Abstract
The thiazolylpeptides are a family of >50 bactericidal antibiotics that block the initial steps of bacterial protein synthesis. Here, we report a biosynthetic gene cluster for thiocillin and establish that it, and by extension the whole class, is ribosomally synthesized. Remarkably, the C-terminal 14 residues of a 52-residue peptide precursor undergo 13 posttranslational modifications to give rise to thiocillin, making this antibiotic the most heavily posttranslationally-modified peptide known to date.
Keywords: biosynthesis, natural products, thiazolylpeptides
Many clinically-used antibiotics are small, highly-modified peptides produced by microbes, including penicillins (1), cephalosporins (1), glycopeptides (2), and lipopeptides (3). These peptidic natural products are synthesized by nonribosomal peptide synthetases, multidomain proteins that function as enzymatic assembly lines (4, 5). However, several classes of peptidic natural products arise from unusual posttranslational modifications of ribosomally-synthesized peptides. These include lantibiotics (6), in which serine and threonine residues are converted to dehydroalanine (Dha) and dehydrobutyrine (Dhb) residues; and the cyanobactin/microcin B17 family (7–10), in which serine, threonine, and cysteine residues are converted to 5-membered oxazole and thiazole rings.
Micrococcin (compounds 1 and 2), the founding member of a class of >50 peptidic antibiotics known as the thiazolylpeptides (11, 12), was discovered in 1948 (13) and shown 2 decades later to inhibit the translocation step of ribosomal protein synthesis (14). Stable isotope feeding studies have shown that the nonproteinogenic residues found in thiazolylpeptides (Dha and Dhb, oxazoles and thiazoles, and a pyridine, piperidine, or dehydropiperidine ring) derive from proteinogenic amino acids (15, 16). These results raised the possibility that, despite their abundance of many different kinds of nonproteinogenic residues, the thiazolylpeptides are synthesized by the ribosome and then posttranslationally modified to take on their active form.
Results and Discussion
A Bioinformatic Search for Thiazolylpeptide Structural Genes.
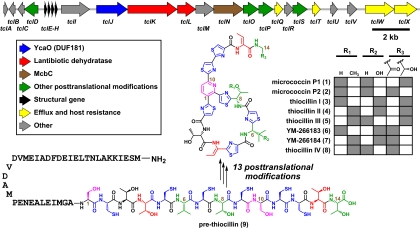
To examine this possibility, we systematically predicted the peptide sequences that would encode known members of the thiazolylpeptide family and used them as queries in tblastn searches against the nonredundant (nr) and whole-genome shotgun (wgs) National Center for Biotechnology Information databases. The highly Cys/Thr/Ser-enriched peptide sequence H2N-SCTTCVCTCSCCTT-CO2H, predicted to encode the thiazolylpeptides micrococcin P1 and P2 (compounds 1 and 2), thiocillin I-III (compounds 3-5), and YM-266183/266184 (compounds 6 and 7) (here collectively termed “thiocillins”), was found embedded in 4 identical, contiguous ORFs in the Bacillus cereus ATCC 14579 genome. Each ORF is predicted to encode a 52-residue peptide comprising the indicated sequence at its C terminus and a 38-residue N-terminal leader sequence. These ORFs are at the 5′ end of a cluster of genes (here termed tcl) that includes homologs of lantibiotic, cyanobactin, and microcin B17 biosynthetic genes (Fig. 1), implicating the tcl gene cluster in the production of the thiocillins.
Fig. 1.
Schematic of the thiocillin gene cluster. A 22-kb gene cluster from B. cereus ATCC 14579 encodes 24 genes responsible for the production of the thiocillins. Four identical structural genes encode a 52-residue peptide, of which the last 14 residues undergo 13 posttranslational modifications of 6 varieties to become the mature thiocillins. Gray squares indicate the R groups in the thiocillin family members. The peptide residues are numbered starting with the first residue after the leader peptide; thus, Ser-39 = Ser-1.
