Abstract
OBJECTIVES
Population-based sequencing of primary/recent HIV infections (PHI) can provide a framework for understanding transmission dynamics of local epidemics. In Quebec, half of PHI represent clustered transmission events. This study ascertained the cumulative implications of clustering on onward transmission of drug resistance.
METHODS
HIV-1 pol sequence datasets were available for all genotyped PHI (<6 months post-seroconversion (n=848 subtype B infections, 1997-2007). Phylogenetic analysis established clustered transmission events, based on maximum likelihood topologies having high bootstrap values (>98%) and short genetic distances. The distributions of resistance to nucleoside and non-nucleoside RT inhibitors (NRTIs and NNRTIs) and protease inhibitors (PIs) in unique and clustered transmissions were ascertained.
Results
Episodic clustering was observed in half of recent/early stage infections from 1997-2008. Overall, 29% and 28% of new infections segregated into small (<5 PHI/cluster, n=242/848) and large transmission chains (≥5 PHI/cluster, n=239/848), averaging 2.8 ± 0.1 PHI/cluster and 10.3 ± 1.0 PHI/cluster, respectively. The transmission of nucleoside analogue mutations and 215 resistant variants (T215C/D/I/F/N/S/Y) declined with clustering (7.9% vs. 3.4% vs. 1.2% and 5.8% vs. 1.7% vs. 1.1% for unique, small and large clustered transmissions, respectively). In contrast, clustering was associated with the increased transmission of viruses harbouring resistance to NNRTIs (6.6% vs. 6.0% vs. 15.5%, respectively).
CONCLUSIONS
Clustering in early/PHI stage infection differentially affects transmission of drug resistance to different drug classes. Public health, prevention and diagnostic strategies, targeting PHI, afford a unique opportunity to curb the spread of transmitted drug resistance.
Introduction
The incidence and prevalence of HIV infections in Western settings have risen in the era of highly active antiretroviral therapy (HAART), particularly among male-sex-male (MSM) populations [1-3]. Comprehensive surveillance strategies are required to monitor emerging trends in HIV transmission and assess the effectiveness of intervention strategies [4-6].
Sequence data acquired from systematic antiretroviral resistance testing programs of primary HIV infections can provide a framework for studying transmission dynamics of local epidemics [7-14]. Accumulated HIV pol sequences allow for phylogenetic reconstruction of possible transmission events. Phylogenetic, mathematical and epidemiological modelling can be combined to better understand factors leading to amplified transmission risk at early/acute disease stages [5-16].
In Quebec, half of early stage infections (n=481/848) are phylogenetically linked to other primary infections, with 28% (n=239) forming large clusters, averaging 10 primary HIV infections (PHI) per cluster [8]. The present study was designed to evaluate the cumulative effects of clustering on forward transmission of viruses containing drug resistance mutations related to nucleoside and non-nucleoside reverse transcriptase inhibitors (NRTIs and NNRTIs), as well as protease inhibitors (PIs).
Methods
Study Population
Our PHI study population was drawn from the provincial genotyping program, at either of two Quebec reference laboratories (2001-2007, n=645) and the Quebec PHI cohort study (1997-2007, n=249). The inclusion criterion for the genotyping program, (PHI< 6 months), was based on laboratory requisitions completed by prescribing physicians. These patients have been infected for an average of 4.9 months [17]. The Quebec PHI cohort study provides demographic data and a serologic testing algorithm for recent seroconversions (STARHS) [2,8].
Sequence data were compiled using non-nominative identifiers and cross-identifiers to ensure confidentiality while controlling for repeat sampling. In all, 848 unique PHI (subtype B infections) were identified. Ethics approval was obtained from individual study sites, the Laboratoire de santé publique du Québec, and the Quebec Ministry of Health committee on confidentiality and access of information.
