Abstract
1-Deoxy-d-xylulose 5-phosphate (DXP) reductoisomerase (DXR, also known as methyl-d-erythritol 4-phosphate (MEP) synthase) is a NADPH-dependent enzyme, which catalyzes the conversion of DXP to MEP in the non-mevalonate pathway of isoprene biosynthesis. Two mechanisms have been proposed for the DXR-catalyzed reaction. In the α-ketol rearrangement mechanism, the reaction begins with deprotonation of the C-3 hydroxyl group followed by a 1,2-migration to give methylerythrose phosphate, which is then reduced to MEP by NADPH. In the retroaldol/aldol rearrangement mechanism, DXR first cleaves the C3-C4 bond of DXP in a retroaldol manner to generate a three-carbon and a two-carbon phosphate bimolecular intermediate. These two species are then reunited by an aldol reaction to form a new C-C bond, yielding an aldehyde intermediate. Subsequent reduction by NADPH affords MEP. To differentiate these mechanisms, we have prepared [3-2H]- and [4-2H]-DXP and carried out a competitive secondary kinetic isotope effect (KIE) study of the DXR reaction. The normal 2° KIEs observed for [3-2H]- and [4-2H]-DXP provide compelling evidence supporting a retroaldol/aldol mechanism for the rearrangement catalyzed by DXR, with the rate-limiting step being cleavage of the C3-C4 bond of DXP.
Terpenoids are a large family of secondary metabolites, consisting of more than 55,000 members, that are widely distributed in nature and rich in biological activities.1,2 Terpenoids are biosynthesized starting with two 5-carbon isoprene units, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which have long been established to be derived from acetate in a pathway involving mevalonic acid as the key intermediate.3 However, a new mevalonate-independent isoprene source has recently been discovered in eubacteria, archeabacteria, algae, and in the plastids of plants.4–6 Since this pathway is absent in mammals but is essential for many pathogens, including Plasmodium falciparum7 and Mycobacterium tuberculosis,8 all enzymes in this pathway are potential antibacterial targets.9
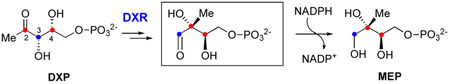
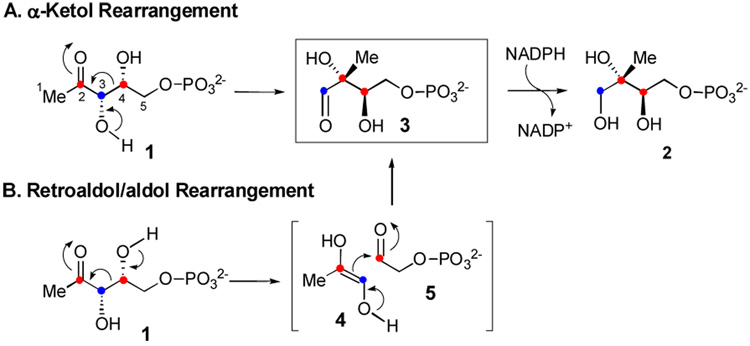
The first committed step of this non-mevalonate pathway is the conversion of 1-deoxy-d-xylulose 5-phosphate (DXP, 1) to methyl-d-erythritol 4-phosphate (MEP, 2), catalyzed by the NADPH-dependent enzyme, DXP reductoisomerase (DXR, also known as MEP synthase, see Scheme 1).10 Since MEP is the first metabolite specific to this pathway, this biosynthetic route is commonly referred to as the MEP pathway. Two mechanisms have been proposed for the DXR-catalyzed reaction (Scheme 1). In the α-ketol rearrangement mechanism (route A), the reaction begins with deprotonation of the C-3 hydroxyl group followed by a 1,2-(C4-to-C2)-migration to give methylerythrose phosphate (3), which is then reduced to MEP (2) by NADPH. In the retroaldol/aldol mechanism (route B), DXR first cleaves the C3-C4 bond of 1 in a retroaldol manner to generate a three-carbon (4) and a two-carbon phosphate (5) bimolecular intermediate. These two species are then reunited by an aldol reaction to form a new C-C bond, yielding the same aldehyde intermediate 3. Subsequent reduction of 3 by NADPH affords MEP (2).
Scheme 1.
Despite much effort,11–15 the catalytic mechanism of DXR remains elusive. To gain direct evidence to differentiate between these mechanisms, we have prepared [3-2H]-DXP (11) and [4-2H]-DXP (17) and carried out a competitive secondary kinetic isotope effect (KIE) study of the DXR reaction. Reported herein are the results and mechanistic implications.
