Abstract
FENs (flap endonucleases) play essential roles in DNA replication, pivotally in the resolution of Okazaki fragments. In eubacteria, DNA PolI (polymerase I) contains a flap processing domain, the N-terminal 5′→3′ exonuclease. We present evidence of paralogous FEN-encoding genes present in many eubacteria. Two distinct classes of these independent FEN-encoding genes exist with four groups of eubacteria, being identified based on the number and type of FEN gene encoded. The respective proteins possess distinct motifs hallmarking their differentiation. Crucially, based on primary sequence and predicted secondary structural motifs, we reveal key differences at their active sites. These results are supported by biochemical characterization of two family members - ExoIX (exonuclease IX) from Escherichia coli and SaFEN (Staphylococcus aureus FEN). These proteins displayed marked differences in their ability to process a range of branched and linear DNA structures. On bifurcated substrates, SaFEN exhibited similar substrate specificity to previously characterized FENs. In quantitative exonuclease assays, SaFEN maintained a comparable activity with that reported for PolI. However, ExoIX showed no observable enzymatic activity. A threaded model is presented for SaFEN, demonstrating the probable interaction of this newly identified class of FEN with divalent metal ions and a branched DNA substrate. The results from the present study provide an intriguing model for the cellular role of these FEN sub-classes and illustrate the evolutionary importance of processing aberrant DNA, which has led to their maintenance alongside DNA PolI in many eubacteria.
Keywords: DNA polymerase I, exonuclease IX, flap endonuclease (FEN), metallonuclease, replication
Abbreviations: DTT, dithiothreitol; ExoIX, exonuclease IX; EcExoIX, Escherichia coli ExoIX; FEN, flap endonuclease; H3TH, helix-three-turn helix; Inv, invader; PolI, polymerase I; PsY, pseudo-Y; SaFEN, Staphylococcus aureus FEN
INTRODUCTION
Classical biochemistry conducted by Kornberg and co-workers revealed that Escherichia coli DNA PolI (polymerase I) contained two readily separable enzymatic activities within the same polypeptide [1,2]. The C-terminal ‘Klenow’ fragment exhibited DNA polymerase and 3′→5′ ‘proofreading’ activity, whereas the N-terminal domain accounted for 5′→3′ exonuclease activity. The 5′→3′ exonuclease domain of PolI falls within a wider class of essential metallonucleases, the FEN (flap endonuclease) family [3]. FENs demonstrate both 5′→3′ exonuclease and structure-specific endonuclease activity, playing important roles in DNA replication, repair and recombination [4–6]. These enzymes are ubiquitous and highly conserved in all three kingdoms of life (Eubacteria, Archaea and Eukaryota), presumably due to their role in these crucial biological processes.
The importance and biological similarity of FENs has been demonstrated in several cross-species studies. In bacteriophage T4, disruption of the rnh gene (which encodes a FEN, known historically as T4 RNase H) resulted in slower, less accurate DNA replication. Furthermore, no measurable progeny were recovered from a host carrying a deletion in the PolI 5′→3′ exonuclease domain [7], clearly implicating the essential requirement of an active FEN for viral replication. The lethal phenotype of Δrad27 (encoding another FEN homologue) in Saccharomyces cerevisiae was suppressed by the PolI 5′→3′ exonuclease domain of E. coli [8]. In higher organisms, heterozygous (Fen1+/−) mice rapidly developed lymphoma and thymic abnormalities, whereas homozygous knockouts (Fen1−/−) displayed the embryonic lethal phenotype [9]. Moreover, mice with a single point mutation (E160D) in one of the two divalent metal ion-binding sites present in the FEN enzyme were reported to be predisposed to autoimmunity, chronic inflammation and cancers [10].
FENs recognize substrates with a DNA duplex and a single-stranded 5′ overhang such as might arise during strand displacement synthesis. The principle site of hydrolytic phosphodiester backbone cleavage is one nucleotide into the DNA duplex, at the bifurcation of single- and double-stranded DNA [11]. In addition, FENs that play a role in nucleotide excision repair (e.g. XPG in humans) can cleave ‘bubble’ substrates that have no free 5′ end at the bifurcation of single- and double-stranded DNA. As exonucleases, FENs cleave free 5′ termini and nicked DNA duplexes, liberating mono-, di- and short poly-nucleotides [12,13]. Both endo- and exo-nucleolytic activities are generally accepted to play a role in Okazaki fragment processing during DNA replication [6].
A number of FEN structures have been elucidated using X-ray crystallography. The structures show striking similarities, with a core β-sheet adorned with a number of surrounding α-helices. The FEN domain of Thermus aquaticus DNA PolI, T4RNaseH and T5FEN all reveal the presence of at least two divalent metal ion-binding sites [14–16]. Experimental evidence based on isothermal titration calorimetry and mutagenesis studies support the presence of two metal-binding sites [17,18]. Metal site I is required for generation of an attacking hydroxyl nucleophile during phosphodiester backbone hydrolysis [19]. Metal site II is required for substrate binding, particularly during exonucleolytic cleavage [17]. In T5FEN, a key structural motif is formed from α-helices bridging above the active site has been described as a ‘helical arch’ [15] or ‘helical clamp’ [20,21]. This feature has been observed in a number of enzymes, and based on biochemical studies, has been proposed to play a role in substrate binding [20–23]. Recently, a 3 Å (1 Å=0.1 nm) X-ray diffraction structure of T4 RNase H in complex with a forked DNA substrate was solved [24]. These results were in close agreement with models of FEN substrate interaction previously proposed from site-directed mutagenesis and substrate-binding studies [25,26].
