The LADI-III diffractometer at the Institut Laue–Langevin has been used to carry out the first neutron crystallographic study of a DNA oligonucleotide in the A conformation. The crystal size was 0.06 mm3, the smallest ever used successfully for a study of this type. The results provide evidence of unexpected base protonation and illustrate the opportunities that now exist for nucleic acid crystallography using both hydrogenated and perdeuterated oligonucleotides.
Keywords: neutron crystallography, A-DNA, LADI-III
Abstract
The LADI-III diffractometer at the Institut Laue–Langevin has been used to carry out a preliminary neutron crystallographic study of the self-complementary DNA oligonucleotide d(AGGGGCCCCT)2 in the A conformation. The results demonstrate the viability of a full neutron crystallographic analysis with the aim of providing enhanced information on the ion–water networks that are known to be important in stabilizing A-DNA. This is the first account of a single-crystal neutron diffraction study of A-DNA. The study was carried out with the smallest crystal used to date for a neutron crystallographic study of a biological macromolecule.
1. Introduction
DNA is capable of extensive structural polymorphism (Fuller et al., 2004 ▶). The role of water and ions in stabilizing DNA conformations and in mediating structural transitions between them is well known. Natural (mixed-sequence) DNA can adopt the A, B and C conformations depending on hydration and the prevailing conditions of ionic strength. A number of repetitive-sequence DNAs that may be of biological importance are capable of adopting other conformations such as the left-handed Z conformation and the tightly wound D conformation. Water-mediated transitions between these structures can be followed in real time using synchrotron X-ray sources (Mahendrasingam et al., 1986 ▶; Forsyth et al., 1986 ▶) and crystallographic approaches have been used to trap DNA oligonucleotides in a range of structures that relate to the A–B structural change (Vargason et al., 2001 ▶).
In the continuous polymer, A-DNA is an 11-fold right-handed double helix having a deep major groove and heavily tilted bases that are displaced away from the axis towards the minor groove. The molecule has an open centre of approximately 6 Å diameter around the axis. This type of structure is adopted by RNA double helices (see, for example, Shi et al., 1999 ▶) and DNA–RNA hybrid molecules (Fedoroff et al., 1993 ▶; Xiong & Sundaralingam, 2000 ▶) and there has been speculation that the A conformation is involved in transcription. In naturally occurring DNA and throughout a large range of synthetic DNA polymers, the A conformation is capable of structural transitions to the C and the B conformations. A-type conformations are also found in oligonucleotide–protein complexes (Kim, Nikolov et al., 1993 ▶; Kim, Geiger et al., 1993 ▶) and are known to bind to a number of anticancer antibiotics (e.g. Welch et al., 1994 ▶).
A fair amount of information on DNA hydration is available from X-ray crystallography (Egli et al., 1998 ▶; Kennard et al., 1986 ▶; Malinina et al., 1998 ▶), neutron fibre diffraction (Forsyth et al., 1989 ▶; Langan et al., 1992 ▶; Shotton et al., 1997 ▶) and, more recently, neutron crystallography (Arai et al., 2005 ▶; Chatake et al., 2005 ▶). These approaches are highly complementary in the information that they provide. High-resolution X-ray methods have provided considerable detail on the location of structured water in oligonucleotide duplexes. However, it is frequently difficult to use X-ray diffraction methods to extract complete information on the location of water and H atoms, particularly for atoms that have high thermal displacement parameters (Tereshko et al., 2001 ▶). High-angle neutron fibre-diffraction experiments that exploit the difference in coherent scattering of H2O and D2O provide unique information that relates to the continuous polymer, in a situation where crystal packing effects are minimized but where the resolution of the observed data is typically poorer than that obtained for single-crystal studies. More recently, neutron Laue methods have been developed at both steady-state (Blakeley, Ruiz et al., 2008 ▶) and spallation (Langan & Greene, 2004 ▶) neutron sources. Since neutron diffraction studies of oligonucleotide single crystals are sensitive to the location of H atoms, they may provide additional detail on the orientation of water molecules and hence on the nature of hydrogen-bond interactions.
