Abstract
Summary: Aromatic compounds belong to one of the most widely distributed classes of organic compounds in nature, and a significant number of xenobiotics belong to this family of compounds. Since many habitats containing large amounts of aromatic compounds are often anoxic, the anaerobic catabolism of aromatic compounds by microorganisms becomes crucial in biogeochemical cycles and in the sustainable development of the biosphere. The mineralization of aromatic compounds by facultative or obligate anaerobic bacteria can be coupled to anaerobic respiration with a variety of electron acceptors as well as to fermentation and anoxygenic photosynthesis. Since the redox potential of the electron-accepting system dictates the degradative strategy, there is wide biochemical diversity among anaerobic aromatic degraders. However, the genetic determinants of all these processes and the mechanisms involved in their regulation are much less studied. This review focuses on the recent findings that standard molecular biology approaches together with new high-throughput technologies (e.g., genome sequencing, transcriptomics, proteomics, and metagenomics) have provided regarding the genetics, regulation, ecophysiology, and evolution of anaerobic aromatic degradation pathways. These studies revealed that the anaerobic catabolism of aromatic compounds is more diverse and widespread than previously thought, and the complex metabolic and stress programs associated with the use of aromatic compounds under anaerobic conditions are starting to be unraveled. Anaerobic biotransformation processes based on unprecedented enzymes and pathways with novel metabolic capabilities, as well as the design of novel regulatory circuits and catabolic networks of great biotechnological potential in synthetic biology, are now feasible to approach.
INTRODUCTION
After carbohydrates, aromatic compounds, found as lignin components, flavonoids, quinones, aromatic amino acids, or constituents of fossil fuels, are the most widely distributed class of organic compounds in nature. Indeed, lignin is the second most abundant polymer in nature after cellulose (3). Moreover, a significant number of xenobiotics, human-made compounds that likely did not exist in the biosphere in significant quantities prior to the industrial revolution (e.g., polychlorinated biphenyls and dioxins and nitroaromatics, etc.), belong to this family of compounds (305). The thermodynamic stability of the benzene ring due to its resonance structure has contributed to the widespread production and industrial usage of natural and xenobiotic aromatic compounds but at the same time has contributed to the persistence of these compounds, many of which are toxic, after their release into the environment, posing a major environmental problem (238). Microorganisms play an essential role in recycling carbon and maintaining the health of the biosphere (76). Bacteria have evolved to degrade most naturally occurring organic compounds, including the persistent aromatics. Moreover, the promiscuity of the catabolic enzymes allows bacteria to degrade, at least partially, xenobiotics that share similar structures with naturally occurring aromatic compounds (84, 238).
The bacterial catabolism of aromatic compounds involves a wide variety of peripheral pathways that activate structurally diverse substrates into a limited number of common intermediates that are further cleaved and processed by a few central pathways to the central metabolism of the cell (142, 238, 284). There are two major strategies to degrade aromatic compounds depending on the presence or absence of oxygen. In the aerobic catabolism of aromatics, oxygen is not only the final electron acceptor but also a cosubstrate for two key processes, i.e., the hydroxylation and oxygenolytic ring cleavage of the aromatic ring, carried out by oxygenases (270, 356). In contrast, the anaerobic catabolism of aromatic compounds uses a completely different strategy, based on reductive reactions, to attack the aromatic ring (116, 128, 142). While the aerobic catabolism of aromatic compounds has been studied for several decades (238), the anaerobic degradation of aromatics is a more recently discovered microbial capacity that still awaits a deeper understanding despite the fact that microbial metabolism in the absence of oxygen is the most ancient of all life processes. The biochemical and genetic bases of the anaerobic degradation of aromatic compounds are not very well established yet due mainly to the difficulties in routinely growing and genetically manipulating the aromatic-degrading microorganisms (152).
The extensive work carried out on the aerobic processes for the degradation of aromatics might lead, to a certain extent, to an overestimation of their ecological importance for the removal and mineralization of these compounds in natural environments. In fact, many habitats containing large amounts of aromatic compounds are often anoxic, e.g., aquifers, aquatic sediments and submerged soils, sludge digesters, and intestinal contents, and at aerobic sites with high carbon concentrations, molecular oxygen is more rapidly consumed than replenished (227). Within hydrocarbon-contaminated aquifers, anoxic contaminant plumes with distinct redox compartments are formed, and local microbial communities capable of using locally available electron donors and acceptors can perform biodegradation processes at different rates according to the particular redox parameters (226, 227). Recent findings also showed that the biodegradation of crude oil in deep subsurface petroleum reservoirs, an important alteration process that generates heavy oil with major economic consequences, is carried out by anaerobic microorganisms through mainly syntrophic hydrocarbon degradation coupled to hydrogenotrophic methanogenesis (2, 147, 175). Thus, anoxic conditions dominate in many natural habitats and contaminated sites, and the anaerobic catabolism of aromatic compounds by microorganisms becomes crucial for the biogeochemical cycles and for the sustainable development of the biosphere (110, 128, 149, 226, 227, 374). Moreover, due to the low chemical reactivity of aromatic compounds, their anaerobic biodegradation requires unusual biochemical reactions that are of great interest not only from a biochemical and evolutionary point of view but also for their potential biotechnological applications. Some examples are the novel enzymatic reactions involved in anaerobic benzoate ring reduction (biological Birch reduction) and the anaerobic carboxylation of phenol (biological Kolbe-Schmitt carboxylation), which were previously thought to exist only in the field of organic chemistry (116).
The mineralization of aromatic compounds by facultative or obligate anaerobic bacteria (and some archaea) can be coupled to anaerobic respiration with a variety of electron acceptors, e.g., nitrate, sulfate, iron(III), manganese(II), and selenate, with each one conserving different yields of energy. The greatest energy conservation is reached when nitrate is the final electron acceptor, followed by ferric ion. The energy conservation when sulfate is the electron acceptor is much more limited (110, 128, 153, 261, 320, 374). Fermentative bacteria can also use aromatic compounds, but usually, complete biodegradation becomes energetically feasible when accompanying methanogens or sulfate reducers use the metabolic end products, such as hydrogen, that are generated by the fermenters (106, 107, 168, 288, 289). Anoxygenic phototrophs avoid the energetic constraints of the heterotrophs since all the energy is derived from light, and therefore, the aromatic compounds are catabolized to generate building blocks (e.g., acetyl-coenzyme A [CoA]) used in biosynthetic reactions (128). Since the redox potential of the electron-accepting system in the anaerobic breakdown of aromatic compounds dictates the biochemical strategy that is applied for the degradation of such compounds, there is wide biochemical diversity among anaerobic aromatic degraders. Thus, the same aromatic compound can follow a more energy-demanding biochemical pathway in facultative anaerobes, such as many denitrifying bacteria, than in obligate anaerobes, e.g., sulfate reducers or fermenters, which show a poor bioenergetic balance. For instance, resorcinol and ethylbenzene are degraded via initial hydroxylation reactions in denitrifying bacteria, but they are reductively dearomatized and activated by the addition of fumarate, respectively, in sulfate reducers (149, 190, 320). On the other hand, aromatic compounds can participate in anaerobic metabolism by serving as electron acceptors rather than electron donors, generally with accompanying modifications of ring substituents that do not perturb the benzene nucleus itself (e.g., chlorinated aromatics as electron acceptors in dehalorespiration, reduction of the acrylate side chain of aromatic compounds, and reduction of nitroaromatics, etc.) (45, 88, 105, 108, 109, 128, 160, 169, 338).
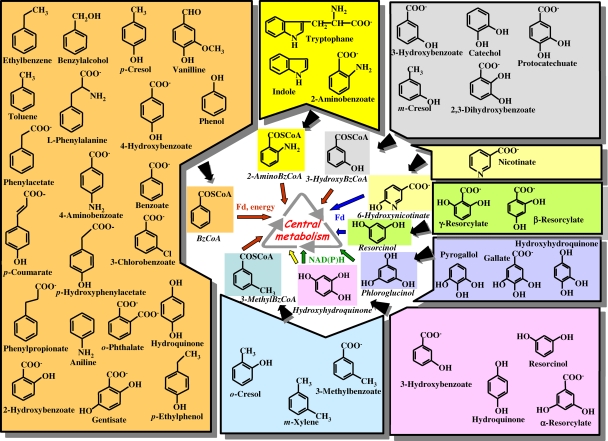
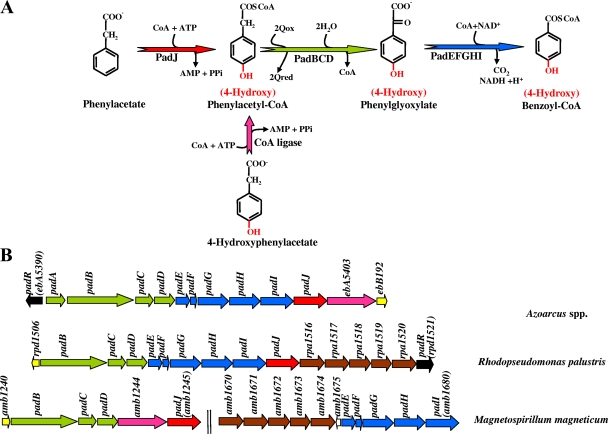
As indicated above, different peripheral pathways involved in the anaerobic activation of a wide variety of (monocyclic) aromatic compounds lead to a few central aromatic intermediates, e.g., resorcinol (1,3-dihydroxybenzene), phloroglucinol (1,3,5-trihydroxybenzene), hydroxyhydroquinone (HHQ) (1,2,4,-trihydroxybenzene), 6-hydroxynicotinate, hydroxybenzoyl-CoA, methylbenzoyl-CoA, aminobenzoyl-CoA, and benzoyl-CoA, with the latter being the most common and studied intermediate (Fig. 1) (4, 116, 128, 142, 151). In aerobic pathways, phenolic hydroxyl groups are introduced by oxygenases to activate the aromatic ring previous to the oxygenolytic ring cleavage. In contrast, in anaerobic catabolism, phenolic hydroxyl groups are often reductively removed, and the aromatic ring is also reduced, leading to alicyclic compounds. The biochemistry of some of these peripheral and central pathways for the anaerobic degradation of aromatic compounds has been studied to some extent, and several excellent reviews have been published (21, 30, 31, 33, 52, 110, 111, 116, 128, 142, 144, 149, 151, 153, 176, 320, 345, 374, 386). However, the genetic determinants of all these processes and the mechanisms involved in their regulation are much less studied. Recent advances in genome sequencing have led to the complete genetic information for five bacterial strains that are able to anaerobically degrade aromatic compounds using different electron acceptors and that belong to different subgroups of proteobacteria, i.e., two alphaproteobacteria, the phototroph Rhodopseudomonas palustris strain CGA009 (213) and denitrifying “Magnetospirillum magneticum” strain AMB-1 (236); a betaproteobacterium, denitrifying Azoarcus sp. strain EbN1 (proposed to be renamed “Aromatoleum aromaticum” strain EbN1) (293); and two obligate anaerobic deltaproteobacteria, the iron reducer Geobacter metallireducens GS-15 (48) and the fermenter Syntrophus aciditrophicus strain SB (237) (Table 1). This review focuses on the recent findings that standard molecular biology approaches together with the new high-throughput technologies (e.g., genome sequencing, transcriptomics, proteomics, and metagenomics) have provided for the genetics, regulation, ecology, and evolution of anaerobic aromatic degradation pathways.
FIG. 1.
The anaerobic catabolic funnel for monoaromatic compounds. A broad range of aromatic compounds funnel through a wide variety of peripheral pathways (black arrows) into a limited number of aromatic central intermediates, e.g., benzoyl-CoA (BzCoA), 2-aminobenzoyl-CoA (2-AminoBzCoA), 3-hydroxybenzoyl-CoA (3-HydroxyBzCoA), 3-methylbenzoyl-CoA (3-MethylBzCoA), 6-hydroxynicotinate, resorcinol, phloroglucinol, and HHQ, which are then dearomatized and channeled by the cognate central pathways (thin arrows) to the central metabolism of the cell. The dearomatization of the central intermediates can involve a ferredoxin (Fd) and energy (e.g., ATP) (red arrows), a ferredoxin (blue arrows), or NAD(P)H (green arrows) as electron donors. An oxidative dearomatization of HHQ has also been described (yellow arrow). It should be noted that the same aromatic compound can be degraded following different peripheral and central pathways depending on the particular redox potential of the final electron-accepting system in the host cell.
TABLE 1.
Comparison of some relevant features of completely sequenced anaerobic biodegraders
| Organism | Taxonomical classification | Habitat | Main electron acceptor(s) | Optimal growth temp (°C) | Cellular shape | Motility | Some aromatic carbon sources under anaerobic conditions | Chromosome size (bp) | Plasmid size (kb) | G+C content (%) | No. of predicted ORFs | No. of IS-like elements |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R. palustris CGA009a | Alphaproteobacteria | Aquatic | O2, phototrophic | 25-30 | Rod | Yes | p-Coumarate, cinnamate, ferulate, caffeate, 3-phenylpropionate, 5-phenylvalerate, mandelate, 4- hydroxybenzoate, 3-hydroxybenzoate, benzoate | 5,459,213 | 8.4 | 65 | 4,836 | 16 |
| M. magneticum AMB-1b | Alphaproteobacteria | Aquatic | O2 (microaerophilic), NO3− | 30 | Spiral | Yes | Benzoate, phenylacetate | 4,967,148 | 65.1 | 4,559 | 33 | |
| Azoarcus sp. strain EbN1c | Betaproteobacteria | Freshwater sediments | O2, NO3−, NO2− | 31 | Rod | No | Toluene, ethylbenzene, phenylacetate, anthranilate, phenol, p-cresol, p-ethylphenol, phenylalanine, benzyl alcohol, 3-hydroxybenzoate, 4-hydroxybenzoate, benzoate | 4,296,230 | 207/223 | 65.1 | 4,603 | 237 |
| G. metallireducens GS-15d | Deltaproteobacteria | Multiple | Fe(III), Mn(IV), NO3−, NO2−, Co(III), U(VI), Cr(VI) | 30 | Rod | Yes | Toluene, phenol, p-cresol, benzyl alcohol, benzaldehyde, 4-hydroxybenzoate, benzoate | 3,997,420 | 13.7 | 59.5 | 3,532 | |
| S. aciditrophicus SBe | Deltaproteobacteria | Multiple | Fermenting | 35 | Rod | No | Benzoate | 3,179,300 | 51.4 | 3,169 | 37 |
Data taken from data reported previously by Larimer et al. (213) and Harwood and Gibson (143) and from the R. palustris CGA009 genome project at the National Center for Biotechnology Information (NCBI).
Data taken from data reported previously by Matsunaga et al. (236) and from the M. magneticum AMB-1 genome project at the NCBI.
Data taken from data reported previously by Rabus et al. (293), Rabus (290), and Wöhlbrand et al. (379) and from the Azoarcus sp. strain EbN1 genome project at the NCBI.
Data taken from data reported previously by Butler et al. (48) and from the G. metallireducens GS-15 genome project at the NCBI.
Data taken from data reported previously by McInerney et al. (237) and from the S. aciditrophicus SB genome project at the NCBI.
GENE CLUSTERS ENCODING CENTRAL CATABOLIC PATHWAYS
As indicated in the Introduction, the general strategy of anaerobes is to convert, via different peripheral pathways, the multitude of different aromatic growth substrates into a few key intermediates, which are then substrates for the corresponding dearomatizing reductases. The major key intermediates formed during the anaerobic catabolism of aromatic compounds with a benzene-based structure (homocyclic aromatics) are benzoyl-CoA (and its 2-amino, 3-hydroxy, and 3-methyl derivatives), phloroglucinol, HHQ, and resorcinol (Fig. 1) (116, 128, 142, 151). The latter three compounds posses keto/enol tautomeries, which largely weaken their aromatic character, and they can be reduced (dearomatized) in exergonic reactions with common physiological reductants, i.e., NAD(P)H in the case of phloroglucinol and HHQ, and ferredoxin in case of resorcinol, that serve as electron donors for the corresponding reductases (30, 31, 151, 320). In contrast, due to the fully aromatic character of the benzene ring in benzoyl-CoA (and its methyl, hydroxyl, and amino derivatives), its reduction is mechanistically difficult to achieve, and it requires not only a low-potential electron donor, ferredoxin, but also a complex reductase system coupled to an input of energy (30, 31, 33, 116). A similar reductive strategy is thought to occur during the anaerobic dearomatization of the polycyclic intermediates, e.g., naphthoyl-CoA, generated during the anaerobic catabolism of polycyclic aromatic compounds (8, 374). Although the anaerobic degradation of heterocyclic aromatic compounds has been less studied than that of the homocyclic compounds, and it usually involves central intermediates of low aromaticity that are subject to a hydrolase-type ring cleavage reaction (21, 74, 111, 176, 178, 366), some intermediates, e.g., hydroxynicotinate (Fig. 1), require a reductive attack of the aromatic ring carried out by reductases that use ferredoxin as an electron donor but require no ATP hydrolysis (4, 111, 176).
In summary, according to the key intermediate that is formed from a particular aromatic substrate and the particular redox potential of the electron-accepting system that dictates the energy balance in the host cell (320), different central pathways that funnel the key intermediates into the central metabolism have evolved (Fig. 1).
Benzoate Catabolism: the Benzoyl-CoA Degradation Pathway
Benzoate has been routinely used as the model compound to study the major central pathway for the anaerobic degradation of aromatic compounds, i.e., the benzoyl-CoA degradation pathway. The anaerobic catabolism of benzoate via benzoyl-CoA has been studied at the molecular level in some facultative anaerobes, i.e., in the denitrifying bacteria Thauera aromatica, Azoarcus spp. (betaproteobacteria) (39, 142, 224), and Magnetospirillum spp. (alphaproteobacteria) (225, 335); in the photosynthetic bacterium R. palustris (alphaproteobacteria) (104); and in the strictly anaerobic Fe(III)-reducing (G. metallireducens) and fermentative (S. aciditrophicus) deltaproteobacteria (237, 377). In all these bacteria, benzoate degradation involves a one-step peripheral pathway that activates benzoate to benzoyl-CoA by the action of an ATP-dependent benzoate-CoA ligase (Fig. 2A). Benzoyl-CoA then becomes degraded to acetyl-CoA and CO2 following a series of reactions that constitute the benzoyl-CoA biodegradation pathway. This central pathway is arranged into two major metabolic blocks: (i) the upper benzoyl-CoA pathway that converts benzoyl-CoA into an aliphatic C7-dicarboxyl-CoA compound and (ii) the lower benzoyl-CoA pathway that converts the C7-dicarboxylic CoA ester to acetyl-CoA and CO2 (52) (Fig. 2A). The genes that encode equivalent enzymatic reactions of the benzoyl-CoA pathway have been named differently in most of the anaerobes studied so far, and these gene designations are compared and summarized in Table 2.
FIG. 2.
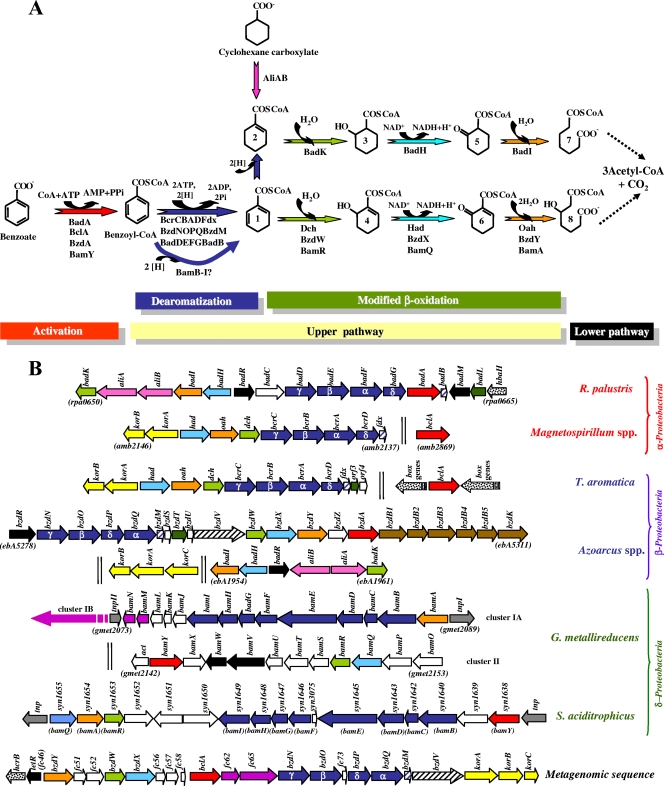
Pathway for the anaerobic catabolism of benzoate in anaerobic bacteria: the benzoyl-CoA biodegradation pathway. (A) Enzymatic reactions of the pathway. The cyclohexane carboxylate convergent pathway is also shown. The reaction steps are indicated at the bottom: I, activation of benzoate to benzoyl-CoA by benzoate-CoA ligase (red arrow); II, upper benzoyl-CoA pathway involving benzoyl-CoA dearomatization by benzoyl-CoA reductase (dark blue arrows) and modified β-oxidation via hydratation (green arrows), dehydrogenation (light blue arrows), and hydrolytic ring cleavage (orange arrows); III, lower benzoyl-CoA pathway (dotted arrows) that generates three molecules of acetyl-CoA and one CO2 (see also Fig. 3). The activation of cyclohexane carboxylate by the cognate CoA ligase (AliA) and its subsequent dehydrogenation by the cyclohexanecarboxyl-CoA dehydrogenase (AliB) are also shown (pink arrow). The metabolites are as follows: cyclohex-1,5-diene-1-carbonyl-CoA (1), cyclohex-1-ene-1-carbonyl-CoA (2), 2-hydroxycyclohexane-1-carbonyl-CoA (3), 6-hydroxycyclohex-1-ene-1-carbonyl-CoA (4), 2-ketocyclohexane-1-carbonyl-CoA (5), 6-ketocyclohex-1-ene-1-carbonyl-CoA (6), pimelyl-CoA (7), and 3-hydroxypimelyl-CoA (8). The enzymes are also indicated, and the names correspond to those of the corresponding genes. (B) Organization of the gene clusters involved in the anaerobic catabolism of benzoate in Rhodopseudomonas palustris CGA009 (GenBank accession number NC_005296), Magnetospirillum spp. (M. magneticum AMB-1 [accession number NC_007626], M. magnetotacticum MS-1 [accession number AAAP00000000], and Magnetospirillum sp. strain TS-6 [accession numbers AB167726 and AB243675]), Thauera aromatica (accession number AJ224959), Azoarcus spp. (Azoarcus sp. strain CIB [accession number AF515816], A. evansii [accession number AJ428529], and Azoarcus sp. strain EbN1 [accession number NC_006513]), Geobacter metallireducens GS-15 (accession number NC_007517), Syntrophus aciditrophicus SB (accession number NC_007759), and a metagenomic sequence (accession number CR931837). The names of the genes in the different organisms are also summarized in Table 2. Genes are represented by arrows: red, genes encoding benzoate-CoA ligases; dark blue, genes encoding the subunits of the benzoyl-CoA reductase; blue hatched, genes encoding ferredoxins associated with benzoyl-CoA reductases; yellow, KGOR-encoding genes; black hatched, genes encoding a putative NADPH:ferredoxin oxidoreductase; light green, genes encoding enoyl-CoA hydratases; light blue, genes encoding NAD-dependent hydroxyacyl-CoA dehydrogenases; orange, genes encoding oxoacyl-CoA ring cleavage hydrolases; black, regulatory genes; dark green, genes encoding putative acyl-transferases; brown, putative transport genes; pink, genes encoding cyclohexanecarboxylate-CoA ligase (aliA) and cyclohexanecarboxyl-CoA dehydrogenase (aliB); violet, putative lower-pathway genes; dotted, genes from other aromatic catabolic pathways; gray, transposase-encoding genes; white, genes of unknown function. Two vertical lines mean that the genes are not adjacent in the genome.
TABLE 2.
Genes encoding benzoate-CoA ligase and the upper benzoyl-CoA pathway in different anaerobes
| Enzyme | Gene
|
|||||
|---|---|---|---|---|---|---|
| Rhodopseudomonas palustris | Magnetospirillum spp. | Thauera aromatica | Azoarcus spp. | Geobacter metallireducens | Syntrophus aciditrophicus | |
| Benzoate-CoA ligase | badA | bclA | bclA | bzdA | bamY | bamY |
| BCRa α-subunit | badF | bcrA | bcrA | bzdQ | ||
| BCR β-subunit | badE | bcrB | bcrB | bzdO | ||
| BCR γ-subunit | badD | bcrC | bcrC | bzdN | ||
| BCR δ-subunit | badG | bcrD | bcrD | bzdP | ||
| Ferredoxin | badB | fdx | fdx | bzdM | ||
| BCRoab | bamBCDEFGHI? | bamBCDEFGHI? | ||||
| Enoyl-CoA hydratase | badK | dch | dch | bzdW | bamR | bamR |
| Hydroxyacyl-CoA dehydrogenase | badH | had | had | bzdX | bamQ | bamQ |
| Oxoacyl-CoA hydrolase | badI | oah | oah | bzdY | bamA | bamA |
BCR from facultative anaerobes.
BCR from obligate anaerobes.
Activation of benzoate.
As mentioned above, the anaerobic degradation of benzoate starts with a one-step peripheral pathway that involves the activation of benzoate to benzoyl-CoA via an ATP-dependent benzoate-CoA ligase that releases AMP and PPi (Fig. 2A). A CoA thioesterification reaction also occurs in many other anaerobic degradation pathways, and it is a general strategy of anaerobes not only for the activation of such compounds to be suitable substrates for further reduction reactions but also for accelerating the accumulation of these aromatic compounds inside the cells (116, 151, 364). Nevertheless, the use of a CoA transferase rather than a CoA ligase (ATP-consuming) reaction for substrate activation has already been reported, e.g., in the 3-hydroxybenzoate degradation pathway from the fermenting bacterium Sporotomaculum hydroxybenzoicum (259) or in the peripheral toluene degradation pathway (218) (see below).
The genes encoding benzoate-CoA ligases from phototrophic (badA in R. palustris) and denitrifying (bclA in T. aromatica and Magnetospirillum spp. and bzdA in Azoarcus spp.) bacteria have been described, and some of these gene products have been biochemically characterized (100, 224, 330). The substrate specificity of benzoate-CoA ligases can differ depending on the host strain, but it is common that these enzymes can recognize and activate halobenzoates (5, 123, 181, 224, 330), and in some cases, they are also responsible for the initial step in the anaerobic catabolism of benzoate derivatives such as 2-aminobenzoate (331). On the other hand, other aromatic/alicyclic acid-CoA ligases can also recognize benzoate as a substrate, and for instance, R. palustris cells synthesize at least three different enzymes, i.e., benzoate-CoA ligase (BadA), 4-hydroxybenzoate (4-HBA)-CoA ligase (HbaA), and cyclohexanecarboxylate-CoA ligase (AliA), that can catalyze the activation of benzoate to benzoyl-CoA during anaerobic growth with benzoate (100).
The aerobic degradation of benzoate in some facultative anaerobes also starts with the activation of this aromatic acid via a benzoate-CoA ligase (box pathway) that shows similarity to the anaerobic benzoate-CoA ligases (124, 125, 238). Interestingly, these types of aerobic hybrid pathways that are initiated through the formation of CoA thioesters (although they still make use of oxygen to introduce hydroxyl groups into the aromatic ring) have been considered to be an adaptation of some denitrifying facultative aerobes to fluctuating oxic/anoxic conditions, since both oxic and anoxic types of catabolism share the same initial aryl-CoA intermediates as substrates (116). The existence of a range of oxygen concentrations that can induce both the aerobic and anaerobic catabolic pathways might explain why some bacteria, such as T. aromatica (330) and Magnetospirillum strains (181), have evolved a single benzoate-CoA ligase gene whose product is shared by the aerobic and anaerobic benzoate degradation pathways.
