sci. 2005;14:1171–1180. doi: 10.1110/ps.041097505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat Struct Mol Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 41.Eneqvist T, Andersson K, Olofsson A, Lundgren E, Sauer-Eriksson AE. The β-slip: A novel concept in transthyretin amyloidosis. Mol Cell. 2000;6:1207–1218. doi: 10.1016/s1097-2765(00)00117-9. [DOI] [PubMed] [Google Scholar]
- 42.Kinch LN, Grishin NV. Evolution of protein structures and functions. Curr Opin Struct Biol. 2002;12:400–408. doi: 10.1016/s0959-440x(02)00338-x. [DOI] [PubMed] [Google Scholar]
- 43.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 44.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 46.Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:29–32. 51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 47.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 48.Letunic I, et al. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong W, et al. Structure of a biological oxygen sensor: A new mechanism for heme-driven signal transduction. Proc Natl Acad Sci. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheuermann TH, et al. Artificial ligand binding within the HIF2α PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci USA. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.