Phylogenetic Analysis
Genotyping was carried out as previously described to generate sequences spanning the protease and reverse transcriptase regions [8]. All sequences were aligned to consensus HXB2 sequences, removing gaps and cutting to identical sequence lengths using BioEdit software [18]. Phylogenetic interrelationships among viral sequences were estimated using Neighbour Joining (NJ) trees and Maximum Likelihood methods using BioEdit and MEGA2 integrated analysis software [18,19]. Clustering was based on the robust statistical criterion of high bootstrap values (>98%) and short genetic distances [8,11,12].
Sequence data identified minor and major resistance mutations [20]. For this study, the standardized list of the HIV-1 PR and RT mutations established by the WHO HIV Drug Resistance Surveillance Programme (V. 07-08-05) was used for comparative analysis of the epidemiology of transmitted resistance [21]. Differences in drug resistance motifs among clustered and non-clustered primary transmissions and chronically infected groups were ascertained using contingency tests [22].
Cell culture-based phenotypic assays, were used to assess drug susceptibilities of select transmitted resistant variants [23-25]. Four G190A isolates from a large transmission cluster (n=27), (GenBank accession numbers EU375798-EU375801), were assessed for baseline susceptibility to nevirapine (NVP), efavirenz (EFV), etravirine (ETV) and TMC-120.
Results
PHI Study Population
Between January 1998 and July 2007, sequence data was available from 848 persons, harbouring subtype B infections having clinical indications of PHI <6 months. Temporal changes in baseline demographics, clinical features, cluster dynamics and transmitted resistance within our PHI cohort are summarized in Table 1. As illustrated, the extent of PHI surveillance improved in 2001 with the introduction of the provincial genotyping program. Our cohort is representative of infections in Quebec [26].
Table 1.
Baseline clinical features, viral load, clustering subgroupings and acquired drug resistance of primary/recent HIV infections (PHI) diagnosed between 1997-2007.
| Baseline Characteristics | Sub Group | Year of PHI diagnosisa | |||
|---|---|---|---|---|---|
| 1997-2001 (n=112) |
2002-2003 (n=233) |
2004-2005 (n=275) |
2006-2007 (n=228) |
||
| Gender (% PHI) | Male | 92.9 | 87.9 | 93.0 | 92.9 |
| Female | 7.0 | 12.0 | 7.0 | 7.1 | |
| Age (yr) | Unique | 38.4 ± 1.0 | 38.4 ± 0.9 | 37.4 ± 0.9 | 39.9 ± 1.1 |
| Cluster | 36.3 ± 1.3 | 37.1 ± 0.8 | 36.9 ± 0.7 | 36.1 ± 0.8 | |
| Viral load (log copies/ml) | Unique | 4.6 ± 0.1 | 4.7 ± 0.1 | 4.5 ± 0.1 | 4.7 ± 0.1 |
| Cluster | 4.5 ± 0.2 | 4.7 ± 0.1 | 4.7 ± 0.1 | 4.8 ± 0.1 | |
| Risk group ( % PHI)b | MSM | 47.0 | 77.0 | 82.5 | 88.8 |
| IDU | 45.0 | 11.0 | 6.3 | 7.4 | |
| Heterosexual | 7.8 | 11.1 | 11.1 | 3.7 | |
| Cluster subgroup A c All PHI (% PHI) |
Unique | 50.4 | 46.4 | 42.8 | 43.0 |
| Small clusters (>5 PHI) |
30.9 | 24.6 | 28.0 | 28.5 | |
| Large clusters (≥5 PHI) |
18.5 | 28.8 | 28.4 | 28.1 | |
| Cluster subgroup B c Male only, non- IDU (% PHI)) |
Unique PHI | 50.0 | 49.3 | 49.8 | 50.0 |
| Small Clusters (mean PHI ± sem) |
26.9 (2.9 ± 0.3) |
23.7 (2.9 ± 0.1) |
24.2 (2.9 ± 0.1) |
25.6 (2.8 ± 0.1) |
|
| Large clusters (mean PHI ± sem) |
23.0 | 26.9 | 25.9 | 24.3 | |
| (11.9 ± 1.6) | (10.6 ± 0.7) | (14.3 ± 0.9) | (13.7 ± 1) | ||
| Drug Resistance (% PHI) |
Unique | 15.7 | 16.5 | 21.2 | 13.1 |
| Clustered | 33.9 | 16.8 | 13.0 | 18.5 | |
Baseline clinical characteristics of all genotyped PHI have been stratified according to year of diagnosis.