Considering the strict energy constraints in strictly anaerobic bacteria, it is surprising that an ATP-consuming (AMP-forming) benzoate-CoA ligase is present in iron reducers (377) as well as in fermentative (9) and sulfate-reducing (278) bacteria (Fig. 2). Moreover, comparison analyses revealed that the AMP-forming benzoate-CoA ligase encoded by the bamY gene from the Fe(III)-reducing organism G. metallireducens shares significant amino acid sequence identity with benzoate-CoA ligases from facultative anaerobes (377). The genome of S. aciditrophicus contains four different genes (syn1638, syn2417, syn2896, and syn2898) that are predicted to encode AMP-forming ligases that could activate aromatic or alicyclic compounds (237). Out of these four genes, syn1638 is the only one that is located in a gene cluster (Sa2) that also contains genes which have been shown to be involved in the reaction steps after benzoyl-CoA reduction (206, 277, 279), and therefore, it is assumed to be the bamY ortholog (bamYSyn) in S. aciditrophicus (Fig. 2B). All these data suggest that the ATP-mediated activation to benzoyl-CoA is a general feature in the anaerobic catabolism of benzoate regardless of the redox potential of the electron-accepting system (116). Nevertheless, in some obligate anaerobes, such as the benzoate-fermenting bacterium Syntrophus gentianae, the hydrolysis of the PPi formed by benzoate-CoA ligase has been shown to be coupled to energy conservation by a membrane-bound proton-translocating pyrophosphatase, which allows the synthesis of one-third of one ATP and thus partially compensates for the two ATP molecules consumed in the activation reaction (327).
The genes encoding benzoate-CoA ligases are of biotechnological interest since they can be used for the heterologous overproduction of enzymes that generate CoA thioesters, which are an important class of activated intermediates in various biological pathways. In plants, benzoyl-CoA is reported to be the substrate in various enzymatic benzoylations for the biosynthesis of compounds such as cocaine, taxol, dianthramide B, benzoylated glucosinolate esters, or benzylbenzoate (22). In bacteria, benzoyl-CoA also serves as a starter unit for the biosynthesis of the polyketides enterocin and soraphen. Consistent with this, a gene, ecnN, encoding a biosynthetic benzoate-CoA ligase in “Streptomyces maritimus” has been characterized (380). Interestingly, the badA gene from R. palustris has already been used for the enzymatic synthesis of aromatic-CoA esters (22). On the other hand, the benzoate-CoA ligase genes can also be used for engineering gene expression systems that respond to benzoyl-CoA as an inducer molecule (14).
The upper benzoyl-CoA pathway.
The anaerobic degradation of benzoyl-CoA to yield an aliphatic C7-dicarboxyl-CoA derivative is called the upper benzoyl-CoA pathway, and it involves two major metabolic steps: (i) the dearomatization of benzoyl-CoA to cyclohex-(di)ene-carbonyl-CoA and (ii) a modified β-oxidation of the latter to form the C7-dicarboxylic CoA ester (Fig. 2A).
(i) Dearomatization.
The key step in the anaerobic degradation of benzoyl-CoA is the dearomatization of the benzene ring by benzoyl-CoA reductase (BCR), the only oxygen-sensitive enzyme within the benzoyl-CoA pathway. Benzoyl-CoA reduction seems to follow a Birch-like mechanism, that is, a sequential transfer of single electrons and protons at extremely low redox potentials (30, 31, 33). This reduction is greatly facilitated by the thioester group, which explains why the anaerobic pathways generally use CoA thioesters throughout (116). Whereas benzoyl-CoA is reduced to a cyclohexadienecarbonyl-CoA intermediate in T. aromatica, Azoarcus spp., G. metallireducens, and S. aciditrophicus, a further reduction of the dienoyl-CoA to a monoenoyl-CoA was reported for R. palustris (Fig. 2A) (30, 31, 52, 104, 129, 142, 274, 279).
So far, BCR has been isolated only from T. aromatica (BCRTa); it is an αβγδ heterotetramer (BcrABCD) that overcomes energetic limitations by using low-potential electron donor ferredoxin (Fdx) containing two [4Fe-4S]+1/+2 clusters and by coupling electron transfer to benzoyl-CoA to stoichiometric ATP hydrolysis (1 ATP/electron) in a manner analogous to, but mechanistically different from, the well-characterized dinitrogen reduction by nitrogenases (30-33, 35, 354). BCRTa is composed of two functionally different modules: (i) the electron activation module that is composed of the αδ subunits (BcrAD) and that harbors two ATP-binding sites and the electron entry [4Fe-4S]+1/+2 cluster and (ii) the benzoyl-CoA reduction module formed by the βγ subunits, which coordinate two further [4Fe-4S]+1/+2 clusters and bind a single benzoyl-CoA molecule (30, 31, 245). A two-subunit ferredoxin-reducing enzyme, T. aromatica 2-oxoglutarate:ferredoxin oxidoreductase (KGORTa), directly regenerates reduced ferredoxin in T. aromatica (33, 94). The bcrABCD and fdx genes, encoding BCRTa and its associated ferredoxin, respectively, are organized in a gene cluster that also includes the korAB genes, encoding the KGORTa enzyme (Fig. 2B) (39, 94). A similar gene cluster has been identified by genome in silico search and reverse transcription (RT)-PCR experiments using Magnetospirillum magnetotacticum MS-1 (225), Magnetospirillum sp. strain TS-6 (335), and M. magneticum AMB-1 (Fig. 2B).
The BCR from R. palustris (BCRRp) has been assayed using crude extracts, since for unknown reasons, no one has succeeded in obtaining an active benzoyl-CoA reductase preparation from this phototroph. Although two cyclohexadienecarboxylates have been extracted from R. palustris cells grown anaerobically on benzoate, thus indicating a two-electron reduction, a cyclohex-3-ene-1-carboxylate product was detected at a 20- to 40-times-higher concentration (129). The further two-electron reduction of the initial cyclohexadienecarbonyl-CoA product to the monoene carbonyl-CoA, which should be energetically easy to accomplish, could be catalyzed by its own BCRRp or by an additional enzyme that is yet unknown (30, 31, 129, 142, 274). The badF, badE, badD, and badG genes encode the αβγδ BCRRp heterotetramer, and these gene products, together with the badB-encoded ferredoxin, show a significant amino acid sequence identity (64 to 76%) with the corresponding bcrA, bcrB, bcrC, bcrD, and fdx gene products from T. aromatica and Magnetospirillum strains (Fig. 2B) (31, 39, 52, 104, 142, 225, 335).
For Azoarcus evansii strains and Azoarcus sp. strain CIB, a BCR (BCRAs) that reduces benzoyl-CoA to a cyclohexadienecarbonyl-CoA intermediate, like in T. aromatica, has been described (99, 224). However, the bzdQ, bzdO, bzdN, and bzdP gene products, corresponding to the αβγδ BCRAa subunits, respectively, and the bzdM-encoded ferredoxin differ significantly (22 to 43% identity) from their equivalent bad and bcr gene products (31, 52, 224, 290, 293). Furthermore, the system that regenerates the electron donor of BCRAs differs from that acting on BCRTa. Thus, the primary electron donor of BCRAs, the BzdM ferredoxin, is regenerated by the combined action of a three-subunit NADP-dependent 2-oxoglutarate:ferredoxin oxidoreductase (KGORAs) encoded by the korABC genes and an inducible NADPH:ferredoxin oxidoreductase (99) that might be encoded by the bzdV gene within the bzd cluster (Fig. 2B) (52, 224, 293).
Based on amino acid sequence comparison analyses, two types of ATP-dependent BCR enzymes that might share a common ancestor have been proposed, i.e., the bcr-type BCR present in T. aromatica, R. palustris, and Magnetospirillum strains and the bzd-type BCR present in Azoarcus strains (30, 343). Despite the possibility that BCRTa and BCRRp may have evolved from a common ancestor, these two enzymes might differ in their biochemical properties, which could account for the single and the two successive two-electron reduction steps of benzoyl-CoA, respectively (Fig. 2A) (30, 31, 52, 104, 128, 142, 274, 279). The absence of korAB and korABC orthologs in the genome of R. palustris (213) suggests that the BCR electron donor-regenerating system in this phototroph is different from that characterized for denitrifying bacteria. It is worth noting here that the badC gene located upstream of the BCR-encoding badDEFG genes (Fig. 2B) encodes a protein that resembles an NADPH:quinone oxidoreductase whose inactivation leads to the lack of growth of the R. palustris badC mutant in benzoate (102). Whether the BadC protein may be involved in the BCRRp electron donor-regenerating system or may participate in the second two-electron reduction of the dienoyl-CoA product is still unknown (142). Nevertheless, korAB orthologs have been found to be linked to the BCR genes in other R. palustris strains whose genomes have been recently sequenced (see below). This observation indicates that the electron donor-regenerating systems of BCRRp and BCRTa do not differ significantly or, more unlikely, that different BCRs (95 to 99% amino acid sequence identity) with different biochemical properties are present in different R. palustris strains. In any case, more studies need to be carried out to understand the diversity and the structural-functional relationships of BCR enzymes.
Since the reduction of benzoyl-CoA is an energetically expensive reaction, it has been argued that it should be different in phototrophs and nitrate-reducing organisms, which use more favorable electron acceptors, than in obligate anaerobes such as Fe(III)-reducing, sulfate-reducing, and fermenting bacteria, which can hardly afford a stoichiometric ATP-dependent benzene ring reduction (31, 128, 142, 328). As observed for sulfate-reducing bacteria like Desulfococcus multivorans, in which the specific induction of selenocysteine-containing proteins occurs when the cells use benzoate as the sole carbon source (278), benzoate degradation by G. metallireducens is dependent strictly on the presence of selenium, and molybdenum (or tungstate) also appears to stimulate growth on benzoate (377). Notably, no molybdenum- and/or selenocysteine-containing enzyme is involved in the benzoate metabolism of facultative anaerobes, and no BCR activity was observed in extracts from G. metallireducens cells grown in benzoate when using the established anaerobic radioactive assay and a wide variety of different electron donors (377). Accordingly, the completion of the genomes of G. metallireducens and S. aciditrophicus has confirmed the lack of the classical ATP-dependent BCR found in facultative anaerobes (48, 237). All these data strongly suggest that obligate anaerobes use different enzymes for benzene ring dearomatization than those reported for facultative anaerobes (116, 377).
Based on sequence comparison analyses, it was suggested that all or some of the benzoate-induced proteins and the corresponding bamBCDEFGHI genes within bam cluster IA from G. metallireducens would be responsible for the benzoyl-CoA dearomatization (Fig. 2B) (377). The bamDE and the bamCF gene products may form a complex that serves as the electron transfer machinery involved in benzene ring reduction. A predicted selenocysteine residue located near the N terminus of BamF would explain the observed selenium dependence during growth on aromatic compounds. The bamGHI gene products might be involved in complex formation with the BamCDEF components, transferring the electrons from NAD(P)H for enzymatic dearomatization. The bamB gene product shows similarities to tungsten- or molybdenum-containing aldehyde:ferredoxin oxidoreductases, and it is predicted to represent the active-site-containing component of the BCR, thus explaining the molybdenum-dependent stimulation of G. metallireducens growth when benzoate is used. According to these data, unlike the stoichiometric ATP hydrolysis used for benzene ring reduction in facultative anaerobes, a new type of BCR is proposed for strictly anaerobic bacteria where electron transfer might be driven by a membrane potential (377). The discovery of benzoate-induced paralogs of bamB and bamC outside of the bam cluster in G. metallireducens (48) adds further complexity to the benzoyl-CoA dearomatization step in this Fe(III)-reducing bacterium and stresses the necessity to characterize this enzymatic reaction further.
In the genome of S. aciditrophicus, two gene clusters have related synteny and high similarity (>50% identity at the amino acid level) to the bamB-bamI gene cluster of G. metallireducens (237). Each of these two gene clusters contains a gene predicted to encode an aldehyde oxidoreductase similar to BamB, which may function as the active-site-containing component of the benzoyl-CoA reductase complex (BamB to BamI) as well as genes likely to code for selenium-containing heterodisulfide reductase subunits and NADH:quinone oxidoreductase-like components (237). Interestingly, one of these gene clusters, Sa2, also contains the putative benzoate-CoA ligase gene (bamYSyn) and genes that have been shown to be involved in the reaction steps after benzoyl-CoA reduction (Fig. 2) (206, 279). The presence of these genes in the genome of S. aciditrophicus suggests that, like in G. metallireducens, the membrane potential rather than ATP hydrolysis may drive the electron transfer needed for benzoyl-CoA ring reduction by as-yet-unknown membrane components (237).
Thermodynamic considerations suggested that the energetically more favorable four-electron reduction of benzoyl-CoA forming cyclohexenecarbonyl-CoA should take place in strictly anaerobic bacteria (128, 328). However, recent works have revealed the existence of highly specific dienoyl-CoA hydratases in the benzoyl-CoA pathway from G. metallireducens, S. aciditrophicus, and D. multivorans (see below), strongly suggesting that dienoyl-CoA, the two-electron reduction product of benzoyl-CoA, is also the product of a BCR activity in obligate anaerobes (Fig. 2A) (279, 377). Therefore, although totally different BCR enzymes are present in facultative and obligate anaerobes, the benzoyl-CoA dearomatization product appears to be similar in all nonphotosynthetic anaerobic bacteria, thus showing a highly conserved reaction scheme (modified β-oxidation pathway) for its subsequent metabolism (Fig. 2A).
(ii) Modified β-oxidation.
After the formation of the cyclic (di)enoyl-CoA by BCR, a modified β-oxidation pathway involving the addition of water to a double bond (acyl-CoA hydratase), a dehydrogenation reaction (hydroxyacyl-CoA dehydrogenase), and a hydrolytic ring fission (oxoacyl-CoA hydrolase) generates an aliphatic C7-dicarboxyl-CoA compound. Two different β-oxidation sets, the Rhodopseudomonas type and the Thauera type, have been reported (Fig. 2A) (52, 142, 144). The Thauera-type β-oxidation pathway was first described for T. aromatica, and it uses the cyclic dienoyl-CoA product of the BCR as a substrate, generating 3-hydroxypimelyl-CoA as the final product (142, 210, 212). The Rhodopseudomonas-type β-oxidation pathway for R. palustris has been described, and it uses the cyclic monoenoyl-CoA product of the benzoyl-CoA reduction as a substrate, generating pimelyl-CoA (142, 273, 274). As a consequence, the enzymes responsible for the Thauera-type pathway show different substrate specificities compared with those acting on the Rhodopseudomonas-type pathway, and these differences also parallel two different types of catabolic genes. Whereas the Rhodopseudomonas-type genes (badK, badH, and badI) are found only in R. palustris CGA009 (and other recently sequenced Rhodopseudomonas strains) (104), orthologs of the Thauera-type genes (dch, had, and oah) (39) are found in other nitrate-reducing bacteria (e.g., Azoarcus and Magnetospirillum strains) (224, 225, 293, 335) and in Fe(III)-reducing bacteria (Geobacter strains) (48) (Fig. 2B). Although enzyme activities for the conversion of cyclohexene carboxyl-CoA to pimelyl-CoA were detected in cell extracts of the benzoate fermenter S. aciditrophicus (106), genes homologous to those encoding these proteins in R. palustris were not detected in the genome of S. aciditrophicus (237). In contrast, the genome of S. aciditrophicus contains three genes (syn1653, syn1654, syn1655) within cluster Sa2 (Fig. 2B) whose products show high sequence similarity to the hydratase (BamRSyn), dehydrogenase (BamQSyn), and ring-opening hydrolase (BamASyn) of the β-oxidation pathway from Azoarcus spp. (224, 237, 293). The cloning and heterologous expression of the genes encoding the presumed dienoyl-CoA hydratases from S. aciditrophicus (bamRSyn) and G. metallireducens (bamRGmet) confirmed that these genes code for highly specific dienoyl-CoA hydratases catalyzing the next step after benzoyl-CoA reduction in these two obligate anaerobes (279). The presence of benzoate-induced dienoyl-CoA hydratases has also been shown for cell extracts from sulfate-reducing bacteria such as D. multivorans (279). Recently, the characterization of the ring-opening hydrolases from G. metallireducens (BamA) and S. aciditrophicus (BamASyn) as well as the BamQ dehydrogenase from G. metallireducens further confirmed that the Thauera-type β-oxidation pathway is present in both facultative and obligate anaerobes (Fig. 2) (206). The observation that the Rhodopseudomonas type is restricted to photosynthetic anaerobes may reflect an evolutionary adaptation of phototrophs to carry out the catabolism of aromatic and alicyclic acids through the same β-oxidation pathway (see below).
Sequence comparison analyses of the dienoyl-CoA hydratases and ring-opening hydrolases suggested the existence of two phylogenetic groups, one including the enzymes from T. aromatica and G. metallireducens and the deduced gene products from Magnetospirillum strains and a second group that includes the enzymes from S. aciditrophicus, the deduced gene products from Azoarcus species, and the hydrolase from the sulfate-reducing bacterium D. multivorans (206, 279). This observation suggests that these two groups represent neither the phylogenetic relationship nor a common overall energy metabolism of the organisms (facultative versus obligate anaerobes) but rather points to the fact that they might have been acquired by the corresponding bacteria through horizontal gene transfer events (206, 279).
The lower benzoyl-CoA pathway.
The further degradation of the aliphatic C7-dicarboxyl-CoA derivative generates three acetyl-CoAs and CO2 (Fig. 3) (118, 140). Usually, bacteria contain many genes that might participate in the metabolism of the dicarboxyl-CoA intermediates formed during the anaerobic metabolism of aromatic (and alicyclic) acids as well as in the catabolism of dicarboxylic acids.
FIG. 3.
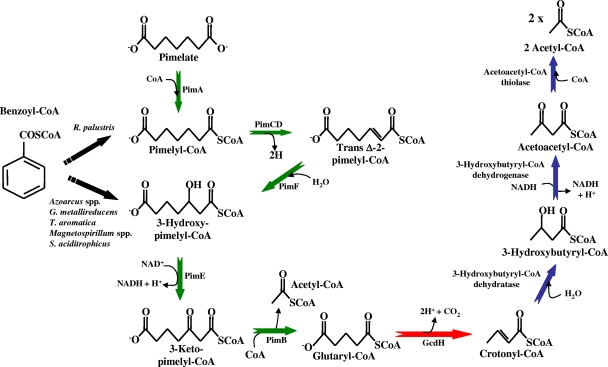
Lower benzoyl-CoA pathway. Benzoyl-CoA becomes converted into pimelyl-CoA (R. palustris) or 3-hydroxypimelyl-CoA (other bacteria) via the central benzoyl-CoA pathway (Fig. 2). Pimelate is another carbon source that funnels into this lower pathway. The C7-dicarboxyl-CoA compounds are further degraded into two molecules of acetyl-CoA and one CO2 through a series of reactions that involve a dicarboxylic acid β-oxidation pathway (green arrows), a glutaryl-CoA dehydrogenase (red arrow), and a short-chain fatty acid β-oxidation pathway (blue arrows). The enzymes indicated are as follows: PimA, acyl-CoA ligase; PimCD, flavin-containing acyl-CoA dehydrogenase; PimF, enoyl-CoA hydratase; PimE, hydroxyacyl-CoA dehydrogenase; PimB, acyl-CoA acetyltransferase (β-ketothiolase); GcdH, glutaryl-CoA dehydrogenase.
The pimFABCDE genes were shown to constitute an operon specifically induced when R. palustris cells grow anaerobically in benzoate (or pimelate), and they were predicted to encode all the enzymes that would be required for the β-oxidation of odd-chain dicarboxylic acids to glutaryl-CoA (Fig. 3) (140, 213, 271, 362). The Pim proteins are not the only R. palustris enzymes that can catalyze dicarboxylic acid degradation, and it is likely that they act in combination with other putative β-oxidation enzymes, some of which have increased levels of expression when the cells are grown in benzoate (362). In the genome of Azoarcus sp. strain EbN1, multiple clusters encoding putative dicarboxylic acid β-oxidation pathways have also been identified by in silico analysis (293).
In the genome of G. metallireducens, there are also multiple clusters encoding putative dicarboxylic acid β-oxidation pathways (48). In G. metallireducens, benzoate-induced gene cluster IB (Fig. 2B) has been proposed to be involved in the lower pathway for anaerobic benzoate degradation. The gene products of cluster IB include acyl-CoA/3-hydroxyacyl-CoA dehydrogenases (acd), enoyl-CoA hydratases (ech), and thiolases (act). Further benzoate-induced genes of cluster IB were annotated as a thioesterase (the), a sodium-dependent symporter (ssy), and some putative regulatory proteins (tre and rbsU) (377). Nevertheless, the gene with the greatest increase in transcript levels during the growth of G. metallireducens in benzoate is that encoding a 50-fold-induced Act thiolase (bamN) (48). This gene together with the bamM gene, which might encode glutaryl-CoA dehydrogenase (see below), do not belong to cluster IB, although they are located in its close vicinity at the left end of cluster IA (Fig. 2B) (377). Interestingly, the lower pathway in G. metallireducens might also involve electron-transferring flavoproteins (etfAB gene products) whose quinone oxidoreductases appear to be replaced by a membrane-bound 4Fe/4S oxidoreductase (oxr gene product), and they are induced in cluster IB when cells grow in benzoate (48, 377). The participation of these enzymes in β-oxidation reactions as electron acceptors of acyl-CoA dehydrogenases, with the latter introducing a double bond in thiol esters of saturated fatty acids, has been postulated (377). All these data suggest that the lower benzoyl-CoA pathway in strict anaerobes can involve enzyme systems more complex than those found in facultative anaerobes.
In most organisms, the oxidation and decarboxylation of glutaryl-CoA is catalyzed by a bifunctional glutaryl-CoA dehydrogenase that forms glutaconyl-CoA as an enzyme-bound reaction intermediate (115, 134). A glutaryl-CoA dehydrogenase was significantly produced in crude extracts of bacteria growing anaerobically in aromatic compounds (and cyclohexane carboxylate), which suggests that glutaryl-CoA was indeed an intermediate formed during the catabolism of these compounds via the pimelyl-CoA/3-hydroxypimelyl-CoA β-oxidation pathway (28, 106, 118, 140, 141, 153, 328). Recently, the gcdH gene, encoding the bifunctional glutaryl-CoA dehydrogenase enzyme, has been identified and characterized for Azoarcus sp. strain CIB (29). In contrast to the redundancy observed for the genes encoding the dicarboxylic acid β-oxidation pathway, the gcdH gene is usually present as a single chromosomal copy. The disruption of gcdH impaired the anaerobic growth of Azoarcus sp. strain CIB in benzoate and in other aromatic and alicyclic compounds as well as in dicarboxylic acids of odd chain length, such as pimelate and glutarate. This finding indicates that the gcdH gene is the only one that encodes a glutaryl-CoA dehydrogenase in Azoarcus sp. strain CIB (29).
In some organisms of limited energetic budget, such as fermenting bacteria, glutaryl-CoA is oxidized by a NAD-dependent glutaryl-CoA dehydrogenase and then decarboxylated by the action of a membrane-bound multicomponent glutaconyl-CoA decarboxylase, which is sodium dependent and couples decarboxylation with the translocation of a sodium ion across the membrane, leading to ATP synthesis (89). The conversion of glutaryl-CoA to crotonyl-CoA with the formation of ATP can be regarded as an additional means of energy conservation imposed by the strict energy constraints of syntrophic metabolism (107, 328). In silico analysis of the S. aciditrophicus genome revealed the existence of a gene cluster (syn479 to syn481) likely encoding a glutaryl-CoA dehydrogenase with low similarity to the classical GcdH enzymes and two products that are similar to two of the four subunits of the glutaconyl-CoA decarboxylase from Acidaminococcus fermentans (44). Nevertheless, whether these genes are involved in glutaryl-CoA metabolism in S. aciditrophicus requires further experimental confirmation.
The enzyme activities responsible for crotonyl-CoA metabolism to acetyl-CoA, i.e., 3-hydroxybutyryl-CoA dehydratase, 3-hydroxybutyryl-CoA dehydrogenase, and acetoacetyl-CoA thiolase (Fig. 3), have been detected in several bacteria (10, 106, 107, 118, 328), and genes predicted to encode these enzymes have been proposed after the in silico analysis of the genomes of some anaerobic biodegraders (140, 290).
Comparative genetics among benzoyl-CoA gene clusters.
Most of the genes involved, or proposed to be involved, in the anaerobic catabolism of benzoate to the aliphatic dicarboxyl-CoA derivative, such as the bad genes from R. palustris, the bcr genes from T. aromatica and Magnetospirillum strains, the bzd genes from Azoarcus strains and S. aciditrophicus, and the bam genes from G. metallireducens, are arranged in a single large cluster (Fig. 2B). However, in some organisms, such as T. aromatica and Magnetospirillum strains, the benzoate-CoA ligase gene is not clustered with the benzoyl-CoA pathway genes (Fig. 2B). In these bacteria, the aerobic degradation of benzoate also starts with the activation of this aromatic acid via a benzoate-CoA ligase (box pathway) that shows similarity to benzoate-CoA ligases involved in anaerobic degradation. Whereas in Azoarcus strains, there are two different benzoate-CoA ligases, one (bzdA) located in the bzd anaerobic cluster (Fig. 2B) and the other (bclA) located within the box cluster for the aerobic degradation of benzoate (124, 125, 224, 293), T. aromatica and Magnetospirillum strains use the same benzoate-CoA ligase (bclA) for the aerobic and anaerobic pathways (181, 330). While the bclA gene from T. aromatica is located in the box cluster for aerobic benzoate degradation (330), the bclA gene in Magnetospirillum strains is located within a gene cluster, which also encodes an ABC transporter, that is not linked to the bcr and box clusters (181). In contrast to what has been observed for the benzoate-CoA ligase-encoding genes, the kor genes, encoding the BCR electron donor-regenerating system, are physically associated with the benzoyl-CoA gene cluster in T. aromatica and Magnetospirillum strains but not in Azoarcus strains (39, 225, 293) (Fig. 2B).
Since the expression of the genes encoding aromatic catabolic pathways is usually induced by the substrate and/or an intermediate of the pathway, regulatory genes are frequently present in the corresponding catabolic clusters (53, 86, 352). Thus, in the bzd and bad clusters from Azoarcus and Rhodopseudomonas strains, the bzdR gene and the badR and badM genes (Fig. 2B), respectively, encode the corresponding specific transcriptional regulators (see below) (14, 102, 276). Whereas in the bam cluster from G. metallireducens, some regulatory genes, such as the putative two-component regulatory system encoded by the bamVW genes (Fig. 2B), have been proposed (377), in the bcr clusters from T. aromatica and Magnetospirillum strains and in the proposed benzoyl-CoA cluster from S. aciditrophicus, a gene encoding a typical transcriptional regulator has not been found (Fig. 2B). Remarkably, the benzoyl-CoA gene clusters from different anaerobes usually contain some genes of unknown function, some of which, e.g., bzdT in Azoarcus spp., badL in R. palustris, and orf3 in T. aromatica, are conserved and might encode acyl-transferases (Fig. 2B). Although further work needs to be carried out to unravel the function of these orphan genes, the fact that they are present in different bacteria suggests that they may also play an important role, e.g., posttranscriptional regulation, in the anaerobic catabolism of benzoyl-CoA.