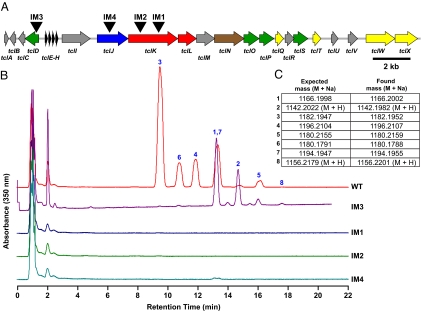
Detection of Eight Thiazolylpeptides from B. cereus ATCC 14579 and Insertional Mutagenesis of the tcl Gene Cluster.
Although micrococcins P1 and P2 (17), thiocillins I-III (18), and YM-266183/266184 (19) had been isolated separately from other strains of B. cereus, they had never been isolated from B. cereus ATCC 14579. To test whether B. cereus ATCC 14579 produces the thiocillins, we assayed extracts from its cell material and cell-free culture fluid by liquid chromatography (LC)/MS. We observed a set of 8 compounds with UV/visible absorption spectra consistent with the thiocillins, which we purified by preparative HPLC and analyzed by NMR (compounds 3 and 6) and high-resolution MS (compounds 1–8) (Fig. 2 and Fig. S1). Seven of these compounds had NMR and/or high-resolution mass spectra consistent with reported values for compounds 1–7, and the eighth was a new compound whose high-resolution mass spectrum was consistent with structure 8.
Fig. 2.
Characterization of thiocillins by HPLC and MS. (A) Schematic of the thiocillin gene cluster, showing the plasmid integration sites of insertional mutants IM1, IM2, and IM4, and control strain IM3. (B) HPLC stack plot showing methanolic extracts of cell material from cultures of B. cereus ATCC 14579, insertional mutants IM1, IM2, IM4, and control strain IM3. Peaks corresponding to compounds 1–8 are labeled. Thiocillin production is abolished in the insertional mutants, whereas in IM3, flux is shifted toward the nonhydroxylated thiocillins 1, 2, 5, and 8. (C) High-resolution MS data for compounds 1–8.
To test whether the tcl gene cluster is involved in thiocillin production, we performed insertional mutagenesis on B. cereus ATCC 14579 by using 3 plasmids that integrated into 2 of its ORFs, tclJ and tclK, predicted to catalyze heterocycle formation and Ser/Thr dehydration, respectively. Thiocillin production was abolished in all 3 mutants, whereas a control strain in which the same plasmid was integrated upstream of the structural genes retained the ability to produce thiocillins, suggesting that the tcl gene cluster is responsible for thiocillin production (Fig. 2).
Bioinformatic Analysis of the tcl Gene Cluster.
The tcl gene cluster harbors a remarkable array of encoded catalysts for the posttranslational processing of thiazolylpeptides, including an unexpected merger of biosynthetic strategies from the lantibiotic and cyanobactin/microcin B17 families. TclJ and TclN are homologous to enzymes in the cyanobactin and microcin B17 gene clusters, respectively, that convert Ser, Thr, and Cys residues to 5-membered oxazole and thiazole rings, altering the connectivity of the peptide backbone (12, 20). These enzymes, which are also homologous to the recently-described streptolysin S maturation enzymes (21), likely catalyze the conversion of all 6 cysteines in the thiocillin backbone to thiazole rings, including the 3 thiazoles surrounding the central pyridine that are characteristic of the thiazolylpeptide family. Although heterocycles in nonribosomal peptides are formed by the same cyclization/dehydration-oxidation sequence (22), the NRPS domain that catalyzes the first step (cyclization/dehydration) is unrelated to its counterparts in the thiocillin and cyanobactin gene clusters, suggesting that the formation of 5-membered heterocycles from β-nucleophilic peptide residues may have evolved convergently in nonribosomal and ribosomal peptide biosynthetic systems. TclK and TclL are homologous to lantibiotic dehydratases (6), and accordingly they are predicted to dehydrate 4 residues by a phosphorylation-elimination reaction sequence (23): Ser-1 and Ser-10, which are predicted by isotope labeling studies to be the precursors of the pyridine ring (11), and Thr-4 and Thr-13, which persist as Dhb residues in the final product. Dhb residues are also found in nonribosomal peptides like syringomycin (24) and nodularin (25), although their biosynthetic origin is not yet known. The nonheme iron-dependent dioxygenase TclD likely hydroxylates Val-6 and the S-adenosylmethionine-dependent methyltransferase TclO is predicted to O-methylate Thr-8. Consistent with the predicted function of TclD, insertional mutagenesis of tclD results in a shift of the product flux to the nonhydroxylated thiocillins 1, 2, 5, and 8 (Fig. 2).