An early interrogation of the incomplete E. coli genome sequence identified a putative gene (later termed xni [27]) whose product shared significant homology to the PolI 5′→3′ exonuclease domain and other known FENs [4,28]. Shafritz et al. [27] reported the purification and characterization of the product of the xni gene. Surprisingly, given the significant homology to a family of enzymes demonstrating 5′→3′ exonuclease activity, the authors suggested that the xni product was a 3′→5′ exonuclease and designated the xni gene product ExoIX (exonuclease IX) [27]. Recently we have shown that exonuclease III (a 3′→5′ exonuclease) co-purifies with ExoIX over a multiple-stage purification process [29]. Therefore, we suggested that the 3′→5′ exonuclease activity described by Shafritz et al. [27] in ExoIX was due to co-purifying exonuclease III.
Here we show that a number of prokaryotes encode a second FEN paralogue, in addition to that present as an N-terminal domain of PolI. Based on amino acid sequence analysis, these single-domain eubacterial FEN homologues can be delineated into two distinct groups. One group appears to possess binding sites for two divalent metal ions, as observed in all FENs characterized to date. The second group, exemplified by E. coli ExoIX, seems to lack a number of normally conserved amino acid residues that act as metal-binding ligands. We demonstrate that, in contrast with ExoIX, which apparently lacks activities expected of a FEN when assayed in isolation, a discrete FEN homologue encoded by Staphylococcus aureus has both FEN and 5′γ3′ exonuclease activities. We compare and contrast these two representatives of the cryptic single-domain eubacterial FEN homologues and speculate as to their biological roles.
EXPERIMENTAL
Alignments and phylogenetics
DNA PolI paralogues were identified by TBLASTN searches [30] and multiple sequence alignments prepared with CLUSTALW [31], edited in Jalview [32] and annotated by hand. Identified primary motifs were analysed with Pattinprot [33]. A phylogenetic tree was constructed from the alignment using Phylip algorithms [34]. These were defined using default parameters in Protdist and Neighbor-joining clustering [35], followed by radial plots, generated using the Phylemon server (http://phylemon.bioinfo.cipf.es) [36].
Secondary and tertiary structural predictions
Structural predictions for SaFEN (Staph. aureus FEN) were generated using FUGUE (http://www-cryst.bioc.cam.ac.uk/∼fugue/prfsearch.html) [37] and Phyre (http://www.sbg.bio.ic.ac.uk/∼phyre/) [38]. The quality of the derived models was checked by eye in the first instance, for the local geometry of strictly conserved residues. A model of SaFEN, docked with a branched substrate, was prepared by superimposition of the Phyre Apo SaFEN model with bacteriophage T4 RNase H in complex with DNA (PDB code 2IHN), using an all atom best-fit algorithm in Deepview (http://www.expasy.org/spdbv/) [39], followed by manual rigid body docking of DNA within a 1.5 Å constraint and final remodelling of single-stranded DNA regions to facilitate best fit. Final models were submitted to PROCHECK [40], hosted on the JCSB server (http://www.jcsg.org/scripts/prod/validation/sv_final.cgi) for assessment of their stereochemical quality. MolProbity (http://molprobity.biochem.duke.edu/) was used for additional checks on the validity of single-stranded DNA remodelling, in comparison with the original DNA template (chain CD, of PDB protein number 2IHN [24]).
Bacterial strains and plasmids
E. coli strain M72 pJONEX4ch was provided by Dr C. E. Meadows (Department of Infection and Immunity, University of Sheffield, Sheffield, U.K.) and purified genomic DNA from Staph. aureus strain 8325-4 was a gift from Professor Simon Foster (Molecular Biology and Biotechnology, University of Sheffield, U.K.). Recombinant bacteriophage T5 FEN and E. coli ExoIX were overexpressed and purified as described previously in [12,29].
Cloning recombinant Staph. aureus fen
PCR was performed using Staph. aureus strain 8325-4 genomic DNA template with primers for MW1329 (forward, 5′-TATTGAATTCAAAGAAGGAACGTTTAAATGCCT-3′; reverse, 5′-TTTATACGGATCCAAAAATGGGATGAAATATATTTTCC-3′), with the restriction site underlined, using the Expand High Fidelity PCR System, according to the manufacturer's instructions (Roche Diagnostics GmbH). The gel-purified product was digested with BamHI and EcoRI, ligated into pJONEX4ch (a gift from Dr C. E. Meadows) and transformed into electrocompetent E. coli M72. Positive clones were confirmed by capillary sequencing on an ABI 3730 instrument (Genetics Core Facility, University of Sheffield). The resulting expression plasmid was designated pJONEX4/fench. This plasmid encoded the Staph. aureus FEN-like protein as a C-terminal polyhistidine fusion linked via an enterokinase cleavage motif, resulting in the addition of the sequence DDDDKHHHHHH to the C-terminus of the protein.
Preliminary inductions
Culture medium {30 ml 2YT/C [2YT: 1.6% (w/v) tryptone, 1% (w/v) yeast extract and 0.5% (w/v) NaCl; with 100 μg ml−1 carbenicillin]} was inoculated with 1/100 volume (300 μl) of an overnight culture of M72 pJONEX4/fench. The culture was grown to an A595 of 0.5 and induced at 42 °C for 0.5, 1, 1.5 or 2 h duration, followed by incubation at 30 °C, for up to a total of 2 h post-induction (i.e. the 2 h 42 °C culture received no further incubation). A control strain containing the empty vector was grown with continuous heat-shock or without temperature-induction. Cells were harvested and analysed by SDS/10% PAGE using either Coomassie Blue stain or DNase substrate gel assays [41].