Since the hydride transfer step is not rate-limiting for the E. coli DXR,16 the observed KIE should result only from the rearrangement event. If the reaction proceeds via the α-ketol rearrangement mechanism, incubation with [3-2H]- and [4-2H]-DXP is expected to yield normal and unit KIEs, respectively. This is because C3 undergoes sp3 to sp2 rehybridization during the rearrangement, whereas C4 remains sp3 through out the reaction. In contrast, sp3 to sp2 rehybridization occurs at both C3 and C4 in the retroaldol reaction, with C4 converting back to sp3 in the subsequent aldol condensation. Therefore, [3-2H]-DXP and [4-2H]-DXP should both display normal 2° KIEs if DXR follows the retroaldol/aldol rearrangement mechanism.
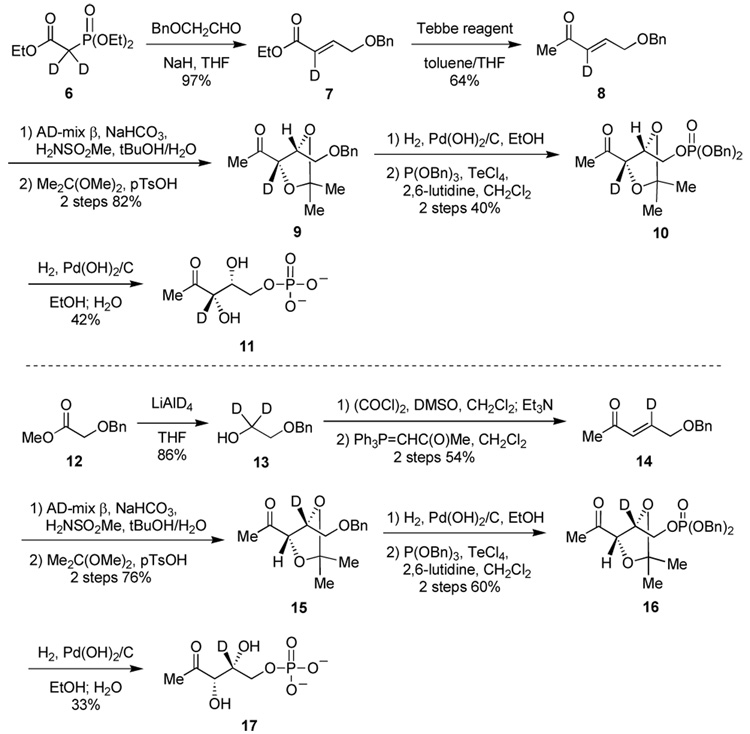
The two labeled compounds were synthesized according to the reaction sequence depicted in Scheme 2.17 To determine the KIEs, we initially turned to the standard noncompetitive method of measuring the respective kcat and Km values. However, it was observed that the progress curves resulting from the incubation of DXR with chemically synthesized [3-2H]-DXP and [4-2H]-DXP are curved, while that for the enzymatically derived DXP is linear (Figure S1).17 This phenomenon may be attributed to the presence of a contaminant (or contaminants) in the synthetic DXP samples, as the nonlinear progress curves are suggestive of slow-binding inhibition.18 As shown in Scheme 2, the key step of the synthesis is the dihydroxylation of deuterated 5-benzyloxy-pent-3-en-2-one (8 and 14) using an AD-mix β mediated protocol,19 which gave a mixture of enantiomers (er > 20:1).17 This step is likely the source of the contaminant(s), because the C4 epimer of 1 is known to be an inhibitor of DXR.12 Further attempts to purify the labeled samples failed to eradicate the non-linear behavior of the progress curves. This situation complicates the mechanistic interpretation of the KIE values. During the course of this investigation, a similar study was published in which inverse DVmax and D(Vmax/Km) KIEs were measured for [3-2H]-DXP (0.56 and 0.92) and [4-2H]-DXP (0.62 and 0.86), respectively.20 The authors concluded that the DXR catalyzed reaction proceeds via the retroaldol/aldol mechanism and that the inverse KIEs indicate that the aldol condensation step (which involves sp2 to sp3 rehybridization) is rate limiting. However, since C3 and C4 of DXP are both sp3 hybridized in the reactant state, D(Vmax/Km) should be normal for both [3-2H]- and [4-2H]-DXP. In addition, the inverse DVmax’s measured for [3/4-2H]-DXP (though possible) are much larger than one would predict. Since a similar synthetic strategy was used by this group and DXP obtained had ee values ranging between 74–84%, depending on the amount of chiral auxiliary used in the dihydroxylation step,21 if the ee for their unlabeled DXP was lower than the isotopically labeled DXPs, inverse KIEs would artificially be determined.