The oligonucleotide sequence d(ApGpGpGpGpCpCpCpCpT) is of interest because of its potential significance for understanding the marked stability of A-type DNA structures in guanine–cytosine homopolymer repeats such as poly-d(G)·poly-d(C). It is known to crystallize in two different space groups, P212121 and P6122, and previous work on similar sequences (Gao et al., 1995 ▶) has demonstrated that while there is a clear influence of the packing on the molecular conformation, the main features of A-DNA are maintained in both symmetries. X-ray structures of the P212121 (at 1.1 Å resolution) and P6122 (at 1.5 Å resolution) forms of d(AGGGGCCCCT)2 have been described by Gao et al. (1999 ▶). Here, we describe preliminary results from the first neutron crystallographic study of an A-type conformation of an oligonucleotide. The study builds on a growing body of results from the LADI-III diffractometer at the Institut Laue–Langevin (ILL) (Blakeley, 2009 ▶; Teixeira, Zaccai et al., 2008 ▶).
2. Methods
2.1. Crystallization
HPSF grade (High Purity Salt Free) oligonucleotides were obtained from MWG Biotech. The oligonucleotides were further purified by cation-exchange chromatography (Pharmacia MonoQ HR 10/10), dialysis and a final desalting step using a Biorad EconoPac column. The oligonucleotides were flash-frozen and lyophilized prior to solubilization in water to obtain the final stock concentration. The concentration of the samples was determined by UV spectroscopy. The samples were annealed from 368 K prior to crystallization.
Crystals were obtained at room temperature in hydrogenated buffer using the sitting-drop vapour-diffusion method. Crystallization conditions were optimized from the previously published protocol (Gao et al., 1999 ▶) to obtain crystals that grew in a few days in 10–40 µl drops containing a 1:1 mixture of DNA and 40 mM sodium cacodylate buffer pH 6, 10 mM cobalt hexammine, 120 mM magnesium chloride, 80 mM lithium chloride, 10% 2-methyl-2,4-ethylpentanediol (MPD). The drops were equilibrated against a reservoir consisting of 35% MPD. The DNA concentration was optimized at 2 mM, which systematically yielded a smaller number of larger crystals. In order to further optimize the growth of large crystals, vapour diffusion was limited by the application of a layer of Al’s oil (a 1:1 mixture of parafin and silicone oil) onto the surface of the MPD in the reservoir (Chayen et al., 1997 ▶). This compensated for the different diffusion rates that were particularly obvious at drop volumes above 30 µl and where the incidence of multiple crystals was otherwise higher. Approximately four weeks prior to data collection, the crystals were mounted on quartz capillaries and left to equilibrate by vapour diffusion against deuterated crystallization buffer to reduce neutron incoherent scattering from exchangeable hydrogen in the crystal sample.
2.2. Neutron diffraction
Neutron Laue diffraction data were recorded at room temperature using the LADI-III diffractometer at the ILL. The LADI-III instrument, which uses a large neutron image-plate detector that completely encircles the sample, is a recent replacement for the LADI-I instrument that was used to collect data for human aldose reductase (Blakeley et al., 2006 ▶; Blakeley, Ruiz et al., 2008 ▶), xylose isomerase (Meilleur et al., 2006 ▶), concanavalin A (Blakeley et al., 2004 ▶; Ahmed et al., 2007 ▶), rasburicase (Budayova-Spano et al., 2006 ▶) and endothiapepsin/hydroxyethylene (Coates et al., 2006 ▶). More recently, the instrument has been used to study the hydration and protonation states of thaumatin (Teixeira, Blakeley et al., 2008 ▶). The improved design of LADI-III provides a twofold to threefold gain in neutron detection (Wilkinson et al., 2007 ▶) and the larger radius of the instrument aids the signal-to-noise ratio of recorded reflections and decreases spatial overlap.