Risk group for participants in the PHI cohort study (n=249).
Cluster subgroup information is presented for all PHI (Cluster group A n=850), as well as for the male non-IDU subpopulation (n=730).
MSM, male-sex-male,; IDU-intravenous drug user
Clustering represented a steady ∼50% of PHI transmission events between 1997-2008. This is evident for the male-sex-male (MSM) population, i.e. the male only population that excludes intravenous drug users (Table 1). The accumulation time for clusters averaged 16.5 ± 9.8 months (mean ± sd), using STARSH for the PHI cohort. However, the majority of transmissions within clusters occurred over shorter 6-12 month periods [8, 10].
Overall, phylogenetic analysis identified 368 unique PHI, 89 small clusters (<5 PHI/cluster) and 30 large clusters (≥5 PHI/cluster) representing 43.3%, 28.5% and 28.1% of transmission events, respectively. There was an increase in the number and size of existent clusters between December 2005 and July 2007. The numbers of infections in small clusters and large clusters increased from 2.7 ± 0.2 to 3.4 ± 0.2 and from 8.8 ± 1.0 to 10.3 ± 1.2 PHI/cluster, respectively (mean ± sem).
It should be noted that non-B subtype infections, including subtypes C, A/AG, AG, G, and complex recombinant forms, introduced into Quebec (n=410) through recent immigration have been excluded from our analysis. Clustering among non-B subtypes was low (13.7%), largely restricted to heterosexual partners, and mostly involved wild-type infections.
Clustering of Transmissions Harbouring Drug Resistance
The prevalence of single drug class and multidrug resistance (MDR) in our PHI cohort was 14.9% (n=126/848) and 2.1% (n=18/848), respectively. No significant trend in the prevalence of transmitted resistance was obvious (Table 1). The overall incidence of PHI harbouring mutations conferring resistance to NRTIs (n=58), NNRTIs (n=65), and PIs (n=42) was 6.8%, 7.6%, and 5.0%, respectively.
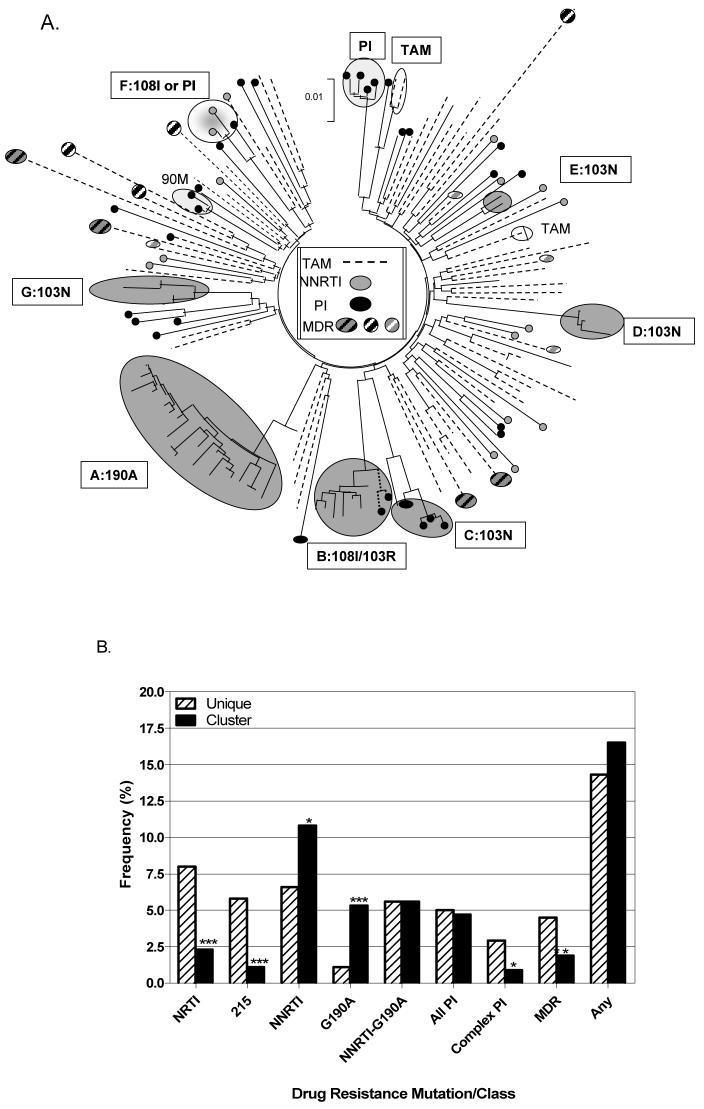
The overall frequency of drug-resistant viruses in non-clustered and clustered transmissions was similar, 14.3% and 16.5%, respectively. However, clustering had a significant impact on the relative distribution of resistance mutations for different drug classes. This is illustrated in the phylogenetic tree in Figure 1A, that includes the 144 cases of transmitted resistance in our treatment-naïve, PHI population.