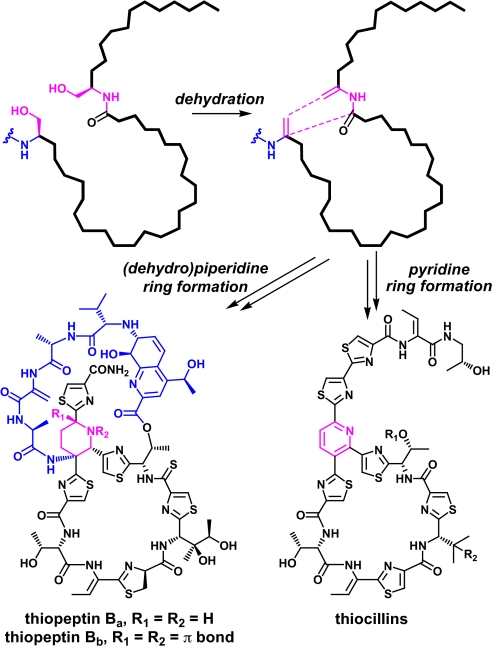
It is more challenging to predict which tcl-encoded enzymes catalyze the 2 most unusual modifications: the decarboxylation of Thr-14 to an aminoisopropanol group, which persists as the aminoacetone intermediate in compounds 2, 6, 7, and 8 and the remarkable conversion of Ser-1 and Ser-10 to a pyridine ring (Fig. 3). In total, 13 of the 14 proteinogenic amino acids in pre-thiocillin (compound 9) undergo 1 of 6 different posttranslational modifications, leaving Thr-3 as the single unmodified amino acid in the product. Not only does such a high degree of modification inhibit proteolysis and rigidify the peptide backbone, but it also provides unique binding moieties like the trithiazolylpyridine, which is remarkably distinct from any chemical group found among the 20 proteinogenic amino acids and forms a rigid recognition element for the thiocillin binding site on the 50S subunit of the ribosome.
Fig. 3.
Cryptic dehydration in the proposed mechanism for pyridine and dehydropiperidine ring formation. Dehydration of Ser-1 and Ser-10 yields Dha residues at both positions. Subsequent cyclization leads to cleavage of the leader peptide and formation of the fully aromatic pyridine in the thiocillins, whereas a portion of the leader peptide is retained and the nonaromatic piperidine or dehydropiperidine is formed in the thiopeptins. The pyridine and (dehydro)piperidine rings are shown in pink, and the leader peptide fragment and leader peptide/quinaldic acid loop are shown in blue.
Conversion of the C-Terminal 14 Residues of the Structural Peptide to Mature Thiazolylpeptides.