Overexpression and purification of SaFEN
A fermenter vessel containing approx. 5 litres of 2×YT/C with 0.01% (v/v) antifoam (Sigma) was seeded with a 1/15 inoculum of an overnight culture of M72 pJONEX4/fench. The culture was grown to an absorbance of 0.6 (A595) at ≤30 °C, followed by heat-shock induction at 42 °C for 2 h. The culture was cooled to 25 °C and incubated for a further 4.5 h prior to harvesting. The cell pellet was recovered by centrifugation at 3000 g for 30 min at 4 °C, washed with 50 mM Tris/HCl, pH 8.0, containing 0.9% (w/v) NaCl and stored in aliquots at −80 °C. Cell pellet (7.5 g) was resuspended in 40 ml of EQ buffer [25 mM potassium phosphate buffer, pH 7.4, containing 20% (w/v) glycerol, 500 mM NaCl and 30 mM imidazole]. Lysozyme (200 μg/ml) and sodium deoxycholate (500 μg/ml) were added, and the reaction was incubated on ice for up to 1 h, followed by sonication at 20% maximum amplitude in 10 s bursts. Insoluble material was removed via centrifugation at 35000 g for 30 min and the supernatant cleared by filtration (Whatman No. 1) to remove residual precipitate, before loading onto a 1 ml HisTrap column (GE Healthcare), equilibrated with EQ buffer at ≤0.5 ml/min. The column was washed in an excess of 30 ml EQ buffer (returning A280 to baseline), followed by elution over a 15 ml 40–500 mM imidazole gradient, collecting 0.5 ml fractions. Fractions were analysed by SDS/PAGE using Coomassie Blue and zymogram assays for exonuclease activity [41]. Fractions of greatest purity were equilibrated against a 100-fold excess of LSQ buffer [25 mM Tris/HCl, pH 8.0, containing 5% (w/v) glycerol, 1 mM EDTA and 5 mM DTT (dithiothreitol)], adjusted to 30% (w/v) ammonium sulphate and incubated for 15 min at room temperature (22 °C). Precipitated material was collected via centrifugation at 4000 g for 10 min and the buffer adjusted to LSQ Buffer, containing 50% (w/v) glycerol, for storage at −20 °C.
Nuclease assays
Structure-specific nuclease assays were performed using oligonucleotides synthesized by MWG Biotech AG (Table 1) and purified by butanol precipitation [42]. Oligonucleotides were radiolabelled in a reaction (30 μl) containing 5 pmol DNA, [1×] kinase buffer (70 mM Tris/HCl, pH 7.6, 10 mM MgCl2, 5 mM DTT), 6 pmol [γ-32P]ATP and 30 units of polynucleotide kinase at 37 °C for 1 h, and quenched by the addition of an equal volume of stop-mix [95% deionised formamide, 10 mM EDTA and 0.05% (w/v) Bromophenol Blue. Labelled oligonucleotides were purified using a QIAquick Nucleotide Removal Kit (Qiagen Ltd) and quantified by comparison with a serial dilution of [γ-32P]ATP using a BioRad Personal FX phosphorimager. Substrates for structure-specific assays were formed in annealling reactions (200 μl) containing 450 pM [γ-32P]-labelled DNA and 13.5 nM unlabelled complementary oligonucleotide(s), in a buffer comprising 25 mM Tris/HCl, pH 8.0 and 500 mM KCl, incubated at 80 °C for 5 min and passively cooled to ambient temperature. Standard nuclease reactions (10 μl) were performed with 3 μl of annealed labelled oligonucleotide, 5 μl of 2×reaction buffer (50 mM Tris/HCl, pH 8.0, 200 mM KCl, 20 mM MgCl2 and 0.6 mg/ml acetylated BSA), and 2 μl of enzyme prepared in enzyme dilution buffer [25 mM Tris/HCl, pH 8.0, containing 10% (w/v) glycerol, 1 mM EDTA, 1 mM DTT and 1 mg/ml acetylated BSA]. The final mixture, which contained 100 nM enzyme and 135 pM radiolabelled oligonucleotide was incubated at 37 °C for 30 min before the reaction was quenched by the addition of an equal volume of stop-mix. Reaction products were analysed by denaturing PAGE (15% 19:1 acrylamide: bisacrylamide) [42]. In addition to the structure-specific nuclease assays described above, an exonuclease assay was deployed based on that described by Fraser [43]. This assay used high-molecular-mass DNA as a substrate. Exposure to exonuclease releases acid soluble mononucleotides and polynucleotides from acid-precipitable substrate. Release of acid-soluble products was monitored by UV spectroscopy after incubation with perchloric acid and centrifugation as described in [29].
Table 1. Oligonucleotides used in structure-specific nuclease assays, presented in Figure 4.
Substrates were prepared as described in the Experimental section. In brief, unlabelled oligonucleotides were present at a 30-fold excess (13.5 nM) when annealed to the complementary radiolabelled strand (450 pM). Radiolabelled oligonucletides are italicised and the labelled 5′ terminal nucleotides are indicated in bold typeface.