Transport genes are also usually present in aromatic catabolic clusters (269). The uptake of benzoate in aerobic bacteria has been suggested to proceed via a proton-symporter major facilitator superfamily (MFS) transporter (BenK) (67). Although nothing is known about benzoate uptake in anaerobic degraders, at the 3′ end of the bzd gene cluster from Azoarcus spp., there are five genes (bzdB1, bzdB2, bzdB3, bzdB4, and bzdB5) encoding a putative ABC transporter of the branched-chain amino acid uptake family and an additional gene, bzdK, encoding a putative MFS transporter, that are likely involved in an efficient uptake of benzoate (Fig. 2B) (293; J. F. Juárez, unpublished data). The hbaEFGH genes from R. palustris (Fig. 2B) may also encode a putative ABC transporter of benzoate/4-HBA (104). Since the bcr clusters from T. aromatica and Magnetospirillum strains are lacking typical regulatory and transport genes, it is likely that these regulatory and transport elements are located in another region of the genome, for instance, linked to the corresponding bclA genes.
A gene cluster that is likely to be involved in the anaerobic degradation of benzoate (and other aromatic compounds) has been identified in a metagenomic library from a microbial mat of the Black Sea (201). The 79-kb FC1 contig contained all genes required for the central benzoyl-CoA pathway, including those encoding the benzoate-CoA ligase (bclA), the four subunits of Azoarcus-type BCR (bzdNOPQ), ferredoxin (bzdM), and the regenerating system (korABC and bzdV), and the genes involved in the modified β-oxidation pathway (bzdWXY) that generates 3-hydroxypimelyl-CoA. Some genes likely encoding a thiolase (fc62) and a hydroxyacyl-CoA dehydrogenase (fc65), which might participate in the lower pathway, are also present (Fig. 2B). Although the phylogenetic affiliation of the source organism remains unclear, the presence of an anaerobic benzoate degradation pathway that resembles that of denitrifying bacteria points to the fact that a member of the latter group could be the likely host organism (201).
Interestingly, in R. palustris, the bad genes are clustered with the ali genes (Fig. 2B), which are involved in the anaerobic degradation of cyclohexane carboxylate (104), and all of them are induced in cells cultivated anaerobically on benzoate (267, 362). The aliA and aliB genes encode the CoA ligase that activates cyclohexane carboxylate to cyclohexanecarboxyl-CoA and the dehydrogenase responsible for the conversion of the latter to cyclohexenecarbonyl-CoA, respectively (Fig. 2) (104, 207, 274). Since cyclohexenecarbonyl-CoA is also the product of the benzoyl-CoA reduction, the metabolic linkage between the pathways for the anaerobic catabolism of benzoate and cyclohexane carboxylate may account for the genomic linkage of the bad and ali genes in R. palustris. Moreover, it can be argued that BCRRp and the Rhodopseudomonas-type β-oxidation pathway arise as an evolutionary adaptation to channel, through the same catabolic pathway, the cyclohexenecarbonyl-CoA generated in the anaerobic degradation of both cyclohexane carboxylate and benzoate (274). In contrast, some denitrifying bacteria, such as Azoarcus sp. strain EbN1, contain an additional gene cluster harboring aliAB-badHIK orthologs but lack the badDEFG orthologs that encode BCRRp (Fig. 2B) (293), which suggests that this bacterium might degrade cyclohexane carboxylate through a Rhodopseudomonas-type β-oxidation pathway that generates pimelyl-CoA and that differs from the benzoyl-CoA pathway (bzd gene cluster) used for anaerobic benzoate catabolism.
The analysis of the aromatic catabolic clusters from different R. palustris strains whose genomes have recently been sequenced, e.g., strains BisB5 (GenBank accession number NC_007958), BisB18 (accession number NC_007925), BisA53 (accession number NC_008435), and HaA2 (accession number NC_007778), revealed that the ali-bad clustering is also conserved in all these strains. However, the gene arrangements of the ali-bad cluster differ from one strain to another. Thus, strain BisA53is the only one that contains a gcdH ortholog (encodes the glutaryl-CoA dehydrogenase from the lower pathway) located downstream of the badK gene. In R. palustris GCA009, the gcdH ortholog (RPA1094) is not linked to the bad genes (140). On the other hand, the hbaEFGH transport genes that are located between the bad and hbaABCD genes (the latter encoding the peripheral 4-HBA pathway) (see below) are lacking in strain BisB5. As shown above, BCRRp belongs to the Thauera type rather than to the Azoarcus type, and therefore, a KorAB-dependent electron donor-regenerating system should be predicted to exist in R. palustris strains. Consistent with this, although korAB orthologs were not found in the genome of the well-studied strain CGA009, they could be found downstream of the badB gene in strains BisB5 and BisB18. In summary, these genomic analyses reveal, therefore, that the continuous effort to sequence different isolates of a particular bacterial species can bring a more detailed reconstruction of the evolutionary history of the genes.
In G. metallireducens, the genes encoding the benzoyl-CoA pathway (bam genes) are arranged into two clusters (clusters IA and II) comprising 44 open reading frames (ORFs) and located ca. 60 kb apart in the chromosome (Fig. 2B) (48, 377). RT experiments and proteomics (377) as well as whole-genome microarray analyses (48) confirmed that the expression of these genes is specifically induced in the presence of benzoate (and 4-HBA) and indicated that adjacent genes in the same orientation might form a transcriptional unit (377). These two clusters are part of a 300-kb catabolic island that also includes the genes responsible for other aromatic catabolic pathways (see below) and the lower benzoyl-CoA pathway (cluster IB) (Fig. 2B) (48, 377).
In general, the genes encoding the benzoyl-CoA degradation pathway in G. metallireducens lack synteny with the genes of benzoyl-CoA clusters from facultative anaerobes (Fig. 2B). Thus, a global analysis of all gene clusters reported so far in facultative anaerobes reveals that the genes involved in the modified β-oxidation pathway are adjacent or in close proximity to each other, and they are associated with BCR-encoding genes (Fig. 2B). In contrast, in G. metallireducens, the bamA gene (cluster IA) is not linked to the bamQ and bamR genes (cluster II) (Fig. 2B). As genes encoding putative transposases flank cluster IA, one could argue that a transposition event separated the bam genes into two clusters, producing cluster IA, which became linked to genes involved in the lower pathway (cluster IB) (Fig. 2B). Another feature of the benzoyl-CoA pathway from G. metallireducens is the duplication of some genes, such as bamA and the bamB/bamC genes, which were found to have paralogs in two chromosomal regions outside of the aromatic catabolic island. Like the bamA and bamB/bamC genes, the bamA2 and bamB2/bamC2 paralogs show large increases in levels of expression during the growth of G. metallireducens in benzoate (48).
Two gene clusters in the S. aciditrophicus genome (Sa1 and Sa2) contain a putative aromatic acid-CoA ligase gene, and they have related synteny and high similarity (>50% identity at the amino acid level) to the bamB-bamI gene cluster in G. metallireducens (237). The location of the cluster Sa2 in the close vicinity of the bzdXYWSyn (bamQARSyn) genes strongly suggests that this extended cluster may encode the benzoyl-CoA degradation pathway in this bacterium (Fig. 2B). Interestingly, cluster Sa2 (including the bzdXYWSyn genes) is flanked by transposases (Fig. 2B), which suggests its acquisition in the S. aciditrophicus genome by horizontal gene transfer events.
Although most benzoyl-CoA gene clusters are chromosomally encoded, there is at least one report of such a gene cluster located in a transmissible plasmid. Thus, in T. aromatica strain PN-1 (formerly Alcaligenes xylosoxidans strain PN-1), the genes involved in the anaerobic degradation of benzoate were reported to be encoded in a small plasmid, pCBI (17.4 kb), that conferred the ability to catabolize benzoate anaerobically to heterologous hosts such as Pseudomonas aeruginosa and Pseudomonas stutzeri strains (27). The location of the anaerobic catabolic clusters in mobile genetic elements, such as plasmids, or in catabolic islands (as in G. metallireducens) may facilitate the adaptation of bacteria to use aromatic compounds as carbon sources and stresses the role of horizontal gene transfer in the acquisition and evolution of such aromatic catabolic pathways (358).
3-Hydroxybenzoate Catabolism
Hydroxybenzoic acids are formed during the degradation of other phenolic compounds; e.g., m-cresol is oxidized to 3-hydroxybenzoate via the addition of fumarate to the methyl group (258). Whereas 2-hydroxybenzoate (salicylate), 4-HBA, and some dihydroxybenzoates such as 2,5-dihydroxybenzoate (gentisate) have been suggested to become dehydroxylated and channeled to the central benzoyl-CoA pathway in some bacteria (37, 135), the anaerobic catabolism of 3-hydroxybenzoate follows a devoted central route in some anaerobes (Fig. 1).
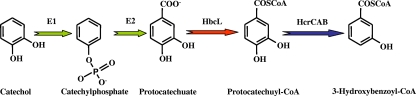
The anaerobic catabolism of 3-hydroxybenzoate has been studied in T. aromatica (211). The capacity to metabolize 3-hydroxybenzoate is induced only when T. aromatica cells are grown in this substrate. A relatively specific and substrate-induced CoA ligase (HbcL, as named previously by Rabus et al.) (293) catalyzes the first committed step, 3-hydroxybenzoyl-CoA formation (Fig. 4). Sequence comparison revealed that the 3-hydroxybenzoate-CoA ligase shows the highest similarity with 4-HBA-CoA ligase (HbaA) from R. palustris (see below) (211). However, 4-HBA-CoA ligase from T. aromatica does not act on 3-hydroxybenzoate, and benzoate-CoA ligase (BclA) does not act on either of the two hydroxy analogs, which could explain why cells grown with benzoate or 4-HBA are not adapted to grow on 3-hydroxybenzoate (211). The hbcL gene corresponds to orf5 of a gene cluster that contains at least six genes, orf1 to orf6, some of which were shown to be induced by 3-hydroxybenzoate (Fig. 4) (211). The hbcL gene from T. aromatica has been cloned and expressed in Escherichia coli cells, and the purified HbcL protein was shown to act not only on 3-hydroxybenzoate but also on protocatechuate, 4-HBA, benzoate, and gallate (3,4,5-trihydroxybenzoate). Thus, HbcL appears to be a promiscuous aromatic acid-CoA ligase (90).
FIG. 4.
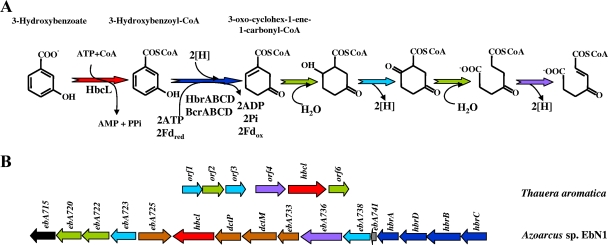
Proposed pathway for anaerobic catabolism of 3-hydroxybenzoate in T. aromatica and Azoarcus sp. strain EbN1. (A) Enzymatic reactions of the 3-hydroxybenzoate catabolic pathway. Enzymes are 3-hydroxybenzoate-CoA ligase (HbcL) and 3-hydroxybenzoyl-CoA reductase (putative HbrABCD in Azoarcus sp. strain EbN1 and BcrABCD in T. aromatica). Further reactions of the pathway are predicted solely from the presence of the corresponding genes in the cluster (the color code of the reactions corresponds to that of the genes in B) (211, 293). (B) Organization of the gene clusters involved in the anaerobic catabolism of 3-hydroxybenzoate in T. aromatica (GenBank accession number AJ278289) (211) and Azoarcus sp. strain EbN1 (accession number NC_006513) (293). Genes are represented by arrows, and their (predicted) functions are indicated by colors as follows: red, 3-hydroxybenzoate-CoA ligases; dark blue, subunits of 3-hydroxybenzoyl-CoA reductase; green, enoyl-CoA hydratases; violet, acyl-CoA dehydrogenases; light blue, hydroxyacyl-CoA dehydrogenases; brown, transporters; gray, gene encoding a BzdT-like protein; black, TetR-like transcriptional regulator.
In T. aromatica, 3-hydroxybenzoyl-CoA is reduced to a cyclic dienoyl-CoA in a two-electron step, a reaction which is coupled to the hydrolysis of two ATP molecules (Fig. 4) Although the reduction of 3-hydroxybenzoyl-CoA can be carried out by the purified BCRTa enzyme, the presence of closely related isoenzymes for benzoyl-CoA and 3-hydroxybenzoyl-CoA reduction in T. aromatica cannot be completely ruled out (211). Although it remains to be shown which isomeric form of the dienoyl-CoA is formed, the Dch enzyme of the benzoyl-CoA pathway cannot recognize the ring reduction product of 3-hydroxybenzoyl-CoA (211). Therefore, it is tempting to speculate that the 3-hydroxybenzoate-induced gene cluster might be responsible for the further catabolism of the 3-hydroxybenzoyl-CoA reduction product. Putative enoyl-CoA hydratases (Orf2 and Orf6), hydroxyacyl-CoA dehydrogenases (Orf1 and Orf3), and acyl-CoA dehydrogenase (Orf4) could be involved in modified β-oxidation (293). The involvement of BCRTa in the 3-hydroxybenzoate degradation pathway would imply that the overall 3-hydroxybenzoate pathway combines the reductase (BCRTa) and the electron donor ferredoxin (Fdx) of the general benzoyl-CoA pathway for ring reduction with β-oxidation-like enzymes that are specific for the 3-hydroxybenzoate pathway (211).
In the genome of Azoarcus sp. strain EbN1, there is a gene cluster that contains orthologs of the 3-hydroxybenzoate-induced genes from T. aromatica, also including two putative 3-hydroxybenzoate uptake systems (293) (Fig. 4). Since most of the proteins encoded by this gene cluster have been shown to be specifically induced when Azoarcus sp. strain EbN1 grows anaerobically in 3-hydroxybenzoate, it is assumed that they are indeed involved in the anaerobic degradation of 3-hydroxybenzoate (378). Interestingly, this 3-hydroxybenzoate gene cluster contains paralogs (hbrABCD genes) of the bzdNOPQ genes that code for the BCR, and therefore, they have been suggested to encode a specific 3-hydroxybenzoyl-CoA reductase in Azoarcus sp. strain EbN1 (Fig. 4). Thus, the in silico analysis of the Azoarcus sp. strain EbN1 genome (293) together with our preliminary data on the genome of Azoarcus sp. strain CIB strongly suggest that the 3-hydroxybenzoate degradation pathway in Azoarcus strains involves a whole set of genes responsible for the activation, dearomatization, and ring cleavage of 3-hydroxybenzoyl-CoA that differs from those encoding the regular benzoyl-CoA pathway. Nevertheless, the genes encoding the ferredoxin that transfers the electrons to the reductase as well as those involved in the reactivation of such ferredoxin, are not present in the 3-hydroxybenzoate degradation cluster in Azoarcus. Consistent with this finding, it was shown that an insertional disruption of the bzd gene cluster, encoding the central benzoyl-CoA pathway in Azoarcus sp. strain CIB, leads to a lack of anaerobic growth of the mutant strain on 3-hydroxybenzoate (224), suggesting that the BzdM ferredoxin and some other bzd gene products, such as the putative BzdV ferredoxin oxidoreductase, could be also shared by the 3-hydroxybenzoate degradation pathway.
The aliphatic CoA-derivative generated in the 3-hydroxybenzoyl-CoA degradation pathway is then subjected to β-oxidation reactions that should lead to the formation of glutaryl-CoA. Accordingly, cells grown in 3-hydroxybenzoate showed glutaryl-CoA dehydrogenase activity (150), and Azoarcus sp. strain CIBdgcdH, lacking a functional gcdH gene, was unable to grow anaerobically in 3-hydroxybenzoate as the sole carbon source (29).
In fermenting bacteria such as S. hydroxybenzoicum, 3-hydroxybenzoate was proposed to be degraded by the reductive elimination of the hydroxyl group via benzoyl-CoA as a central intermediate. As the first step in the degradation pathway, 3-hydroxybenzoate was activated to 3-hydroxybenzoyl-CoA in a CoA transferase-mediated reaction with acetyl-CoA or butyryl-CoA as the CoA donor. The use of a CoA transferase rather than a CoA ligase (ATP-consuming) reaction for substrate activation has important energetic implications for this fermenting bacterium and constitutes an important exception to the general principle of the activation of aromatic acid compounds by CoA ligases (259). A third pathway for anaerobic 3-hydroxybenzoate degradation that involves hydroxylation reactions to form HHQ has been reported for the denitrifying bacterium strain BoNHB (320). However, the genes involved in these two alternative 3-hydroxybenzoate degradation pathways have not been reported so far.
3-Methylbenzoate Catabolism
The anaerobic catabolism of 3-methylbenzoate involves its activation to 3-methylbenzoyl-CoA. This benzoyl-CoA analog can also be formed in the anaerobic degradation of m-xylene (Fig. 1) (199). o-Cresol is also metabolized in Azoarcus buckelii strain U120 via 3-methylbenzoyl-CoA as a central intermediate. A carboxylation similar to that reported for phenol or catechol degradation (see below) may occur with o-cresol, which is converted to 4-hydroxy-3-methylbenzoate. This intermediate is activated to its CoA thioester and reductively dehydroxylated to 3-methylbenzoyl-CoA (308). Further metabolism of 3-methylbenzoyl-CoA most likely involves ATP-driven ring reduction by a BCR that is different from that involved in the reduction of benzoyl-CoA. The modified β-oxidation of the alicyclic-CoA compound might also be encoded by a set of genes that are different from those involved in the benzoyl-CoA pathway (Juárez, unpublished). The methyl substituent of the aliphatic product generated after the β-oxidation step may require subsequent steps catalyzed by enzymes that are not required for the catabolism of the 3-hydroxypimelyl-CoA generated in the benzoyl-CoA pathway. Thus, an Azoarcus sp. strain CIB mutant harboring a disrupted gcdH gene is still able to use m-xylene anaerobically (29), which suggests that a devoted methylglutaryl-CoA dehydrogenase different from the glutaryl-CoA dehydrogenase encoded by the gcdH gene is involved in the lower 3-methylbenzoyl-CoA pathway.
2-Aminobenzoate (Anthranilate) Catabolism: a Devoted Central Pathway?
Nitro- and amino-aromatic compounds are produced in large amounts by chemical industries, e.g., trinitrotoluene. Frequently, these compounds can be used as electron acceptors for respiration with the subsequent reduction and removal of the nitro substituents (108, 109). However, some nitrogen-containing xenobiotics are finally channeled to single-amino-aromatic compounds such as anthranilate (2-aminobenzoate). Anthranilate also plays an important role in the synthesis and degradation of many N-heterocyclic aromatic compounds such as tryptophan, indole, indoleacetic acid, and derived compounds (Fig. 1) (21, 111, 331). In A. evansii, the aerobic degradation of 2-aminobenzoate involves the initial activation to 2-aminobenzoyl-CoA carried out by specific aerobic CoA ligases. 2-Aminobenzoyl-CoA is subsequently hydroxylated and reduced to a nonaromatic product by the flavoenzyme 2-aminobenzoyl-CoA monooxygenase/reductase (331). Whereas two gene clusters responsible for the hybrid pathway for the aerobic degradation of 2-aminobenzoate have been characterized for A. evansii (331), most of the genes involved in the anaerobic 2-aminobenzoate degradation pathway still remain unknown.
The anaerobic catabolism of 2-aminobenzoate also starts with its activation to 2-aminobenzoyl-CoA. In T. aromatica, a single benzoate-CoA ligase (BclA) is used for the anaerobic (and aerobic) activation of benzoate and 2-aminobenzoate (330). In contrast, in Azoarcus sp. strain EbN1, the anaerobic benzoate-CoA ligase (BzdA) is 4.5-fold decreased in abundance in cells grown with 2-aminobenzoate. However, the HbcL enzyme, which was predicted to be specific for 3-hydroxybenzoate activation (see above), revealed a marked increase in abundance (17.2-fold), suggesting that this enzyme could be involved in the first step of the anaerobic 2-aminobenzoate pathway in Azoarcus sp. strain EbN1 (378). In A. evansii, however, cells grown anaerobically in 2-aminobenzoate show three different CoA ligases: the anaerobic BzdA enzyme (which also shows some activity with 2-aminobenzoate), an anaerobic 2-aminobenzoate-CoA ligase (which also efficiently activates benzoate but not 3-hydroxybenzoate and therefore, is not the HbcL enzyme), and a minor aerobic 2-aminobenzoate enzyme (5).
Two different pathways for the anaerobic degradation of 2-aminobenzoyl-CoA were proposed, i.e., direct ring reduction and reductive deamination to benzoyl-CoA (150, 220). At present, the only enzyme present in cells of T. aromatica grown anaerobically on 2-aminobenzoate that would reduce 2-aminobenzoyl-CoA to a nonaromatic alicyclic CoA thioester product was purified and shown to be BCR (245, 330). In Azoarcus sp. strain EbN1, the HbrB and HbrC subunits of the putative 3-hydroxybenzoyl-CoA reductase are significantly induced when the cells were grown in 2-aminobenzoate. Thus, the dearomatization of 2-aminobenzoyl-CoA is most likely performed by the same BCR enzyme proposed for the 3-hydroxybenzoate pathway (378). A putative ferredoxin (ebA5004), which was strongly induced in 2-aminobenzoate-adapted cells of strain EbN1, may function as an electron donor for reductive deamination or dearomatization, and it appears to also accept electrons from a phenylacetaldehyde oxidoreductase during the catabolism of phenylalanine (see below) (378). The subsequent catabolism of the 2-aminobenzoyl-CoA reduction product remains unknown, and it might involve some enzymes of the 3-hydroxybenzoate degradation pathway, such as the putative acyl-CoA dehydrogenase (ebA736) (Fig. 4), enzymes from the modified β-oxidation of the benzoyl-CoA pathway, or additional enzymes of a still-unknown 2-aminobenzoate-specific pathway (378).
Resorcinol Catabolism: the Central Oxidative Hydroxyhydroquinone Pathway
Resorcinol (1,3-dihydroxybenzene) is produced and utilized in large amounts by industry. Moreover, roots of aquatic plants exude resorcinol in considerable amounts into the aquatic environment (80), and resorcinol is also generated during the anaerobic catabolism of other aromatic compounds such as 3,5-dihydroxybenzoate (α-resorcylate), 2,4-dihydroxybenzoate (β-resorcylate), and 2,6-dihydroxybenzoate (γ-resorcylate) (Fig. 1) (189). Anaerobic resorcinol degradation has been documented for various microorganisms such as sulfate-reducing bacteria (325), fermenting bacteria (353), and denitrifying bacteria (136, 189, 346). Notably, none of these bacteria use the benzoyl-CoA pathway in resorcinol catabolism. Two major catabolic strategies for the degradation of resorcinol have been described, i.e., the classical reductive dearomatization in sulfate-reducing or fermenting bacteria (see below) and the oxidative HHQ pathway, which is energetically favorable only for denitrifying bacteria (283, 320).
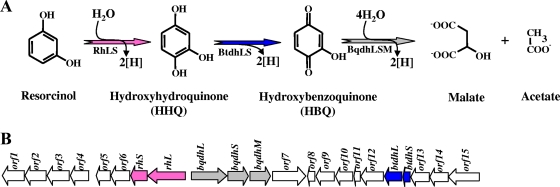
The obligate anaerobe Azoarcus anaerobius degrades resorcinol through two consecutive oxidative reactions using nitrate as the final electron acceptor (Fig. 5) (283). Resorcinol is hydroxylated at position 4 of the aromatic ring to form the key intermediate HHQ in a reaction catalyzed by resorcinol hydroxylase (283). HHQ is also a common intermediate in the aerobic degradation of various aromatic compounds, and it usually becomes degraded through the β-ketoadipate pathway (145). However, in A. anaerobius, HHQ is subsequently oxidized to 2-hydroxy-1,4-benzoquinone (HBQ) in a reaction catalyzed by HHQ dehydrogenase (Fig. 5) (283). Both oxidative reactions were measured in the membrane fraction of cells grown in resorcinol. A ring cleavage enzyme acting on HBQ leads to the formation of malate and acetate (Fig. 5). The electrons from resorcinol and HHQ oxidation can directly enter the denitrification process and may allow energy conservation by proton translocation (283). A similar oxidative strategy is carried out by T. aromatica strain AR1, which hydroxylates α-resorcylate to 3,5,6-trihydroxybenzoate that is then decarboxylated to HHQ (119). Thus, T. aromatica AR1 and A. anaerobius harbor, next to the classical benzoyl-CoA pathway for benzoate degradation, a second mechanistically distinct central pathway with HHQ as the central intermediate for the anaerobic degradation of certain aromatic compounds (e.g., resorcinol, α-resorcylate, 3-hydroxybenzoate, gentisate, and, perhaps, hydroquinone) (Fig. 1) (320).
FIG. 5.
Proposed pathway for anaerobic catabolism of resorcinol in Azoarcus anaerobius. (A) Enzymatic reactions of the HHQ oxidative pathway. The enzymes involved are as follows: RhLS, putative resorcinol hydroxylase; BtdhLS, putative HHQ dehydrogenase; BqdhLSM, putative enzyme complex involved in the ring cleavage of HBQ (77). (B) Organization of the gene cluster involved in anaerobic catabolism of resorcinol in A. anaerobius (GenBank accession number EF078692) (77). Genes are represented by arrows: pink, rhLS genes, encoding the putative resorcinol hydroxylase; blue, btdhSL genes, encoding the putative HHQ dehydrogenase; gray, bqdhLSM genes, encoding the HBQ ring cleavage enzyme complex; white, genes of unknown function.
The only gene cluster described so far for the anaerobic degradation of resorcinol and HHQ is that from A. anaerobius. This 30-kb gene cluster is organized into three operon-like units (Fig. 5) (77). Sequence comparison analysis allowed the tentative assignation of functions to 20 out of the 22 gene products, including high-affinity nutrient acquisition systems (three transporters), detoxification (one glutathione S-transferase), catabolism (10 oxidoreductases), and components with regulatory functions (σ factors, regulators, and a stress response system) (77). Transconjugants of T. aromatica strains that were unable to use resorcinol but that were harboring cosmid R+ (contains the cloned 30-kb gene cluster) were able to grow with resorcinol at a rate similar to that of A. anaerobius. Accordingly, resorcinol hydroxylase activity, HHQ dehydrogenase activity, as well as an HBQ ring cleavage enzyme activity with the formation of malate and acetate (Fig. 5) could be detected in the transconjugants, suggesting that the whole resorcinol degradation pathway is encoded by cosmid R+. Moreover, the expression of the genes is inducible when the cells grow in the presence of resorcinol or some analogs such as α-resorcylate (77). This report constitutes one of the few examples of vertical pathway expansion, i.e., increasing the catabolic potential by transferring a new central pathway, in anaerobes that degrade aromatic compounds.
The mapping of transposon insertions in cosmid R+ derivatives of those mutants impaired in resorcinol utilization revealed that eight genes, rhLS, bqdhLS, btdhLS, orf13, and orf14, encode enzymes specific for resorcinol degradation in A. anaerobius (77). The rhL and rhS genes encode the large and small subunits of the resorcinol hydroxylase, respectively (Fig. 5). The best match for the rhL and rhS gene products are the large and small subunits of the heterodimeric pyrogallol- phloroglucinol transhydroxylase from Pelobacter acidigallici, a member of the dimethyl sulfoxide reductase family (244). As in the case of the transhydroxylase, the RhL and RhS subunits contain putative binding sites for two molybdopterin guanine dinucleotide cofactors and two iron-sulfur clusters, respectively (77). Although resorcinol hydroxylase is predicted to be soluble from the amino acid sequence, it was measured almost exclusively in the membranes of A. anaerobius cells and T. aromatica transconjugants, suggesting the existence of an as-yet-unknown membrane anchor in its molecule (77).