Figure 1. Distribution of sequenced primary infections harbouring drug resistance.
A. The phylogenetic tree shows clustering patterns of sequenced PHI (n=144/848) harbouring resistance to thymidine analogue mutations (TAMs), non-nucleoside RT inhibitors (NNRTIs), as well as protease inhibitors (PIs). This includes seven NNRTI clusters, one TAM, and two PI clusters. Boxes list relevant NNRTI mutations. B. The mean frequency of PHI harbouring mutations to NRTIs, NNRTIs, PIs, T215 revertants, and multidrug resistant (MDR) viruses, as well as the G190A mutation.
As shown, viral variants harbouring mutations to NRTIs, including revertants at codon 215 (T215C/D/I/N/S), thymidine analogue mutations, and M184V, as well as to PIs were less frequent in clustered transmissions. In marked contrast, seven transmission clusters (Clusters A-G) harboured mutations, e.g. K103N, V108I, G190A, associated with resistance to NNRTIs (Fig. 1A). In addition, cluster C represents an MDR transmission network, wherein all four PHIs harboured K103N and three of the four also harboured L10I, I54V, A71V, V82A/I/T, I84I/V, and L90M. These PHIs, dating from Oct 2002 to May 2003, were resistant to the PIs available at that time.
Cluster A is noteworthy in that it represents the largest in our cohort, including 24 PHI and three chronic asymptomatic patients. Clustering was episodic; 6 patients were infected in 2004, 8 patients within a 3-month interval in 2005, and 8 within a 6-month interval in 2006. All 27 patients harboured the G190A and A98S mutations. Cell-based phenotypic assays on four isolates revealed >100-fold NVP resistance, sensitivity to EFV and ETV, and hypersensitivity to TMC-120. This resistance profile was confirmed for these and two other isolates based on Virco Antivirogram phenotypic analysis.
Effect of Clustering on Transmissibility of Resistance
The overall frequency of viral species harbouring resistance to different classes of drugs in unique and clustered transmissions is summarized in Fig. 1B. There was a marked diminution in the incidence of viruses harbouring NRTI mutations, 215 revertants, and MDR variants in clustered transmissions (X2= 15.4, 15.6, and 4.8, p< 0.0001, 0.0001, and 0.05, respectively).
In contrast, clustering led to increased frequencies of NNRTI resistance in large clusters, compared to unique and small cluster transmissions (X2= 4.8 and 11.4, p< 0.05 and 0.001, respectively). Such forward transmission of NNRTI resistance in clusters was independent of the G190A mutation (Fig, 1).
There appeared to be no significant impact of clustering on the incidence of viral variants harbouring mutations associated with resistance to PIs. Most PI transmitted resistance was restricted to infections harbouring single mutations, e.g. L90M, V82I, that have limited impact on drug susceptibility and viral replicative capacity. The transmission of complex PI mutational patterns, conferring measureable phenotypic resistance, was lower in clustered transmissions (X2= 5.2, p< 0.05) (Figure 1A).