| Sequence | Reference | Used in Figure 4 panel |
|---|---|---|
| 5′-d(*GATCCTAACTGGGGTTCAGAATCC) | The present study | A–G |
| 5′-d(GGATTCTGAACCCCAGTTAGGATC) | The present study | B |
| 5′-d(GGATTCTGAACCCCAGTTAGGATCTCGGATCCTGTA) | The present study | C |
| 5′-d(TCGGATCCTGTAGGATTCTGAACCCCAGTTAGGATC) | The present study | D |
| 5′-d(TACAGGATCCGA) | The present study | C and D |
| 5′-d(GGATTCTGAACCCC) | The present study | E |
| 5′-d(GGATTCTGAACCCCGACGGTAGTCAACGTG) | The present study | F and G |
| 5′-d(CACGTTGACTACCGTC) | The present study | G |
| 5′-d(*AGTCTAGACTGCAGTTGAGTCCTTGCTAGGACGGATCCCT) | [49] | H, I and J |
| 5′-d(AGGAATTCAACCACCGCTCAACTCAACTGCAGTCTAGACT) | [49] | I and J |
| 5′-d(AGGGATCCGTCCTAGCAAGGGGCTGCTACCGGAAGCTTCT) | [49] | H and J |
| 5′-d(AGAAGCTTCCGGTAGCAGCCTGAGCGGTGGTTGAATTCCT) | [49] | H and I |
RESULTS AND DISCUSSION
Delineation of eubacterial FENs
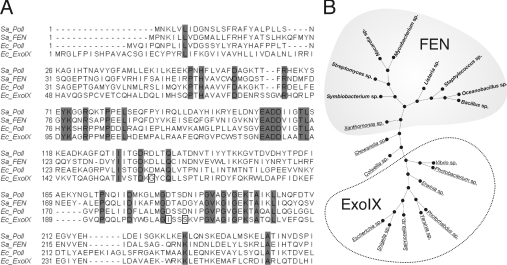
We originally identified the gene now known as xni by homology searches of the sequence databases in 1994 [4,28]. The xni gene encodes ExoIX, a paralogue of the N-terminal FEN domain of DNA polymerase I. The defining feature of all the eubacterial FEN family is a highly conserved cluster of carboxylate ligands at the putative active site (Supplementary Figure S1 at http://www.BiochemJ.org/bj/418/bj4180285add.htm). In EcExoIX (E. coli ExoIX), these are Asp10, Asp51, Glu103, Asp105 and Asp128. A key feature of the EcExoIX polypeptide sequence is the absence of certain aspartic acid residues, conserved in previously characterized FENs (Figure 1A). These conserved residues form the ligands that co-ordinate one of two divalent metal ions, directly observed in several FEN crystal structures [14–16]. EcExoIX possesses the conserved aspartates which make up the high affinity catalytic metal-binding site, known as site I [17]. However, the multiple sequence alignment (Figure 1) reveals that key aspartate residues, normally involved in binding the second divalent metal ion in classical FENs, have been replaced by the small side chains of Gly130, Ser175, and the aliphatic residue Ile178, in EcExoIX (boxed residues in Figure 1A). These residues are unable to chelate a divalent metal ion. Indeed, when only two of the analogous mutations (D201I and D204S) were introduced into T5FEN, the resulting mutant lost its ability to bind metal at site II [17] as well as much of its potency as an exonuclease. However, the structure-specific endonuclease activity of the T5FEN D201I/D204S mutant remained intact. The metal site II substitutions form a striking motif that delineates the ExoIX subset from the wider FEN family of polypeptides (Figure 1B and Supplementary Figure S1).
Figure 1. Evaluation of eubacterial DNA PolI 5′-3′ exonuclease paralogues.
(A) ClustalW multiple sequence alignment of 5′-3′-like nucleases with their related Pol I paralogues. Both classes of eubacteria contain two putative paralogous FEN-like proteins, one associated with the Klenow fragment in the form of DNA Pol I (polA), and the other encoded autonomously. The autonomous FEN-like proteins can be divided into those which retain all the putative ligands to bind two metal ions (e.g. SaFEN) and those which retain the ligands to bind a single divalent metal ion (EcExoIX). Perfectly conserved residues are boxed in grey. The amino acid substitutions affecting binding at the second metal site in ExoIX are shown in clear boxes. (B) Phylip radial phylogenetic tree, derived from ClustalW alignment of 19 secondary FENs from eubacteria (Supplementary Figure S1). A clear differentiation of ExoIX and FEN classes is evident. Eubacterial paralogous FENs fall into two discrete classes with predicted affinity for one (hatched area) or two (grey area) metal ions. This broadly, but not exclusively, correlates with Gram-positive and -negative classification (bold and underlined type respectively).
Our sequence analysis revealed a further class of FEN paralogues. We observed a subset encoding short FEN-like proteins that retain all the conserved aspartic acid residues associated with both metal sites I and II. This subset encoded FEN-like proteins that were independent of the N-terminal FEN domains found in DNA PolI orthologues. Figure 1(A) shows exemplars of the two subsets of autonomous FEN-like proteins. The SaFEN represents those containing all the conserved features of the FENs, whereas EcExoIX represents those lacking the aspartates necessary for binding metal site II. Phylogenetic analysis reveals that ExoIX and FEN fall broadly, though not exclusively, in classification based on Gram type (Figure 1B). Here we analysed 19 previously identified PolI FEN paralogues [44], performing ClustalW multiple alignments (Supplementary Figure S1) to generate a phylogenetic tree (Figure 1B).
In our analysis we also observed another intriguing class of PolI FEN paralogues in the marine bacteria Shewanella sp. and Colwellia sp. These organisms appear to fall at the divergent branch point between autonomous eubacterial FEN and ExoIX subsets (Figure 1B). They retain more conserved residues than EcExoIX (Colwellia sp., Gly150, Asp195 and Asn198) in place of typical FEN site II aspartate ligands. Based on site-directed mutagenesis in T4 RNase H and other FENs, we suggest these gene products would be likely to exhibit diminished enzymatic activity [22].
We tentatively suggest that prokaryotes can be classified into four groups based on the number and type of FEN homologue that they encode: (i) a FEN only as an N-terminal domain of DNA PolI, as exemplified in Streptococcus pneumoniae; (ii) N-terminal DNA PolI with a separate FEN paralogue with one divalent metal-binding site, e.g ExoIX in E. coli; (iii) N-terminal DNA PolI with a separate FEN paralogue with two divalent metal-binding sites, e.g. Staph. aureus; and (iv) a Klenow-like PolI domain (with no attached FEN domain) and a separate FEN domain with two divalent metal ion-binding sites, as exists in Mycoplasma genitalium (NP_072928). Accordingly, we therefore propose a more precise nomenclature for prokaryotic FEN-like genes, to allow for this simple but important distinction. The gene name xni should be used to specify the subset containing only the site I carboxylate ligands (i.e. those genes encoding ExoIX orthologues). The gene fen should specify all other homologues, whether carried alongside polA or as the sole chromosomal copy. We support this proposition with the following biochemical analysis of the Staph. aureus fen gene product.
