Abstract
The human forkhead box O3A gene (FOXO3A) encodes an evolutionarily conserved key regulator of the insulin–IGF1 signaling pathway that is known to influence metabolism and lifespan in model organisms. A recent study described 3 SNPs in the FOXO3A gene that were statistically significantly associated with longevity in a discovery sample of long-lived men of Japanese ancestry [Willcox et al. (2008) Proc Natl Acad Sci USA 105:13987–13992]. However, this finding required replication in an independent population. Here, we have investigated 16 known FOXO3A SNPs in an extensive collection of 1,762 German centenarians/nonagenarians and younger controls and provide evidence that polymorphisms in this gene were indeed associated with the ability to attain exceptional old age. The FOXO3A association was considerably stronger in centenarians than in nonagenarians, highlighting the importance of centenarians for genetic longevity research. Our study extended the initial finding observed in Japanese men to women and indicates that both genders were likely to be equally affected by variation in FOXO3A. Replication in a French centenarian sample generated a trend that supported the previous results. Our findings confirmed the initial discovery in the Japanese sample and indicate FOXO3A as a susceptibility gene for prolonged survival in humans.
Keywords: aging, forkhead box O3A, genetic association study, long-lived individuals
Life expectancy in humans is influenced by various environmental and genetic factors. Approximately 25–32% of the overall variation in adult lifespan is accounted for by genetic differences that become particularly important for survival after the age of 60 (1–5). The mechanisms influencing lifespan have been intensively studied in Caenorhabditis elegans, Saccharomyces cerevisiae, or Drosophila melanogaster, and hundreds of genetic variants that lead to life extension in model systems have been identified (6–8). The success in finding lifespan-control genes in lower organisms has also motivated efforts to search for corresponding genes in humans. However, to date variation in only 1 gene, which codes for apolipoprotein E (APOE), has been found to be consistently associated with survival in various populations. Although numerous case-control candidate studies have been performed and associations of the longevity phenotype with biologically plausible genes have been described, results from these experiments have proven difficult to validate (5). These findings emphasize the importance of conducting large-scale studies with adequate replication to identify variants that are likely to exhibit only a weak or moderate effect.
The human forkhead box O3A gene (FOXO3A) is one of the homologues of daf-16 in C. elegans. The DAF-16 protein is a transcription factor and an evolutionarily conserved key regulator of the insulin–IGF1 signaling (IIS) pathway that influences metabolism and lifespan in model organisms (9–11). These aspects also render FOXO3A a very likely candidate for genetic longevity studies in humans. Recently, Willcox et al. (12) described 3 SNPs in the FOXO3A gene that were statistically significantly associated with longevity and different aging phenotypes in a discovery sample of long-lived Americans of Japanese ancestry. However, this finding required replication in an independent population. Here, we have investigated 16 known SNPs, which capture the majority of the variation in FOXO3A via its common haplotypes, in an extensive collection of 1,762 German centenarians, nonagenarians, and younger controls and provide evidence that polymorphisms in this gene are indeed associated with the ability to attain exceptional old age. Our findings confirmed the initial discovery in the Japanese sample and thus support FOXO3A as a susceptibility gene for prolonged survival in humans.
Results
In the present study, 16 polymorphisms in FOXO3A were analyzed for association with the human longevity phenotype (Tables 1 and 2). The tested SNPs are spaced across the entire gene region, including the promoter (Fig. 1) and capture the majority of its allelic variation by haplotype tagging. All SNPs were in Hardy-Weinberg equilibrium (HWE) in the control population. For the association analyses, we applied an established longevity study design (13, 14) by comparing German long-lived individuals (LLI; subset A; n = 1,031; aged 95–110 years) and a centenarian subset (subset B; n = 388) to appropriately matched younger controls (n = 731; aged 60–75 years). All markers were subjected to allelic case-control comparisons (CCA) by using the entire LLI sample (subset A) and the centenarian subset (subset B). For subset A, single-marker analysis revealed 4 SNPs with nominally significant PCCA values (Table 1). For the centenarians (subset B), 11 SNPs showed significant association (Table 2). Although subset B is smaller in size and therefore expected to have less power than the overall LLI sample, the significance level was more pronounced in the centenarians and revealed a stronger effect as reflected in the odds ratios (ORs) (Tables 1 and 2). The 3 top-ranking FOXO3A markers in subset B (rs3800231, rs9400239, and rs479744) passed correction for multiple testing (Bonferroni-adjusted significance threshold = 0.0016; for 2 × 16 tests). Because this adjustment did not take into account the strong linkage disequilibrium (LD) between the investigated markers (Fig. 1), the obtained threshold must be regarded as conservative. The results from the comparison of the genotypic data (CCG) are presented as additional information but they were not included in the initial statistical assessment (Tables 1 and 2). Because the age of the study participants seemed to influence the strength of the associations, we looked at the SNP allele frequencies of the nonagenarians separately (n = 643; aged 95–99 years; mean age 96.5 years). As expected, their frequencies were intermediate between those of the controls and the whole LLI sample (Table 3).