The timing of the various posttranslational modifications in this complex biosynthetic scheme remains unclear. As with the maturation of microcins (26) and lantibiotics (27), the 38-residue thiocillin leader peptide likely serves as a recognition element for some of the posttranslational modification catalysts. In total, 10 of the 14 residues are predicted to undergo heterocyclization or dehydration; 1 modification type may precede the other, or both may occur distributively. The most probable mechanism for pyridine ring formation (17) (Fig. 3), which is consistent with stable isotope labeling studies (15, 16), begins with the cryptic dehydration of Ser-1 and Ser-10 to Dha moieties, implying that dehydration precedes pyridine ring formation. Likewise, because pyridine ring formation necessarily involves the cleavage of the leader peptide, any leader peptide-dependent modifications must occur before the pyridine ring is fully formed. Importantly, not all thiazolylpeptides harbor a pyridine ring; some, like the thiopeptins (28) (Fig. 3), have a piperidine or dehydropiperidine ring instead. Although the mechanism of cyclization to the 6-membered heterocyclic ring is not yet known, the presence of (dehydro)piperidine moieties in thiazolylpeptides suggests that pyridine formation is a multistep process that likely involves piperidine intermediates and can be diverted down multiple pathways by redox control of an initial dihydropyridine condensation product. Interestingly, a 4-residue fragment of the leader peptide remains attached to the reduced heterocycle in the (dehydro)piperidine subclass of thiazolylpeptides, forming an additional loop that is esterified to the primary macrocycle via a quinalidic acid moiety. This structural difference suggests that for pyridine-containing thiazolylpeptides, the final desaturation to the pyridine involves cleavage of the peptide chain upstream of Ser-1, releasing the leader peptide. The 2- or 4-electron reduction that is likely required to reach the dehydropiperidine or piperidine oxidation state may stabilize this leader peptide ring substituent, because its elimination no longer leads to an aromatic ring.
Micrococcin, the best-studied member of the thiocillin family, inhibits protein synthesis by a novel mechanism: dysregulating the interaction between ribosomal proteins L7 and L11, thereby perturbing the binding of the translation factor EF-G to the ribosome (29). TclQ and TclT each encode a variant copy of ribosomal protein L11, the wild-type copy of which is a direct binding partner of the thiocillins in the ribosome. It remains to be seen whether TclQ and TclT prevent the thiocillins from binding to the ribosome, thereby playing a role in the resistance of B. cereus ATCC 14579 to its own antibiotic.
Identification of Additional Gene Clusters Predicted to Encode Novel Thiazolylpeptides.
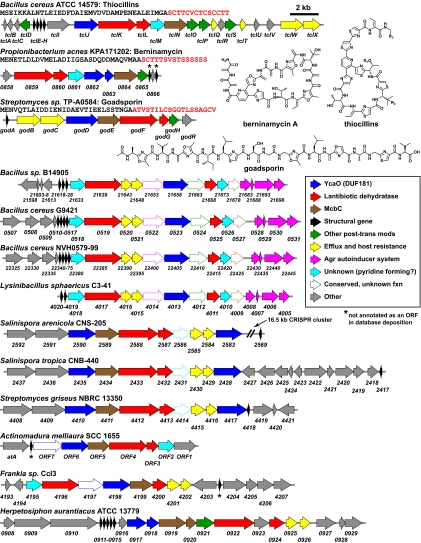
The predicted roles of the tcl genes in thiazolylpeptide maturation are further supported by a second gene cluster identified by our bioinformatic search for thiazolylpeptide-encoding genes (Fig. 4). This 11-gene cluster from Propionibacterium acnes KPA171202 harbors homologs of tclJ-N and encodes a 47-residue peptide, of which the 15 C-terminal residues putatively encode the thiazolylpeptide berninamycin (30). Further bioinformatic searches for gene clusters harboring homologs of both the heterocycle-forming (tclJ/tclN) and the Dha/Dhb-forming (tclK/tclL) genes uncovered 11 additional gene clusters (Fig. 4), including one responsible for producing goadsporin (31), a linear molecule containing heterocycles and Dha moieties. The remainder of the gene clusters we identified are likely to produce novel thiazolylpeptides, suggesting that this class will be much larger than the currently known set of >50 members. Further genetic and biochemical analysis will be required to determine how differences in the structural peptide and the complement of posttranslational modification catalysts determine the product's topology, including the presence or absence of a pyridine or (dehydro)piperidine.