Discussion
Our findings are consistent with other studies showing that early/PHI infections play a key role in the spread of HIV [6-8,16]. Our new study provides additional data that confirm that clustering of early/recent infections in Quebec may contribute to ∼50% of transmissions [8]. High rates of clustering in our cohort reflect a concentrated MSM epidemic in which persons unaware of their HIV status may engage in high risk behaviours [27]. Universal access to resistance testing allows for in depth population sampling. Our results are also consistent with models that suggest that early chronic infection contributes to HIV spread [28]. Universal access to antiretroviral drugs is an obvious factor in transmission of resistant quasispecies.
This study illustrates the potential benefits of phylogenetics in understanding factors that govern HIV transmission and the spread of drug resistance. The implications of PHI/early infection and viral fitness in transmission networks of drug resistance is underscored by the distribution of resistant drug classes in early and chronic infection [23,29-33]. The frequencies of mutations among Quebec patients failing HAART are NRTI (64%) > PI (42%) > NNRTI (38%) [31]. The reverse order is observed in transmitted resistance, i.e. NNRTI (11%/7%) > PI (5%/5%) > NRTI (2%/ 8%) in clustered and nonclustered transmissions, respectively.
Viruses harbouring NRTI and MDR resistance may be replicatively unfit, creating a bottleneck for forward transmission of such variants [29,30,32]. In contrast, clustering may facilitate spread of viruses harbouring NNRTI resistance mutations. This finding is highlighted by a transmission chain of 27 G190A-containing infections and a transmission network of NNRTI/PI dual resistant infections (n=4). In general, NNRTI mutations do not impact on viral replicative fitness to the same extent as either NRTI or PI mutations [29,32,33]. The elevated frequency of acquired NNRTI resistance reported here is consistent with data from other PHI cohorts [34,35]. In contrast, clustering did not impact on the frequency of transmitted drug-resistant infections in the Swiss cohort [36].
Our findings further underscore the recommendation that all newly infected persons undergo drug resistance testing [37,38]. Resistance to antiretroviral drugs is present 10 to 25% of the time in PHI [8,34-36]. Transmitted drug resistance persists over time, can affect disease course in drug-naïve patients, and limit strategies for antiretroviral therapy [39-44]. Although MDR variants may be less replicatively fit than wild-type archival species, the generation of resistant and MDR strains that may circumvent replicative transmission barriers is of growing concern [39]. Observations of superinfection and coinfections with resistant and MDR viruses show that resistant viruses may become more virulent and transmissible over time [33,39]. Public health strategies that target early/PHI stage infection, including the introduction of rapid testing and routine genotyping, may be of significant benefit toward reducing the incidence and spread of HIV, including that of drug resistant variants [6-,8, 39,44].
Acknowledgements
We thank the patients and attending staff from all centres participating in the Quebec genotyping program and the Montreal PHI cohort studies. Members of the Quebec Genotyping Program/Montreal HIV Prevention Study Group: INSQ (Joe Cox, Gilles Lambert, M Couillard), R Rousseau (Séro-Zéro), J Flores, R Lavoie (COCQ), G Emond (Concordia University); J Otis (CHUM). Members of the Quebec PHI Study Group: M Legault (PHI cohort coordinator); R Leblanc, RG Lalonde, N Gilmore, M Klein, J MacLeod, G Smith, J Allan, C Tsoukas, M Potter, J Falutz, J Cox (McGill University Health Center); D Rouleau, C Tremblay, J Bruneau, C Fortin, A de Pokomandy (CHUM); R Thomas, B Trottier, F Asselin, M Boissonnault, L Charest, H Dion, S Lavoie, D Legault, D Longpré, PJ Maziade, ME Morin, D Murphy, VK Nguyen, R O’Brien, S Vézina (Clinique médicale l’Actuel); JG Baril, P Côté, S Dufresne, P Junod, F Laplante, D Poirier, Y Parent, MA Charron, B Lessard, D Tessier, É Sasseville, A Talbot, MS Joyal (Clinique médicale du Quartier Latin); N Lapointe (Hôpital Ste-Justine); A Dascal (Jewish General Hospital); M Munoz (CLSC Cote des Neiges).