Staph. aureus fen encodes an active nuclease
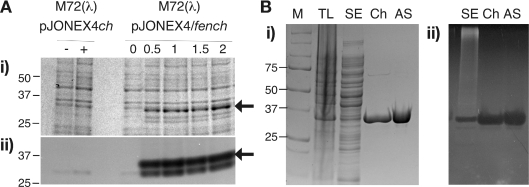
Based on the retention of a full complement of metal-binding aspartyl ligands, we reasoned that the newly identified Staphylococcal enzyme should display similar structure-specific endo- and exo-nucleolytic activities to previously identified FENs. We cloned and expressed Staph. aureus fen under the control of the temperature-inducible bacteriophage λ PL promoter in the pJONEXCH expression vector [45]. The recombinant protein was expressed with a C-terminal polyhistidine tag. Figure 2(A) shows the properties of the recombinant SaFEN protein in preliminary inductions. A distinct band of the anticipated molecular mass (35 kDa) was induced upon heat-shock (0.5–2 h) but was absent in control lysates (−/+,0 h). Furthermore, using an in situ zymogram assay we observed a co-induced nuclease activity, under conditions of heat shock (Figure 2A, panel ii), with some evidence of proteolysis. The protein was purified to >95% homogeneity (Figure 2B, panel i). Zymogram assays confirmed that the DNase activity shared chromatographic and physicochemical properties with the induced SaFEN protein (Figure 2B, panel ii). These data show that the novel Staph. aureus fen gene encodes a bona fide nuclease. The nuclease activity required divalent metal ions as expected and was not adversely affected by the C-terminal fusion (results not shown). We also obtained very similar zymogram data for an untagged version of the SaFEN protein, but it proved difficult to purify without the affinity tag, unlike the untagged T5FEN and ExoIX (results not shown).
Figure 2. SDS/10% PAGE analysis of recombinant Staph. aureus FEN.
(A) Heat-shock induction (0–2 h at 42 °C) of recombinant SaFEN demonstrates protein of the anticipated molecular mass (35 kDa; black arrow); i) stained with Commassie Blue or ii) zymogram for DNase activity. (B) Fractions taken during the purification of SaFEN were analysed by SDS/10% PAGE and stained with i) Coomassie Blue or ii) a zymogram assay of DNase activity. A replicate gel was prepared with 40 μg/ml of Type XIV herring sperm DNA, and assayed for in situ DNase activity, furnished with 10 mM MgCl2 as cofactor [41]. M, Marker (BioRad precision unstained), molecular mass of standards is given in kDa; TL, total lysate; SE, soluble extract; Ch, pooled Ni-chelate eluate; AS; ammonium sulphate precipitated fraction (final 30% saturated).
Comparison of SaFEN with other FEN family members
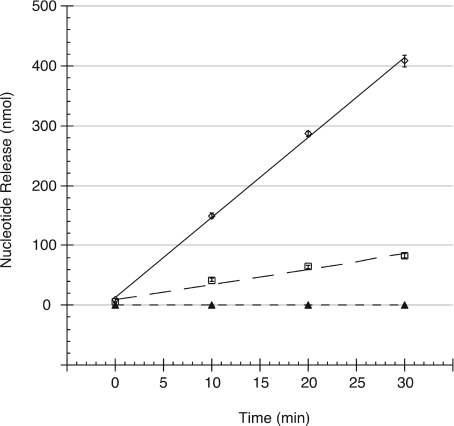
Using a standard spectrophotometric assay, we quantified the exonucleolytic activity of purified SaFEN against the well-characterized control FEN T5FEN (Figure 3). This has proven to be a simple, highly reproducible and robust assay, in the characterization of other FENs [12,23,29]. Under similar experimental conditions (pH 8.0 at 37 °C), the specific activity for T5FEN, calculated in a previous study (610 units/μg) [29] was similar to that reported in the present study (420±10 units/μg), The specific activity of SaFEN (90±5 units/μg) was approx. 5-fold lower than that of T5FEN, equivalent to a turnover rate of 106 min−1. T5FEN is a highly processive exonuclease, which serves as a stringent control. By comparison, the reported turnover of E. coli Pol I FEN under similar assay conditions is 37 min−1 (37 °C, pH 7.4) [2]. Thus, the activity of SaFEN is approx. 3-fold greater than E. coli PolI FEN activity under similar conditions, suggesting that the activity of the novel polypeptide is biologically relevant. As reported previously, ExoIX showed no detectable activity under any assay conditions tested [29].
Figure 3. Comparison of exonucleolytic properties of prokaryotic FEN homologues.
Release of acid-soluble nucleotides (nmol) in a standard reaction (1 ml) at pH 8.0, containing an excess of type XIV herring sperm DNA substrate, measured spectrophotometrically and adjusted to 1 μg of protein to aid comparison: ExoIX (▲), SaFEN (□), T5FEN (◊). Mean values from three independent readings were plotted, shown ±S.E.M (error bars). Nucleotide release was determined as described elsewhere [12].
Structure-specific activity of SaFEN and EcExoIX
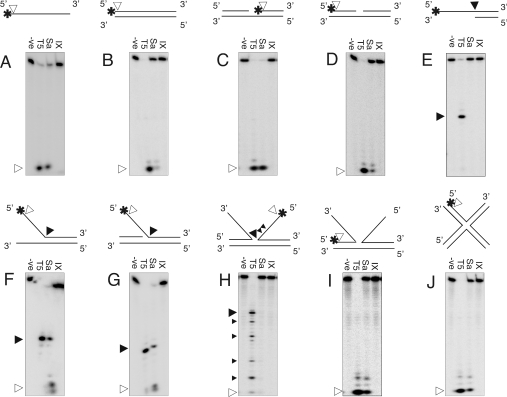
We hypothesized that the alterations in the predicted active sites of the two classes of divergent FENs would lead to pronouncedly different catalytic activity. To test this, we employed a substrate screen formed from partial or completely complementary oligonucleotides (Table 1). The catalytic activity of the proteins was examined for exo- and endo-nucleolytic cleavage under denaturing PAGE (Figure 4).
Figure 4. Comparison of enzymes' structure-specific nucleolytic cleavage on a selection of exo- and endo-nuclease substrates.