Fig. 4.
Thiazolylpeptide and related gene clusters. The gene clusters for thiocillin and goadsporin and the predicted gene cluster for berninamycin are shown at the top; the structural peptide sequence is shown below the strain name, with the small molecule-encoding C-terminal sequence highlighted in red. Ten other gene clusters harboring homologs of both the heterocycle-forming (tclJ/tclN) and the Dha/Dhb-forming (tclK/tclL) genes are shown.
Eight Products from Three Incomplete Posttranslational Modifications.
It was not known that thiazolylpeptides 1–8 comprise a family of compounds from the same biosynthetic system. Although thiazolylpeptides 1–7 had been discovered, they were isolated in sets of 2 or 3 (17–19) from 3 different strains of B. cereus, whereas the eighth, thiocillin IV (compound 8), is a new molecule. Although a set of core posttranslational modifications (thiazole formation, Thr dehydration, and pyridine ring formation) occur in all family members, the differences among thiazolylpeptides 1–8 arise from a set of 3 auxiliary modifications that occur on only a subset of the molecules: C-hydroxylation of Val-6, O-methylation of Thr-8, and the reduction of the presumed Thr-14 decarboxylation product (an aminoacetone) to an aminoisopropanol. Each of the 23 = 8 possible products is formed, highlighting the biosynthetic potential of using incompletely penetrant posttranslational modifications to generate natural combinatorial diversity.
Materials and Methods
See Tables S1–S5 for primers, plasmids, and strains used in this work.
Materials, General Methods, and Instrumentation.
Escherichia coli TOP10 competent cells were purchased from Invitrogen. Plasmid pKM082 was generously provided by David Rudner (Harvard Medical School). Oligonucleotide primers were synthesized by Integrated DNA Technologies. PfuTurbo DNA polymerase was purchased from Stratagene. TaqDNA polymerase was purchased from Roche. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. All other chemicals were purchased from Sigma–Aldrich. Analytical RP-HPLC was performed on a Beckman System Gold HPLC system by using a Phenomenex Luna 5-μm C18(2) 100-Å 100 × 4.6-mm column at 1 mL/min. Preparative RP-HPLC was performed on a Beckman System Gold HPLC system by using a Phenomenex Luna 5-μm C18(2) 100-Å 250 × 10-mm column. LC/MS analysis was carried out on an Agilent LC/MS by using a Phenomenex Luna 5-μm C18(2) 100-Å 100 × 4.6-mm column at 1 mL/min. High-resolution MS was performed by the FT-ICR MS Resource of the Keck Biotechnology Resource Laboratory (Yale University, New Haven, CT) and the University of Illinois Mass Spectrometry Laboratory (Urbana, IL). 1H NMR spectra were recorded on a Varian Unity INOVA 400 (400 MHz) spectrometer. Chemical shifts are reported in ppm from tetramethylsilane with the solvent resonance resulting from incomplete deuteration as the internal standard (CDCl3: δ 7.26). Data are reported in table form as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, br = broad, m = multiplet), integration, and coupling constants. All PCRs were performed by using PfuTurbo or Taq according to the manufacturer's instructions. The identities of all DNA fragments amplified by PCR were confirmed by DNA sequencing (DNA Sequencing Core Facility, Massachusetts General Hospital).
Plasmid Construction and Transformation.
To construct the plasmids for insertional mutagenesis, fragments of ≈1 kb from the thiocillin gene cluster were PCR-amplified with specific primer pairs (LCW012/LCW013; LCW010/LCW011; LCW001/LCW002; LCW014/LCW015) from the genomic DNA of B. cereus ATCC 14579. The PCR products were digested with the restriction enzymes specified in Table S1 and ligated into the BamHI and SalI restriction sites of pKM082, yielding plasmids pLW106, pLW105, pLW111, and pLW107. B. cereus ATCC 14579 was transformed with plasmids pLW106, pLW105, pLW111, and pLW107 as described (32).