A nickel/titanium multi-layer bandpass filter was used to select a neutron wavelength range (Δλ/λ ≃ 25%) extending from 3.1 to 4.2 Å and centred at 3.7 Å. Data were recorded in a series of 18 Laue images with a step separation of 5° about the vertical rotation (ϕ) axis of the detector. Neutron Laue data were indexed and integrated using the LAUEGEN software suite (Helliwell et al., 1989 ▶; Campbell, 1995 ▶) modified for the cylindrical geometry of the LADI-III detector (Campbell et al., 1998 ▶). The LSCALE program (Arzt et al., 1999 ▶) was used to derive the wavelength-normalization curve using the intensities of symmetry-equivalent reflections measured at different wavelengths. SCALA (Collaborative Computational Project, Number 4, 1994 ▶) was used to combine and merge the observed data. There was no appreciable radiation damage to the sample and no explicit absorption correction was applied.
2.3. X-ray diffraction
Two X-ray data sets were collected in connection with this work. Since an important goal of this study was to be able to carry out a joint X-ray/neutron refinement, room-temperature data were collected at medium resolution from the same crystal as used for the neutron data collection. This was performed with an in-house diffractometer using Cu Kα radiation (λ = 1.5418 Å) equipped with a MAR345 image-plate detector. The data were integrated and reduced using MOSFLM (Leslie, 1992 ▶) and SCALA and TRUNCATE from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). Observations were rejected during scaling if they deviated by more than five standard deviations from the mean. Immediately after the room-temperature data had been recorded, the crystal was cryocooled in a goniometer loop and data to a resolution of 1.3 Å were recorded on beamline ID23-1 at the ESRF.
3. Results
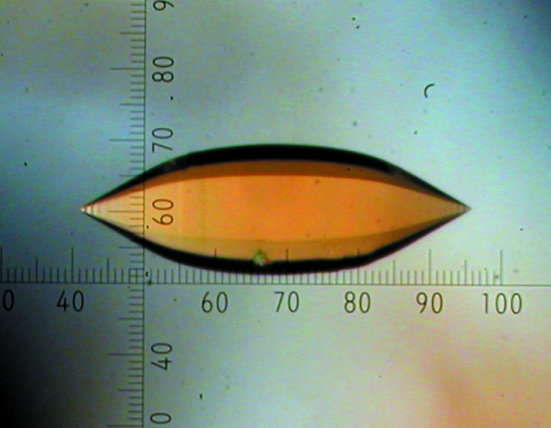
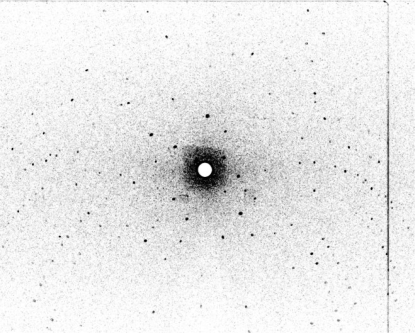
Fig. 1 ▶ shows a photograph of the crystal used for the preliminary neutron work described here. The crystal was 1.12 mm long and 0.36 mm wide and was hexagonally symmetric about its long axis. It was mounted in a thin-walled quartz capillary. The volume of the crystal was calculated to be around 0.06 mm3. Fig. 2 ▶ shows a representative example of the neutron Laue diffraction patterns recorded from this sample. Table 1 ▶ shows the relevant neutron and (room-temperature) X-ray data-collection statistics. There were 6724 observed reflections recorded for the neutron data collection. Interestingly, preliminary density maps based on phases derived from the published X-ray model suggest the presence of D atoms at some of the guanine 7 positions (data not shown).
Figure 1.
The oligonucleotide crystal used for this neutron diffraction study on LADI-III. The crystal volume was estimated to be 0.06 mm3.
Figure 2.
Neutron Laue diffraction pattern recorded from the hydrogenated oligonucleotide crystal shown in Fig. 1 ▶ using the LADI-III instrument at the ILL. The exposure time was 12 h.
Table 1. Data-collection statistics for the oligonucleotide crystal.