Table 1.
Association statistics for 16 FOXO3A SNPs in German LLI (subset A)
| No. | dbSNP ID | Min AF controls, n = 731 | Min AF LLI, n = 1,031 | PCCA | PCCG | OR | 95% C.I. |
|---|---|---|---|---|---|---|---|
| 1 | rs2274776 | 0.490 | 0.485 | 0.747 | 0.807 | 0.98 | 0.85–1.12 |
| 2 | rs1571631 | 0.465 | 0.463 | 0.893 | 0.987 | 0.99 | 0.87–1.13 |
| 3 | rs6911407 | 0.378 | 0.415 | 0.030 | 0.101 | 1.16 | 1.01–1.34 |
| 4 | rs768023 | 0.382 | 0.415 | 0.051 | 0.134 | 1.15 | 1.00–1.32 |
| 5 | rs2802288 | 0.385 | 0.417 | 0.058 | 0.146 | 1.14 | 1.00–1.31 |
| 6 | rs2883881 | 0.099 | 0.084 | 0.129 | 0.019 | 0.83 | 0.66–1.05 |
| 7 | rs12200646 | 0.118 | 0.141 | 0.050 | 0.141 | 1.22 | 1.00–1.50 |
| 8 | rs2802290 | 0.387 | 0.417 | 0.081 | 0.185 | 1.13 | 0.99–1.30 |
| 9 | rs13220810 | 0.263 | 0.248 | 0.308 | 0.125 | 0.92 | 0.79–1.08 |
| 10 | rs7762395 | 0.153 | 0.175 | 0.090 | 0.191 | 1.17 | 0.98–1.41 |
| 11 | rs9400239 | 0.295 | 0.331 | 0.025 | 0.033 | 1.18 | 1.02–1.37 |
| 12 | rs3800231 | 0.287 | 0.327 | 0.011 | 0.025 | 1.21 | 1.04–1.40 |
| 13 | rs1268170 | 0.347 | 0.377 | 0.073 | 0.171 | 1.14 | 0.99–1.31 |
| 14 | rs473268 | 0.337 | 0.366 | 0.075 | 0.196 | 1.14 | 0.99–1.31 |
| 15 | rs479744 | 0.199 | 0.229 | 0.033 | 0.082 | 1.20 | 1.01–1.41 |
| 16 | rs519007 | 0.186 | 0.183 | 0.816 | 0.796 | 0.98 | 0.82–1.16 |
LLI, entire sample; 95–110 years. Min AF, minor allele frequency; PCCA, P value obtained from an allele-based case-control comparison, using a χ2 test with 1 df; PCCG, P value obtained from a genotype-based case-control comparison, using a χ2 test with 2 df; OR, for attaining old age with the minor allele in controls as reference allele; 95% C.I. is for OR.
Table 2.