An excess of enzyme (100 nM) was incubated with preformed 5′-labelled substrate (135 pM) in the presence of 10 mM MgCl2, at 37 °C for 30 min, and the reactions analysed on denaturing 15% PAGE. Filled triangles (▼) denote structure-specific endonucleolytic cleavage products and open triangles (∇) exonucleolytic cleavage products. -ve, negative control (BSA carrier, no enzyme); T5, bacteriophage T5 FEN; Sa, SaFEN; IX; EcExoIX. See Table 1 for explanation of substrates used in A J.
Single-stranded and fully duplexed DNA substrates were cleaved exonucleolytically by SaFEN, liberating products of the same electrophoretic mobility as those released from T5FEN, albeit to a lesser extent (Figures 4A and 4B). The enzyme also liberated exo-product (≤4 mer) from the 5′ terminus of the downstream strand of a nicked DNA strand. Under the conditions tested, all substrate was degraded (Figure 4C). The nicked substrate (36 mer) was larger than the duplex DNA (24 mer), though this difference is unlikely to account for the heightened activity on the former substrate. This is supported by the similar substrate turnover of the strand upstream of a nick, compared with purely duplex DNA (compare Figure 4D with 4B). These data suggest that SaFEN favours a free 3′ terminus, upstream of the free 5′ end, which perhaps facilitates loading of the enzyme onto the DNA substrate. In this assay the activity of SaFEN was apparently equivalent to T5FEN. Such activity correlates with the involvement of this class of proteins in nick translation [46].
Although SaFEN did not endonucleolytically hydrolyse a 5′ DNA overhang, 5′ PsY (pseudo-Y) and 3′ Inv (invader) bifurcations were cleaved by the staphylococcal enzyme (Figures 4E–4G). On PsY and Inv substrates, SaFEN exhibited both endo- and exo-nucleolytic cleavage, the products of which corresponded to decamers (closed triangle) and tetramers (open triangle). Interestingly, a free 5′ flap of a partially formed junction (Figure 4H) was not hydrolysed by SaFEN. This structure resembled the double flap substrate used in the early characterization of human FEN [47]. In those assays, human FEN cleaved the 5′ flap of double flap structures efficiently, when 3′ overlaps of up to 10 nucleotides were tested. T5FEN cut the large double flap substrate at a number of sites, the most prominent product of which was a 20 mer, formed from hydrolysis at the bifurcation junction. The complex cleavage pattern with this substrate is probably due to transient base pairing, causing the formation of substrates with different free 5′ arms.
The length of duplex DNA also appears to play a role in cleavage efficiency. This hypothesis is supported by the cleavage properties on the double-stranded DNA region of larger (20 bp) Holliday junction intermediates (Figures 4I and 4J). Duplex DNA regions of partially formed junctions were hydrolysed more efficiently than shorter substrates (3′ PsY and 5′ Inv substrates, results not shown), with comparable activity to all duplex DNA (Figure 4B). SaFEN activity was limited to exonucleolytic degradation of duplex DNA in Holliday junction intermediates.
In contrast to SaFEN, ExoIX did not demonstrate FEN activity when incubated with a range of substrates (Figure 4). Therefore, ExoIX (when assayed in isolation) did not possess either 5–3′ exonuclease or 5′ FEN activity that would be expected of a FEN enzyme. We hypothesize that the lack of observable characteristic FEN activity is due to the absence of a divalent metal ion-binding site II in this protein.
Modelling cofactor and substrate interactions of SaFEN
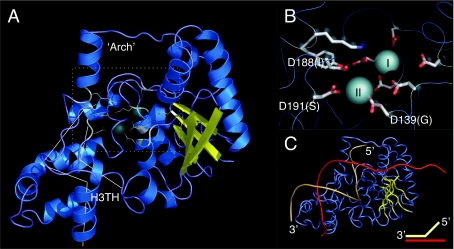
We used a structural bioinformatics approach to gain insight into the catalytic differences observed in the different FEN homologues characterized above (Figure 5). Using a co-ordinated approach of different structural prediction algorithms, we were able to ascertain with confidence the probable structure of the newly identified staphylococcal enzyme (FUGUE z-score 43.6, where ≥6 represents 99% confidence [37]). A Phyre-generated model was used for analysis based on its favourable structural constraints [38]. The model bore striking resemblance to many experimentally derived FEN structures, most notably at the DNA-binding H3TH (helix-three-turn helix) motif and the putative active site (Figures 5A and 5B). The model reveals that both divalent metal-binding sites are present in SaFEN, whereas metal site II ligands are absent in ExoIX. We postulate this accounts for the different catalytic activities observed in our screen when the Staphylococcal enzyme was compared to ExoIX. Comparison of this threaded model with other FEN family members revealed a smaller arch domain than that found in T5FEN. The first co-crystal structure of a FEN in complex with a bifurcated substrate has recently been solved [24]. We sought to use this as a model to understand the behaviour of SaFEN in its activity on a similar PsY substrate assayed above (Figure 5C; compare with Figure 4F). The restricted arch of the SaFEN model could still allow DNA to thread though upon interaction with a branched substrate, positioning the scissile phosphate diester close to the site I metal binding site, which is known to be essential for catalysis in the FENs [17,18]. The more restricted arch present in SaFEN may account for the slightly different cleavage pattern it exhibits compared to the T5FEN (Figure 4).
Figure 5. Homology modelling SaFEN in complex with M2+ cofactor and a branched PsY DNA substrate.
Bacteriophage T5FEN (PDB 1UT8) served as a template for the Phyre-threaded model of SaFEN [38], confirmed by two independent algorithms and structural validation (refer to Experimental section). (A) Cartoon representation of SaFEN. α-helices are blue and β-sheets are yellow. The DNA-binding H3TH motif is illustrated in white and the predicted active site region is boxed, shown in detail in (B). Perfectly conserved eubacterial FEN residues include tyrosine and lysine, in addition to a cluster of carboxyl ligands at the active site, here modelled with two Mn2+ ions (cyan spheres). The ligand substitutions affecting site II in the ExoIX sub-class are in brackets. (C) A branched DNA substrate was modelled by rigid body docking, based on structural superimposition of the SaFEN model with PDB protein number 2IHN, and steric clashes are reduced to within 1.5 Å. The free 5′ end (in gold) fits best with a threading model, as originally proposed for T5FEN [15], although the arch is disordered and more spatially constricted in SaFEN.