Values in parentheses are for the highest resolution shell.
| Data set | Neutron | X-ray |
|---|---|---|
| Beamline | LADI-III | Rotating anode |
| Wavelength (Å) | 3.1–4.2 | 1.54 |
| No. of images | 18 | 89 |
| Oscillation angle (°) | 5 | 1 |
| Exposure time per image (min) | 720 | 1 |
| Space group | P6122 | |
| Unit-cell parameters (Å) | a = b = 33.021, c = 78.817 | |
| Resolution range (Å) | 26.88–2.30 (2.42–2.30) | 26.88–1.92 (2.02–1.92) |
| No. of unique reflections | 1082 (119) | 2218 (265) |
| Completeness (%) | 82.4 (66.6) | 97.7 (84.5) |
| Rmerge† | 0.18 (0.25) | 0.043 (0.383) |
| Multiplicity | 6.2 (2.8) | 8.7 (7.3) |
| Mean I/σ(I) | 11.8 (2.1) | 19.3 (4.1) |
| Wilson B factor (Å2) | 32.1 | 36.0 |
R
merge = 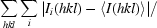
 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of I(hkl) for all i measurements.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of I(hkl) for all i measurements.
4. Discussion
The final stages of data collection are currently under way; this will involve further data collection to improve resolution and completeness and will be used to pursue a full analysis of d(AGGGGCCCCT)2 in the A conformation with the aim of providing detailed structural characterization inclusive of hydration and protonation. This analysis will exploit the availability of both neutron and X-ray data sets from the same sample and a joint structure refinement will be carried out using the nCNS (Langan & Mustyakimov, 2009 ▶) and PHENIX (Afonine et al., 2005 ▶) programs. The results will provide further information on the factors that stabilize this type of DNA conformation.
This study highlights a number of technical issues that are of general interest for neutron crystallography. Firstly, the work was carried out using a crystal that is the smallest ever described to date for a neutron crystallographic study of a biological macromolecule. In making such comparisons, one should of course consider the ratio between the sample volume and the volume of the asymmetric unit (Blum et al., 2007 ▶; Helliwell, 1992 ▶), which in this case is 49 (Blakeley, Langan et al., 2008 ▶) and is the third smallest ratio to date after the study of aldose reductase by Hazemann et al. (2005 ▶) and that of dihydrofolate reductase (Bennett et al., 2006 ▶). This is a tribute to recent developments at the LADI-III diffractometer at the ILL and emphasizes the major gains possible in the scope of neutron macromolecular crystallography as this instrument is optimized. Current instrument developments, together with the imminent move of LADI-III to a site that will provide a major flux gain, are likely to yield a future efficiency gain of ∼10. Secondly, while in this experiment hydrogen incoherent scattering was substantially reduced by the use of a crystal containing deuterated buffer, the oligonucleotide itself was not perdeuterated and a large amount of hydrogen remains attached to the C atoms of the furanose sugar groups and the bases. Further major gains in terms of the minimum usable sample volume can be expected with the use of perdeuterated oligonucleotide samples. In neutron protein crystallography, perdeuteration is usually associated with an effective gain factor of ∼10 in the minimum required crystal volume. Given the crystal volumes typically attainable in oligonucleotide or oligonucleotide–ligand systems, such a gain, in tandem with the developments planned for LADI-III, would dramatically widen the scope of nucleic acid crystallography. Thirdly, this type of work on nucleic acids is likely to be complemented in the future by studies in which other neutron and X-ray scattering techniques are brought to bear on situations in which crystallization is not possible or where parametric studies are envisaged. For example, Miles et al. (2009 ▶) have recently characterized using small-angle scattering the variation of DNA quadruplex structure in response to changes in ionic strength and temperature. This work has also illustrated how selective deuteration of DNA oligonucleotides could be heavily exploited.
Acknowledgments
RMFL was supported by a studentship jointly funded by ESRF, the ILL and Keele University. SCMT and VTF acknowledge support from the EPSRC under grant EP/C015452/1 and from the EU under contract RII3-CT-2003-505925. We acknowledge the ILL for provision of beamtime, the ESRF for test time on beamline ID14-2 and the EMBL for partial support of the LADI-III diffractometer. We thank the staff of the ILL–EMBL Deuteration Laboratory, Joanne McCarthy for help with ESRF instrumentation and Hassan Belhrali for assistance with in-house data collection. We are also grateful to Sax Mason for useful discussion and for help with tests on ILL’s D19 diffractometer.
References
- Afonine, P. V., Grosse-Kunstleve, R. W. & Adams, P. D. (2005). CCP4 Newsl.42, 8.