Association statistics for 16 FOXO3A SNPs in German centenarians (subset B)
| No. | dbSNP ID | Min AF controls, n = 731 | Min AF centenarians, n = 388 | PCCA | PCCG | OR | 95% C.I. |
|---|---|---|---|---|---|---|---|
| 1 | rs2274776 | 0.490 | 0.477 | 0.536 | 0.373 | 0.95 | 0.79–1.13 |
| 2 | rs1571631 | 0.465 | 0.461 | 0.872 | 0.543 | 0.99 | 0.83–1.17 |
| 3 | rs6911407 | 0.378 | 0.438 | 0.006 | 0.012 | 1.28 | 1.07–1.53 |
| 4 | rs768023 | 0.382 | 0.440 | 0.007 | 0.010 | 1.27 | 1.07–1.52 |
| 5 | rs2802288 | 0.385 | 0.445 | 0.006 | 0.007 | 1.28 | 1.07–1.53 |
| 6 | rs2883881 | 0.099 | 0.078 | 0.100 | 0.010 | 0.77 | 0.56–1.05 |
| 7 | rs12200646 | 0.118 | 0.146 | 0.064 | 0.157 | 1.28 | 0.99–1.65 |
| 8 | rs2802290 | 0.387 | 0.444 | 0.010 | 0.008 | 1.26 | 1.06–1.51 |
| 9 | rs13220810 | 0.263 | 0.218 | 0.020 | 0.053 | 0.78 | 0.63–0.96 |
| 10 | rs7762395 | 0.153 | 0.199 | 0.005 | 0.012 | 1.38 | 1.10–1.73 |
| 11 | rs9400239 | 0.295 | 0.366 | 0.0007 | 0.0009 | 1.38 | 1.14–1.66 |
| 12 | rs3800231 | 0.287 | 0.362 | 0.0002 | 0.0003 | 1.42 | 1.18–1.70 |
| 13 | rs1268170 | 0.347 | 0.397 | 0.020 | 0.051 | 1.24 | 1.03–1.48 |
| 14 | rs473268 | 0.337 | 0.380 | 0.044 | 0.078 | 1.21 | 1.01–1.45 |
| 15 | rs479744 | 0.199 | 0.258 | 0.001 | 0.002 | 1.40 | 1.14–1.73 |
| 16 | rs519007 | 0.186 | 0.176 | 0.550 | 0.799 | 0.93 | 0.74–1.17 |
For abbreviations see legend to Table 1.
Fig. 1.
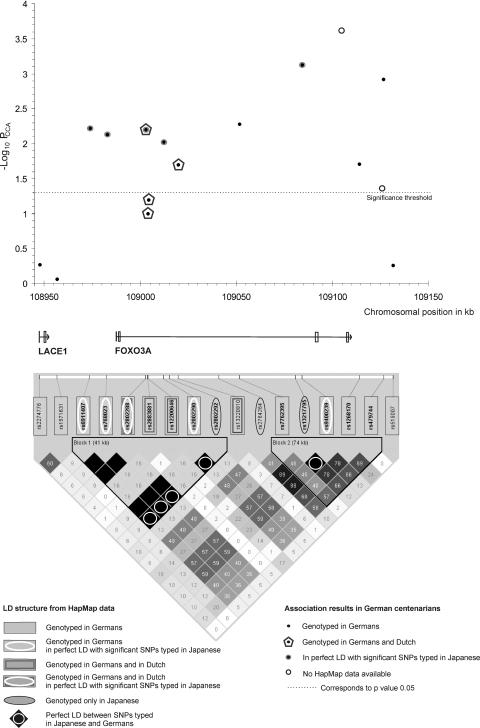
Fine mapping of the FOXO3A region on chromosome 6 (German centenarian sample). The physical position (in kb) of all 16 genotyped SNPs with their allele-based P values (−log10 PCCA, German centenarian sample) and a schematic representation of the gene structure NM_201559 are shown. Coordinates refer to National Center for Biotechnology Information genome assembly build 36. Plotted association results (PAR) of SNPs that have previously been typed in the Dutch sample are indicated by a pentagon; PAR of SNPs that are in perfect LD with SNPs typed in the Japanese sample (according to CEU HapMap data) are indicated by a gray curb; PAR with no available HapMap data are presented as a black circle. The LD plot of the locus is based on the measure r2 and was generated with Haploview by using CEU HapMap data. The markers typed in the German sample are indicated by a gray box; markers typed in Germans that are in perfect LD with the significant SNPs typed in the Japanese (according to CEU HapMap data) are indicated by a white circle; markers typed in the German and Dutch samples are indicated by a double gray box; markers typed in the German and Dutch samples that are in perfect LD with the significant SNPs typed in the Japanese (according to CEU HapMap data) are indicated by a white circle and a double gray box; markers only genotyped in the Japanese sample with no corresponding PCCA values in the German sample are indicated by a black circle; perfect LD between the SNPs typed in the Japanese and the German samples (according to CEU HapMap data) is indicated by a white circle in the lower LD plot.
Table 3.