Purification of Thiocillins from B. cereus ATCC 14579.
Three hundred microliters of Luria–Bertani medium was inoculated with 100 μL of a glycerol stock of B. cereus ATCC 14579, the cells were suspended by vortexing, and 3 mL of the suspension was transferred into each of 80 culture tubes. After incubating with shaking at 28 °C for 68 h, the cultures were combined and centrifuged. Twenty milliliters of methanol was added to the cell material and vortexed, and the resulting suspension was dried with solid Na2SO4, filtered, and concentrated to give a yellow residue. This residue was suspended in 4 mL of solvent B [0.1% trifluoroacetic acid (TFA)/CH3CN], and an additional 4 mL of solvent A (0.1% TFA/H2O) was added. The mixture was syringe-filtered and purified by preparative HPLC (30–60% solvent B in solvent A; 3.5 mL/min for 60 min, with monitoring at 220 and 350 nm).
Assay of Thiocillin Production by B. cereus ATCC 14579 and Mutant Strains.
A 3-mL culture of B. cereus ATCC 14579 in Luria–Bertani broth was incubated with shaking at 28 °C for the specified period. The culture was centrifuged, 1 mL of methanol was added to the cell material and vortexed, and the resulting suspension was dried with solid Na2SO4, filtered, and concentrated to give a yellow residue. This residue was suspended in 100 μL of solvent B, and an additional 100 μL of solvent A was added. The solution was analyzed by analytical HPLC (30–55% solvent B in solvent A; 1 mL/min for 20 min, with monitoring at 220 and 350 nm).
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants CA24487 and CA59021 (to J.C.) and GM20011 and GM49338 (to C.T.W.). M.A.F. is supported by the Department of Molecular Biology and the Center for Computational and Integrative Biology at Massachusetts General Hospital. L.C.W.B. is supported by a Scientific Planning and Allocation of Resounces Committee grant from the Broad Institute of Massachusetts Institute of Technology and Harvard.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900008106/DCSupplemental.
References
- 1.Baldwin JE, Abraham E. The biosynthesis of penicillins and cephalosporins. Natural Product Rep. 1988;5:129–145. doi: 10.1039/np9880500129. [DOI] [PubMed] [Google Scholar]
- 2.Donadio S, Sosio M, Stegmann E, Weber T, Wohlleben W. Comparative analysis and insights into the evolution of gene clusters for glycopeptide antibiotic biosynthesis. Mol Genet Genomics. 2005;274:40–50. doi: 10.1007/s00438-005-1156-3. [DOI] [PubMed] [Google Scholar]
- 3.Baltz RH. Biosynthesis and genetic engineering of lipopeptide antibiotics related to daptomycin. Curr Top Med Chem. 2008;8:618–638. doi: 10.2174/156802608784221497. [DOI] [PubMed] [Google Scholar]
- 4.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides 1. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 5.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 6.Willey JM, van der Donk WA. Lantibiotics: Peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 7.Donia MS, et al. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 8.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: Microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 12.Hughes RA, Moody CJ. From amino acids to heteroaromatics–thiopeptide antibiotics, nature's heterocyclic peptides. Angew Chem Int Ed. 2007;46:7930–7954. doi: 10.1002/anie.200700728. [DOI] [PubMed] [Google Scholar]
- 13.Su TL. Micrococcin, an antibacterial substance formed by a strain of Micrococcus. Br J Exp Pathol. 1948;29:473–481. [PMC free article] [PubMed] [Google Scholar]