The HHQ dehydrogenase that converts HHQ into HBQ appears to be encoded by the btdhL and btdhS genes (Fig. 5). Whereas the btdhL gene product is similar to putative β-hydroxy acid dehydrogenases, the btdhS gene product seems to be a membrane protein, which would explain why the HHQ dehydrogenase activity was measured in the membrane fraction (77). The further conversion of HBQ to acetate and malate requires the splitting of two C-C bonds and releases two electrons. Such an oxidative cleavage reaction is compatible with an enzyme system analogous to a pyruvate dehydrogenase complex. Consistent with this, the bqdhLSM gene products show similarity to the E1, E2, and E3 components of multicomponent enzymes, such as the pyruvate dehydrogenase, and they have been proposed to be involved in the ring cleavage reaction of HBQ in A. anaerobius (Fig. 5) (77). Although a tentative pathway for anaerobic resorcinol degradation in A. anaerobius has been postulated based on the putative function of the key gene products (Fig. 5), further studies are needed to confirm the proposed catabolic scheme and the role of some genes, such as orf13 and orf14, that were shown to be essential for this catabolism (77).
The Central Phloroglucinol, Hydroxyhydroquinone, and Resorcinol Reductive Pathways
So far, three types of dearomatizing reductases, which use dihydroxylated (resorcinol) or trihydroxylated (phloroglucinol and HHQ) benzenes, have been described (Fig. 1). The aromaticity of these compounds is substantially weakened with two or more meta-positioned hydroxyl groups, which allows the tautomerization of the enol form to the keto form, generating an isolated double bond that can be easily reduced without a coupling to ATP hydrolysis (30, 31, 320).
A fermenting Clostridium sp. uses reductive biochemistry to convert resorcinol to cyclohexanedione (189, 353), which is further hydrolyzed to 5-oxohexanoate (320). The resorcinol reductase of this bacterium consists of subunits of 49.5 kDa and contains flavin adenine dinucleotide (FAD), but iron-sulfur centers have not been detected. A reduced ferredoxin has been proposed to be the in vivo electron donor (30, 320). No resorcinol reductase activity has been found in denitrifying bacteria. In the latter organisms, which are endowed with an electron-accepting system of higher redox potential than that of sulfate reducers and fermenters, the catabolism of resorcinol follows an alternative pathway that involves two oxidative reactions and HHQ as a central intermediate (see above) (30, 31, 320).
The central phloroglucinol pathway in fermenting bacteria such as Eubacterium oxidoreducens and P. acidigallici has been studied. Phloroglucinol is reduced by an NADPH-dependent phloroglucinol reductase to dihydrophloroglucinol, which finally renders three acetate molecules (30, 320). Phloroglucinol is a central intermediate formed during the anaerobic catabolism of trihydroxybenzoates. Thus, gallate (3,4,5-trihydroxybenzoate), which originates from oak gall and as a degradation product of lignin or tannins, is decarboxylated to pyrogallol (Fig. 1), which then becomes isomerized to phloroglucinol through a transhydroxylation reaction catalyzed by the cytoplasmic pyrogallol-phloroglucinol transhydroxylase (320). The amino acid sequence and three-dimensional structure of this transhydroxylase are known (11, 244). The heterodimeric protein contains a molybdenum ion coordinated to two molybdopterin guanine dinucleotide cofactors in the large α-subunit and three [4Fe-4S] clusters in the small β-subunit. During the transhydroxylation reaction, the transferred hydroxyl group does not originate from water; instead, a cosubstrate such as 1,2,3,5-tetrahydroxybenzene is used without apparent electron transfer (244).
The central HHQ reductive pathway in sulfate reducers has been described. In Desulfovibrio inopinatus, HHQ is dearomatized by a two-electron reaction catalyzed by an NADH-dependent reductase (299). In fermenting bacteria, however, HHQ is isomerized to phloroglucinol by three subsequent transhydroxylation reactions analogous to pyrogallol-phloroglucinol transhydroxylation (43).
No genes encoding these reductases have been described so far, and therefore, future work should be carried out to identify and characterize the gene clusters responsible for all these central reductive pathways.
Central Pathways for the Catabolism of N-Heteroaromatic Compounds: Nicotinate Catabolism
N-heteroaromatic compounds are very abundant in nature, and they play key roles within the cells as electron carriers (pyridine and flavin cofactors, e.g., NAD and FAD), nucleotides (purines and pyrimidines), energy storage molecules (e.g., ATP), toxins, and alkaloids. Moreover, synthetic N-hereteroaromatic compounds are used as industrial solvents, dyes, explosives, pharmaceuticals, and pesticides (e.g., atrazine). N-heteroaromatic compounds usually show good water solubility, low sorption, and, thus, high aqueous mobility, which explains why they are commonly found in anaerobic subsurface environments. The anaerobic degradation of these compounds has been less studied than that of the homocyclic aromatic compounds, and excellent reviews on this topic have been published (21, 111, 176). Usually, the degradation of these aromatic compounds generates central intermediates of low aromaticity that are subject to a hydrolase-type ring cleavage reaction (21, 74, 111, 176, 178, 366). However, the anaerobic catabolism of some N-heteroaromatic compounds, such as pyridines, requires a reductive attack of the aromatic ring carried out by reductases that use ferredoxin as an electron donor (111, 176, 302, 303).
As benzoate has been used as a model compound to study the anaerobic central benzoyl-CoA pathway, nicotinate (3-carboxypyridine) is the analogous N-heterocyclic compound used as a model to study the cognate anaerobic central pathway. Nicotinate is widely distributed in nature as part of pyridine cofactors (e.g., NAD and NADP) and alkaloids (e.g., nicotine and anabasine), and it is essential (vitamin B3) for those organisms that are not able to carry out its synthesis (221). Nicotinate is also a carbon and nitrogen source for different bacteria and some fungi (6, 176), and some of its catabolic intermediates, e.g., 6-hydroxynicotinate, are of pharmacological and agrochemical value (381).
Nicotinate catabolism in all organisms starts with its hydroxylation to 6-hydroxynicotinate by the well-characterized and industrially used enzyme nicotinate dehydrogenase (6). Further catabolism depends on the availability of oxygen in the environment. So far, the only fully elucidated nicotinate degradation pathway is the anaerobic route from the fermenting bacterium Eubacterium barkeri (formerly Clostridium barkeri) (347). A gene cluster containing 17 catabolic genes arranged as a putative operon has been identified (Fig. 6) (4). The ndhF, ndhS, ndhL, and ndhM genes encode the four subunits of the E. barkeri selenium/molybdenum-containing nicotinate dehydrogenase (Fig. 6) (132). High-level sequence identities of NdhS and NdhF were found with the 2× [2Fe-2S]- and FAD-containing subunits/domains of xanthine dehydrogenases, respectively. The NdhL and NdhM proteins correspond to the three molybdopterin domains of the large subunit of other xanthine dehydrogenase-like enzymes (36, 354). The hnr gene encodes the 6-hydroxynicotinate reductase, a homotetrameric [4Fe-4S]-flavoprotein that reduces 6-hydroxynicotinate to 1,4,5,6-tetrahydro-6-oxonicotinate (Fig. 6) (159). The activity of this enzyme is lost when purified in the presence of oxygen. An electron flow from the physiological electron donor ferredoxin to the [4Fe-4S] clusters followed by single-electron transfers to the covalently bound flavin in the active site of Hnr has been proposed. The heterologous expression of the hnr gene in E. coli has not been successful and might require the coexpression of orfC and/or orfE, two ORFs that are also linked to hnr orthologs present in putative nicotinate degradation clusters from different proteobacteria (4).
FIG. 6.
Nicotinate fermentation pathway in E. barkeri. (A) Enzymatic reactions of the pathway according to data described previously by Alhapel et al. (4). The enzymes shown are as follows: NdhFSLM, nicotinate dehydrogenase; Hnr, 6-hydroxynicotinate reductase; Ena, enamidase; Hgd, 2-(hydroxymethyl)glutarate dehydrogenase; Hmd, 2-(hydroxymethyl)glutarate dehydratase; Mgm, 2-methyleneglutarate mutase; Mii, (R)-3 methylitaconate isomerase; DmdAB, (2R,3S)-dimethylmalate dehydratase; Dml, (2R,3S)-dimethylmalate lyase. (B) Organization of the nicotinate gene cluster (GenBank accession number DQ310789) (4). Genes are represented by arrows, and their color code corresponds to that of the encoded reactions shown above (panel A). A putative regulatory gene (nicR) is shown in black; genes of unknown function are indicated in white.
The ena gene encodes the enamidase that catalyzes the hydrolysis of tetrahydro-6-oxonicotinate to ammonia and 2-formylglutarate. Enamidase shares 15 to 25% amino acid sequence identity with members of the α/β-barrel amidohydrolase family (typified by dihydroorotase) and contains a typical metal binding His-X-His pattern at the N-terminal part (4, 161). The 2-formylglutarate is then reduced to 2-(hydroxymethyl)glutarate by the action of a stereospecific 2-(hydroxymethyl)glutarate dehydrogenase that requires NADH, and it is encoded by the hgd gene. The conversion of 2-(hydroxymethyl)glutarate to 2-methyleneglutarate is carried out by a putative [4Fe-4S]-containing dehydratase encoded by the hmd gene (Fig. 6). The mgm and mii genes encode the adenosylcobalamin-dependent carbon skeleton-rearranging enzyme 2-methyleneglutarate mutase and 3-methylitaconate isomerase, respectively (Fig. 6) (4, 15). The analysis of the nicotinate cluster revealed the existence of two genes, dmdAB, located at the 3′ end of the cluster that encode proteins with amino acid sequence identities to members of the aconitase family. The DmdAB proteins are suggested to be the Fe2+-dependent and oxygen-sensitive (2R,3S)-dimethyl-malate dehydratase previously identified in E. barkeri (194). The last step of the nicotinate pathway is carried out by the homotetrameric (2R,3S)-dimethylmalate lyase (Fig. 6), a member of the isocitrate lyase family, encoded by the dml gene, which has been successfully expressed in E. coli cells (4).
The nicotinate cluster from E. barkeri contains two additional genes postulated to encode a 2-methyleneglutarate mutase repair enzyme (mgmL) and a transmembrane protein of unknown function (orfD). The nicotinate operon is flanked by a divergently transcribed gene (nicR) encoding a putative LysR-type transcriptional regulator. The chemical inducer could be the 6-hydroxynicotinate intermediate, which is known to accumulate early in the growth phase (4).
Database searches identified nine proteobacteria that contain gene clusters encoding proteins with significant amino acid sequence identities (about 40%) to the Ndh, Hnr, and Ena proteins from E. barkeri. These observations suggest that these organisms are also nicotinate-catabolizing bacteria that share the first three enzymatic steps with those of the anaerobic nicotinate pathway from E. barkeri. This assumption is supported by preliminary data that show that the heterologously expressed Bradyrhizobium japonicum ena homolog has kinetic parameters similar to that of E. barkeri enamidase (4). Since it is known that the alphaproteobacterium Azorhizobium caulinodans is able to catabolize nicotinate aerobically via 1,4,5,6-tetrahydro-6-oxonicotinate and 2-formylglutarate (187), it could be expected that the first steps of this aerobic pathway might be encoded by ndh, hnr, and ena orthologs. The further catabolism of 2-formylglutarate in the nine identified proteobacteria probably proceeds via the glutarate pathway described for A. caulinodans (187), thus explaining the absence of hgd, hmd, mgm, mii, dmdAB, and dml homologs in such aerobic gene clusters (4).
GENE CLUSTERS ENCODING PERIPHERAL CATABOLIC PATHWAYS
As indicated in the Introduction, the peripheral pathways are those devoted to the removal of functionalities from the aromatic nucleus, leading to the formation of the central intermediates. In the anaerobic catabolism of aromatic compounds, most of the peripheral pathways converge into the benzoyl-CoA central compound (Fig. 1). Although in some cases, the peripheral route involves just one reaction step, e.g., the activation of some aromatic acids (benzoate, 3-hydroxybenzoate, 3-methylbenzoate, and 2-aminobenzoate), to the corresponding aryl-CoA esters (see above), many aromatic compounds are channeled to benzoyl-CoA via different multistep peripheral pathways whose genetic determinants are usually not linked to the benzoyl-CoA gene cluster.
Gene Clusters for Degradation of Aromatic Acids
4-HBA catabolism.
Aromatic compounds derived from lignin, the second most abundant polymer on earth, are often methoxylated or hydroxylated. Methyl groups are removed to leave hydroxyl substituents. The anaerobic degradation of many industrially produced phenolics, including phenol and p-cresol (4-methylphenol), converge at the level of 4-HBA, which is also a common growth substrate for different anaerobic bacteria (38). The peripheral pathway for the anaerobic degradation of 4-HBA involves two catabolic steps, i.e., its CoA-dependent activation and further reductive dehydroxylation to benzoyl-CoA (Fig. 7) (133).
FIG. 7.
Peripheral pathway for anaerobic catabolism of 4-HBA in anaerobic bacteria. (A) Enzymatic reactions of the pathway as described previously for R. palustris (127) and T. aromatica (40). The following R. palustris (Hba) and T. aromatica (Hcr) enzymes are indicated: HbaA/HcrL, 4-HBA-CoA ligase; HbaBCD/HcrCAB, 4-hydroxybenzoyl-CoA reductase. (B) Organization of the gene clusters involved in anaerobic catabolism of 4-HBA in R. palustris (GenBank accession number NC_005296) and T. aromatica (accession number AJ001830) and those proposed for the genomes of M. magneticum AMB-1 (accession number NC_007626), Azoarcus sp. strain EbN1 (accession number NC_006513), G. metallireducens (accession number NC_007517), and a metagenomic sequence (accession number CR931837). Genes are represented by arrows: red, genes encoding 4-HBA-CoA ligase; green, genes encoding the subunits of 4-hydroxybenzoyl-CoA reductase; black, putative transcriptional regulators; brown, genes encoding putative 4-HBA transporters and/or permeases; yellow, genes encoding a putative maturation factor; white, genes of unknown function. Two vertical lines mean that the genes are not adjacent in the genome.
4-HBA-CoA ligase is the enzyme that activates 4-HBA to 4-hydroxybenzoyl-CoA. Enzymes catalyzing this reaction have been purified from R. palustris (126) and from T. aromatica (24). The R. palustris gene encoding this enzyme, hbaA, was disrupted, and the resulting strain was unable to use 4-HBA as a carbon source, indicating that the HbaA enzyme is essential for the anaerobic degradation of this compound (126). The hba gene is flanked by the hbaEFGH genes, encoding a putative 4-HBA transport system related to the branched-chain amino acid uptake family of ABC transporters (312), and the hbaBCD genes, encoding 4-hydroxybenzoyl-CoA reductase (Fig. 7). Moreover, the hba gene cluster is adjacent to the bad genes (Fig. 2B), generating a supraoperonic clustering (104). The hbaA ortholog in T. aromatica is still unknown but is not adjacent to the hcrCBA genes, which encode the 4-hydroxybenzoyl-CoA reductase in this bacterium (Fig. 7) (40).
The oxygen-sensitive 4-hydroxybenzoyl-CoA reductase (dehydroxylating) is a three-subunit protein that catalyzes the removal of the phenolic hydroxyl group of 4-hydroxybenzoyl-CoA by a two-electron reduction, yielding water and the central intermediate benzoyl-CoA. The three-dimensional structure of the heterohexameric dehydroxylating reductase from T. aromatica is known (354). The enzyme is a member of the xanthine oxidase family, and it contains two [2Fe-2S] clusters, one [4Fe-4S] cluster, one FAD, and one molybdopterin cytosine dinucleotide cofactor per monomer (31, 34). However, the 4-hydroxybenzoyl-CoA reductase shows some unique properties, e.g., irreversible reduction of the substrate, among the members of the xanthine oxidase family (172). The amino acid sequences of the three subunits of the T. aromatica (HcrCAB) and R. palustris (HbaBCD) 4-hydroxybenzoyl-CoA reductases showed 47 to 62% identity (40, 127). Interestingly, among all Mo-containing members of the xanthine oxidase family (172), the two enzymes contain a unique extra sequence domain in the HcrB (HbaD) subunit coordinating the [4Fe-4S] cluster, which plays an essential role in mediating two-electron transfer from the low-potential donor, reduced ferredoxin to the other redox centers. The same reduced ferredoxin may serve as an electron donor for 4-hydroxybenzoyl-CoA reductase and BCR (31). It was proposed that in both cases, catalysis involves the stabilization of a highly reactive radical species. However, the electron-donating para-hydroxyl group of 4-hydroxybenzoyl-CoA destabilizes a putative benzene ring radial anion, which makes the redox potential for the first electron transfer rather lower for benzoyl-CoA than for 4-hydroxybenzoyl-CoA. Consequently, BCR catalysis requires the input of external energy (ATP) to promote electron transfer at a physiological rate, whereas 4-hydroxybenzoyl-CoA reductase does not (31). The hbaBCD genes from R. palustris have been cloned and expressed in E. coli cells, but no 4-hydroxybenzoyl-CoA was detected, suggesting that the active enzyme is not easily reconstituted from separately synthesized subunits and/or that some of the enzyme cofactors, such as molybdopterin cytosine dinucleotide, are not synthesized in E. coli (127).
In both T. aromatica and R. palustris, the genes encoding the 4-hydroxybenzoyl-CoA reductase are associated with genes encoding a putative transcriptional regulator of the MarR family (orf1) and a transcriptional activator of the fumarate nitrate reductase (FNR)/cyclic AMP receptor protein (CRP) superfamily (hbaR), respectively (Fig. 7) (40, 103, 104). On the other hand, the hbaEFGH genes, encoding a putative ABC transporter of 4-HBA in R. palustris, are substituted by a gene (orf2) encoding a hydrophobic permease protein that might be involved in the uptake of 4-HBA in T. aromatica (Fig. 7) (40, 104).
The hcr and hba genes have been used to identify orthologs in the genomes of other bacteria. Thus, in the genome of M. magneticum AMB-1 (236), there is also an hcr gene cluster similar to that of T. aromatica and, therefore, is likely to be involved in the anaerobic degradation of 4-HBA. This cluster also contains a putative regulatory gene (amb2018) encoding a MarR-type transcriptional regulator and an additional ORF (amb2022) located downstream of the hcrCAB genes, which might encode a maturation factor of the 4-hydroxybenzoyl-CoA reductase as already reported for other members of the xanthine oxidase family (Fig. 7) (112). As observed for T. aromatica, the gene encoding the 4-HBA-CoA ligase is not linked to the hcr genes in M. magneticum. In Azoarcus sp. strain EbN1, the hcrCAB orthologs are also linked to a gene (ebA3601) encoding a putative maturation factor of the 4-hydroxybenzoyl-CoA reductase. Next to the hcr cluster, there are genes encoding putative molybdenum transport proteins. Like in T. aromatica, the gene encoding 4-HBA-CoA ligase in strain EbN1 is not linked to the hcr cluster (Fig. 7) (293). This gene (here named hcrL) has been cloned and expressed in E. coli cells, and the purified 4-HBA-CoA ligase was shown to act not only on 4-HBA but also on protocatechuate, 3-hydroxybenzoate, and benzoate (90). Although a proteomic study of Azoarcus sp. strain EbN1 grown in 4-HBA did not reveal the specific induction of any of the hcr gene products, a significant induction of the korA2 gene product (α-subunit of the two-component KGOR enzyme) was observed, which might be involved in the regeneration of the ferredoxin of the BCR system (see above), suggesting that such a ferredoxin may also be shared by 4-hydroxybenzoyl-CoA reductase (378). Interestingly, the HcrA and HcrC subunits of a putative 4-hydroxybenzoyl-CoA reductase are found fused and encoded in a single gene within an hcr cluster from a metagenomic sequence of a Black Sea microbial mat. A TetR family transcriptional regulator is likely regulating the expression of the hcr genes in this metagenomic clone (Fig. 7) (201).
The anaerobic catabolism of 4-HBA has also been studied for the obligate anaerobic bacterium G. metallireducens. Orthologs of the genes encoding 4-hydroxybenzoyl-CoA reductase from facultative anaerobes have been identified within the large genomic island of G. metallireducens (48, 277). Thus, the pcmRST genes will encode the three-subunit 4-hydroxybenzoyl-CoA reductase from G. metallireducens (Fig. 7). Notably, although PcmR would be the FAD-containing subunit of 4-hydroxybenzoyl-CoA reductase, it does not contain the [4Fe-4S] cluster binding loop, which suggests a mode of electron transfer to the substrate that is different from that postulated for the classical 4-hydroxybenzoyl-CoA reductases from facultative anaerobes (277). The deduced pcmUVWX gene products show similarities to proteins involved in the maturation of molybdenum-containing enzymes and are proposed to play a role in the molybdenum cofactor assembly of 4-hydroxybenzoyl-CoA reductase. On the other hand, the pcmQ gene may encode a MarR-type transcriptional regulator that could be involved in the expression of other pcm genes (277). Proteomic (277) and transcriptomic (48) results have shown that the pcmRST genes are induced not only when the G. metallireducens cells grow in the presence of 4-HBA but also when they are cultivated with benzoate. However, 4-hydroxybenzoyl-CoA reductase activity was detected in G. metallireducens cells grown in 4-HBA but not in cells grown in benzoate, suggesting a posttranslational regulation of this enzyme activity (277). An in silico search of the G. metallireducens genome indicated that bamY appears to be the only gene coding for a typical aromatic carboxylic acid-CoA ligase (Fig. 2). Since BamY does not recognize 4-HBA as a substrate (377), and a specific 4-HBA-CoA ligase activity was detected in G. metallireducens cells grown in 4-HBA, it should be assumed that the gene encoding the 4-HBA-CoA ligase is still unknown (277).
Halobenzoate catabolism.
Because of acute and chronic toxicity, persistence, and bioaccumulation, the presence of haloaromatic compounds in the environment, usually in anoxic ecosystems, is of major concern and a threat to human and environmental health. Some anaerobic bacteria, many of which are sulfate reducers, are able to reductively dehalogenate aromatic compounds such as chlorobenzenes and chlorobenzoates by using them as terminal electron acceptors. These anaerobes can couple the reductive dehalogenation by specific enzymes to energy conservation via electron transport-coupled phosphorylation in a process termed dehalorespiration (105, 128, 160, 169, 338). However, some proteobacteria (e.g., isolates of the genera Thauera, Pseudomonas, and Ochrobactrum) completely degrade halobenzoates as a carbon source for growth when nitrate is present as a terminal electron acceptor, although the mechanisms involved in the removal of the halide ion are, in most cases, still unknown (339-342).
The benzoate degradation pathway can be used to catabolize some halobenzoates. Thus, the relaxed substrate specificity of the benzoate-CoA ligase allows the activation of some halobenzoates to the corresponding CoA esters. In some cases, such as with 2-fluorobenzoate, the fluoride ion can then be eliminated gratuitously from the corresponding CoA derivative by a regioselective reaction carried out by the BCR (245), and the bacteria can use this haloaromatic acid as a sole carbon source (319).
A devoted peripheral pathway for the catabolism of haloaromatic compounds has been reported for some phototrophs. R. palustris strains often acquire the ability to utilize 3-chlorobenzoate as the sole carbon source after prolonged incubation in medium containing only this haloaromatic compound, suggesting that a limited number of mutations are needed to acquire this function and providing a defined basis for studying this adaptation (263). The peripheral 3-chlorobenzoate pathway in R. palustris strain RCB100 starts with the activation of 3-chlorobenzoate to 3-chlorobenzoyl-CoA by the action of a CoA ligase (AliA100) that was almost identical (only a single-nucleotide change that replaces Ser-208 with Thr-208) to the AliA cyclohexane carboxylate-CoA ligase (AliA009) from the non-3-chlorobenzoate degrader R. palustris strain CGA009 (313). AliA100 was 10-fold more active with 3-chlorobenzoate than AliA009, and it was also more active with several other halogenated substrates including 3-bromobenzoate, 4-fluorobenzoate, and 2-fluorobenzoate than AliA009. An aliA100 disrupted strain grew extremely slowly on 3-chlorobenzoate and cyclohexane carboxylate. Nevertheless, the aliA100 gene did not confer the ability to grow on 3-chlorobenzoate on strain CGA009, indicating that the inability of this strain to degrade this haloaromatic compound is not due solely to the lack of the appropriate CoA ligase activity (313). Consistent with this, benzoyl-CoA has been found as a metabolite formed from 3-chlorobenzoate by whole cells of R. palustris RCB100, indicating that a reductive dehalogenation of 3-chlorobenzoyl-CoA to benzoyl-CoA must occur. Such reductive dehalogenation in cell extracts has not yet been demonstrated, and the R. palustris strain CGA009 genome does not contain any genes that resemble known reductive dehalogenase genes (101). Further work needs to be carried out to identify the reductive dehalogenation genes and/or proteins in halobenzoate-degrading R. palustris strains.
Under anaerobic conditions, only reductive dehalogenation has been described so far (338). However, in Pseudomonas sp. strain CBS3, there are three genes involved in an aerobic 4-chlorobenzoate dehalogenation pathway via CoA derivatives that generate 4-HBA as the final product (hydrolytic dehalogenation). By cloning the dehalogenation genes from strain CBS3 into T. aromatica T1, a strain that anaerobically degrades 4-HBA, it was possible to engineer a recombinant strain that is able to use 4-chlorobenzoate via 4-HBA as the sole carbon source under denitrifying conditions (71). Interestingly, this is one of the few examples of metabolic engineering (horizontal pathway expansion) in anaerobes that degrade aromatic compounds.
Phenylalanine/phenylacetate catabolism.
The aromatic amino acids phenylalanine and tyrosine can serve as the sole source of carbon for a variety of microorganisms. The anaerobic degradation of these aromatic amino acids in fermenting bacteria that are unable to cleave the aromatic ring has been studied, and it involves oxidation and reduction reactions with the formation of phenylacetate and 4-hydroxyphenylacetate from phenylalanine and tyrosine, respectively (13). Some of the genes that are likely to be involved in the fermentation of phenylalanine to phenylacetate via phenyllactate and cinnamate have been identified for some Clostridium strains (87). In contrast to fermenting bacteria, phototrophs and anaerobic respirers can further oxidize the aromatic ring of the phenylacetate and 4-hydroxyphenylacetate generated during the anaerobic catabolism of phenylalanine and tyrosine, respectively (324).
Phenylalanine is a common carbon source for different anaerobic bacteria. Anaerobic phenylalanine degradation was suggested to proceed in T. aromatica via transamination and decarboxylation to phenylacetaldehyde and then via dehydrogenation to phenylacetate (324) (Fig. 8). The initial transamination of phenylalanine to phenylpyruvate is catalyzed by the phenylalanine aminotransferase (Pat). The transaminase uses 2-oxoglutarate as a preferential cosubstrate (324). The second step in phenylalanine degradation is performed by phenylpyruvate decarboxylase (Pdc), which generates phenylacetaldehyde from phenylpyruvate. Phenylacetaldehyde is then oxidized to phenylacetate (324). The predicted proteins of the anaerobic phenylalanine degradation pathway were identified by in silico analysis of the Azoarcus sp. strain EbN1 genome, and the corresponding genes are not organized in a cluster but are rather distributed along the chromosome in at least three different regions (Fig. 8) (293). The putative pat gene is apparently not specifically upregulated during the anaerobic growth of strain EbN1 in phenylalanine, suggesting that either Pat is constitutively formed or an alternative aminotransferase, out of the 16 predicted by in silico analysis of the Azoarcus sp. strain EbN1 genome, catalyzes this initial reaction, and it could not be detected by differential proteomics (378). In contrast, the level of Pdc is significantly increased in phenylalanine-adapted cells, supporting the proposed function of the pdc gene in phenylalanine degradation (Fig. 8) (290, 293, 378). A pdh gene encoding a putative phenylacetaldehyde dehydrogenase (Pdh) that catalyzes the third step of the phenylalanine degradation pathway was identified in the genome of strain EbN1 (293). However, the pdh gene product did not increase in abundance during the anaerobic growth of strain EbN1 in phenylalanine. Instead, the pdh gene product was upregulated when the cells were grown in p-cresol, suggesting that pdh encodes a dedicated function in p-cresol rather than in phenylalanine degradation (see below) (378). The differential protein profiling of strain EbN1 growing in phenylalanine suggested a predicted aldehyde:ferredoxin oxidoreductase (AOR) as a possible substitute for Pdh. The AOR-encoding gene, ebA5005, forms a putative operon with ebA5004, encoding a putative ferredoxin that acts as an electron acceptor of AOR-dependent oxidation (and may also be involved in the reduction and/or deamination of 2-aminobenzoyl-CoA) (see above), and ebA5007, encoding the putative ferredoxin:NADH oxidoreductase that regenerates the oxidized ferredoxin (Fig. 8) (378). An alternative pathway shortcutting the phenylalanine degradation from phenylpyruvate directly to phenylacetyl-CoA, as occurs in fermenting bacteria (87), was previously proposed to involve indolepyruvate:ferredoxin oxidoreductase (IorAB) (293). However, the proteomic data do not support this possibility since IorA was not differentially upregulated in Azoarcus sp. strain EbN1 cells grown in phenylalanine (378).