Biological role of the cryptic FEN-like proteins
The gene now known as xni was initially designated exo by this laboratory. It was speculated at the time that the gene might encode a backup function for the FEN-domain of PolI [28]. This inference was initially undermined by the observation that Strep. pneumoniae polA null mutants are non-viable [48]. However, this organism does not encode an additional FEN-like protein. Recently, Fukushima et al. [44] demonstrated that the FEN-domain encoded by either polA or xni was required for the viability of E. coli. Similarly, they also showed that either polA or ypcP is essential for Bacillus subtilis. The ypcP gene encodes a two-metal-site FEN that shares 48% identity and 64% similarity to SaFEN (Supplementary Figure S1). Thus, it would appear that either subclass of the single domain FEN-like proteins discussed here acts in redundancy with DNA PolI. The actual biochemical activity of the xni-encoded proteins remains elusive, as we were unable to identify any nucleolytic activity when tested in isolation in biochemical assays, despite testing a wide range of substrates and cofactors. Interestingly, amino acid sequence alignments also suggest that the helical arch motif identified as being important in FEN–substrate interactions is truncated in EcExoIX, which may be an additional factor in the lack of expected nuclease activity. Perhaps the ExoIX-type proteins need to be part of a complex of protein(s) to display nuclease activity or require some as yet unidentified cofactor. Alternatively, it could be that they process some form of unusual nucleic acid structure.
However, it is clear from genetic experiments and their widespread occurrence that these small FEN-like proteins are capable of fulfilling a critical biological role. The reasons why each group of organisms has retained FEN-like proteins with differing properties is unclear and will require further investigation in to how these proteins participate in biological processes.
Online data
Acknowledgments
We thank Professor Peter Artymiuk (University of Sheffield) for helpful advice on model building.
FUNDING
This work was supported by the BBSRC (Biotechnology and Biological Sciences Research Council) for funding to J. R. S. [grant number B19466] and studentships for L. M. A. [grant number 99B1B05418] and M. R. G. H. [grant number 02B1B08371].
References
- 1.Setlow P., Brutlag D., Kornberg A. Deoxyribonucleic acid polymerase: two distinct enzymes in one polypeptide. I. A proteolytic fragment containing the polymerase and 3′ leads to 5′ exonuclease functions. J. Biol. Chem. 1972;247:224–231. [PubMed] [Google Scholar]
- 2.Setlow P., Kornberg A. Deoxyribonucleic acid polymerase: two distinct enzymes in one polypeptide. II. A proteolytic fragment containing the 5′ leads to 3′ exonuclease function. Restoration of intact enzyme functions from the two proteolytic fragments. J. Biol. Chem. 1972;247:232–240. [PubMed] [Google Scholar]
- 3.Robins P., Pappin D. J., Wood R. D., Lindahl T. Structural and functional homology between mammalian DNase IV and the 5′-nuclease domain of Escherichia coli DNA polymerase I. J. Biol. Chem. 1994;269:28535–28538. [PubMed] [Google Scholar]
- 4.Lieber M. R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 5.Ceska T. A., Sayers J. R. Structure-specific DNA cleavage by 5′ nucleases. Trends Biochem. Sci. 1998;23:331–336. doi: 10.1016/s0968-0004(98)01259-6. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y., Kao H. I., Bambara R. A. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 7.Hobbs L. J., Nossal N. G. Either bacteriophage T4 RNase H or Escherichia coli DNA polymerase I is essential for phage replication. J. Bacteriol. 1996;178:6772–6777. doi: 10.1128/jb.178.23.6772-6777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X., Wu P., Zheng L., Thrower D., Partikian A., Qiu J., Shen B. Suppression of Saccharomyces cerevisiae rad27 null mutant phenotypes by the 5′ nuclease domain of Escherichia coli DNA polymerase I. Curr. Genet. 2002;41:379–388. doi: 10.1007/s00294-002-0323-x. [DOI] [PubMed] [Google Scholar]
- 9.Kucherlapati M., Yang K., Kuraguchi M., Zhao J., Lia M., Heyer J., Kane M. F., Fan K., Russell R., Brown A. M., et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng L., Dai H., Zhou M., Li M., Singh P., Qiu J., Tsark W., Huang Q., Kernstine K., Zhang X., Lin D., Shen B. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 11.Rossi M. L., Bambara R. A. Reconstituted Okazaki fragment processing indicates two pathways of primer removal. J. Biol. Chem. 2006;281:26051–26061. doi: 10.1074/jbc.M604805200. [DOI] [PubMed] [Google Scholar]
- 12.Sayers J. R., Eckstein F. Properties of overexpressed phage T5 D15 exonuclease. J. Biol. Chem. 1990;265:18311–18317. [PubMed] [Google Scholar]
- 13.Lundquist R. C., Olivera B. M. Transient generation of displaced single-stranded DNA during nick translation. Cell. 1982;31:53–60. doi: 10.1016/0092-8674(82)90404-4. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y., Eom S. H., Wang J. M., Lee D. S., Suh S. W., Steitz T. A. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 15.Ceska T. A., Sayers J. R., Stier G., Suck D. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature. 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 16.Mueser T. C., Nossal N. G., Hyde C. C. Structure of bacteriophage-T4 RNASE-H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 17.Feng M., Patel D., Dervan J. J., Ceska T., Suck D., Haq I., Sayers J. R. Roles of divalent metal ions in flap endonuclease-substrate interactions. Nat. Struct. Mol. Biol. 2004;11:450–456. doi: 10.1038/nsmb754. [DOI] [PubMed] [Google Scholar]