- Ahmed, H. U., Blakeley, M. P., Cianci, M., Cruickshank, D. W. J., Hubbard, J. A. & Helliwell, J. R. (2007). Acta Cryst. D63, 906–922. [DOI] [PubMed]
- Arai, S., Chatake, T., Ohhara, T., Kurihara, K., Tanaka, I., Suzuki, N., Fujimoto, Z., Mizuno, H. & Niimura, N. (2005). Nucleic Acids Res.33, 3017–3024. [DOI] [PMC free article] [PubMed]
- Arzt, S., Campbell, J. W., Harding, M. M., Hao, Q. & Helliwell, J. R. (1999). J. Appl. Cryst.32, 554–562.
- Bennett, B., Langan, P., Coates, L., Mustyakimov, M., Schoenborn, B., Howell, E. E. & Dealwis, C. (2006). Proc. Natl Acad. Sci. USA, 103, 18493–18498. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P. (2009). In the press.
- Blakeley, M. P., Kalb, A. J., Helliwell, J. R. & Myles, D. (2004). Proc. Natl Acad. Sci. USA, 101, 16405–16410. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Langan, P., Niimura, N. & Podjarny, A. (2008). Curr. Opin. Struct. Biol.18, 593–600. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Mitschler, A., Hazemann, I., Meilleur, F., Myles, D. & Podjarny, A. (2006). Eur. Biophys. J.35, 577–583. [DOI] [PubMed]
- Blakeley, M. P., Ruiz, F. R., Cachau, R. C., Hazemann, I., Meilleur, F., Mitschler, A., Ginell, S., Afonine, P., Ventura, O. N., Cousido-Siah, A., Haertlein, M., Joachimiak, A., Myles, D. & Podjarny, A. (2008). Proc. Natl Acad. Sci. USA, 105, 1844–1848. [DOI] [PMC free article] [PubMed]
- Blum, M.-M., Koglin, A., Rüterjans, H., Schoenborn, B., Langan, P. & Chen, J. C.-H. (2007). Acta Cryst. F63, 42–45. [DOI] [PMC free article] [PubMed]
- Budayova-Spano, M., Bonneté, F., Ferté, N., El Hajji, M., Meilleur, F., Blakeley, M. P. & Castro, B. (2006). Acta Cryst. F62, 306–309. [DOI] [PMC free article] [PubMed]
- Campbell, J. W. (1995). J. Appl. Cryst.28, 228–236.
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst.31, 496–502.
- Chatake, T., Tanaka, I., Umino, H., Arai, S. & Niimura, N. (2005). Acta Cryst. D61, 1088–1098. [DOI] [PubMed]
- Chayen, N. E. (1997). J. Appl. Cryst.30, 198–202.
- Coates, L., Erskine, P. T., Mall, S., Gill, R., Wood, S. P., Myles, D. & Cooper, J. B. (2006). Eur. Biophys. J.35, 559–566. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Egli, M., Tereshko, V., Teplova, M., Minasov, G., Joachimiak, A., Sanishvili, R., Weeks, C. M., Miller, R., Maier, M. A., An, H., Cook, P. D. & Manoharan, M. (1998). Biopolymers, 48, 234–252. [DOI] [PubMed]
- Fedoroff, O. Y., Salazar, M. & Reid, B. R. (1993). J. Mol. Biol.233, 509–523. [DOI] [PubMed]
- Forsyth, V. T., Greenall, R. J., Hussain, R., Mahendrasingam, A., Nave, C., Pigram, W. J. & Fuller, W. (1986). Biochem. Soc. Trans.14, 553–557. [DOI] [PubMed]
- Forsyth, V. T., Mahendrasingam, A., Pigram, W. J., Greenall, R. J., Bellamy, K., Fuller, W. & Mason, S. A. (1989). Int. J. Biol. Macromol.11, 236–240. [DOI] [PubMed]
- Fuller, W., Forsyth, V. T. & Mahendrasingam, A. (2004). Philos. Trans. R. Soc. Lond. B Biol. Sci.359, 1237–1247. [DOI] [PMC free article] [PubMed]
- Gao, Y. G., Robinson, H., van Boom, J. H. & Wang, A. H. (1995). Biophys. J.69, 559–568. [DOI] [PMC free article] [PubMed]
- Gao, Y.-G., Robinson, H. & Wang, A. H.-J. (1999). Eur. J. Biochem.261, 413–420. [PubMed]
- Hazemann, I., Dauvergne, M. T., Blakeley, M. P., Meilleur, F., Haertlein, M., Van Dorsselaer, A., Mitschler, A., Myles, D. A. A. & Podjarny, A. (2005). Acta Cryst. D61, 1413–1417. [DOI] [PubMed]
- Helliwell, J. R. (1992). Macromolecular Crystallography with Synchrotron Radiation. Cambridge University Press.