Comparison of FOXO3A association statistics from Japanese, German, French and Dutch samples
| Significantly associated SNPs in Japanese |
SNPs in strong LD with Willcox' SNPs | r2 | Significantly associated SNPs in Germans* |
Significantly associated SNPs in French* |
Significantly associated SNPs in Dutch* |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Min AF controls | Min AF elderly | PCCA | Min AF controls | Min AF centenarians | PCCA† | Min AF LLI | PCCA‡ | Min AF 95–99 years | PCCA§ | Min AF controls | Min AF centenarians | PCCA | Min AF controls | Min AF cohort '87 | PCCA | ||
| rs2802292 | 0.255 | 0.371 | <0.0001 | rs6911407 | 1 | 0.378 | 0.438 | 0.006 | 0.415 | 0.030 | 0.402 | 0.211 | ||||||
| rs768023 | 1 | 0.382 | 0.440 | 0.007 | 0.415 | 0.051 | 0.400 | 0.330 | 0.393 | 0.429 | 0.097 | |||||||
| rs2802288 | 1 | 0.385 | 0.445 | 0.006 | 0.417 | 0.058 | 0.402 | 0.384 | 0.354 | 0.365 | ns | |||||||
| rs2802290 | 1 | 0.387 | 0.444 | 0.010 | 0.417 | 0.081 | 0.401 | 0.458 | ||||||||||
| rs2764264 | 0.248 | 0.347 | 0.0002 | rs9400239 | 0.88 | 0.295 | 0.366 | 0.0007 | 0.331 | 0.025 | 0.311 | 0.389 | ||||||
| rs13217795 | 0.248 | 0.340 | 0.0006 | rs9400239 | 1 | 0.295 | 0.366 | 0.0007 | 0.331 | 0.025 | 0.311 | 0.389 | ||||||
| rs2153960 | 0.88 | 0.281 | 0.293 | ns | ||||||||||||||
Min AF, minor allele frequency; PCCA, P value obtained from an allele-based case-control comparison, using a χ2 test with 1 df; LD, linkage disequilibrium; LLI, entire sample; 95–110 years); ns, not significant. r2, LD values from CEU HapMap data between the markers typed in Japanese men and SNPs typed in Germans, French and Dutch.
*Association statistics for SNPs typed in Germans, French, or Dutch that are in perfect or strong LD with significant SNPs typed in Japanese men.
†PCCA for centenarian sample.
‡PCCA for entire LLI sample.
§PCCA for nonagenarians.
In the following we concentrated our analyses on the centenarians. When we analyzed the 16 SNPs by logistic regression, incorporating APOE status and gender with possible interactions, we obtained only negligible differences to the unadjusted results. There was no significant interaction between any of the SNPs and either APOE status or gender in the impact on longevity.
Three of the SNPs that were associated in the German centenarian sample (rs768023, rs7762395, and rs3800231) were subsequently investigated for replication in an independent collection of 535 French centenarians (mean age: 103.8 years) and 553 younger controls (aged 18–70 years). The 3 SNPs included rs3800231 (the top-ranking marker in Germans); they were spaced evenly across the FOXO3A gene region and located in different haplotype blocks (Fig. 1). No evidence for an association was detected between any of the tested SNPs and the longevity phenotype (Table 4). SNP rs768023 yielded the lowest PCCA value (0.097) and had an allele frequency distribution that was close to that in Germans. When we repeated the analysis with only the older French controls (137 individuals aged 60–70 years), whose age range was comparable with that of the Germans, a similar PCCA value for rs768023 (0.094) and a larger estimated OR (1.26) were observed.
Table 4.
Association statistics for 3 FOXO3A SNPs in French centenarians
| No. | dbSNP ID | Min AF controls, n = 553 | Min AF centenarians, n = 535 | PCCA | PCCG | OR | 95% C.I. |
|---|---|---|---|---|---|---|---|
| 4 | rs768023 | 0.393 | 0.429 | 0.097 | 0.224 | 1.16 | 0.97–1.37 |
| 10 | rs7762395 | 0.176 | 0.177 | 0.928 | 0.981 | 1.01 | 0.81–1.26 |
| 12 | rs3800231 | 0.297 | 0.317 | 0.332 | 0.452 | 1.10 | 0.91–1.32 |
For abbreviations see legend to Table 1.