Sponsorship: Canadian Institutes for Health Research and the Fonds de la Recherche en Santé du Québec-Réseau SIDA (FRSQ-SIDA).
References
- 1.Boulos D, Yan P, Schanzer D, Remis RS, Archilbald CP. Estimates of HIV prevalence and incidence in Canada. Can Commun Dis Rep. 2006;32:165–74. [PubMed] [Google Scholar]
- 2.Fisher M, Pao D, Murphy G, Dean G, McElborough D, Homer G, Jarry PV. Serological testing algorithm shows rising HIV incidence in a UK cohort of men who have sex with men: 10 years application. AIDS. 2007;21:2309–14. doi: 10.1097/QAD.0b013e3282ef9fed. [DOI] [PubMed] [Google Scholar]
- 3.Marcus U, Voss L, Kollan C, Hamouda O. HIV incidence increasing in MSM in Germany: Factors influencing infection dynamics. Eurosurveill. 2006;11:157–160. [PubMed] [Google Scholar]
- 4.Saloman JA, Hogan DR, Stover J, Stanecki KA, Walker N, Ghys PD, Schwartlander B. Integrating HIV prevention and treatment: from slogans to impact. PloS Med. 2005;2(e16):50–55. doi: 10.1371/journal.pmed.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopman JS, Jacquez JA, Welch GW, Simon CP, Foxman B, Pollack SM, et al. The role of early HIV infection in the spread of HIV through populations. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:249–58. doi: 10.1097/00042560-199703010-00009. [DOI] [PubMed] [Google Scholar]
- 6.Cohen MS, Pilcher CD. Amplified HIV transmission and new approaches to HIV prevention. J Infect Dis. 2005;191:1391–93. doi: 10.1086/429414. [DOI] [PubMed] [Google Scholar]
- 7.Pillay D, Fisher M. Primary HIV Infection, phylogenetics, and antiretroviral prevention. J Infect Dis. 2007;195:924–6. doi: 10.1086/512090. [DOI] [PubMed] [Google Scholar]
- 8.Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C, et al. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis. 2007;195:951–9. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 9.Yerly S, von Wyl V, Junier T, Ledergerber B, Boni J, Burgisser P, et al. The contribution of individuals with recent infection to the spread of HIV-1 in Switzerland: A 10-year survey. 15th Conference on Retroviruses and Opportunistic Infections; Boston. February 2008; [Abstract 512] [Google Scholar]
- 10.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh-Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5(e50):392–402. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hue S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004;18:719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- 12.Hue S, Clewley, Cane PA, Pillay D. Investigation of HIV-1 transmission events by phylogenetic analysis; requirement for scientific rigour. AIDS. 2005;19:440–50. doi: 10.1097/01.aids.0000161778.15568.a1. [DOI] [PubMed] [Google Scholar]
- 13.Grant RM, Liegler TI, Hecht FM. Clusters and trends in primary resistance in San Francisco 2001-2003. Antivir Ther. 2004;9:S104–1. [Google Scholar]
- 14.Pao D, Fisher M, Hue S, Dean G, Murphy G, Cane PA, et al. Transmission of HIV-1 during primary infection: relationship to sexual risk and sexually transmitted infections. AIDS. 2005;9:85–90. doi: 10.1097/00002030-200501030-00010. [DOI] [PubMed] [Google Scholar]
- 15.Wawer MJ, Gray RH, Sewankambo NK, Serwadda D, Li X, Laeyendecker O, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–09. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 16.Pilcher CD, Joacki G, Hoffman IF, Martinson FE, Mapanje C, Stewart PW, et al. Amplified transmission of HIV-1: comparison of HIV-1 concentrations in semen and blood during acute and chronic infection. AIDS. 2007;21:1723–30. doi: 10.1097/QAD.0b013e3281532c82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Routy JP, Machouf N, Edwardes MD, Brenner BG, Thomas R, Trottier B, et al. Factors associated with a decrease in the prevalence of drug resistance in newly HIV-1 infected individuals in Montreal. AIDS. 2004;18:2305–12. doi: 10.1097/00002030-200411190-00011. [DOI] [PubMed] [Google Scholar]