Scheme 2.
To circumvent this problem, we opted to determine the KIEs using the equilibrium perturbation method developed by Cleland and coworkers.22 DXR is an excellent system for applying this method because the reaction is freely reversible, and the progress of the reaction can be followed by monitoring the formation/consumption of NADPH.17 The experiment is initiated by adding DXR to a solution of NADPH, NADP+, [3/4-2H]-DXP (11 or 17), and MEP (2) at chemical equilibrium. After enzyme addition, labeled DXP is processed to give labeled MEP, in the forward direction, with the concomitant oxidation of NADPH to NADP+. In the reverse direction, the unlabeled MEP is converted to unlabeled DXP, initiated by NADP+ reduction. If there is a normal KIE on the reaction, there will be a temporary increase in the concentration of NADPH because the reverse (unlabeled) reaction is faster than the forward (labeled) reaction. If there is an inverse isotope effect, then there will be a temporary decrease in the concentration of NADPH. Since the labeled and unlabeled substrates are competing for the same enzyme active site, any enzyme sequestered by an inhibitor should affect both forward and reverse reactions equally. Thus, any putative contaminant inhibitor(s) can slow down the overall turnover, but will not have an effect on the magnitude of the KIE.22,23
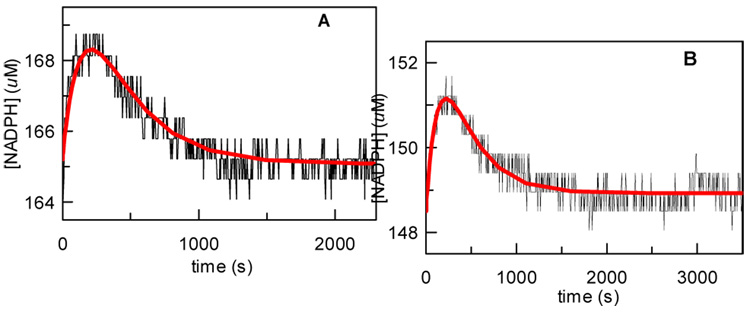
The results of the equilibrium perturbation experiments are shown in Figure 2. Importantly, for both [3-2H]-DXP and [4-2H]-DXP a temporary increase in the concentration of NADPH was observed before returning to equilibrium, corresponding to a normal 2° KIE for both compounds. The observed perturbation patterns strongly favor the retroaldol/aldol rearrangement mechanism. The corresponding KIE values were determined by computer simulation using the KinTek Explorer program and are listed in Table 1. While these values are smaller than the 2° KIE values for muscle aldolase (1.21–1.28),24 which catalyzes a similar reaction, they may simply reflect that either the retroaldol reaction is only partially rate-limiting, or that it has an early transition state where little rehybridization has occurred. The size of the KIE for [4-2H]-DXP is slightly larger than that of [3-2H]-DXP. One possible explanation for this observation is that C3 has less s-character than C4 in the transition state, because the negative charge on C3 is not fully delocalized onto the C2-carbonyl in the transition state.
Figure 2.
Experimental (•) and simulated (−) changes in [NADPH] during the equilibrium perturbation experiments with [3/4-2H]-DXP. Reaction conditions: A) 165 µM NADPH, 1 mM [3-2H]-DXP (11), 1.1 mM MEP, 3.1 mM NADP+, and 2 mM MgCl2; B) 148.5 µM NADPH, 900 µM [4-2H]-DXP (17), 990 µM MEP, 2.79 mM NADP+, and 1.8 mM MgCl2. The reaction was initiated by the addition of 1.7 µM DXR.12
Table 1.
Summary of kinetic data.
| Substrate | kcat (s−1) | Dkchema |
|---|---|---|
| DXP | 16.1 ± 0.7 | -- |
| [3-2H]-DXP | 13.2 ± 0.7 | 1.04 ± 0.02 |
| [4-2H]-DXP | 13.7 ± 0.5 | 1.11 ± 0.02 |
This KIE is determined by computer simulation of the equilibrium perturbation data and is the ratio of k3 to k3* (see Scheme S1).
In conclusion, the 2° KIEs measured in the equilibrium perturbation experiments with [3-2H]- and [4-2H]-DXP provide, for the first time, direct evidence supporting a retroaldol/aldol mechanism for the DXR catalyzed rearrangement of DXP (1) to MEP (2). This mechanistic information will assist in future design of specific inhibitors for DXR, which offers much promise for the development of new antibacterial agents.