Recently, Willcox et al. (12) reported an association of 3 FOXO3A SNPs with longevity in Japanese–American men from the island of Oahu (Hawaii). Unfortunately, there is no overlap between the 16 polymorphisms tested here and the 3 investigated in the Hawaii sample as the genotyping in the German and French samples had been conducted before publication of the Willcox study. However, based on LD data from HapMap individuals of European ancestry (CEU), the SNPs described by Willcox et al. were in perfect or very high LD with 5 highly significant markers in our study (Fig. 1 and Table 3). In addition, the FOXO3A LD structure is comparable in Europeans and Japanese (HapMap data for CEU and JPT) (see Fig. S1). Although the allele frequencies differed between Germans and Japanese, it is noteworthy that in both populations the frequency distribution followed the same trend, with a significant increase of the minor allele in the LLI relative to the younger controls (Table 3). In the Japanese sample, only SNP rs2802292 was investigated for further analyses. The OR of this SNP was larger for the homozygote of the minor allele than for the heterozygote (when the homozygote of the major allele was taken as the reference genotype), suggesting an additive effect (12) (Table 5). For the German centenarian sample (subset B), rs2802288, which is in perfect LD with Willcox et al.'s rs2802292 (according to CEU HapMap data), revealed no evidence for an additive effect (Table 5). Comparisons of the homozygote, the heterozygote, and carriers of the minor allele with the homozygote of the major allele all yielded similar ORs of ≈1.5 (P = 0.025, 0.0027, and 0.0016, respectively), whereas there was no difference between the heterozygote and the homozygote of the minor allele (OR = 1.01, P = 0.97) (Table 5). These findings suggest a dominant effect for the minor allele of rs2802288 in the German population. However, because the corresponding confidence intervals (C.I.) reported by us (Table 5) and Willcox et al. are rather large, the available data do not allow us to ascertain whether there is an additive or a dominant effect.
Table 5.
Effect comparison between Japanese and German samples
| Comparison | Japanese sample,rs2802292 |
German centenarian sample,rs2802288 |
||||
|---|---|---|---|---|---|---|
| OR | 95% C.I. | P | O.R. | 95% C.I. | P | |
| Homozygote of minor vs. homozygote of major allele | 2.75 | 1.51–5.02 | 0.0007 | 1.53 | 1.06–2.21 | 0.025 |
| Heterozygote vs. homozygote of major allele | 1.91 | 1.34–2.72 | 0.0003 | 1.54 | 1.16–2.04 | 0.0027 |
| Carriers of minor vs. homozygote of major allele | * | * | * | 1.53 | 1.18–2.00 | 0.0016 |
| Homozygote of minor allele vs. heterozygote | * | * | * | 1.01 | 0.71–1.42 | 0.970 |
For abbreviations see legend to Table 1.
*No data available.
The influence of FOXO3A variation on longevity had also been examined in the Dutch Leiden 85-plus study using both single-point and haplotype analysis (15). None of the 11 tested SNPs yielded a significant result when 2 elderly cohorts (aged 85 and 88–92 years) were, in a cross-sectional manner, compared with a younger control sample (aged 27–36 years). Four of these SNPs were part of our marker panel (rs2802288, rs2883881, rs12200646, and rs13220810), and in contrast to the Leiden results, 3 of them were significantly associated in German centenarians (Table 2). Furthermore, one of the SNPs tested in the Dutch sample (rs2153960) was in strong LD with the SNP rs13217795 that was highly associated in the Japanese men from Hawaii (P = 0.0006; r2 = 0.88 between rs13217795 and rs2153960 according to CEU HapMap data; Table 3).
The data of the 2 Dutch cohorts were also previously analyzed by applying a longitudinal design, including information on mortality and age-related phenotypes (15). Kuningas et al. (15) observed that carriers of a particular 4-SNP haplotype (GAGC from rs2802288, rs2883881, rs12200646, and rs13220810) had increased risks for all-cause and cardiovascular mortalities. The effects were very small, and the findings did not hold up to multiple testing (15). These 4 SNPs were also subjected to haplotype analysis in our sample collection. Although risk alleles or haplotypes for age-related diseases are expected to decrease in frequency in population strata of increasing age (16), the mortality haplotype identified in the Dutch was not significantly depleted in German centenarians compared with younger controls (0.212 in centenarians versus 0.238 in controls; P = 0.17).