FIG. 8.
Proposed pathway for anaerobic catabolism of l-phenylalanine to phenylacetate in Thauera and Azoarcus strains. (A) Enzymatic reactions of the pathway according to data described previously by Schneider et al. (324) and Wöhlbrand et al. (378). The enzymes involved are l-phenylalanine:2-oxoglutarate transaminase (Pat), phenylpyruvate decarboxylase (Pdc), and phenylacetaldehyde oxidoreductase (AOR). Fdx (ferredoxin) and Fdx:NADH oxidoreductase are predicted to be auxiliary enzymes of AOR (378). (B) Organization of the genes likely to be involved in anaerobic catabolism of phenylalanine to phenylacetate in Azoarcus sp. strain EbN1 (GenBank accession number NC_006513). Genes are represented by arrows: red, pat gene, encoding the putative l-phenylalanine:2-oxoglutarate transaminase; blue, pdc gene, encoding the putative phenylpyruvate decarboxylase; green, genes encoding a putative phenylacetaldehyde oxidoreductase AOR and the ferredoxin and ferredoxin:NADH oxidoreductase enzymes. Two vertical lines mean that the genes are not adjacent in the genome.
Phenylacetate is a free intermediate formed in the anaerobic degradation of phenylalanine but can also be found in the environment as a common carbon source. The catabolism of phenylacetate, under both aerobic and anaerobic conditions, involves the formation of phenylacetyl-CoA as a first intermediate (85, 238, 247, 265, 321). In bacteria where phenylacetate can be degraded either aerobically or anaerobically, e.g., T. aromatica and A. evansii, two different phenylacetate-CoA ligases are induced either aerobically or anaerobically, respectively (150, 246, 247). However, whereas the aerobic degradation of phenylacetyl-CoA involves an oxygenolytic attack of the aromatic ring (167, 238), anaerobic catabolism uses a different strategy based on an α-oxidation of the phenylacetyl-CoA side chain with the formation of benzoyl-CoA as a central intermediate (Fig. 9) (247).
FIG. 9.
Peripheral pathway for anaerobic catabolism of (4-hydroxy)phenylacetate in bacteria. (A) Enzymatic reactions of the pathway in T. aromatica (248, 301, 324) and A. evansii (157, 248). The enzymes involved are phenylacetate-CoA ligase (PadJ), phenylacetyl-CoA:acceptor oxidoreductase (PadBCD), phenylglyoxylate:NAD+ oxidoreductase (PadEFGHI), and a proposed 4-hydroxyphenylacetate-CoA ligase (CoA ligase). (B) Organization of the gene clusters likely to be involved in anaerobic catabolism of phenylacetate in A. evansii (GenBank accession number AJ428571), Azoarcus sp. strain EbN1 (accession number NC_006513), R. palustris BisB5 (accession number NC_007958), and M. magneticum AMB-1 (accession number NC_007626). Genes are represented by arrows: red, padJ genes encoding phenylacetate-CoA ligase; pink, genes encoding a putative 4-hydroxyphenylacetate-CoA ligase; green, padBCD genes likely encoding phenylacetyl-CoA:acceptor oxidoreductase; blue, padEFGHI genes likely encoding phenylglyoxylate:NAD+ oxidoreductase; brown, genes encoding a putative ABC-type transporter; black, genes coding for putative transcriptional regulators; yellow, genes encoding a putative thioesterase; white, gene of unknown function. Two vertical lines mean that the genes are not adjacent in the chromosome.
In the anaerobic pathway for phenylacetate degradation from T. aromatica and A. evansii, the activated phenylacetyl-CoA becomes oxidized at the α-methylene carbon in a four-electron reaction catalyzed by a membrane-bound molybdenum-iron-sulfur enzyme, phenylacetyl-CoA:acceptor oxidoreductase, generating phenylglyoxylate and releasing CoA (301, 323). Ubiquinone is most likely to act as the electron acceptor, and the oxygen atom introduced into the product is derived from water (301). The subsequent oxidative decarboxylation of phenylglyoxylate to benzoyl-CoA and CO2 is catalyzed by an oxygen-sensitive membrane-bound phenylglyoxylate:NAD+ oxidoreductase (CoA benzoylating) (Fig. 9) (157). Phenylglyoxylate oxidation was detected in cell extracts of T. aromatica and A. evansii when cells were grown anaerobically on phenylalanine, phenylacetate, or phenylglyoxylate (157, 301).
An identically organized pad cluster likely encoding the anaerobic phenylacetate degradation pathway has been identified in the genomes of A. evansii (GenBank accession number AJ428571) and Azoarcus sp. strain EbN1 (293) (Fig. 9). Phenylacetate-CoA ligase is likely encoded by the padJ gene, which is located near the 3′ end of the pad cluster. The pad catabolic operon also includes the padBCD genes, encoding the putative phenylacetyl-CoA:acceptor oxidoreductase, and the padEFGHI genes, which may code for phenylglyoxylate:NAD+ oxidoreductase (Fig. 9). The PadJ, PadC, PadG, and PadH proteins were most strongly increased in abundance during the anaerobic growth of Azoarcus sp. strain EbN1 with phenylacetate (and phenylalanine), reinforcing the hypothesis that the pad cluster is indeed responsible for the anaerobic degradation of phenylacetate in Azoarcus strains (378). Two additional genes, located downstream of padJ and likely belonging to the pad catabolic cluster, encode a putative aromatic acid-CoA ligase (ebA5403) and a thioesterase (ebB192) (Fig. 9). The existence of a thioesterase in the aerobic phenylacetate catabolic pathway was also reported, and it was suggested that such enzymes could participate in a salvage reaction to avoid a depletion of the cellular CoA pool when CoA derivatives cannot be metabolized further by any blockage in phenylacetate catabolism (167, 344). The thioesterase could be also involved in the proposed enzymatic release of CoA from the phenylglyoxylyl-CoA intermediate formed in the reaction catalyzed by the phenylacetyl-CoA:acceptor oxidoreductase (301). The product of the padR gene, which is divergently transcribed to the pad catabolic genes (Fig. 9), shows significant similarity to PaaX, a transcriptional repressor that responds to phenylacetyl-CoA and controls the expression of the paa catabolic genes, which are involved in aerobic phenylacetate degradation in different bacteria (53, 82, 85, 86, 287). Thus, it is likely that PadR, a member of the GntR family of transcriptional regulators, regulates the expression of the pad genes, with phenylacetyl-CoA as the inducer molecule. Interestingly, the putative regulatory protein (PaaR) of the aerobic phenylacetate pathway (paa genes) in Azoarcus strains does not show similarity to the PaaX-type regulators, and it rather belongs to the TetR family of transcriptional regulators (53, 86), suggesting different regulatory schemes for the aerobic and anaerobic catabolisms of phenylacetate in these bacteria.
The in silico search for pad orthologs in bacterial genomes revealed the existence of putative pad clusters in two alphaproteobacteria, the denitrifying bacterium M. magneticum strain AMB-1 and the phototroph R. palustris strain BisB5. In R. palustris BisB5, the pad genes are organized into a single cluster that also contains a putative regulatory gene (padR), a putative ABC-type transport system, and a thioesterase-like gene (Fig. 9). In M. magneticum, the predicted pad genes are organized into two clusters located at different regions of the chromosome; i.e., whereas the padBCD genes are linked to padJ and to homologs of the thioesterase and CoA ligase genes, the padEFGHI genes are linked to a putative ABC-type transport system that might be involved in the transport of phenylacetate/phenylglyoxylate (Fig. 9). These results suggest that the anaerobic pathway for phenylacetate degradation that has been characterized for betaproteobacteria such as Azoarcus and Thauera strains is also present in other facultative anaerobes that degrade aromatic compounds.
The anaerobic degradation of some hydroxy derivatives of phenylacetate, such as the 4-hydroxyphenylacetate that can be generated during the metabolism of tyrosine (333), appears to also be carried out through the Pad pathway, although activation to the 4-hydroxyphenylacetyl-CoA intermediate seems to involve a different CoA ligase than that encoded by the padJ gene (247, 248). Consistent with this, the existence of a second CoA ligase-encoding gene next to padJ in some pad clusters (Fig. 9) could be related to the specific 4-hydroxyphenylacetate-CoA ligase necessary for 4-hydroxyphenylacetate catabolism via the Pad pathway, leading to the formation of 4-hydroxybenzoyl-CoA (Fig. 9) (248).
p-Coumarate catabolism.
Lignin and suberin are polymers of phenylpropanoid units, and their biodegradation involves the depolymerization and subsequent catabolism of the derived aromatic monomers such as p-coumarate (4-hydroxycinnamate). p-Coumarate can also be generated from the deamination of tyrosine (42, 272). Two different routes for aerobic p-coumarate catabolism have been described, i.e., a β-oxidative route and a non-β-oxidative pathway. However, both routes generate p-coumaroyl-CoA as the first intermediate, and both could easily function under anaerobic conditions (122, 170, 380).
The genes and proteins involved in the anaerobic degradation of p-coumarate have been studied using R. palustris cells grown in this aromatic acid by integrating transcriptomics and quantitative proteomics data. None of the genes upregulated during p-coumarate degradation appeared to belong to a β-oxidation gene set that could likely be used for p-coumarate catabolism. This global study suggested that the anaerobic degradation of p-coumarate proceeds through a non-β-oxidation route and then through the central benzoyl-CoA pathway (Fig. 10) (267).
FIG. 10.
Proposed peripheral pathway for anaerobic catabolism of p-coumarate in R. palustris. (A) Enzymatic reactions of the pathway (267). The postulated enzymes are p-coumarate-CoA ligase (RPA1787), p-coumaroyl-CoA hydratase/lyase (RPA1786), and p-hydroxybenzaldehyde dehydrogenase (RPA1206). (B) Organization of the gene clusters likely to be involved in peripheral catabolism of p-coumarate in R. palustris CGA009 (GenBank accession number NC_005296). Genes are represented by arrows: red, RPA1787 gene, encoding p-coumarate-CoA ligase; blue, RPA1786 gene, encoding p-coumaroyl-CoA hydratase/lyase; green, RPA1206 gene, encoding p-hydroxybenzaldehyde dehydrogenase; orange and brown, genes encoding putative TRAP and ABC transporters, respectively; black, RPA1794 and RPA1207 genes, encoding putative transcriptional regulators of the MarR and Fis families, respectively; violet, gene encoding a putative aromatic alcohol dehydrogenase; yellow, gene encoding a putative thioesterase; white, genes of unknown function. Two vertical lines mean that the genes are not adjacent in the chromosome.
Among the three loci that likely encode ferulate/p-coumarate-CoA ligases in the R. palustris CGA009 genome, RPA1787 is the only one upregulated under p-coumarate conditions, which makes it the probable gene responsible for the p-coumarate-CoA ligation (Fig. 10). This gene is located within a gene cluster that also contains RPA1786, a gene encoding a protein that is closely related to members of the enoyl-CoA hydratase/isomerase family and therefore likely to be the putative p-coumaroyl-CoA hydratase/lyase that generates p-hydroxybenzaldehyde and an acetyl-CoA molecule (Fig. 10) (122, 272). RPA1782 (putative periplasmic subunit of a TRAP-type transporter), RPA1788 (putative thioesterase), and RPA1789 to RPA1793 (putative ABC-type transporter) are also upregulated in p-coumarate-grown cells, suggesting their involvement in the uptake of this aromatic acid (Fig. 10) (267). The expression of this operon may be controlled by RPA1794, a divergently transcribed gene that encodes a putative ΜarR-type transcriptional regulator (Fig. 10). The genomes of other R. palustris strains, e.g., BisB5, BisA53, and HaA2, also contain a gene cluster similar to that reported for strain CGA009, which is likely to be involved in p-coumarate catabolism.
It is known that R. palustris readily oxidizes 4-hydroxybenzaldehyde to 4-HBA in a CoA-independent manner (143). The RPA1206 gene product is the only annotated aldehyde dehydrogenase with significantly upregulated protein abundance under p-coumarate conditions, suggesting that it could be the p-hydroxybenzaldehyde dehydrogenase of the peripheral p-coumarate pathway in R. palustris. 4-HBA-CoA ligase (HbaA) (see above) is also substantially upregulated when R. palustris cells are grown in p-coumarate, confirming that a large flux of 4-HBA exists during p-coumarate catabolism. The RPA1206 gene is arranged in a putative operon that includes the RPA1205 gene, encoding a putative aromatic alcohol dehydrogenase. The expression of this operon may be controlled by RPA1207, a divergently transcribed gene that encodes a putative σ54-dependent transcriptional regulator of the Fis family (Fig. 10). Orthologous clusters encoding a transcriptional regulator and two catabolic genes that might encode aromatic alcohol and aldehyde dehydrogenases have also been detected in other anaerobes that degrade aromatic compounds, e.g., the putative benzyl alcohol degradation cluster from Azoarcus sp. strain EbN1 (see below).
Some derivatives of p-coumarate, such as cinnamate and ferulate (4-hydroxy-3-methoxycinnamate), are also common bacterial carbon sources derived from lignin degradation. R. palustris is able to anaerobically use these phenylpropenoid compounds as the sole carbon source (Table 1) (143). Whether the anaerobic pathway for the catabolism of p-coumarate could also be used for the catabolism of additional phenylpropenoids in R. palustris is a plausible hypothesis that requires further experimental confirmation.
Gene Clusters for Degradation of Aromatic Alcohols and Phenols
Aromatic alcohols and phenols comprise a large group of natural compounds, and they can also originate as metabolic intermediates in the course of the degradation of aromatic hydrocarbons and lignin (25).
Benzyl alcohol catabolism.
Benzyl alcohol is one of the main aromatic alcohols present in nature, e.g., as a fragrance of some flowers, and it can also originate as a metabolic intermediate in the degradation of aromatic hydrocarbons (25). Benzyl alcohol becomes oxidized, under both aerobic and anaerobic conditions, via benzaldehyde, to benzoate. Whereas the genes responsible for benzyl alcohol and benzaldehyde dehydrogenases in different aerobic bacteria are known (62), they have not been identified yet in anaerobes. A homotetrameric NAD+-dependent benzyl alcohol dehydrogenase that generates benzaldehyde from benzyl alcohol was purified from T. aromatica cells grown anaerobically in benzyl alcohol. The N-terminal amino acid sequence suggests that the enzyme belongs to the long-chain Zn2+-dependent alcohol dehydrogenases, as it occurs with benzyl alcohol dehydrogenases from aerobic bacteria (25).
Analysis of the genome of Azoarcus sp. strain EbN1 suggested the presence of various aromatic alcohol and aldehyde dehydrogenases, some of which were proposed to be involved in benzyl alcohol (ebA3118, ebA3166 [Adh], and ebA4623 [AdhB]) and benzaldehyde (ebA5642 [Ald] and ebA4625 [AldB]) oxidation (Fig. 11) (293, 378). However, a proteomic study with Azoarcus sp. strain EbN1 cells grown in benzyl alcohol or benzaldehyde revealed that most putative benzyl alcohol and benzaldehyde dehydrogenases were not significantly increased in abundance (e.g., Adh) or that they were nonspecifically induced in cells grown with other aromatic compounds such as phenol (e.g., AdhB and Ald), providing no clear evidence for its involvement in benzyl alcohol and benzaldehyde oxidation (378).
FIG. 11.
Peripheral pathway for anaerobic catabolism of benzyl alcohol in Azoarcus sp. strain EbN1. Putative benzyl alcohol and benzaldehyde dehydrogenases are indicated according to data described previously by Rabus et al. (293) and Wöhlbrand et al. (378). Adh, AdhB, Ald, AldB, and PchA are encoded by the ebA3166, ebA4623, ebA5642, ebA4625, and ebA3161 genes, respectively.
Interestingly, the adhB and aldB genes form an operon that shows significant similarity to the RPA1205-RPA1206 operon from R. palustris, which encodes a putative aromatic alcohol dehydrogenase and the putative 4-hydroxybenzaldehyde dehydrogenase, respectively, upregulated in cells grown in p-coumarate (Fig. 10). A similar operon-like structure of genes encoding a benzyl alcohol dehydrogenase and a benzaldehyde dehydrogenase has been described for aerobic bacteria such as A. calcoaceticus (131). However, AldB could not be identified among the upregulated protein spots in Azoarcus sp. strain EbN1 cells grown in benzyl alcohol and benzaldehyde (378). On the other hand, the adh gene is located in the vicinity of pchA, a gene that was initially thought to encode a p-hydroxybenzaldehyde dehydrogenase involved in p-cresol degradation (see below) but that displays significant upregulation during the anaerobic growth of strain EbN1 with benzaldehyde and benzyl alcohol (378), suggesting its implication in the degradation of the latter compounds.
The ald gene is located within a gene cluster that encodes putative acyl-CoA transferases and dehydrogenases as well as a protein (ebA5637) showing the strongest increase in abundance during the anaerobic growth of strain EbN1 with benzyl alcohol and benzaldehyde. Although ebA5637 shows similarity to a mandelate racemase, Azoarcus sp. strain EbN1 cannot utilize either of the two-mandelate isomers, which is in agreement with the absence of genes coding for mandelate and benzylformate dehydrogenase in the genome of this bacterium (378). Therefore, although the operon embracing the ald gene seems to be involved in the anaerobic metabolism of aromatic compounds, its physiological role remains to be determined.
In summary, it appears that Azoarcus sp. strain EbN1 possesses a diverse set of genes located at different regions of the genome and with a multifaceted regulatory pattern, which are likely involved in the peripheral pathways that convert different aromatic alcohols and aldehydes into the corresponding carboxylic acids. Whether a similar situation occurs in other genomes of anaerobic biodegraders remains to be analyzed.
Phenol catabolism.
Phenol is a natural substrate that is formed from a variety of natural compounds. Phenol arises from tyrosine by tyrosine:phenol lyase (tyrosinase) but also arises during the degradation of many secondary phenolic plant constituents, notably in the course of the degradation of lignin and phenylpropanoid compounds. Phenol is also produced in large quantities by industrial activities (322).
Anaerobic phenol catabolism by pure cultures of T. aromatica has been studied in detail, and it proceeds via a two-step process that involves the para-carboxylation of phenol (biological Kolbe-Schmitt carboxylation) (Fig. 12) (262). In the first step, phenol is converted to phenylphosphate in a reaction catalyzed by a phenylphosphate synthase (protein E1) (322). Phenylphosphate synthase consists of three different subunits (proteins 1, 2, and 3) and transfers the β-phosphoryl group from ATP to phenol, generating phenylphosphate, AMP, and Pi. Protein 1, which resembles the central part of E. coli phosphoenolpyruvate synthase, becomes phosphorylated at the conserved His-569 residue in the course of the reaction cycle by ATP. This reaction requires protein 2, which resembles the N-terminal part of phosphoenolpyruvate synthase and is stimulated by an unknown mechanism by protein 3, which contains two cystathionine-β-synthase domains but does not show significant similarity to known proteins. Phosphorylated protein 1 then transfers the phosphoryl group to the phenolic substrate (262, 322). The genes encoding subunits 1 (orf1), 2 (orf2), and 3 (orf3) of phenylphosphate synthase are located adjacent to each other on a large operon that is phenol induced (Fig. 12) (41, 322).
FIG. 12.
Peripheral pathway for anaerobic catabolism of phenol. (A) Enzymatic reactions of the pathway as determined previously for T. aromatica (322, 329). The enzymes are as follows: E1, phenylphosphate synthase; E2, phenylphosphate carboxylase. Brackets indicate an enzyme-bound phenolate intermediate. (B) Organization of the gene cluster involved in anaerobic catabolism of phenol in T. aromatica (GenBank accession number AJ272115), and that of the clusters likely to be involved in anaerobic phenol degradation in Azoarcus sp. strain EbN1 (accession number NC_006513) (288) and G. metallireducens (accession number NC_007517) (329). Genes are represented by arrows: red, genes encoding phenylphosphate synthase; green, genes encoding phenylphosphate carboxylase; black, genes encoding putative XylR/DmpR-like transcriptional regulators; white, genes likely to be involved in a different carboxylation reaction. Numbers underneath the genes in Azoarcus sp. strain EbN1 and G. metallireducens clusters correspond to the ortholog ORF in T. aromatica.
In the second step, phenylphosphate is the substrate of enzyme E2, phenylphosphate carboxylase, which catalyzes the carboxylation of phenylphosphate to 4-HBA (329). The carboxylase enzyme is a member of a new family of carboxylases/decarboxylases that act on phenolic compounds, use CO2 as a substrate, do not contain biotin or thiamine diphosphate, require K+ and a divalent metal cation (Mg2+ or Mn2+) for activity, and are strongly inhibited by oxygen (329). The phenylphosphate carboxylase consists of four proteins (Orf4, Orf5, Orf6, and Orf12) whose genes are located adjacent to each other in the phenol gene cluster (Fig. 12). The orf4 and orf6 products show similarity to UbiD/UbiX aryl decarboxylases, which are involved in ubiquinone biosynthesis. The orf5 product belongs to a hydratase/phosphatase protein family (329).
Whereas the genes located in the 5′ region of the phenol gene cluster from T. aromatica (orf1 to orf6 and orf12) appear to be directly involved in the anaerobic catabolism of phenol to 4-HBA, those arranged in the 3′ half of the cluster consisting of orf7 to orf10 and orf13 to orf15 may play a role in the metabolism (carboxylation or decarboxylation) of other phenolic compounds (329). Consistent with this, the orf7 and orf8 products also show similarity to the UbiD and UbiX proteins (329). Nevertheless, both parts of the gene cluster are coordinately expressed as a single transcript, as indicated by the identification of phenol-induced proteins and RT-PCR experiments (41, 262). A putative σ54-dependent promoter with the consensus −12/−24 region is located upstream of the transcription initiation site (+1) of the catabolic operon. Inverted repeats representing possible binding sites for a σ54-dependent regulatory protein are found about 260 bp upstream of the +1 site. The putative regulatory gene, orf11, is transcribed divergently from the catabolic operon. ORF11 shows similarity to members of the XylR/DmpR family of transcriptional regulators that control gene clusters involved in the aerobic degradation of phenolic compounds (53, 322, 352). As was shown for DmpR, the ORF11 regulator is likely to be activated not only by phenol but also by some (methyl) derivatives whose degradation may need the proteins encoded by the 3′ half of the cluster (322).
G. metallireducens and Azoarcus sp. strain EbN1 are the only bacteria that grow with phenol under anaerobic conditions and whose genomes have been sequenced. These two bacteria contain orthologs of many genes of the phenol gene cluster from T. aromatica, including the DmpR-like (ORF11) regulatory gene (Fig. 12), suggesting that anaerobic phenol metabolism in these bacteria is similar to that found in T. aromatica (293, 329). In G. metallireducens, the putative phenol gene cluster is located within the large aromatic catabolic island between cluster IA, involved in benzoyl-CoA degradation, and the pcm operon for the catabolism of p-cresol (see below) (48), and it contains only two out of the four ppc genes conserved in Thauera and Azoarcus (Fig. 12), which may indicate the existence of a different type of carboxylase in this strict anaerobe.
The phenol degradation genes (pps and ppc genes) in Azoarcus sp. strain EbN1 are organized identically to their orthologs in T. aromatica (Fig. 12) (293), and some of their products have been shown to be upregulated in phenol-grown cells by proteomic studies (378). Several of the associated genes of unknown function are also conserved in strain EbN1, as occurs with the ppcXY genes (which are equivalent to orf7 and orf8 from T. aromatica) that might encode the subunits of another carboxylase/decarboxylase of the UbiD/UbiX family and that could be responsible for the catabolism of phenol derivatives. Consistent with this, the phenol gene cluster from strain EbN1 is located in the close vicinity of the pch gene cluster, which was initially thought to be responsible for p-cresol degradation (293), although recent proteomic experiments suggest that it is involved in the degradation of other aromatic compounds such as benzyl alcohol and benzaldehyde (see above) (378). A paralogous pps operon (ebA5781 to ebA5783) lacking the ppsC gene but containing putative carboxylase-encoding genes is located at a different chromosomal position in strain EbN1. Since this operon is not induced by phenol but rather by p-cresol or phenylacetate, it was suggested to be involved in a hitherto unknown degradation pathway (378). In addition to the Pps and Ppc proteins, several heat shock proteins and proteins related to oxidative stress (e.g., superoxide dismutase [SodB] and glutathione peroxidase [BtuE]), DNA protection (Dps), or the cell envelope revealed increased abundances in phenol-grown cells. Since the levels of the majority of these stress proteins were also increased in the presence of the solvent p-cresol, a common and complex strategy of survival to the toxic nature of organic solvents in Azoarcus sp. strain EbN1 has been suggested (see below) (378). An alternative way of feeding phenol into the catabolic pathway may be provided by a putative tyrosine:phenol lyase (tyrosinase) encoded in the chromosome of strain EbN1. Although the tnaA gene (ebA605) coding for this predicted enzyme is located near the pat gene, encoding the putative phenylalanine:2-oxoglutarate aminotransferase (Fig. 8) (293), it has been reported that Azoarcus sp. strain EbN1 does not use tyrosine as a carbon source under anaerobic conditions (294), which brings into question the physiological role of the ebA605 gene.
Interestingly, in some strict anaerobes that encounter high phenol and CO2 concentrations in their natural habitat, such as the spore-forming heterotrophic bacterium Sedimentibacter hydroxybenzoicus strain JW/Z-1 (formerly Clostridium hydroxybenzoicum), phenol carboxylation appears to be accomplished by an ATP-independent biochemical reaction catalyzed by an oxygen-sensitive and cofactor-independent reversible enzyme with the equilibrium on the side of the decarboxylation reaction (164). This reversible hydroxyarylic acid decarboxylase/phenol carboxylase is encoded by the shdCDB genes, and it is specifically induced in the presence of 4-HBA. Similar shdC (ubiD homolog), shdB (ubiX homolog), and shdD genes have been found in the genomes of other bacteria, and they were suggested to encode a novel enzyme family of reversible, nonoxidative, and cofactor-independent hydroxyarylic acid decarboxylases (230).
p-Cresol catabolism.
Monomethylphenols (cresols) and dimethylphenols are produced in large amounts in the petrochemical industry as constituents of resins, solvents, disinfectants, and wood-preserving chemicals. Among cresols, the catabolism of p-cresol (4-methylphenol) has been studied most intensively. Although p-cresol can also be formed biologically from tyrosine via the decarboxylation of p-hydroxyphenylacetate by some Clostridium species (384), the major part of p-cresol-contaminated wastewaters is derived from industrial activities (277).