- 18.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Series. 2000;41:95–98. [Google Scholar]
- 19.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–5. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 20.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, et al. Update of the Drug Resistance Mutations in HIV-1: Spring 2008. Top HIV Med. 2007;16:62–68. doi: 10.1007/s11750-007-0034-z. [DOI] [PubMed] [Google Scholar]
- 21.Shafer RW, Rhee S-Y, Pillay D, Miller V, Sandstrom P, Schapiro JM, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007;21:215–233. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van de Vijver, Brenner B, Turner D, Sandstorm P, Dunn D, Green H, et al. A systematic approach to identify mutations that can be used in epidemiological studies on transmission of drug resistant HIV. HIV Medicine. 2004;5:196. [Google Scholar]
- 23.Salomon H, Quan Y, Brenner B, Quan Y, Rouleau D, Côté P, et al. Prevalence of HIV-1 resistant to antiretroviral drugs in 81 individuals newly infected by sexual contact or injecting drug use. AIDS. 2000;14:F17–23. doi: 10.1097/00002030-200001280-00003. [DOI] [PubMed] [Google Scholar]
- 24.Brenner B, Routy JP, Petrella M, Moisi D, Oliveira M, Detorio M, et al. Persistence and fitness of multidrug resistant human immunodeficiency virus type 1 acquired in primary infection. J Virol. 2002;76:1753–1761. doi: 10.1128/JVI.76.4.1753-1761.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loemba H, Brenner B, Parniak MA, Ma’ayan S, Spira B, Moisi D, et al. Genetic divergence of human immunodeficiency virus type 1 Ethiopian clade C reverse transcriptase (RT) and rapid development of resistance against nonnucleoside inhibitors of RT. Antimicrob Agents Chemother. 2002;46:2087–2094. doi: 10.1128/AAC.46.7.2087-2094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert G, Cox J, Tremblay F, Gadoury M-A, Frigault C, Tremblay M, et al. [August 2006];ARGUS 2005: Summary of the survey on HIV, viral hepatitis and blood-borne infections, as well as associated rik factors among Montreal men who have sex with men. Montreal Public Health Department, Institut national de santé publique du Québec and the Public Health Agency of Canada. www.argusquebec.ca
- 27.Marks G, Crepaz N, Janssen RS. Estimating sexual transmission of HIV from persons aware and unaware that they are infected with the virus in the USA. AIDS. 2006;18:983–9. doi: 10.1097/01.aids.0000233579.79714.8d. [DOI] [PubMed] [Google Scholar]
- 28.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci USA. 2007;104:17441–6. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner BG, Turner D, Wainberg MA. HIV-1 drug resistance: can we overcome? Exp Opin Biol Ther. 2002;2:751–761. doi: 10.1517/14712598.2.7.751. [DOI] [PubMed] [Google Scholar]
- 30.Turner D, Brenner B, Routy JP, Petrella M, Wainberg MA. Rationale for maintenance of the M184V resistance mutation in human immunodeficiency virus type 1 reverse transcriptase in treatment experienced patients. New Microbiol. 2004;27:31–39. [PubMed] [Google Scholar]
- 31.Brenner B, Wainberg MA, Roger M. Primary HIV infections vs. genotyped chronic infections in the province of Quebec. Data from the Quebec program for HIV drug resistance testing. HIV Strain and Primary Drug Resistance in Canada. :32–36. www.phacaspc.gc.ca/publicat/hiv1Report to -vih1-05/pdf/hiv1-vih1_05_e.pdf Surveillance March 31, 2005. August 2006.