Discussion
Recently, variation in FOXO3A was shown to be associated with longevity in a discovery cohort comprising 213 long-lived men of Japanese ancestry (95–106 years) and 402 younger probands (73–81 years) (12). Our study of 1,762 German centenarians, nonagenarians, and controls provides independent evidence that FOXO3A is a susceptibility gene for prolonged survival. As can be expected for a polygenic trait like longevity, the effect was rather modest (demonstrated by an OR of 1.42 for the top-ranking marker rs3800231; Table 2). However, because the association has been observed in 2 genetically diverse groups of European and Asian descent, our results render FOXO3A a modifier of general relevance that may play a role in many human populations. The analysis in the French centenarian sample generated a trend that supported the findings in the Japanese and German collections, but did not yield a statistically robust replication result. The validation could have been hindered by low levels of undetected population structure, but as demonstrated previously, the French sample shows no evidence of stratification (17). Another explanation might be the younger age of the French controls (18–70 years). This idea is supported by the fact that the estimated OR was larger when only controls aged 60–70 years were used for analysis. The resulting PCCA value still did not reach significance, which is possibly caused by the small number of available older controls (n = 137). The lack of reproducibility presents a well-known problem of case-control studies. To prove the validity of association results, confirmation in independent and larger samples is generally mandatory. However, this requirement may sometimes be difficult to achieve when polygenic traits, such as human longevity, are analyzed for which only modest or weak effects are predicted (5). This problem is aggravated in underpowered experiments where the sizes of appropriate samples are relatively small. The French centenarian-control collection used in this study is one of the largest to date and yet it had only an a priori power of 47% to confirm the association of the FOXO3A SNP rs768023 (assuming the same allele frequencies as in the German centenarian and control samples).
In genetic longevity research, the age of the cases is of utmost importance. In our study, the association of FOXO3A revealed stronger effects in the centenarians than in the nonagenarians or the whole LLI sample (Tables 1–3). The marker rs2802288 showed a continuous increase in the frequency of the minor allele with age. A remarkable rise was observed from the age group 100–104 years (44.1%) to that of 105–110 years (52.4%) (Table 6). However, it should be taken into account that there were only 21 individuals who were 105 years and older. It has recently been shown by computer simulation that samples of nonagenarians need to be 5 times larger than those of centenarians to achieve comparable power and that the performance of association studies is drastically reduced when nonagenarians are considered as cases (18). It can be expected that the power is even more decreased when octogenarians are used, as was done in the Dutch study (15), which may explain why Kuningas et al. did not observe any association with FOXO3A in their cross-sectional comparisons. Our results highlight the relevance of centenarians for genetic longevity research, because they appear more valuable for such studies than octogenarians and nonagenarians. This finding might be attributed to the fact that centenarians represent the top percentiles of their respective birth cohort-specific age distributions and have outlived most of their peers by several decades. Only ≈4% of male and 5.6% of female 90-year-olds and 15–17% of 95-year-olds in Germany are likely to become 100 years themselves (Human Mortality Database, www.mortality.org), indicating that centenarians represent a highly selected phenotype even among LLI. Because the genetic contribution to survival is strongest at very advanced ages (4), centenarians may be particularly enriched for beneficial variants in “longevity assurance genes” (16). Hence, it would seem that centenarians are the more suitable, but rarer, phenotype for genetic longevity studies.
Table 6.
Minor allele frequency distribution of rs2802288 in Germans by age groups
| Age at recruitment, years | n | Minor allele frequency of rs2802288 |
|---|---|---|
| 60–75 | 731 | 0.385 |
| 95–99 | 631 | 0.402 |
| 100–104 | 362 | 0.441 |
| 105–110 | 21 | 0.524 |
SNP rs2802288 is in perfect LD with Willcox et al.'s (12) rs2802292 (according to CEU HapMap data),
The effect (as reflected in the ORs) reported for the Japanese men was larger than that for the German collection of mixed gender. It is possible that this discrepancy could be attributed to an inaccuracy of estimation caused by chance (as reflected in the confidence intervals for the ORs). If the difference is actually real, the question arises as to which factors could have contributed to it. First, it may be caused by some population-specific factors that result in a stronger effect in Japanese in general. Second, the association described by Willcox et al. (12) referred to males only. Men become long-lived less often, and males may depend more on genetic factors than females to attain exceptional old age (19). It would therefore seem plausible that longevity variants are more enriched in men or impact more strongly on male survival. This may be the case for the Japanese men examined (12). However, our study also extended the association finding to females and indicates that genetic variation in FOXO3A is likely to contribute equally to the longevity phenotype in both genders. Third, the 2 investigations differed in the choice of controls. The selection of appropriate controls for genetic longevity research is a matter of concern (5, 14). Willcox et al. (12) used controls (mean age ≈77 years) whose birth cohorts were very close to those of the cases and who had already died before the age of 81. This so-called nested case-control design avoids or minimizes a number of potential pitfalls that may arise when controls are drawn from much more recent birth cohorts than the LLI and are still alive at the time of recruitment (13, 14). The latter strategy was applied for the German association study described here.