The aerobic metabolism of p-cresol involves a periplasmic flavocytochrome p-cresol methylhydroxylase that generates p-hydroxybenzyl alcohol by the hydroxylation of the methyl group with water. Although the oxidation of p-hydroxybenzyl alcohol to p-hydroxybenzaldehyde can also be carried out by p-cresol methylhydroxylase, an NAD+-dependent p-hydroxybenzyl alcohol dehydrogenase was supposed to catalyze this reaction in vivo (75, 171, 183). This oxygen-independent p-cresol hydroxylation has also been described for several denitrifying bacteria such as Thauera and Azoarcus strains (239, 308) as well as for an Achromobacter strain (162). p-Hydroxybenzaldehyde becomes oxidized by a specific NAD+- or NADP+-dependent dehydrogenase to form 4-HBA (Fig. 13). The genes pchFC, coding for the putative α-subunit (flavoprotein) and β-subunit (c-type cytochrome) of a p-cresol methylhydroxylase, respectively, as well as the pchX and pchA genes, encoding a protein of unknown function and a predicted p-hydroxybenzaldehyde dehydrogenase, respectively, are organized into an operon-like structure that is adjacent to the adh gene (ebA3166), which is predicted to encode an alcohol dehydrogenase (see above), in the chromosome of Azoarcus sp. strain EbN1 (Fig. 13) (293). However, both the PchF and PchA proteins did not display significant increases in abundance during the anaerobic growth of strain EbN1 with p-cresol but rather with benzaldehyde and benzyl alcohol (see above) (378). On the contrary, the largest increase in abundance in p-cresol-grown cells was observed for a predicted FAD-dependent oxidase (ebA5380), which displays high similarities to p-cresol methylhydroxylases. The ebA5380 gene (pch) constitutes an operon-like structure with pdh (ebA5381), encoding a formerly predicted phenylacetaldehyde dehydrogenase, and ebA5384, encoding a hypothetical protein that is found to be associated with phenol degradation clusters (Fig. 13) (293). However, since p-cresol functions as a gratuitous inducer for other anaerobic peripheral pathways, such as that for toluene (bbs and bss genes) and the one encoded by the paralogous ebd operon, whose aromatic substrate is still unknown (378), it is difficult to identify the dedicated p-cresol degradation genes among those that become induced when the cells are cultivated in the presence of this compound. Therefore, although the pch and pdh gene products are currently suggested to be involved in anaerobic p-cresol degradation in Azoarcus sp. strain EbN1 (378), a role for PchCF and PchA in p-cresol and/or benzyl alcohol (or other aromatic alcohol) degradation cannot be ruled out so far.
FIG. 13.
Proposed peripheral pathway for oxidation of p-cresol to 4-HBA in different bacteria. (A) Enzymatic reactions of the pathway in denitrifying (308) and iron-reducing (171, 277) bacteria. The enzymes are indicated as follows: Pch and PcmGIJ, p-cresol methylhydroxylase; Adh, p-hydroxybenzyl alcohol dehydrogenase; Pdh, PchA, and PcmO, p-hydroxybenzaldehyde dehydrogenase. (B) Organization of the gene clusters likely to be involved in anaerobic p-cresol oxidation in Azoarcus sp. strain EbN1 (GenBank accession number NC_006513) and G. metallireducens (accession number NC_007517). Genes are represented by arrows: green, genes known or predicted to encode p-cresol methylhydroxylase and a putative p-hydroxybenzyl alcohol dehydrogenase (adh); blue, genes predicted to encode a proposed p-hydroxybenzaldehyde dehydrogenase (or an aromatic aldehyde dehydrogenase); yellow, genes predicted to encode a membrane-bound cytochrome bc1 complex; white, genes of unknown function; black, the pcmQ regulatory gene of the p-hydroxybenzoate degradation cluster (Fig. 7). Two vertical lines mean that the genes are not adjacent in the genome.
In the strictly anaerobic bacterium G. metallireducens, the enzymes involved in p-cresol metabolism clearly differ from those found in aerobic and facultatively anaerobic bacteria. Thus, although p-cresol metabolism is also initiated by methyl hydroxylation in G. metallireducens, this activity was surprisingly found in the membrane fraction of the cell extracts (277). Proteomic studies of the membrane fraction of G. metallireducens cells grown in p-cresol revealed an upregulation of the pcmI and pcmJ gene products, which might represent the two isoforms of the FAD-containing α-subunit of p-cresol methylhydroxylase. On the other hand, sequence comparison analyses indicated that the pcmG gene, located upstream of pcmIJ, might encode the cytochrome c-like β-subunit of p-cresol methylhydroxylase (Fig. 13). The purification and characterization of the soluble PcmIJ and PcmG components confirmed that p-cresol methylhydroxylase from G. metallireducens has a unique αα′β2-subunit composition, with the α subunit (PcmI) harboring the typical FAD cofactor, which is lacking in the catalytically inactive α′-subunit (PcmJ), and the β-subunit (PcmG) representing a c-type cytochrome (171). In contrast to other aerobic and facultatively anaerobic p-cresol-degrading organisms, G. metallireducens appears to use p-cresol methylhydroxylase for both p-cresol and p-hydroxybenzyl alcohol oxidation in vivo. Interestingly, the observed membrane location of the p-cresol methylhydroxylase activity is considered to result from a strong interaction of the PcmIJG components with the p-cresol-induced pcmCDEF gene products (Fig. 13), which share amino acid sequence similarities (30 to 66% identity) with the subunits of membrane-bound bacterial ubiquinol:cytochrome b oxidoreductases (cytochrome bc1 complex) (171, 277). The existence of a p-cresol-induced membrane-bound cytochrome bc1-like complex, which usually mediates electron transfer between quinols and cytochrome c, could also account for the unusual asymmetric architecture of the p-cresol methylhydroxylase with two differing α-subunits. Thus, it has been suggested that the noncatalytic α′-subunit may be involved in mediating both alternative electron transfer routes either to the periplasmic cytochrome c pool (in the case of p-cresol oxidation) or to the menaquinone pool (in the case of p-hydroxybenzyl alcohol oxidation), which would significantly increase the energy yield in an obligately anaerobic organism (171).
The oxidation of the p-hydroxybenzaldehyde generated by the p-cresol methylhydroxylase to form 4-HBA is carried out by an NADP+-dependent soluble dehydrogenase in G. metallireducens. The p-cresol-induced pcmO gene product is assigned as the p-hydroxybenzaldehyde dehydrogenase since it shows high levels of similarities to known or deduced p-hydroxybenzaldehyde or p-hydroxyphenylacetaldehyde dehydrogenases (up to 75% amino acid sequence identity) (Fig. 13) (277). As expected, the 4-hydroxybenzoyl-CoA reductase activity (PcmRST) (Fig. 7) was also induced in G. metallireducens cells grown in p-cresol (277). Thus, the p-cresol (and 4-HBA)-induced pcm gene cluster in G. metallireducens involves a set of genes responsible for the oxidation of p-cresol to finally generate benzoyl-CoA (Fig. 7 and 13).
In sulfate-reducing bacteria, two different pathways for p-cresol oxidation have been described. In the gram-positive bacterium Desulfotomaculum sp. strain Groll, p-cresol appears to be oxidized by the hydroxylation of the methyl group as in aerobic, facultative anaerobic, and iron-reducing bacteria (222). However, in the gram-negative sulfidogenic bacterium Desulfobacterium cetonicum, p-cresol degradation is initiated by the formation of p-hydroxybenzylsuccinate. This reaction, an addition of the methyl group of p-cresol to fumarate, and the proposed metabolism of p-hydroxybenzylsuccinate to 4-hydroxybenzoyl-CoA through a β-oxidation-like scheme, resembles the anaerobic degradation of toluene (see below) and m-cresol in anaerobic bacteria. However, the genes involved in p-cresol degradation in sulfate-reducing bacteria have not been characterized so far (258).
Regarding the two other cresol isomers, i.e., o-cresol and m-cresol, they are usually catabolized via carboxylation/dehydroxylation and fumarate addition/β-oxidation, leading to the formation of 3-methylbenzoyl-CoA and 3-hydroxybenzoyl-CoA, respectively (Fig. 1), but the genes specifically involved have not been identified so far (258, 308).
p-Ethylphenol catabolism.
p-Ethylphenol is present in coal tars and crude oils and is also a prominent constituent of petrochemical wastewaters. The aerobic p-ethylphenol degradation pathway in Pseudomonas putida is initiated by water-dependent hydroxylation carried out by a p-ethylphenol methylhydroxylase similar to the equivalent hydroxylase that acts on p-cresol (see above) (298). The anaerobic degradation of p-ethylphenol in Azoarcus sp. strain EbN1 was recently studied, and a peripheral catabolic pathway has been proposed based on the identification of some intermediates and the upregulation of some proteins when the cells are grown with p-ethylphenol/p-hydroxyacetophenone as the sole carbon source (Fig. 14) (379). The initial hydroxylation of p-ethylphenol to 1-(4-hydroxyphenyl)-ethanol is proposed to be catalyzed by a p-ethylphenol methylhydroxylase encoded by the pchCF gene (379). Here we have named these genes pehCF (Fig. 14) to distinguish them from the homologous pchCF genes that were previously proposed to be involved in p-cresol hydroxylation (Fig. 13). Two predicted alcohol dehydrogenases (ChnA and ebA309) that show significant similarity to the 1-phenylethanol dehydrogenase (Ped) involved in anaerobic ethylbenzene degradation (see below) may catalyze the oxidation of 1-(4-hydroxyphenyl)-ethanol to p-hydroxyacetophenone (Fig. 14) (379). The further catabolic steps proposed are equivalent to those for ethylbenzene degradation, and they may involve (i) the carboxylation of p-hydroxyacetophenone by a putative biotin-dependent carboxylase (XccABC) that does not share any similarity to the acetophenone carboxylase (Apc1 to Apc5) of the ethylbenzene pathway; (ii) the activation of the carboxylation product for its respective CoA-ester by a putative acetoacetyl-CoA synthase-like protein (AcsA), which is not homologous to the benzoylacetate CoA-ligase (Bal) of the ethylbenzene pathway; and (iii) the thiolytic cleavage of the CoA-activated product by the predicted TioL thiolase with the final formation of acetyl-CoA and p-hydroxybenzoyl-CoA (Fig. 14), with the latter presumably being catabolized to benzoyl-CoA through the hcr pathway (Fig. 7). Nevertheless, hydrolytic cleavage of the carboxylation product without prior CoA activation cannot be excluded (379).
FIG. 14.
Proposed peripheral pathway for the oxidation of p-ethylphenol to 4-hydroxybenzoyl-CoA in Azoarcus sp. strain EbN1. (A) Enzymatic reactions of the pathway according to data described previously by Wöhlbrand et al. (379). The enzymes are indicated as follows: PehCF, putative p-ethylphenol methylhydroxylase (this enzyme was named PchCF by Wöhlbrand et al.) (379); ChnA and/or ebA309, putative 1-(4-hydroxyphenyl)-ethanol dehydrogenase; XccABC, putative p-hydroxyacetophenone carboxylase; AcsA, predicted CoA synthetase; TioL, predicted thiolase. (B) Organization of the gene cluster likely involved in anaerobic p-ethylphenol oxidation in Azoarcus sp. strain EbN1 (GenBank accession number NC_006513). Genes are represented by arrows whose color codes correspond to those above (A) for the corresponding encoded enzymes. Genes of unknown function, putative p-ethylphenol/p-hydroxyacetophenone stress-related genes, and regulatory genes are indicated in white, pink, and black, respectively.
All the predicted enzymes involved in anaerobic p-ethylphenol degradation are encoded in a large operon-like structure on the chromosome of Azoarcus sp. strain EbN1. These catabolic genes are divergently transcribed from a gene (ebA324) that may encode a σ54-dependent regulatory protein that belongs to the XylR/DmpR family of transcriptional regulators and that might recognize p-ethylphenol and p-hydroxyacetophenone as inducer molecules (Fig. 14) (379). Interestingly, adjacent to the regulatory ebA324 gene, and transcribed in the opposite orientation, there is an additional operon-like structure that contains the ebA326 to ebA335 genes, some of whose products are also upregulated when Azoarcus sp. strain EbN1 cells are grown on p-ethylphenol/p-hydroxyacetophenone (Fig. 14). Both operon-like structures harbor the −12/−24 consensus sequence of σ54-dependent promoters located around 100 bp upstream of the translational start of the acsA and ebA335 genes, suggesting the transcriptional control of the corresponding promoters by the ebA324 regulator. The participation of ebA327-ebA332-ebA335 in a so-far-uncharacterized solvent-specific efflux system analogous to the RND (resistance nodulation cell division)-type ArcA-ArcB-TolC efflux pump that is known to be involved in solvent tolerance in several gram-negative bacteria has been proposed (379). The fact that the ebA326 gene product shows domains related to the cytoplasmic universal stress protein (UspA) from E. coli is also in agreement with the potential participation of the ebA326 to ebA335 gene products in a specific solvent stress system in p-ethylphenol and p-hydroxyacetophenone metabolism (379).
Catechol catabolism.
Catechol (1,2-dihydroxybenzene) is one of the main central intermediates in the aerobic catabolism of aromatic compounds. Catechol metabolism has also been observed under anoxic conditions, but pure-culture studies are still rare. Catechol is generated, for instance, during the decarboxylation of 2,3-dihydroxybenzoate in fermenting bacteria (266). As established for phenol, growth on catechol requires CO2 (209). It was suggested that the initial steps of catechol metabolism are similar to those of phenol metabolism, i.e., phosphorylation and carboxylation to generate protocatechuate. The further degradation of protocatechuate may yield 3-hydroxybenzoyl-CoA (282). Recently, anaerobic catechol catabolism in T. aromatica was investigated (90). Interestingly, it appears that no enzyme of the pathway was specific for catechol. Rather, the promiscuity of several enzymes and regulators involved in phenol, 3-hydroxybenzoate, 4-HBA, and benzoate metabolism may be sufficient to reconstruct the catechol pathway. Accordingly, the gene products that were induced when T. aromatica cells were grown on catechol are those that were postulated to be involved in these pathways (90).
In the proposed scheme of catechol metabolism in T. aromatica (Fig. 15), degradation is initiated by the carboxylation of catechol to protocatechuate via catechylphosphate as an intermediate. This process is catalyzed by the two enzymes required for phenol carboxylation to 4-HBA, i.e., phenylphosphate synthase (E1) and phenylphosphate carboxylase (E2). As shown above, the genes coding for these two enzymes are located on a large operon that contains genes that are predicted to encode additional carboxylation enzymes and that are also present in phenol degradation clusters from other bacteria (Fig. 12). The involvement of these additional gene products in the carboxylation of phenol derivatives such as catechol should be studied further. Obviously, both phenol and catechol act as inducers of the putative σ54-dependent regulatory protein (ORF11) (90). The activation of protocatechuate to protocatechuyl-CoA requires 3-hydroxybenzoate-CoA ligase, which is encoded in a 3-hydroxybenzoate-induced gene cluster (Fig. 4). Both 3-hydroxybenzoate and protocatechuate may act as inducers of the 3-hydroxybenzoate metabolic genes. Protocatechuyl-CoA is then reductively dehydroxylated, most likely by 4-hydroxybenzoyl-CoA reductase (HcrCAB), generating 3-hydroxybenzoyl-CoA (Fig. 15), which is further metabolized by the central 3-hydroxybenzoate pathway (Fig. 4) (90).
FIG. 15.
Proposed peripheral pathway for the anaerobic catabolism of catechol to 3-hydroxybenzoyl-CoA in T. aromatica. The enzymes involved, according to data described previously by Ding et al. (90), are recruited from the phenol degradation pathway (green arrows), 3-hydroxybenzoate pathway (red arrows), and 4-HBA pathway (blue arrows) The enzymes are as follows: E1 and E2, phenylphosphate synthase and phenylphosphate carboxylase, respectively; HbcL, 3-hydroxybenzoate-CoA ligase; HcrCAB, 4-hydroxybenzoyl-CoA reductase.
The genetics and biochemistry of the catechol pathway of T. aromatica provide an example of catabolic flexibility that enables an organism to utilize a variety of different but chemically related substrates by using a limited set of genes and enzymes, which are even under proper transcriptional control (90). The genetics of catechol degradation in other anaerobes are still unknown.
Other aromatic compounds are also anaerobically degraded via the carboxylation of the aromatic ring. For instance, aniline becomes carboxylated to 4-aminobenzoate, which is then activated to the CoA thioester and further reductively deaminated to benzoyl-CoA in some sulfate-reducing bacteria (326). No information about the genes responsible for these reactions has been reported so far.
Gene Clusters for Degradation of Aromatic Hydrocarbons
Aromatic hydrocarbons are frequently released into the environment, coming from natural sources, e.g., petroleum deposits or tar pits, as a result of human activities. Moreover, smaller quantities of aromatic hydrocarbons are also continuously produced by biological processes in microorganisms, plants, and animals (359). Although the biological decomposition of these compounds was believed for decades to be absolutely dependent on the presence of molecular oxygen as a highly reactive cosubstrate, about 20 years ago, the first pure cultures of bacteria that degraded hydrocarbons under strictly anoxic conditions were reported, implying that there must be different and unknown biochemical principles for hydrocarbon biotransformation (33, 149, 153, 374). Benzene, toluene, ethylbenzene, and the xylene isomers, collectively known as BTEX, are some of the most water-soluble hydrocarbons of crude oil, and therefore, they are frequently found as soil and groundwater contaminants. Thus far, most of the research regarding the anaerobic catabolism of aromatic hydrocarbons has been conducted on toluene and ethylbenzene degradation (33, 59, 116, 149, 153, 345, 374).
Although an addition of the methyl group of toluene to the double bond of fumarate (fumarate addition) was first described during the anaerobic catabolism of toluene in T. aromatica (26), this enzyme reaction has been shown to be a general strategy for all anaerobic toluene degraders. Moreover, additional reactions to fumarate, similar to that described for toluene degradation, represent a common biochemical principle for activating C-H bonds at the initial step of the anaerobic catabolism of other aromatic compounds such as m-xylene (199), m- and p-cresols (33, 59, 258), 2-methylnaphthalene (7), and ethylbenzene (190) and even for the anaerobic degradation of saturated hydrocarbons such as n-alkanes (295) and cycloalkanes (106) in some bacteria.
Toluene catabolism.
Toluene can be degraded by many bacteria with anaerobic respiratory chains that use nitrate, Mn(IV), Fe(III), humic substances, sulfate, and CO2 as terminal electron acceptors, and it can also be assimilated anaerobically as a carbon source by anoxygenic phototrophs. Toluene is considered to be the model compound for studying anaerobic hydrocarbon degradation (60, 149, 383, 385). Most of the available studies of anaerobic toluene catabolism were carried out with denitrifying T. aromatica strains K172 and T1 and with Azoarcus sp. strain T. The genes responsible for the anaerobic catabolism of toluene are organized into two different large clusters, termed bss (tut in T. aromatica strain T1) and bbs, separated by an intervening sequence (Fig. 16) (33, 59, 116, 149, 153, 345, 374).
FIG. 16.
Peripheral pathway for anaerobic catabolism of toluene in bacteria. (A) Enzymatic reactions of the pathway as determined for Thauera and Azoarcus strains. The enzyme names are as follows: BssABC and BssD, subunits and activase of benzylsuccinate synthase, respectively; BbsEF, succinyl-CoA:(R)-benzylsuccinate CoA transferase; BbsG, (R)-benzylsuccinyl-CoA dehydrogenase; BbsH, putative phenylitaconyl-CoA hydratase; BbsCD, putative 2-[hydroxyl(phenyl)methyl]-succinyl-CoA dehydrogenase; BbsAB, benzoylsuccinyl-CoA thiolase; Sdh, succinate dehydrogenase. Chemical intermediates are indicated as follows: 1, (R)-benzylsuccinyl-CoA; 2, (E)-phenylitaconyl-CoA; 3, 2-[hydroxyl(phenyl)methyl]-succinyl-CoA; 4, benzoylsuccinyl-CoA. (B) Organization of the bss and bbs gene clusters in Magnetospirillum sp. strain TS-6 (GenBank accession number AB167725) (335), Azoarcus sp. strain EbN1 (accession number NC_006513) (203), Azoarcus sp. strain T (accession number AY032676) (1), T. aromatica K172 (accession numbers AJ001848 and AF173961) (155, 216), T. aromatica T1 (accession numbers U57900 and AF113168) (68, 72), Thauera sp. strain DNT-1 (accession number AB066263) (336), and G. metallireducens (accession number NC_007517) (48, 202). Genes are represented by arrows: dark blue, genes encoding benzylsuccinate synthase; brown, genes encoding succinyl-CoA:(R)-benzylsuccinate CoA transferase; red, genes encoding (R)-benzylsuccinyl-CoA dehydrogenase (bbsG) and the predicted electron acceptor system; orange, genes predicted to encode phenylitaconyl-CoA hydratase; dark green, genes predicted to encode 2-[hydroxyl(phenyl)methyl]-succinyl-CoA dehydrogenase; yellow, genes predicted to encode benzoylsuccinyl-CoA thiolase; light green, genes encoding a putative benzylsuccinate synthase chaperone; light blue, gene encoding a putative succinate-dehydrogenase flavoprotein; violet, genes encoding putative toluene transport systems, black, genes predicted to encode transcriptional regulators; gray, genes encoding a putative transposase and its helper protein; white, genes of unknown function. The 3′ end of the bssF and bssG genes from Azoarcus sp. strain T and T. aromatica K172, respectively, as well as the region between the bss and bbs clusters of these two bacteria have not been sequenced yet.
The initial step of anaerobic toluene catabolism involves a new enzymatic reaction catalyzed by a strictly anaerobic glycyl radical enzyme (benzylsuccinate synthase) that adds the methyl group of toluene to fumarate to yield (R)-benzylsuccinate (Fig. 16) (18, 26, 200, 217, 252, 332). The genes bssCAB (tutFDG) code for the γ-subunit (BssC or TutF), α-subunit (BssA or TutD), and β-subunit (BssB or TutG) of the heterohexameric benzylsuccinate synthase, respectively (Fig. 16) (19, 72, 199, 200, 217). Gene inactivation studies with Azoarcus sp. strain T and T. aromatica strain T1 revealed that each of the subunits is essential for the function of the benzylsuccinate synthase (1, 69). Site-directed mutagenesis of the tutF, tutD, and tutG genes from T. aromatica strain T1 determined residues that are critical for the activity of benzylsuccinate synthase (23, 72). The bssD (tutE) gene, which is always located upstream of the bssCAB (tutFDG) genes, codes for BssD (TutE) activase, which introduces the glycyl radical at the large subunit (BssA/TutD) of benzylsuccinate synthase in an S-adenosylmethionine-dependent reaction (332). Site-directed mutagenesis of certain cysteine residues of TutE in T. aromatica strain T1 revealed that they are likely to be critical for the radical activation of benzylsuccinate synthase (23). The benzylsuccinate synthase is irreversibly inactivated in the presence of molecular oxygen by the oxygenolytic cleavage of the α-subunit (200, 217). Whereas the bssD and bssCAB genes are cotranscribed in most bacteria, in T. aromatica strain T1, these genes were reported to be organized as two different operons, tutE and tutFDG (68). Two additional genes, bssE (tutH) and bssF, are also conserved in most bss clusters (Fig. 16). The bssE gene product contains a Walker-type ATP/GTP binding-site motif and is similar to an emerging class of chaperone-like ATPases required for the assembly, operation, and disassembly of protein complexes, which suggests that it may be required for either the assembly or activation of benzylsuccinate synthase (33, 155). The tutH gene product in T. aromatica strain T1 has been shown to be necessary for toluene metabolism, and the 52-Gly Lys Ser-54 residues, which are part of the putative ATP/GTP binding domain, are essential for its activity (23). The bssF gene was predicted to be part of the bss operon because genes coding for highly similar proteins are found directly downstream of bssE in all completely sequenced bss clusters (Fig. 16), although the function of BssF in toluene degradation is still unknown (202). However, in Azoarcus sp. strain T, the bssF gene appears to have its own transcriptional start site (1). Comparative analysis reveals that the order of the genes in the bssDCABEF cluster is conserved in all toluene-degrading bacteria analyzed so far (Fig. 16). However, the five bss clusters from betaproteobacteria can be arranged into two subgroups differing in the length of the intergenic distance between the genes bssB and bssE. In the bss clusters of Azoarcus sp. strain EbN1 and T. aromatica strain K172, these genes are separated by a transcribed intergenic region of 122 nucleotides, which may contain a RNA hairpin loop (155). This distance is much shorter (47 nucleotides) in the clusters from Azoarcus sp. strain T, T. aromatica strain T1, and Thauera sp. strain DNT-1. The different operon organizations are correlated with the fact that the respective benzylsuccinate synthases belong to different similarity subgroups. Thus, the bssABC gene products of Azoarcus sp. strain EbN1 are most similar to those of T. aromatica strain K172 (>94% identity) and slightly less similar (around 80% identity) to those of Azoarcus sp. strain T, T. aromatica strain T1, and Thauera sp. strain DNT-1; the sequences from the latter three denitrifying strains are again >95% identical to each other (202). The bssABC gene products from the alphaproteobacterium Magnetospirillum sp. strain TS-6 showed 60 to 80% identity with the orthologs from T. aromatica K172 (335). On the other hand, the bssABC gene products of the phylogenetically more remote bacterium G. metallireducens are only 71 to 73% identical to those of any of the denitrifying strains (177, 202).
In Azoarcus sp. strain EbN1, the bssDCABEF genes are cotranscribed with two closely spaced genes, bssGH (Fig. 16). In agreement with this operon organization, the BssG protein was specifically upregulated in toluene-grown cells (202). Whereas the bssG gene encodes a protein of unknown function, the bssH gene product displays sequence similarity to members of the MFS and the Bcr/CflA subfamily of drug resistance transporters. Since some members of these transporter families are involved in the uptake/efflux of aromatic compounds, and toluene is expected to diffuse freely across the cytoplasmic membrane, one may speculate that BssH functions in the export of toxic levels of toluene from the cytosol rather than in toluene uptake. However, an alternative function of BssH in the transport of toluene-derived metabolites (e.g., benzylsuccinate) cannot be ruled out (202). Although a bssG ortholog is also present immediately downstream of bssF in T. aromatica strain K172, the bss clusters of other denitrifying toluene degraders are not sequenced sufficiently far to detect possible orthologs. Five additional genes are located downstream of bssH (between the bss and bbs clusters) in Azoarcus sp. strain EbN1 (Fig. 16), but the relevance of these genes in anaerobic toluene catabolism and whether they belong to the bss operon are still unknown (202). In the bss cluster of G. metallireducens, bssG and bssH are missing, and the intercalating sequence between the bss and bbs clusters codes for three predicted genes (Fig. 16). The gene following bssF in G. metallireducens (gmet1535) codes for a protein that shows similarity to TodX-like outer membrane proteins, which are involved in toluene uptake in different proteobacteria (148, 368). Even though this TodX-like protein does not show significant sequence similarity with BssH from Azoarcus sp. strain EbN1, the presence of genes coding for potential toluene transporters in (or next to) the bss clusters in both organisms is remarkable. Interestingly, the gmet1533 protein shows similarity to fumarate reductase/succinate dehydrogenase flavoproteins, and it could be involved in the regeneration of fumarate used by benzylsuccinate synthase (Fig. 16).