- 18.Zheng L., Li M., Shan J., Krishnamoorthi R., Shen B. Distinct roles of two Mg2+ binding sites in regulation of murine flap endonuclease-1 activities. Biochemistry. 2002;41:10323–10331. doi: 10.1021/bi025841s. [DOI] [PubMed] [Google Scholar]
- 19.Tock M. R., Frary E., Sayers J. R., Grasby J. A. Dynamic evidence for metal ion catalysis in the reaction mediated by a flap endonuclease. EMBO J. 2003;22:995–1004. doi: 10.1093/emboj/cdg098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosfield D. J., Mol C. D., Shen B., Tainer J. A. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 21.Hwang K. Y., Baek K., Kim H. Y., Cho Y. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol. 1998;5:707–713. doi: 10.1038/1406. [DOI] [PubMed] [Google Scholar]
- 22.Bhagwat M., Meara D., Nossal N. G. Identification of residues of T4 RNase H required for catalysis and DNA binding. J. Biol. Chem. 1997;272:28531–28538. doi: 10.1074/jbc.272.45.28531. [DOI] [PubMed] [Google Scholar]
- 23.Garforth S. J., Ceska T. A., Suck D., Sayers J. R. Mutagenesis of conserved lysine residues in bacteriophage T5 5′-3′ exonuclease suggests separate mechanisms of endo- and exonucleolytic cleavage. Proc. Natl. Acad. Sci. U.S.A. 1999;96:38–43. doi: 10.1073/pnas.96.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devos J. M., Tomanicek S. J., Jones C. E., Nossal N. G., Mueser T. C. Crystal structure of bacteriophage T4 5′ nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J. Biol. Chem. 2007;282:31713–31724. doi: 10.1074/jbc.M703209200. [DOI] [PubMed] [Google Scholar]
- 25.Dervan J. J., Feng M., Patel D., Grasby J. A., Artymiuk P. J., Ceska T. A., Sayers J. R. Interactions of mutant and wild-type flap endonucleases with oligonucleotide substrates suggest an alternative model of DNA binding. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8542–8547. doi: 10.1073/pnas.082241699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allawi H. T., Kaiser M. W., Onufriev A. V., Ma W. P., Brogaard A. E., Case D. A., Neri B. P., Lyamichev V. I. Modeling of flap endonuclease interactions with DNA substrate. J. Mol. Biol. 2003;328:537–554. doi: 10.1016/s0022-2836(03)00351-6. [DOI] [PubMed] [Google Scholar]
- 27.Shafritz K. M., Sandigursky M., Franklin W. A. Exonuclease IX of Escherichia coli. Nucleic Acids Res. 1998;26:2593–2597. doi: 10.1093/nar/26.11.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayers J. R. Computer aided identification of a potential 5′-3′ exonuclease gene encoded by Escherichia coli. J. Theor. Biol. 1994;170:415–421. doi: 10.1006/jtbi.1994.1202. [DOI] [PubMed] [Google Scholar]
- 29.Hodskinson M. R., Allen L. M., Thomson D. P., Sayers J. R. Molecular interactions of Escherichia coli ExoIX and identification of its associated 3′-5′ exonuclease activity. Nucleic Acids Res. 2007;35:4094–4102. doi: 10.1093/nar/gkm396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Clamp M., Cuff J., Searle S. M., Barton G. J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 33.Combet C., Blanchet C., Geourjon C., Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 34.Retief J. D. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 36.Tarraga J., Medina I., Arbiza L., Huerta-Cepas J., Gabaldon T., Dopazo J., Dopazo H. Phylemon: a suite of web tools for molecular evolution, phylogenetics and phylogenomics. Nucleic Acids Res. 2007;35:W38–W42. doi: 10.1093/nar/gkm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J., Blundell T. L., Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 38.Bennett-Lovsey R. M., Herbert A. D., Sternberg M. J., Kelley L. A. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 39.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. A., Moss D. S., Thornton J. M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 41.Rosenthal A., Lacks S. A. Nuclease detection in SDS-polyacrylamide gel electrophoresis. Anal. Biochem. 1977;80:76–90. doi: 10.1016/0003-2697(77)90627-3. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J., Fritsch E. F., Manniatis T. New York: Cold Spring Harbor Laboratory Press; 1989. Cloning: A Laboratory Manual. [Google Scholar]
- 43.Fraser M. J. Purification and properties of Neurospora crassa endo-exonuclease, an enzyme which can be converted to a single-strand specific endonuclease. Methods Enzymol. 1980;65:255–263. doi: 10.1016/s0076-6879(80)65035-6. [DOI] [PubMed] [Google Scholar]
- 44.Fukushima S., Itaya M., Kato H., Ogasawara N., Yoshikawa H. Reassessment of the in vivo functions of DNA polymerase I and RNase H in bacterial cell growth. J. Bacteriol. 2007;189:8575–8583. doi: 10.1128/JB.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sayers J. R., Eckstein F. A single-strand specific endonuclease activity copurifies with overexpressed T5 D15 exonuclease. Nucleic Acids Res. 1991;19:4127–4132. doi: 10.1093/nar/19.15.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kornberg A., Baker T. A. New York: W.H. Freeman; 1992. DNA Replication. [Google Scholar]
- 47.Harrington J. J., Lieber M. R. DNA structural elements required for FEN-1 binding. J. Biol. Chem. 1995;270:4503–4508. doi: 10.1074/jbc.270.9.4503. [DOI] [PubMed] [Google Scholar]
- 48.Díaz A., Lacks S. A., López P. The 5′ to 3′ exonuclease activity of DNA polymerase I is essential for Streptococcus pneumoniae. Mol. Microbiol. 1992;6:3009–3019. doi: 10.1111/j.1365-2958.1992.tb01759.x. [DOI] [PubMed] [Google Scholar]
- 49.White M. F., Lilley D. M. The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction. J. Mol. Biol. 1997;266:122–134. doi: 10.1006/jmbi.1996.0795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.