- Helliwell, J. R., Habash, J., Cruickshank, D. W. J., Harding, M. M., Greenhough, T. J., Campbell, J. W., Clifton, I. J., Elder, M., Machin, P. A., Papiz, M. Z. & Zurek, S. (1989). J. Appl. Cryst.22, 483–497.
- Kennard, O., Cruse, W. B., Nachman, J., Prangé, T., Shakked, Z. & Rabinovich, D. (1986). J. Biomol. Struct. Dyn.3, 623–647. [DOI] [PubMed]
- Kim, J. L., Nikolov, D. B. & Burley, S. K. (1993). Nature (London), 365, 520–527. [DOI] [PubMed]
- Kim, Y., Geiger, J. H., Hahn, S. & Sigler, P. B. (1993). Nature (London), 365, 512–520. [DOI] [PubMed]
- Langan, P., Forsyth, V. T., Mahendrasingam, A., Pigram, W. J., Mason, S. A. & Fuller, W. (1992). J. Biomol. Struct. Dyn.10, 489–503. [DOI] [PubMed]
- Langan, P. & Greene, G. (2004). J. Appl. Cryst.37, 253–257.
- Langan, P. & Mustyakimov, P. (2009). In preparation.
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Mahendrasingam, A., Forsyth, V. T., Hussain, R., Greenall, R. J., Pigram, W. J. & Fuller, W. (1986). Science, 233, 195–197. [DOI] [PubMed]
- Malinina, L., Tereshko, V., Ivanova, E., Subirana, J. A., Zarytova, V. & Nekrasov, Y. (1998). Biophys. J.74, 2482–2490. [DOI] [PMC free article] [PubMed]
- Meilleur, F., Myles, D. & Blakeley, M. P. (2006). Eur. Biophys. J.35, 611–620. [DOI] [PubMed]
- Miles, S., Callow, P. & Forsyth, V. T. (2009). In preparation.
- Shi, K., Wahl, M. & Sundaralingam, M. (1999). Nucleic Acids Res.27, 2196–2201. [DOI] [PMC free article] [PubMed]
- Shotton, M. W., Pope, L. H., Forsyth, V. T., Langan, P., Denny, R. C., Giesen, U., Dauvergne, M. T. & Fuller, W. (1997). Biophys. Chem.69, 85–96. [DOI] [PubMed]
- Teixeira, S. C. M., Blakeley, M. P., Leal, R. M. F., Mitchell, E. P. & Forsyth, V. T. (2008). Acta Cryst. F64, 378–381. [DOI] [PMC free article] [PubMed]
- Teixeira, S. C. M., Zaccai, G. et al. (2008). Chem. Phys.345, 133–151.
- Tereshko, V., Wilds, C. J., Minasov, G., Prakash, T. P., Maier, M. A., Howard, A., Wawrzak, Z., Manoharan, M. & Egli, M. (2001). Nucleic Acids Res.29, 1208–1215. [DOI] [PMC free article] [PubMed]
- Vargason, J. M., Henderson, K. & Ho, P. S. (2001). Proc. Natl Acad. Sci. USA, 98, 7265–7270. [DOI] [PMC free article] [PubMed]
- Welch, J. J., Rauscher, F. J. & Beerman, T. A. (1994). J. Biol. Chem.269, 31051–31058. [PubMed]
- Wilkinson, C., Blakeley, M. P. & Dauvergne, F. (2007). ILL Internal Report, p. ILL07WI02T.
- Xiong, Y. & Sundaralingam, M. (2000). Nucleic Acids Res.28, 2171–2176. [DOI] [PMC free article] [PubMed]