- 32.Turner D, Brenner B, Routy JP, Moisi D, Rosberger Z, Roger M, Wainberg MA. Diminished representation of HIV-1 variants containing select drug resistance-conferring mutations in reverse transcriptase in primary HIV infection. J Acquir Immunodefic Syndr. 2004;37:1627–1631. doi: 10.1097/00126334-200412150-00017. [DOI] [PubMed] [Google Scholar]
- 33.Brenner, Routy JP, Quan Y, Moisi D, Turner D, Wainberg MA. Persistence of multidrug-resistant HIV-1 in primary infection leading to superinfection. AIDS. 2004;18:1653–60. doi: 10.1097/01.aids.0000131377.28694.04. [DOI] [PubMed] [Google Scholar]
- 34.Shet A, Berry L, Mohri H, Mehandru S, Chung C, Kim A, et al. Tracking the prevalence of transmitted antiretroviral drug-resistant HIV-1: A decade of experience. J Acquir Immune Def Syndr. 2006;41:439–46. doi: 10.1097/01.qai.0000219290.49152.6a. [DOI] [PubMed] [Google Scholar]
- 35.SPREAD programme Transmission of drug-resistant HIV-1 in Europe remains limited to single classes. AIDS. 2008;22:625–35. doi: 10.1097/QAD.0b013e3282f5e062. [DOI] [PubMed] [Google Scholar]
- 36.Yerly S, von Wyl V, Ledergerber B, Boni J, Shupbach J, Burgisser P, et al. Transmission of HIV-1 drug resistance in Switzerland: A 10-year molecular epidemiology survey. AIDS. 2007;21:2223–9. doi: 10.1097/QAD.0b013e3282f0b685. [DOI] [PubMed] [Google Scholar]
- 37.Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vézinat F, Clotet B, Hammer SM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Med. 2008;47:266–85. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 38.Smith D, Moini N, Pesano R, Cachay F, Aiem H, Lie Y, et al. Clinical utility of HIV standard genotyping among antiretroviral-naive individuals with unknown duration of infection. Clin Infect Dis. 2007;44:456–8. doi: 10.1086/510748. [DOI] [PubMed] [Google Scholar]
- 39.Shet A, Markowitz M. Transmitted multidrug resistant HIV-1: new and investigational therapeutic approaches. Curr Opin Investig Drugs. 2006;7:709–20. [PubMed] [Google Scholar]
- 40.Chaix ML, Desquilbet L, Descamps D, Costagliola D, Deveau C, Galimand J, et al. Response to HAART in French patients with resistant HIV-1 treated at primary infection: ANRS Resistance Network. Antiviral Ther. 2007;12:1305–10. [PubMed] [Google Scholar]
- 41.Fox J, Dustan S, McClure M, Weber J, Fidler S. Transmitted drug-resistant HIV-1 in primary HIV-1 infection; incidence, evolution and impact on response to antiretroviral therapy. HIV Med. 2006;7:477–483. doi: 10.1111/j.1468-1293.2006.00412.x. [DOI] [PubMed] [Google Scholar]
- 42.Pillay D, Bhaskaran K, Jurriaans S, Prins M, Masqualier B, Dabis F, et al. The impact of transmitted drug resistance on the natural history of HIV infection and response to first-line therapy. AIDS. 2006;20:21–28. doi: 10.1097/01.aids.0000196172.35056.b7. [DOI] [PubMed] [Google Scholar]
- 43.Smith DM, Wong JK, Shao H, Hightower GK, Mai SH, Moreno JM, et al. Long-term persistence of transmitted HIV drug resistance in male genital tract secretions: implications for secondary transmission. J Inf Dis. 2007;196:356–60. doi: 10.1086/519164. [DOI] [PubMed] [Google Scholar]
- 44.Ghosn J, Pellegrin I, Goujard C, Deveau C, Viard JP, Galimand J, et al. HIV-1 resistant strains acquired at the time of primary infection massively fuel the cellular reservoir and persist for lengthy periods of time. AIDS. 2006;20:159–70. doi: 10.1097/01.aids.0000199820.47703.a0. [DOI] [PubMed] [Google Scholar]