The further degradation of (R)-benzylsuccinate proceeds via β-oxidation to benzoyl-CoA and succinyl-CoA. The reaction cycle is completed by succinate dehydrogenase (Sdh), which recycles the fumarate cosubstrate of benzylsuccinate synthase from succinate (Fig. 16). Based on the N-terminal sequences of toluene-induced proteins, nine genes that form the bbs cluster (bbsA to bbsI) were identified in T. aromatica strain K172 (216). The bbsE and bbsF genes code for the two subunits of a succinyl-CoA:(R)-benzylsuccinate CoA-transferase that activates benzylsuccinate to its CoA thioester as the first step of benzylsuccinate oxidation (218). The bbsG gene codes for the second enzyme of the pathway, a benzylsuccinyl-CoA dehydrogenase that generates (E)-phenyl-itaconyl-CoA (219). The next three enzymes of the pathway are encoded by bbsH (putative phenyl-itaconyl-CoA hydratase), bbsCD (two subunits, or two isoenzymes, of a putative hydroxyacyl-CoA dehydrogenase), and bbsB (a putative benzoylsuccinyl-CoA thiolase) (Fig. 16). The bbsA gene product might be connected to the function of the benzoylsuccinyl-CoA thiolase (202, 216). bbsI is the only toluene-induced gene unaccounted for in the bbs cluster of T. aromatica strain K172. All the bbs genes, with the exception of bbsI, were also identified in Azoarcus sp. strain EbN1, and they show high sequence similarity (Fig. 16) (202). Although the genetic organization of the bbs clusters in strains EbN1 and K172 differ in the presence of bbsJ (encodes a toluene-induced protein of unknown function) between bbsF and bbsG in strain EbN1 (203), there is recognizable nucleotide similarity between the rather large bbsFG intergenic gap of strain K172 and the last quarter of bbsJ from strain EbN1. This similarity can be interpreted as that the bbs cluster of strain K172 has evolved from a bbsJ-containing operon by the deletion of most of this gene and the subsequent modification of the remaining intergenic region (202). Despite the fact that an extensive transcriptional analysis of the bss operon has been accomplished (see above), no transcriptional analysis of the bbs operon has been carried out so far for any organism.
Interestingly, the sequence of the bbs cluster of G. metallireducens codes only for orthologs of bbsA to bbsH but does not contain genes encoding a BbsI- or BbsJ-like protein. Moreover, this gene cluster starts with the bbsEFGH genes, followed by three inserted genes not present in the bbs clusters from denitrifying bacteria, and ends with the bbsABCD genes (Fig. 16) (202). Two of the three additional genes in the bbs cluster from G. metallireducens code for the α-subunit (etfA gene) and β-subunit (etfB gene) of a putative electron transfer ring flavoprotein (ETF), which is expected to serve as the physiological electron acceptor in the reaction catalyzed by the benzylsuccinyl-CoA dehydrogenase (219). The third gene encodes a protein (gmet1527) that contains iron-sulfur clusters and may also be involved in electron transfer to the respiratory chain (202). Therefore, in contrast to the well-conserved bss cluster, the genetic arrangement of the bbs cluster from the iron reducer G. metallireducens differs significantly from that of the bbs cluster from denitrifying bacteria.
The known flanking genes of the bss and bbs clusters differ among T. aromatica strain K172, Azoarcus sp. strain EbN1 (a similar gene arrangement is found in Azoarcus sp. strain T), and G. metallireducens (Fig. 16). Thus, upstream of the bbs cluster in Azoarcus sp. strain EbN1, there is an insertion element, ISE1, which contains the istA and istB genes (Fig. 16), whose products are highly similar to transposases/cointegrases and correlated helper proteins, respectively, found in mobile elements of the IS21 family. Such an insertion element appears to be absent in either flanking region of the bbs clusters from G. metallireducens and T. aromatica strain K172 (202). On the other hand, upstream of the bss clusters in Azoarcus and Thauera strains, there are two adjacent genes, tdiR (tutB1) and tdiS (tutC1) (Fig. 16), that code for a putative two-component regulatory system that was suggested to control the expression of the bss and bbs genes (see below) (1, 202). Orthologs of tdiSR are lacking in G. metallireducens, and putative xylR-like and tetR-like regulatory genes are flanking the bss and bbs clusters, respectively (Fig. 16). Recently, an additional toluene-induced gene cluster (ebA1926 to ebA1938) located upstream of the bbs genes was identified in Azoarcus sp. strain EbN1, and it was suggested that it plays a role in coping with toxic toluene concentrations, as pointed out previously for the BssH protein (203). In summary, genetic comparisons revealed that the conserved clusters involved in anaerobic toluene metabolism reflect a common degradation strategy, but they seem to be embedded in quite different genomic contexts in various bacterial species. The transcriptional organizations of the bss and bbs gene clusters also differ among different toluene degrader species and even between different strains of the same species, e.g., T. aromatica strains T1 and K172 (see below). These observations may well reflect that the bss and bbs genes have been subjected to extensive horizontal gene transfer events and that they have been acquired by different host bacteria as an adaptative response to toluene-contaminated habitats.
Ethylbenzene catabolism.
Based on physiological and biochemical studies of the two closely related strains Azoarcus sp. strains EbN1 and EB1, a complete pathway that allows the anaerobic conversion of ethylbenzene to benzoyl-CoA in denitrifying bacteria has been described (Fig. 17). In contrast to the anaerobic catabolism of toluene, which is based on the addition of fumarate to the methylene carbon, the anaerobic ethylbenzene (and probably other alkylbenzenes with carbon chain length of at least 2) degradation involves a direct oxidation of the methylene carbon via (S)-1-phenylethanol to acetophenone. The enzymes of the first two reactions, ethylbenzene dehydrogenase and (S)-1-phenylethanol dehydrogenase, have been extensively characterized, and they constitute the upper part of the ethylbenzene degradation pathway (12, 174, 191, 192, 291, 294). Ethylbenzene dehydrogenase is unique in that it catalyzes a stereoselective direct hydroxylation of a hydrocarbon in the absence of molecular oxygen, a reaction previously unknown in biochemistry. The enzyme is a heterotrimeric soluble molybdenum/iron-sulfur/heme b protein with a periplasmic location in strain EbN1. Gene sequence comparisons and the elucidation of a high-resolution structure indicated that ethylbenzene dehydrogenase is a member of the dimethyl sulfoxide reductase family of molybdenum bis-molybdopterin-guanine dinucleotide enzymes (188). The enzyme catalyzes the water-dependent stereospecific hydroxylation of ethylbenzene to 1-(S)-phenylethanol (Fig. 17). The reaction is apparently driven by exergonic electron transfer from the substrate to a high-potential c-type cytochrome that acts as the natural electron acceptor (174, 191). The ebdABC genes, encoding the α-subunit (EbdA), β-subunit (EbdB), and γ-subunit (EbdC) of ethylbenzene dehydrogenase, respectively, have been identified in strains EB-1 and EbN1, and they are almost identical (173, 292). The ebdABC genes may form an operon that also includes the ethylbenzene-induced ebdD gene (Fig. 17) (203, 292). Sequence comparison analyses suggested that EbdD may function as an enzyme-specific chaperone required for the incorporation of the molybdenum cofactor. EbdA was suggested to contain a twin-arginine leader peptide sequence (173). Accordingly, ethylbenzene dehydrogenase (EbdABC) may be assembled in the cytoplasm in an EbdD-dependent process prior to translocation and proteolytic processing by the Sec-independent twin-arginine translocation pathway (292).
FIG. 17.
Proposed peripheral pathway for anaerobic catabolism of ethylbenzene in Azoarcus strains. (A) Enzymatic reactions of the pathway according to data described previously by Rabus et al. (292) and Johnson et al. (173). The enzymes shown are as follows: EbdABC, ethylbenzene dehydrogenase; Ped, (S)-1-phenylethanol dehydrogenase; Apc1 to Apc5, acetophenone carboxylase; Bal, benzoylacetate-CoA ligase. (B) Organization of the gene cluster involved in anaerobic catabolism of ethylbenzene in Azoarcus sp. strain EbN1 (GenBank accession number NC_006513) (203, 292). Genes are represented by arrows: orange, genes encoding ethylbenzene dehydrogenase; pink, gene encoding a putative ethylbenzene dehydrogenase-specific chaperone; red, gene encoding (S)-1-phenylethanol dehydrogenase; blue, genes predicted to encode acetophenone carboxylase; green, gene predicted to encode benzoylacetate-CoA ligase; brown, gene predicted to encode a transport system; black, genes encoding putative transcriptional regulators; gray, genes encoding putative transposases or transposase fragments; white, genes of unknown function.
The product of the initial reaction, (S)-1-phenylethanol, is further stereospecifically oxidized to acetophenone by an NAD+-dependent alcohol dehydrogenase (Fig. 17) that also catalyzes the biotechnologically interesting reverse reaction (191). 1-Phenylethanol dehydrogenase from strain EbN1 is a tetramer whose three-dimensional structure has been solved. This novel enzyme belongs to the short-chain alcohol dehydrogenase/aldehyde reductase family, and it shows highly cooperative catalysis and strong regulation of its activity by elevated concentrations of substrates and products as well as by the wrong enantiomer of 1-phenylethanol (158). The ped gene, encoding 1-phenylethanol dehydrogenase, is located directly downstream of the ebdABCD genes (Fig. 17), suggesting that they are cotranscribed (292), and it is induced when Azoarcus sp. strain EbN1 grows in the presence of ethylbenzene (203).
The further degradation of acetophenone (lower part of the ethylbenzene pathway) has been suggested to proceed via carboxylation to benzoylacetate (3-oxo-3-phenylpropionate) by acetophenone carboxylase (Fig. 17), which has been confirmed indirectly by demonstrating a CO2 dependence of ethylbenzene and acetophenone degradation (12, 63). Subsequent reactions have been suggested to involve benzoylacetate-CoA ligase, forming benzoylacetyl-CoA, and a benzoylacetyl-CoA thiolase, thiolytically cleaving the substrate to form acetyl-CoA and benzoyl-CoA (Fig. 17). N-terminal sequences of several proteins that are specifically formed during growth of Azoarcus sp. strain EbN1 on ethylbenzene or acetophenone were separated by two-dimensional gel electrophoresis (63), and they matched the amino acid sequences translated from the genes apc5, apc4, apc3, and apc1 (292). Since sequence analyses indicated significant similarities of Apc4 to the α-subunit, Apc1 and Apc3 to the β-subunit, and Apc2 to the γ-subunit of ATP-dependent acetone carboxylases, it was suggested that the apc4, apc3, apc2, and apc1 gene products constitute the four subunits of the postulated acetophenone carboxylase. Since apc5 is apparently cotranscribed with the other four ORFs, the apc5 gene product might constitute another subunit of this enzyme. Hence, acetophenone and acetone carboxylases may differ with respect to subunit composition (five versus three) of the holoenzymes despite the fact that they catalyze similar enzymatic reactions (292). The apc genes are arranged into an operon-like structure that also contains the bal gene, encoding a putative benzoylacetate-CoA ligase (292). The role of the apc and bal genes in the anaerobic degradation of ethylbenzene and acetophenone is also supported by the observed specific upregulation of these genes (transcriptomics) and their products (proteomics) in Azoarcus sp. strain EbN1 cells grown in ethylbenzene (203).
Similarly to the genes involved in anaerobic toluene catabolism, those of anaerobic ethylbenzene catabolism in Azoarcus sp. strain EbN1 are apparently organized into two operons, i.e., the upper (ebdABCD ped) and lower (apc12345 bal) operons, separated by a 16-kb DNA sequence (Fig. 17) (292). Nevertheless, transcriptomic analyses carried out with Azoarcus sp. strain EbN1 cells grown in ethylbenzene revealed a specific upregulation of additional genes that are also likely to be involved in anaerobic ethylbenzene degradation. Thus, the orf57 and orf68 genes flanking the ped and ebd genes, respectively, become highly upregulated in cells adapted to ethylbenzene. Similarly, orf92, orf90, orf87, and orf84 are also highly upregulated in response to ethylbenzene, and they may extend the lower operon beyond the bal gene (Fig. 17) (203). Although the function of most of these ORFs remains elusive, the product of orf90 shows similarity to transport proteins, and it can be speculated that Orf90 may transport some intermediate of ethylbenzene catabolism, such as (S)-1-phenylethanol produced in the periplasmic space by ethylbenzene dehydrogenase (292). The intercalating sequence between the putative upper and lower operons contains genes coding for two different two-component regulatory systems, Tcs2/Tcr2 and Tcs1/Tcr1 (Fig. 17), which are probably involved in the sequential regulation of both operons according to the presence of their substrates, ethylbenzene and acetophenone, respectively (see below) (292). The lower operon and the tcs genes are flanked by genes encoding putative transposases and transposase fragments (Fig. 17), suggesting that this catabolic segment may have been recruited in the vicinity of the upper operon by horizontal gene transfer processes. Strain EbN1 harbors a second operon (ebA5789 to ebA5795), located at about a 2-Mb distance on the chromosome, containing genes paralogous to those of the ethylbenzene upper operon (ebd and ped genes) (293). A gene encoding a thiolase is located immediately following the genes of the second upper operon. Since the benzoylacetyl-CoA thiolase involved in the last step of the ethylbenzene degradation pathway is the only enzyme of the metabolic pathway whose gene is missing in the previously known ethylbenzene gene cluster (Fig. 17), the thiolase encoded in the second gene cluster may well be the putative benzoylacetyl-CoA thiolase (293). Nevertheless, this second paralogous operon is not induced by ethylbenzene but rather by p-cresol and is located adjacent to a second pps operon and to some putative carboxylase genes that are also induced by p-cresol (see above), which may provide a first hint regarding the nature of the true, so-far-unknown, substrate that would follow a degradation scheme similar to that of ethylbenzene (378).
It should be noted that under poor bioenergetic conditions, ethylbenzene becomes degraded via the addition of fumarate, as in the case of toluene. This alternative ethylbenzene degradation mechanism in sulfate reducers, where the redox potential of the ethylbenzene/phenylethanol couple is too positive for an electron transfer to the final electron acceptors, has been reported, but the genes involved are still unknown (190).
Catabolism of other aromatic hydrocarbons.
The anaerobic catabolism of the other two BTEX compounds, i.e., benzene and xylenes, as well as that of some polycyclic aromatic hydrocarbons (PAHs) have also been studied, and some genes were identified.
(i) Catabolism of benzene.
Of the BTEX compounds, benzene appears to be the most recalcitrant one under anaerobic conditions. The recalcitrance of benzene is enhanced by the presence of ethanol added to gasoline to replace methyl tert-butyric ether as a fuel oxygenate. The high electron acceptor demand exerted by ethanol at sites contaminated with reformulated gasoline rapidly induces methanogenic conditions that decrease the thermodynamic feasibility of benzene degradation (78, 79).
Anaerobic benzene biodegradation has been shown to occur under iron-reducing (204, 229), nitrate-reducing (46, 64, 179, 180), sulfate-reducing (228), and methanogenic (182) conditions. However, despite the enrichment of a large number of different cultures under many different terminal electron-accepting conditions, only a few pure cultures (denitrifying Azoarcus and Dechloromonas species) were obtained (64, 180), and the pathways involved remain essentially unknown (65).
Apart from the environmental significance of benzene as a contaminant with carcinogenic potential and high toxicity, the anaerobic activation of benzene is probably one of the most interesting reactions in microbial degradation today because the activation in the absence of molecular oxygen of a nonsubstituted aromatic ring, which is endowed with the highest C-H bond dissociation energy among all hydrocarbons, constitutes an unprecedented biochemical reaction. Phenol and benzoate, which have been detected in the culture medium of methanogenic, sulfate-reducing, and iron-reducing mixed cultures, are thought to be potential metabolites of anaerobic benzene biodegradation. A direct methylation of benzene to toluene has also been proposed (49, 65, 365). Interestingly, the anaerobic nitrate-dependent benzene degradation by Dechloromonas aromatica strain RCB appears to involve an initial hydroxylation with the formation of phenol. Phenol would then be the subject of further carboxylation and dehydroxylation to form benzoate (61). Contrary to previous reports on methanogenic benzene degradation, in which the source of the hydroxyl group of phenol was suggested to be water (365), in strain RCB, phenol formation was suggested to be mediated through a reaction with a hydroxyl free radical that does not originate from water but is formed on the outer membrane or in the periplasm of the organism (60). Although the genome of D. aromatica strain RCB has been finished, the genes responsible for anaerobic benzene degradation are still unknown, and the enzymes involved in the proposed pathway need to be characterized. Moreover, although D. aromatica strain RCB also appears to be able to grow anaerobically with toluene, ethylbenzene, and xylene isomers (60), no genes coding for any of the expected key enzymes involved in the anaerobic degradation of these compounds can be identified in the genome of this bacterium, which is phylogenetically related to the Azoarcus/Thauera group.
Recently, the abiotic phenol formation in anaerobic media with benzene was described, suggesting that the appearance of phenol as a putative degradation product of benzene has to be interpreted with caution (204). These studies confirmed, however, that benzoate was a true intermediate of benzene degradation, at least in an enriched ferric iron-reducing culture, and they strongly support a direct carboxylation of benzene as the initial activation reaction (204).
(ii) Catabolism of xylenes.
Out of the three xylene isomers, the catabolism of m-xylene is the best-studied one. The anaerobic degradation of m-xylene resembles that of toluene since it is also initiated by the addition of a fumarate to the methyl group of the hydrocarbon, yielding (3-methylbenzyl)succinate (199). A bssA null mutant strain and biochemical studies revealed that the same benzylsuccinate synthase enzyme (BssABCD) activates toluene and m-xylene in Azoarcus sp. strain T (1, 363). Differences in the substrate specificities of the benzylsuccinate synthases from different bacteria may explain, at least partially, the degradative capabilities of these bacteria. Thus, the benzylsuccinate synthase from Azoarcus sp. strain T converts toluene and m-xylene to the corresponding succinate adducts, whereas the equivalent enzyme in T. aromatica strain K172, which grows on toluene but not in m-xylene, is active with toluene but not with m-xylene (363). Although it was initially thought that the same bbs pathway used for toluene degradation would also be used for m-xylene degradation, leading finally to the formation of 3-methylbenzoyl-CoA (199), biochemical studies have shown that cells of Azoarcus sp. strain T grown in m-xylene contained about twofold-higher levels of succinyl-CoA:(R)-benzylsuccinate CoA transferase and benzylsuccinyl-CoA dehydrogenase activities than the same cells grown in toluene, which suggests that there are separated Bbs isoenzymes for some steps of the toluene and m-xylene catabolic pathways (218, 219). A genetic analysis carried out using Azoarcus sp. strain CIB, a strain that degrades toluene and m-xylene anaerobically, also points to the existence of an additional cluster that could be involved in some reactions of the peripheral m-xylene pathway as well as in the catabolism of the central intermediate generated, 3-methylbenzoyl-CoA, (Juárez, unpublished). Moreover, an Azoarcus sp. strain CIB mutant with a disrupted gcdH gene, encoding the glutaryl-CoA dehydrogenase of the lower benzoyl-CoA pathway (see above), did not grow in toluene, but it grew in m-xylene (29). For this reason, it can be concluded that the anaerobic degradation of m-xylene through 3-methylbenzoyl-CoA generates an intermediate whose further catabolism does not require the GcdH enzyme. Whether a different enzymatic system or a different GcdH isoenzyme acting on methylated glutaryl-CoA is required for the anaerobic catabolism of m-xylene is still unknown. In summary, although the anaerobic degradation of toluene and m-xylene share identical activation steps, some of the peripheral reactions, as well as the central and lower pathways for the catabolism of the benzoyl-CoA and 3-methylbenzoyl-CoA generated, appear to be different and specific for each one of these two hydrocarbons. Further work needs to be carried out to unravel the m-xylene-specific enzymes and genes.
Whereas several organisms that can completely mineralize o-xylene coupled to the reduction of nitrate (294) or sulfate (253) have been described, the anaerobic degradation of p-xylene has been reported only for an undefined nitrate-reducing enrichment culture, and there is no organism available in pure culture that can anaerobically mineralize this compound (59). Some reports indicated that the peripheral pathway for o- and p-xylene degradation very likely starts with activation by the addition of fumarate and further proceeds via stepwise β-oxidation to methylbenzoyl-CoA, as is known for toluene and m-xylene (251). Nevertheless, the enzymes and the genes responsible for o- and p-xylene degradation are still unknown, and more efforts should be invested in the future to characterize these peripheral pathways.
(iii) Catabolism of PAHs.
PAHs, also including O-, S-, and N-heterocyclic compounds, are widespread pollutants in the environment, and they are present as complex mixtures of many substances. In most cases, they originate from the natural and anthropogenic pyrolysis of organic material, such as in forest fires, coal-refining processes, and the oil industry. Although many studies have examined the microbial degradation of PAHs under aerobic conditions (57), detailed information on anaerobic degradation of PAHs under sulfate-reducing and nitrate-reducing conditions is scarce. Pathways have been partially described for the anaerobic degradation of naphthalene and 2-methylnaphthalene; very little is known about the degradation of phenanthrene, and some details are available for the anaerobic degradation of heterocyclic aromatic compounds such as benzothiophene, dibenzothiophene, benzofuran, quinoline, and indole (111, 186, 240, 260, 310, 383).
Regarding the anaerobic catabolism of naphthalene, early reports showed that this compound was initially activated by carboxylation to 2-naphthoic acid (387). However, results from further experiments carried out with a highly enriched sulfate-reducing freshwater culture indicated that in the anaerobic degradation of naphthalene, 2-naphthoic acid is not a direct product of carboxylation. In contrast, naphthalene is rather methylated to 2-methylnaphthalene, the methyl group originating from CO2 reduction via the acetyl-CoA pathway. 2-Methylnaphthalene then becomes oxidized to the central metabolite, 2-naphthoic acid or naphthoyl-CoA, by a fumarate addition reaction analogous to that of anaerobic toluene degradation (311). Although all three enzymes involved in the anaerobic degradation of 2-methylnaphthalene to 2-naphthoic acid could be measured in vitro (311), the genes responsible for such activities, as well as those responsible for the assumed methylation of naphthalene to 2-methylnaphthalene, are still unknown in sulfate-reducing freshwater cultures.
Three marine sulfate-reducing naphthalene-degrading pure cultures (NaphS2, NaphS3, and NaphS6) have been isolated so far, and they belong to the Desulfobacteriaceae (deltaproteobacteria). These bacteria also mineralize 2-methylnaphthalene and 2-naphthoate, and two of them (NaphS2 and NaphS3) also degrade benzoate (120, 260). The nmsA gene, encoding the catalytic subunit of the 2-methylnaphthalene-activating enzyme, 2-naphthylmethyl-succinate synthase, from strains NaphS2, NaphS3, and NaphS6 has been cloned and sequenced. The predicted NmsA protein contains the consensus radical-bearing glycine, and it constitutes a phylogenetic subbranch different from those of the benzylsuccinate synthases (for toluene degradation) and 1-methylalkyl-succinate synthases (for n-alkane degradation) (260). The apparent absence of the NmsA protein band in cells grown in naphthalene suggests that naphthalene degradation in marine strains is unlikely to occur via 2-methylnaphthalene. Moreover, substrate adaptation experiments and the isotopic differentiation of metabolites also point to the fact that the activation of naphthalene through methylation is unlikely in marine strains (260) and leave open the possibility for naphthalene activation by carboxylation to 2-naphthoate as initially proposed (387).
Based on the detection of several intermediates in the culture medium, an anaerobic degradation pathway for 2-naphthoic acid or naphthoyl-CoA has been proposed. This pathway involves a series of reduction steps where hydrogen is added to the unsubstituted ring first, generating a tetrahydro-2-naphthoic acid intermediate, which is also the central intermediate of other anaerobic pathways such as that for tetralin degradation. Further hydrogenation of the tetrahydro-2-naphthoic acid finally generates a decalin-2-carboxylic acid. After the cleavage of the first ring of the bicyclic carboxylic acid, the degradation pathway proceeds through saturated intermediates with a cyclohexane ring structure and two carboxylic acid side chains and, therefore, does not generate monoaromatic compounds (8). Whereas the genes for cyclohexane carboxylic acid degradation are known for some bacteria (see above), those responsible for the conversion of 2-naphthoic acid to such cyclohexane carboxylic acids are still unknown.
ANAEROBIC GENE CLUSTERS: COMPARATIVE GENOMICS
Genomic analyses of different aerobic degraders of aromatic compounds have revealed that some of them, such as P. putida and E. coli (85, 170), contain the aromatic gene clusters spread along the chromosome, whereas other bacteria, e.g., Acinetobacter baylyi ADP1, show a supraoperonic clustering of many of these genes in a particular region of the chromosome (357, 382). Among the anaerobic degraders of aromatic compounds, there are also examples of organisms in which the gene clusters involved in anaerobic aromatic degradation are scattered in the chromosome, e.g., Azoarcus sp. strain EbN1 (293) and M. magneticum AMB-1 (236), or are organized into supraoperonic clusters, e.g., R. palustris (104), that may even constitute a catabolic island, e.g., G. metallireducens (48) (Fig. 18). Two major selective forces have been suggested to account for the clustering of catabolic genes: (i) horizontal gene transfer that allows the exchange of the ability to utilize growth substrates among different bacteria and (ii) coamplification that allows an increase in the copy number both for enhancing protein synthesis and exchange or, perhaps more significantly, for the repair of genes in nearby bacteria responding to an environmental challenge. Available evidence suggests that gene amplification is a driving force in the evolution of functionally related gene clusters and in the vertical evolution of catabolic islands. (297, 382). Tandem duplications (amplicons) provide multiple gene copies upon which evolution forces can play, facilitating the genetic rearrangements within the clusters (297, 382). For instance, interspecies, and even intraspecies, comparisons of the benzoyl-CoA gene cluster reveal that whereas the catabolic genes are conserved, their order is not (Fig. 2B).
FIG. 18.
Comparative distribution of the gene clusters involved in anaerobic catabolism of aromatic compounds in different bacterial chromosomes. Characterized or predicted gene clusters for the anaerobic degradation of different aromatic (and cyclohexane carboxylate) compounds are indicated with a different color code. The supraoperonic clustering (26 kb) and the catabolic island (300 kb) of R. palustris (RPA0650 to RPA0673) and G. metallireducens (gmet2037 to gmet2284), respectively, are boxed and expanded. The gene arrangements within the clusters are shown in detail in Fig. 2 to 17.
In R. palustris, the bad genes for the anaerobic degradation of benzoate are clustered with the ali and hba genes, which are involved in the anaerobic degradation of cyclohexane carboxylate and 4-HBA, respectively (Fig. 2 and 18) (104), and all of them are induced in cells cultivated anaerobically on benzoate (267, 362). Since the ali- and hba-encoded pathways converge at two different steps of the central benzoyl-CoA pathway, i.e., the cyclohexenecarbonyl-CoA intermediate and benzoyl-CoA, respectively (Fig. 2 and 7), the arrangement of the ali, hba, and bad genes as a supraoperonic gene cluster in the chromosome of R. palustris may reflect the functional linkage of the corresponding gene products to constitute the benzoyl-CoA catabolon. The analysis of the aromatic catabolic clusters from different R. palustris strains whose genomes have been recently sequenced, e.g., strains BisB5 (GenBank accession number NC_007958), BisB18 (accession number NC_007925), BisA53 (accession number NC_008435), and HaA2 (accession number NC_007778), revealed that the supraoperonic gene cluster composed of bad, ali, and hba previously reported for strain CGA009 is generally conserved in all the new strains, with the exception of strain HaA2, where it is missing. The fact that the genes flanking the supraoperonic gene cluster composed of bad, ali, and hba are different in all four strains reinforces the previous assumption that these genes have evolved together and were acquired independently by different Rhodopseudomonas strains to confer the ability to degrade anaerobically aromatic and alicyclic acids.