- 14.Pestka S, Brot N. Studies on the formation of transfer ribonucleic acid-ribosome complexes. XV. Effect of antibiotics on steps of bacterial protein synthesis: Some new ribosomal inhibitors of translocation. J Biol Chem. 1971;246:7715–7722. [PubMed] [Google Scholar]
- 15.Mocek U, et al. Biosynthesis of the modified peptide antibiotic nosiheptide in Streptomyces actuosus. J Am Chem Soc. 1993;115:7557–7568. [Google Scholar]
- 16.Mocek U, et al. Biosynthesis of the modified peptide antibiotic thiostrepton in Streptomyces azureus and Streptomyces laurentii. J Am Chem Soc. 1993;115:7992–8001. [Google Scholar]
- 17.Bycroft BW, Gowland MS. Structures of highly modified peptide antibiotics micrococcin P1 and P2. J Chem Soc Chem Commun. 1978:256–258. [Google Scholar]
- 18.Shoji J, et al. Isolation of three new antibiotics, thiocillins I, II, and III, related to micrococcin P. J Antibiot. 1976;29:366–374. doi: 10.7164/antibiotics.29.366. [DOI] [PubMed] [Google Scholar]
- 19.Suzumura K, et al. YM-266183 and YM-266184, novel thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge II. Structure elucidation. J Antibiot. 2003;56:129–134. doi: 10.7164/antibiotics.56.129. [DOI] [PubMed] [Google Scholar]
- 20.Milne JC, et al. Cofactor requirements and reconstitution of microcin B17 synthetase: A multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry. 1999;38:4768–4781. doi: 10.1021/bi982975q. [DOI] [PubMed] [Google Scholar]
- 21.Lee SW, et al. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh CT, et al. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr Opin Chem Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee C, et al. Lacticin 481 synthetase phosphorylates its substrate during lantibiotic production. J Am Chem Soc. 2005;127:15332–15333. doi: 10.1021/ja0543043. [DOI] [PubMed] [Google Scholar]
- 24.Scholz-Schroeder BK, Soule JD, Lu SE, Grgurina I, Gross DC. A physical map of the syringomycin and syringopeptin gene clusters localized to an approximately 145-kb DNA region of Pseudomonas syringae pv. syringae strain B301D. Mol Plant Microbe Interact. 2001;14:1426–1435. doi: 10.1094/MPMI.2001.14.12.1426. [DOI] [PubMed] [Google Scholar]
- 25.Moffitt MC, Neilan BA. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl Environ Microbiol. 2004;70:6353–6362. doi: 10.1128/AEM.70.11.6353-6362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madison LL, Vivas EI, Li YM, Walsh CT, Kolter R. The leader peptide is essential for the posttranslational modification of the DNA-gyrase inhibitor microcin B17. Mol Microbiol. 1997;23:161–168. doi: 10.1046/j.1365-2958.1997.2041565.x. [DOI] [PubMed] [Google Scholar]
- 27.Patton GC, Paul M, Cooper LE, Chatterjee C, van der Donk WA. The importance of the leader sequence for directing lanthionine formation in lacticin 481. Biochemistry. 2008;47:7342–7351. doi: 10.1021/bi800277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hensens OD, Albers-Schonberg G. Total structure of the highly modified peptide antibiotic components of thiopeptin. J Antibiot. 1983;36:814–831. doi: 10.7164/antibiotics.36.814. [DOI] [PubMed] [Google Scholar]
- 29.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Abe H, Kushida K, Shiobara Y, Kodama M. The structures of sulfomycin-I and berninamycin-A. Tetrahedron Lett. 1988;29:1401–1404. [Google Scholar]
- 31.Onaka H, Nakaho M, Hayashi K, Igarashi Y, Furumai T. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiology. 2005;151:3923–3933. doi: 10.1099/mic.0.28420-0. [DOI] [PubMed] [Google Scholar]
- 32.Turgeon N, Laflamme C, Ho J, Duchaine C. Elaboration of an electroporation protocol for Bacillus cereus ATCC 14579. J Microbiol Methods. 2006;67:543–548. doi: 10.1016/j.mimet.2006.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.