Another difference between the 2 investigations may be the type of the effect. The ORs in the Japanese suggested an additive mechanism (12), whereas in the German sample the data implicated a dominant-recessive effect. However, the large confidence intervals for the ORs in both studies imply some uncertainty. The future challenge is to investigate whether the observed discrepancies in the strength and kind of the FOXO3A effect can be verified and, if so, to clarify whether they are caused by study design, population- and/or gender-specific differences, or some other factors.
There is growing evidence for an important role of the FOXO3A protein in healthy human aging. It has been shown to control insulin sensitivity and influence coronary heart disease, type 2 diabetes, and longevity. These functions are indicative of a “master regulator” in the IIS pathway, and allelic variation in the transcription factor FOXO3A may modulate a broad array of downstream targets that could exhibit larger effects on extending lifespan (12). The association findings in 2 independent populations of diverse origin confirm FOXO3A as a genetic susceptibility factor for human longevity.
Materials and Methods
Study Population.
A total sample of 1,031 unrelated German LLI was studied that were recruited from different geographic regions throughout Germany. Nonagenarians and centenarians were between 95 and 110 years of age at the time of recruitment (mean age: 98.4 years). The gender ratio was 74.1% females vs. 25.9% males. A subset comprised 388 centenarians (mean age: 101.6 years). The 731 German younger control subjects were between 60 and 75 years of age (mean age: 67.2 years). There are no mortality data available for the controls. However, based on current predictions only 1.5% of all 60-year-old and 1.8% of all 75-year-old German females will become 100 years. For males, the probability is even lower (Human Mortality Database; www.demogr.mpg.de). Hence, we can estimate that only ≈13 of our younger controls (of 731) will become centenarians themselves, a negligible proportion that does not affect power. In Germany genetic differentiation in population structure is considered to be very low (20). Moreover, the recruited controls match the LLI as closely as possible in terms of ancestry, gender, and geographical origin within the country (13), thus minimizing any systematic genetic differences between the samples that might arise because of very low levels of undetected population structure. The good matching is reflected in a genomic inflation factor λ of 1.02 (centenarian group) and 1.00 (LLI sample), respectively, using PLINK version 1.01 (21) (http://pngu.mgh.harvard.edu/purcell/plink), based on 290 randomly chosen, genomewide SNPs. Approval for the project was received from the Ethics Committee of the University Hospital Schleswig–Holstein, Campus Kiel and local data protection authorities.
The French sample consisted of 535 centenarians (mean age: 103.8 years) from different regions throughout France (Île-de-France, Northeast, Northwest, Southeast, Southwest) (22). The centenarians were matched for sex and geographic origin with healthy control individuals (553 younger controls ranged from 18 to 70 years; mean 51.2 years). The gender ratio of the sample was 83.6% females vs. 16.4% males. To test for population stratification, 57 microsatellites from the Applied Biosystems LMS-MD10 panel that were located on 6 different chromosomes (chromosomes 2, 9, 10, 11, 17, and 18) were tested in all cases and controls. χ2 values were calculated from the allele counts. The obtained mean χ2 of the G test statistics for the markers was 1.00, which reflected the good matching of the French sample collection (17). All German and French subjects gave informed, written consent before participation. The study was approved by the Ethics Committee of the Hospital Saint-Antoine in Paris.
Genotyping.
Genotyping of the German and French samples was performed with Taqman SNP Genotyping Assays (Applied Biosystems) on an automated platform (23).
Statistical Analysis.
Single marker case-control analyses on allele and genotype frequency data were performed with χ2 statistics by using the open-source software GENOMIZER (24) (www.ikmb.uni-kiel.de/software.html). P < 0.05 was considered significant. All analyzed SNPs had a minimal overall call rate of 98% and were tested for HWE in controls before inclusion in the analyses (PHWE > 0.05). The LD structure was determined from HapMap data (CEU and JPT; www.hapmap.org) with the Haploview v4.0 program (http://www.broad.mit.edu/mpg/haploview) (25). The Haploview program was also used for selection of tagging SNPs based on the HapMap genotypes of Europeans with the pairwise tagging option (pairwise r2 > = 0.8; PHWE > 0.01) and for the haplotype analysis.
Power calculations were performed by using the PS Power and Sample Size Program (26) (http://medipe.psu.ac.th/episoft/pssamplesize). Single-marker association analyses with adjustment for gender and APOE, and OR statistics were conducted by logistic regression in R, version 5.2.1 (ref. 27 and www.R-project.org).
Supplementary Material
Acknowledgments.