Supplementary Material
Experimental procedures (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgement
We would like to thank Prof. Kenneth Johnson (University of Texas at Austin) for helpful discussion and the use of the KinTeK Explorer program. This work is supported in part by a grant from the Welch Foundation (F-1511) and a NIH Fellowship (GM082085) awarded to S.O.M.
References
- 1.Thulasiram HV, Erickson HK, Poulter CD. J. Am. Chem. Soc. 2008;130:1966–1971. doi: 10.1021/ja0771282. [DOI] [PubMed] [Google Scholar]
- 2.Sacchettini JC, Poulter CD. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 3.Bochar DA, Freisen JA, Stauffacher CV, Rodwell VW. Biosynthesis of mevalonic acid from acetyl-CoA. In: Cane DE, editor. Comprehesive Chemistry of Natural Products. Vol. 2. Amsterdam: 'Ed.'^'Eds.' Elsevier Science B.V.; 1999. pp. 15–44. [Google Scholar]
- 4.Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk MH, Bacher A. Chem. Biol. 1998;5:R221–R233. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 5.Rohmer M. Nat. Prod. Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- 6.Kuzuyama T, Seto H. Nat. Prod. Rep. 2003;20:171–183. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 7.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 8.Argyrou A, Blanchard JS. Biochemistry. 2004;43:4375–4384. doi: 10.1021/bi049974k. [DOI] [PubMed] [Google Scholar]
- 9.Eisenreich W, Bacher A, Arigoni D, Rohdich F. Cell Mol Life Sci. 2004;61:1401–1426. doi: 10.1007/s00018-004-3381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi S, Kuzuyama T, Watanabe H, Seto H. Proc. Natl. Acad. Sci. U S A. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong A, Munos JW, Devasthali V, Johnson KA, Liu HW. Org. Lett. 2004;6:3625–3628. doi: 10.1021/ol048459b. [DOI] [PubMed] [Google Scholar]
- 12.Phaosiri C, Proteau PJ. Bioorg. Med. Chem. Lett. 2004;14:5309–5312. doi: 10.1016/j.bmcl.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 13.Fox DT, Poulter CD. J. Org. Chem. 2005;70:1978–1985. doi: 10.1021/jo048022h. [DOI] [PubMed] [Google Scholar]
- 14.Hoeffler JF, Tritsch D, Grosdemange-Billiard C, Rohmer M. Eur. J. Biochem. 2002;269:4446–4457. doi: 10.1046/j.1432-1033.2002.03150.x. [DOI] [PubMed] [Google Scholar]
- 15.Munos JW, Pu X, Liu HW. Bioorg. Med. Chem. Lett. 2008;18:3090–3094. doi: 10.1016/j.bmcl.2007.11.062. [DOI] [PubMed] [Google Scholar]
- 16.Fox DT, Poulter CD. Biochemistry. 2005;44:8360–8368. doi: 10.1021/bi047312p. [DOI] [PubMed] [Google Scholar]
- 17.See supporting information for details.
- 18.Sculley MJ, Morrison JF, Cleland WW. Biochim. Biophys. Acta. 1996;1298:78–86. doi: 10.1016/s0167-4838(96)00118-5. [DOI] [PubMed] [Google Scholar]
- 19.Kolb HC, VanNieuwenhze MS, Sharpless KB. Chem. Rev. 1994;94:2483–2547. [Google Scholar]
- 20.Wong U, Cox RJ. Angew. Chem. Int. Ed. Engl. 2007;46:4926–4929. doi: 10.1002/anie.200700647. [DOI] [PubMed] [Google Scholar]
- 21.Cox RJ, de Andrés-Gómez A, Godfrey CR. Org. Biomol. Chem. 2003;1:3173–3177. doi: 10.1039/b307314a. [DOI] [PubMed] [Google Scholar]
- 22.Schimerlik MI, Rife JE, Cleland WW. Biochemistry. 1975;14:5347–5354. doi: 10.1021/bi00695a020. [DOI] [PubMed] [Google Scholar]
- 23.Cleland WW. Measurement of isotope effects by the equilibrium perturbation method; Isotope effects on enzyme-catalyzed reactions : proceedings of the Sixth Annual Harry Steenbock Symposium; 1976. p. 303. [Google Scholar]
- 24.Biellmann JF, O'Connell EL, Rose IA. J. Am. Chem. Soc. 1969;91:6484–6488. doi: 10.1021/ja01051a053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental procedures (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.