We thank all study participants for their cooperation and the laboratory personnel of the Institute of Clinical Molecular Biology, the members of the Popgen Biobank, and the staff of the Biological Resource Center of the Fondation Jean Dausset–Centre d'Étude du Polymorphisme Humain for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft Excellence Cluster Inflammation at Interfaces, the German Federal Ministry of Science and Education through an Explorative Project of the National Genome Research Network, the French Ministère de l'Enseignement Supérieur et de la Recherche, and the Genetics of Healthy Aging Consortium, 6. Framework Program of the European Commission.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809594106/DCSupplemental.
References
- 1.McGue M, Vaupel JW, Holm N, Harvald B. Longevity is moderately heritable in a sample of Danish twins born 1870–1880. J Gerontol. 1993;48:B237–B244. doi: 10.1093/geronj/48.6.b237. [DOI] [PubMed] [Google Scholar]
- 2.Herskind AM, et al. The heritability of human longevity: A population-based study of 2,872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 3.Skytthe A, et al. Longevity studies in GenomEUtwin. Twin Res. 2003;6:448–454. doi: 10.1375/136905203770326457. [DOI] [PubMed] [Google Scholar]
- 4.Hjelmborg JvB, et al. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119:312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- 5.Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: Challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 7.Guarente L, Kenyon C. Genetic pathways that regulate aging in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 8.Partridge L, Gems D. Mechanisms of aging: Public or private? Nat Rev Genet. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- 9.White MF. Insulin signaling in health and disease. Science. 2003;302:1710–1711. doi: 10.1126/science.1092952. [DOI] [PubMed] [Google Scholar]
- 10.Gems D, McElwee JJ. Aging: Microarraying mortality. Nature. 2003;424:259–261. doi: 10.1038/424259a. [DOI] [PubMed] [Google Scholar]
- 11.Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Willcox BJ, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nebel A, et al. No association between microsomal triglyceride transfer protein (MTP) haplotype and longevity in humans. Proc Natl Acad Sci USA. 2005;102:7906–7909. doi: 10.1073/pnas.0408670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nebel A, Schreiber S. Allelic variation and human longevity. Sci Aging Knowledge Environ. 2005;2005:pe23. doi: 10.1126/sageke.2005.29.pe23. [DOI] [PubMed] [Google Scholar]
- 15.Kuningas M, et al. Haplotypes in the human Foxo1a and Foxo3a genes; Impact on disease and mortality at old age. Eur J Hum Genet. 2007;15:294–301. doi: 10.1038/sj.ejhg.5201766. [DOI] [PubMed] [Google Scholar]
- 16.Perls T, Kunkel LM, Puca AA. The genetics of exceptional human longevity. J Am Geriatr Soc. 2002;50:359–368. doi: 10.1046/j.1532-5415.2002.49283.x. [DOI] [PubMed] [Google Scholar]
- 17.Geesaman BJ, et al. Haplotype-based identification of a microsomal transfer protein marker associated with the human lifespan. Proc Natl Acad Sci USA. 2003;100:14115–14120. doi: 10.1073/pnas.1936249100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan Q, Zhao JH, Zhang D, Kruse TA, Christensen K. Power for genetic association study of human longevity using the case-control design. Am J Epidemiol. 2008;168:890–896. doi: 10.1093/aje/kwn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franceschi C, et al. Do men and women follow different trajectories to reach extreme longevity? Italian Multicenter Study on Centenarians (IMUSCE) Aging (Milan) 2000;12:77–84. doi: 10.1007/BF03339894. [DOI] [PubMed] [Google Scholar]
- 20.Steffens M, et al. SNP-based analysis of genetic substructure in the German population. Hum Hered. 2006;62:20–29. doi: 10.1159/000095850. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanché H, Cabanne L, Sahbatou M, Thomas G. A study of French centenarians: Are ACE and APOE associated with longevity? C R Acad Sci III. 2001;324:129–135. doi: 10.1016/s0764-4469(00)01274-9. [DOI] [PubMed] [Google Scholar]
- 23.Hampe J, et al. An integrated system for high throughput TaqMan-based SNP genotyping. Bioinformatics. 2001;17:654–655. doi: 10.1093/bioinformatics/17.7.654. [DOI] [PubMed] [Google Scholar]
- 24.Franke A, et al. GENOMIZER: An integrated analysis system for genomewide association data. Hum Mutat. 2006;27:583–588. doi: 10.1002/humu.20306. [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Dupont WD, Plummer WD., Jr Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19:589–601. doi: 10.1016/s0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 27.R Development Core Team. A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.