In contrast to the supraoperonic clustering observed in R. palustris strains, the gene clusters for anaerobic benzoate, cyclohexane carboxylate, and 4-HBA degradation are located at different chromosomal regions in denitrifying bacteria such as Azoarcus sp. strain EbN1 and M. magneticum (Fig. 18). In Azoarcus sp. strain EbN1, the aromatic gene clusters, including those encoding aerobic pathways, are generally distributed across the chromosome and in some cases, such as the genes encoding the peripheral phenylalanine pathway, are not even organized into a cluster (293). However, it is worth noting that those clusters likely responsible for the anaerobic conversion of phenolic compounds (phenol and p-cresol) and aromatic alcohols appear to be organized in supraoperonic structures, one of them including the benzoate and phenylacetate clusters, in two defined regions of the genome (Fig. 18). A gene cluster likely involved in the anaerobic degradation of benzoate and other aromatic compounds has been identified in a metagenomic library from a microbial mat of the Black Sea (201). The 79-kb FC1 contig contained all genes required for the central benzoyl-CoA pathway of denitrifying bacteria, some genes that might participate in the lower benzoyl-CoA pathway (Fig. 2B), genes similar to those involved in the initial reactions of anaerobic phenol degradation (ubi-like genes), as well as orthologs of the genes encoding the 4-hydroxybenzoyl-CoA reductase (hcr genes) (Fig. 2 and 7). Thus, this metagenomic sequence represents another example of a supraoperonic clustering from an organism, likely a denitrifying bacterium, which may be able to use phenol, benzoate, and 4-HBA anaerobically through the classical central benzoyl-CoA pathway (201).
The highest clustering of aromatic catabolic genes is that found in the iron-reducing bacterium G. metallireducens, where most aromatic catabolic clusters, with the sole exception of the toluene catabolic genes (bss and bbs genes), are located in a chromosomal region that shows many of the hallmarks of a genomic island (48, 92). This 300-kb catabolic island contains 244 genes (gmet2037 to gmet2284) including those responsible for the benzoyl-CoA central (clusters IA and II) and lower (cluster IB) pathways and those predicted to be responsible for the catabolism of phenol, p-cresol, 4-hydroxybenzaldehyde, 4-HBA, benzyl alcohol, and benzaldehyde (benzoyl-CoA catabolon) (Fig. 18) (48, 277, 377). The catabolic island contains genes that are species specific since they do not have orthologs in the closely related species G. sulfurreducens, which cannot degrade aromatic compounds. The genes are encoded in large, discrete units flanked by genes with atypical nucleotide compositions, tRNA genes, and phage-like integrases. Since the region contains insertion sequence (IS) elements and repetitive sequences, it shows substantial evidence of potential genetic mobility. However, the aromatic catabolic genes do not have atypical nucleotide composition (GC and dinucleotide content) compared to that of the rest of the genome, implying that if there were a horizontal gene transfer event(s) from distantly related organisms, this was not a recent transfer, and the genes have ameliorated since this event occurred (48). The association of aromatic catabolic clusters with IS elements, transposons, and/or phage-like integrases in the genomes of other anaerobes, such as Azoarcus sp. strain EbN1, has also been observed, and it has been pointed out that this general feature indicates a major role of horizontal gene transfer in the metabolic evolution and catabolic versatility of the aromatic degraders (293, 358).
In some cases, certain genes of aromatic catabolic clusters have paralogs located in other regions of the bacterial genome. Thus, in G. metallireducens, the bamA and bamB/bamC genes of the benzoyl-CoA cluster have benzoate-induced paralogs (bamA2 and bamB2/bamC2) outside of the aromatic catabolic island (48). In Geobacter sp. strain FRC-32, an isolate from a hydrocarbon-contaminated field, bam genes similar to those described for G. metallireducens have been identified. However, in strain FRC-32, eight genes downstream of the bamC-bamI cluster, there are two bamB homologs and a second, identically organized, bamC-bamI gene cluster (48). The role of these paralogous genes in Geobacter strains remains to be determined.
A paralogous bzdNOPQ gene cluster encoding a putative BCR enzyme has also been shown for Azoarcus sp. strain EbN1. However, in this case, this second bzdNOPQ gene cluster (hbrCBDA) has been suggested to encode the enzyme that dearomatizes 3-hydroxybenzoyl-CoA (Fig. 4) (293, 378). Interestingly, in addition to two sets of korCAB genes (encoding the putative three-component BCRAs ferredoxin-regenerating system), the genome of Azoarcus sp. strain EbN1 also contains a korAB operon (encoding a putative two-component BCRTa-type ferredoxin-regenerating system), which is located adjacent to the gene cluster involved in phenol degradation and whose function is still unknown (293). Azoarcus sp. strain EbN1 also harbors genes paralogous to those of the ethylbenzene upper operon (ebd and ped genes) and phenol operon (pps and putative carboxylase genes) (293). It was suggested that these additional gene clusters could be involved in the catabolism of hydrocarbons and phenolic compounds other than ethylbenzene and phenol, respectively (378).
Another example of genetic redundancy is found in the genome of S. aciditrophicus, where five different gene clusters (Sa1 to Sa5) contain putative BCR-encoding genes (237). Although cluster Sa2 is thought to be the benzoyl-CoA gene cluster (Fig. 2B), the role of the other Sa clusters in anaerobic benzoate degradation cannot be ruled out. Nevertheless, these additional Sa clusters may well be involved in the metabolism of cyclohexane carboxylate in S. aciditrophicus. Consistent with this, a biosynthetic pathway that forms cyclohexane carboxylate from crotonate by using benzoyl-CoA as an intermediate has been reported (256). The reversal of the pathway used to synthesize cyclohexane carboxylate may by used by S. aciditrophicus to degrade this alicyclic compound, which would explain that cell extracts of syntrophically grown cells contain the enzyme activities for the conversion of cyclohexene-carbonyl-CoA to pimelyl-CoA (106, 256).
In prokaryotes, genetic redundancy is considered to confer a certain degree of robustness to organisms because they can maintain a stable phenotype under hereditary (e.g., genetic mutations) or environmental changes. In functional terms, the existence of genomic redundancy can be explained as the consequence of some selective processes, i.e., elevated protein dosage (giving rise to identical, duplicated genes), protein diversification (giving rise to standard paralogs devoted to different specialized functions), and adaptation to environmental variations (giving rise to ecoparalogs that perform the same cellular function under different ecological conditions) (314). The existence of ecoparalogs in anaerobic aromatic degraders that inhabit multiple niches, e.g., aerobic and anaerobic lifestyles as in facultative anaerobes, or that are exposed to many external fluctuations might be a successful strategy to maintain essential catabolic functions over a wider range of environmental conditions. Further research to confirm these assumptions should be carried out in the future.
AUXILIARY FUNCTIONS FOR ANAEROBIC METABOLISM OF AROMATIC COMPOUNDS
Auxiliary Genes for Anaerobic Catabolism of Aromatic Compounds
The transcriptomic and proteomic approaches carried out with some anaerobic biodegraders, such as R. palustris (362), Azoarcus sp. strain EbN1 (378), and G. metallireducens (48), have provided information on genes induced by aromatic growth substrates and that have not been assigned a function in aromatic catabolism. Although many of these genes encode proteins of unknown function, this global gene expression profiling revealed so-far-unknown cellular activities that are also related (directly or indirectly) to the anaerobic degradation of aromatic compounds.
Metabolic genes.
As shown above, in the central benzoyl-CoA pathway, one molecule of benzoyl-CoA finally yields three molecules of acetyl-CoA and one CO2 (Fig. 2 and 3). In photosynthetic bacteria, energy is derived from light, and acetyl-CoA is used in biosynthetic reactions (128, 142, 213). Since the aromatic compounds are electron rich relative to cell material, they cannot be fully assimilated into a biomass unless an external electron acceptor like carbon dioxide is available. Accordingly, the cbb genes, encoding the two forms of ribulose bisphosphate carboxylase (RPA1559 to RPA1560 and RPA4641), along with their associated genes for carbon dioxide fixation (RPA1561 and RPA4642 to RPA4645), were induced when R. palustris was cultivated in the presence of benzoate or p-coumarate. Therefore, the main carbon dioxide-assimilating enzymes of the Calvin cycle were expressed at higher abundances to serve as a reducing equivalent sink during the growth of R. palustris cells on aromatic compounds (267, 362).
In contrast to what has been observed for phototrophs, in heterotrophic bacteria, most of the acetyl-CoA is fully oxidized to CO2 in a modified tricarboxylic acid (TCA) cycle with nitrate, Fe(III), sulfate, or some other compound acting as the final electron acceptor in an anaerobic respiratory chain (128, 142). In denitrifying bacteria, one can estimate that approximately two-thirds of the acetyl-CoA molecules are oxidized and that one-third is used for biosynthesis via the glyoxylate bypass (99). In Thauera and Azoarcus cells growing in aromatic compounds, the 2-oxoglutarate dehydrogenase complex acting under aerobic conditions in the TCA cycle becomes replaced by a KGOR, which is encoded by the kor genes, generates succinyl-CoA, and acts as the reducing enzyme of the low-potential ferredoxin that donates electrons to the BCR (see above) (33, 94, 99). The further metabolism of succinyl-CoA in Geobacter species grown on acetate, such as G. sulfurreducens, involves an acetate:succinyl-CoA transferase that converts succinyl-CoA to succinate coupled to the activation of acetate to acetyl-CoA in the TCA cycle (121). However, the ortholog of the acetate:succinyl-CoA transferase-encoding gene in G. metallireducens (gmet3044) had a decreased abundance during benzoate oxidation (48). On the contrary, three set of genes (including the scsAB genes of cluster IB within the catabolic island), each one predicted to encode a two-subunit ATP-yielding succinyl-CoA synthetase enzyme from the TCA cycle, were shown to have increased transcript abundance during benzoate degradation (48, 377). Thus, in G. metallireducens, the enzyme that catalyzes the conversion of succinyl-CoA to succinate is changed when benzoate is used as the electron donor. This result is in agreement with the fact that the product of benzoate oxidation is directly acetyl-CoA and not acetate, and therefore, there would be no need for an acetate:succinyl-CoA transferase. Moreover, the involvement of a succinyl-CoA synthetase instead of an acetate:succinyl-CoA transferase in G. metallireducens cells grown in benzoate would allow the generation of an ATP by substrate-level phosphorylation for each of the acetyl-CoA molecules generated from benzoate oxidation (48).
Stress genes.
Recently, the existence of a newly discovered benzoate-induced, benzoyl-CoA-forming, 1,5-dienoyl-CoA:acceptor oxidoreductase activity in different facultative (T. aromatica, A. evansii, and R. palustris) and obligate (G. metallireducens, D. multivorans, and S. aciditrophicus) anaerobic bacteria grown on benzoate has been shown. The corresponding enzyme was characterized in T. aromatica as being a monomeric flavoprotein/FeS protein that catalyzes the irreversible oxidation of the 1,5-dienoyl-CoA product of BCR, using O2 as the final electron acceptor and generating benzoyl-CoA. Therefore, this new enzyme is referred to as 1,5-dienoyl-CoA oxidase (DCO) (349). Although the gene encoding this new enzyme activity is still unknown, mass spectrometric analysis of tryptic digests of the purified enzyme from T. aromatica revealed significant similarities with 2,4-dienoyl-CoA reductases, NADH oxidases, and “old yellow” enzymes. The best matches were obtained with the ebA2099 gene product from Azoarcus sp. strain EbN1, which is annotated as an NADH oxidase (NoxB-2) (349). It was proposed that DCO provides protection for the extremely oxygen-sensitive BCR enzyme when facultative anaerobes, like Thauera and Azoarcus strains, thrive in narrow oxygen gradients and must be able to adapt rapidly to changing oxygen concentrations. DCO could behave as an oxygen-scavenging enzyme since it has a very high affinity for dioxygen, keeping oxygen levels as low as possible and, therefore, providing an attractive enzymatic tool to remove molecular oxygen rapidly. Furthermore, DCO becomes activated by a redox-dependent switch that guarantees that no futile 1,5-dienoyl-CoA aromatization occurs when the redox potential is low and BCR is active, but it enables a rapid activation of DCO during oxidative stress. The copurification of BCR and DCO suggests an association of both enzymes, which would facilitate the use of the product formed by BCR for oxygen detoxification (349). The requirement for keeping oxygen levels as low as possible when an oxygen-sensitive enzyme is synthesized has been well documented for the protection of nitrogenase against oxygen damage under several conditions (20). In S. aciditrophicus, benzoate formation was observed during crotonate fermentation, which suggests the presence of a rearomatizing activity in vivo (256). It is worth noting that the putative DCO from Azoarcus sp. strain EbN1, encoded by the ebA2099 gene, is located in the close vicinity of the katA gene (ebA2102), encoding a catalase enzyme. Moreover, the purified DCO from T. aromatica was also enriched with an additional protein that was identified as being an aconitase-like protein (349). Since both catalase and aconitase enzymes, which are classical oxidative stress-related proteins, have been shown to be upregulated when anaerobes are grown in the presence of several aromatic compounds (351), the hypothesis that the production of DCO may represent an additional strategy to cope with the oxidative stress conditions generated during the anaerobic catabolism of aromatic compounds (see below) sounds plausible.
Physiological adaptation experiments combined with global expression profiling (DNA microarrays and proteomics) revealed that gene products in addition to those encoded by the bss and bbs clusters are likely to be involved in toluene degradation in Azoarcus sp. strain EbN1. Thus, 7.2 kb upstream of the bbs cluster, there are five genes (ebA1926 to ebA1936) organized in an operon-like structure, two of which (ebA1932 and ebA1936) encode proteins that are highly induced when the cells were grown in toluene and whose putative promoter region shows significant similarity to that of the bbs and bss operons, agreeing well with its toluene-specific induction (203). Within this operon-like structure, the ebA1926 gene may encode a universal stress-like protein, and the ebA1928-ebA1932-ebA1936 genes may encode a putative toluene-specific RND-type efflux pump involved in coping with toxic toluene concentrations (203, 379). Interestingly, ebA1932 and ebA1936 are highly similar to the products of orf1 and orf2 from T. aromatica K172. In the latter organism, these two ORFs are located adjacent to the tdiSR genes (Fig. 16). Since the new toluene-related gene cluster of Azoarcus sp. strain EbN1 is flanked by ISE1 and a transposon gene fragment (tnpF4) at one end and by another transposon gene fragment (tnpF) at the other end, one may speculate that these new genes have been translocated in the genome of strain EbN1 compared to their organization in T. aromatica K172 (203). The chromosome of strain EbN1 harbors two additional gene clusters, which display high sequence similarity to the gene cluster comprising the ebA1926 to ebA1936 cluster, and they exhibit the same gene order: (i) the cluster comprising the ebA326 to ebA335 cluster, which is likely involved in anaerobic p-ethylphenol and p-hydroxyacetophenone metabolism (see above), and (ii) the cluster comprising ebA5762 to ebA5768, which is located near to the catabolic genes involved in a putative phenolic compounds degradation pathway (see above). Interestingly, these three paralogous gene clusters are specifically expressed in the presence of different aromatic compounds, which correlate well with the genomic content in each case (379). The products of the ebA326 to ebA335, ebA1926 to ebA1936, and ebA5762 to ebA5768 genes were specifically formed during anaerobic growth on p-ethylphenol (379), toluene (203), and phenol/p-cresol (378), respectively. The formation of ebA5762 to ebA5768 was also observed in succinate-utilizing cells suddenly stressed with phenolic compounds (351). These similar regulatory patterns support the predicted solvent-related function of the encoded proteins and point to a fine-tuned system of solvent stress tolerance in Azoarcus sp. strain EbN1 (see below) (379).
Transport and chemotaxis genes.
In R. palustris, several methyl-accepting chemotaxis proteins (RPA0139, RPA0142, RPA1678, RPA3185, RPA4302, and RPA4639) were induced under conditions of anaerobic growth in benzoate and other aromatic compounds such as p-coumarate. Since R. palustris is a motile bacterium, these upregulated proteins could serve as chemoreceptors that enable cells to sense and swim toward plant-derived aromatic compounds (267, 268). A significant number of membrane transporters were also induced under conditions anaerobic growth in aromatic compounds, and some of them may facilitate the uptake of these carbon sources and/or the reuptake of some catabolic intermediates transiently excreted into the growth medium (267). Transmembrane trafficking of substrates and metabolic intermediates generated during the anaerobic catabolism of aromatic compounds can therefore play a pivotal role when these compounds are used as a carbon source.
Recently, one of the genes (rpaI) induced during p-coumarate degradation in R. palustris was shown to encode a new acyl-homoserine lactone synthase that generates a new class of homoserine lactone quorum-sensing signals using environmental p-coumarate rather than fatty acids from cellular pools. Although the expression of none of the genes thought to be involved in p-coumarate degradation is controlled by the new p-coumaroyl-homoserine lactone, and an R. palustris rpaI mutant grows normally on p-coumarate, this new signal molecule raises fundamental questions about quorum sensing within the context of environmental signaling (317).
Aromatic Compounds as Stressors under Anaerobic Conditions
A significant number of aromatic compounds simultaneously serve as potential nutrients to be metabolized by bacteria but also serve as stressors since they are membrane-damaging toxic compounds, and their metabolism can also generate stress signals, e.g., oxidative stress, within the cell. The toxicity of some aromatic compounds, such as BTEX and phenolic (phenol and cresols, etc.) compounds, generally correlates with their hydrophobicity, which is described by the logarithm of their partition coefficients in a mixture of n-octanol and water (log PO/W). Compounds with a log PO/W of between 1 and 4 are cytotoxic because they preferentially dissolve in biological membranes. Consequently, membrane fluidity increases, which leads to a loss of ions, ATP, and other cellular metabolites. Moreover, the dissipation of the proton motive force and the denaturation of membrane proteins (e.g., respiratory complexes or nutrient transporters) result in severe energetic problems (117, 337). Some aromatic hydrocarbons, such as toluene and xylene, are sensed by the aerobic degrader P. putida firstly as stressors rather than as nutrients, and the cells display a short-term response that activates stress tolerance genes at a minimal cost in terms of energy. The short-term response to such toxic compounds involved a loss of motility functions and the inhibition of large portions of the basic metabolic machinery (93). Nevertheless, after the short-term stress responses, a metabolic program for the degradation of toxic compounds has been observed. Thus, the adaptative solution to the biodegradation-versus-stress dilemma appears to be temporally subordinating the expression of biodegradative genes (metabolic program) to adaptation to physicochemical stresses (stress program) in order to not compromise survival and/or metabolic fitness (361).
Bacterial solvent tolerance has been studied mainly in aerobic pseudomonads, and it involves solvent efflux pumps, heat shock proteins, and modifications of the cytoplasmic membrane (117, 154). It has been reported that anaerobically grown T. aromatica, D. multivorans, and G. sulfurreducens bacteria are more sensitive to organic solvents than are aerobic bacteria (96). However, in Azoarcus sp. strain EbN1, the semi-inhibitory alkylbenzene concentrations determined for cells grown in succinate were at least 2.5-fold higher than those reported for other anaerobic biodegraders and similar to those reported for aerobic P. putida strains (351). It was shown that during anaerobic growth with 0.32 mM ethylbenzene or 0.74 mM toluene, the corresponding degradation pathways were operative in strain EbN1. However, when cells grown in succinate were suddenly exposed to 0.5 mM ethylbenzene, 1.2 mM toluene, 3.0 mM p-cresol, and 6.5 mM phenol, applied as single stressors or as a mixture (total solvent concentration, 2.7 mM), the corresponding degradation pathways were not operative, and complete growth inhibition was observed (351). Thus, aromatic compound degradation seems not to contribute to the solvent tolerance of strain EbN1 (351). Remarkably, the toluene tolerance of P. putida DOT-T1E also did not change when toluene degradation was disabled by the deletion of one subunit of toluene dioxygenase (255). Considering its markedly high solvent tolerance (e.g., growth with <0.48 mM ethylbenzene and <0.86 mM toluene), Azoarcus sp. strain EbN1 should be able to survive and proliferate in most contaminated environments (139) and represents a promising model organism for in situ anaerobic removal processes.
During the anaerobic growth of Azoarcus sp. strain EbN1 with increasing concentrations of solvents, nitrate consumption and the turnover of the intermediary nitrite formed decelerated. Accordingly, the abundance of several denitrification enzymes changed in response to solvent stress. Thus, the periplasmic cytochrome cd1 nitrite reductase (NirS) and nitrous oxide (NO) reductase (NosZ) as well as the reductase-related chaperone (NorQ) were upregulated (351, 378). It could be argued that increased NirS, NosZ, and NorQ concentrations might compensate for solvent-induced NirS and NosZ inactivation in strain EbN1 (351). On the other hand, alkylbenzene-utilizing cultures also displayed polyhydroxybutyrate (PHB) accumulation up to 10% of the cell dry weight. One may speculate that in alkylbenzene-utilizing cells of strain EbN1, alkylbenzene-derived acetyl-CoA is rerouted from oxidation via the TCA cycle to PHB synthesis, which is accompanied by a decrease in the NAD(P)H pool and recycling of free CoA. Thus, PHB would behave as a sink for reducing equivalents, ensuring continuous alkylbenzene degradation under conditions where the potential electron supply provided by the organic substrate exceeded the electron-accepting capacity of nitrate (351).
Oxidative stress responses in aerobic degraders challenged with aromatic solvents, where highly reactive oxygen species can be generated from impaired oxygen respiration, have been reported (93). Even though dioxygen and superoxide are not present in anoxic media, highly reactive NO compounds can be generated during denitrification (166, 351). Consistent with this, the observed increased abundance of superoxide dismutase (SodB) during anaerobic growth with toluene as well as the increased abundance of catalase (KatA), a DNA binding protein related to oxidative stress (Dps), and the NO-detoxifying flavorubredoxin (NorVW) in succinate-utilizing Azoarcus sp. strain EbN1 cultures shocked with a solvent mixture may have been due to the presence of such reactive NO species (351). It is known that there is a coordinated regulation of antioxidative defense and cellular iron homeostasis in bacteria (350). Accordingly, the oxidative stress observed in Azoarcus sp. strain EbN1 cells utilizing alkylbenzenes anaerobically is paralleled by large decreases in the abundances of two predicted iron uptake proteins, ebA1861 and ebA4918. On the other hand, the downregulation of these two proteins might result in a reduced intracellular iron availability, which would agree with the upregulation of aconitases A and A2 (351).
Some general stress-related proteins were also upregulated in succinate-utilizing, solvent-shocked Azoarcus sp. strain EbN1 cultures. Thus, betaine aldehyde dehydrogenase (BetB), catalyzing the last biosynthetic reaction of osmotically active betaine, was induced during growth with toluene. The WbjB protein, likely involved in the biosynthesis of some components of lipopolysaccharide, was upregulated in cells shocked with ethylbenzene (351). Differences in solvent tolerance have already been correlated with lipopolysaccharide structure or composition in other microorganisms (370). The upregulation of several heat shock proteins, e.g., HtpG, GrpE, ebB88, and ebA2730, and the chaperone ClpB was also observed in succinate-utilizing Azoarcus sp. strain EbN1 cells shocked with most aromatic solvents (351). In alkylbenzene-utilizing cells, the ClpB chaperone is constitutively formed, which was also observed during the aerobic and anaerobic growth of strain EbN1 with other aromatic compounds (378). Finally, some porins, such as OmpC, were also upregulated specifically in succinate-utilizing cells shocked with phenolic compounds (phenol and p-cresol), probably to stabilize the outer membrane (351). As indicated above, three paralogous gene clusters specifically expressed when Azoarcus sp. strain EbN1 cells are grown anaerobically in the presence of solvents such as toluene, p-ethylphenol, phenol, and p-cresol have been suggested to be involved in a solvent stress response (379).
Whether an aromatic-induced stress program similar to that found in Azoarcus sp. strain EbN1 is also present in other anaerobic biodegraders is still unknown, and further studies concerning the short-term response of these bacteria toward aromatic compounds as stressors under anaerobic conditions should be undertaken.
REGULATION OF GENE EXPRESSION OF ANAEROBIC CATABOLIC CLUSTERS
The catalytic performance of a bacterial cell to degrade aromatic compounds is dependent not only on the presence of the adequate catabolic genes that encode the transporters and enzymes able to catalyze the degradation of the compounds but also on their expression where and when needed. The efficient expression of genes depends not only on the presence and/or absence of the aromatic compound but also on a wide range of diverse environmental signals (55, 227). Although regulation can be carried out at different levels (transcription, translation, and posttranslation), transcriptional regulation appears to be the most common, or at least the most studied, mechanism for the control of gene expression of aromatic catabolic clusters (53, 86, 334, 352).
A global analysis of the specific regulatory proteins that control the catabolism of aromatic compounds revealed a great divergence in their evolutionary origins and suggests that catabolic and regulatory genes have evolved independently (55, 81). One regulatory problem, i.e., inducing the expression of an operon in the presence of a given aromatic compound, can be solved through different types of regulators and mechanisms of transcriptional control. One strategy to respond to similar signals in different ways is to combine similar input domains (small-molecule binding motifs) with sets of different DNA binding domains. Moreover, even regulators that show similar whole-domain architectures might be responsible for different biological effects depending on the locations of their binding sites (operator regions) in the cognate promoters. Thus, the same regulator can activate some genes when it binds upstream of the RNA polymerase (RNAP) binding sites while it represses others when it binds downstream of such RNAP binding regions. Taken together, all these possibilities endow regulatory networks with an extraordinary degree of plasticity and adaptability (53, 55, 86, 352).
Whereas most of the current information on regulatory issues comes from aerobic catabolic pathways, there is increasing evidence that similar principles of regulation apply to the anaerobic catabolism of aromatic compounds. Thus, although only a few effector-specific regulators have been characterized so far, a number of potential regulatory proteins that belong to different families of transcriptional regulators and that may control the expression of different aromatic catabolic genes can be identified in the currently described catabolic clusters (Table 3).
TABLE 3.
Some regulatory proteins involved in anaerobic catabolism of aromatic compounds
| Regulator | Familyb | Microorganism | Activity | Pathway | GenBank accession no. |
|---|---|---|---|---|---|
| Effector specific | |||||
| BzdR | BzdR | Azoarcus sp. strain CIB | Repressor | Benzoate | AAQ08805 |
| ORF11a | XylR/DmpR | T. aromatica K172 | Unknown | Phenol | CAC12685 |
| PdeRa | XylR/DmpR | Azoarcus sp. strain EbN1 | Unknown | Phenol | Q5P474 |
| Gmet1542a | XylR/DmpR | G. metallireducens GS-15 | Unknown | Toluene | ABB31776 |
| EbA324 | XylR/DmpR | Azoarcus sp. strain EbN1 | Unknown | p-Ethylphenol | Q5P8R7 |
| TutC1/B1 | TCRb | T. aromatica T1 | Activator | Toluene | AAD12187/AAD12186 |
| TdiS/Ra | TCR | T. aromatica K172 | Activator | Toluene | CAA05048/CAA05049 |
| TdiS/Ra | TCR | Azoarcus sp. strain EbN1 | Activator | Toluene | CAI07156/CAI07155 |
| TdiS/Ra | TCR | Azoarcus sp. strain T | Activator | Toluene | AAK50369/AAK50368 |
| Tcs2/Tcr2a | TCR | Azoarcus sp. strain EbN1 | Unknown | Ethylbenzene | CAI07438/CAI07439 |
| Tcs1/Tcr1a | TCR | Azoarcus sp. strain EbN1 | Unknown | Acetophenone | CAI07436/CAI07437 |
| BamV/Wa | TCR | G. metallireducens GS-15 | Unknown | Benzoate | ABB32375/ABB32374 |
| HbaR | FNR/CRP | R. palustris CGA009 | Activator | 4-Hydroxybenzoate | AAF04013 |
| CprK | FNR/CRP | D. hafniense | Activator | o-Chlorophenol dehalogenation | AAL87770 |
| GcdR | LysR | Azoarcus sp. strain CIB | Activator | Lower benzoyl-CoA pathway | ABM69269 |
| NicRa | LysR | E. barkeri | Unknown | Nicotinate | ABC88392 |
| PadRa | GntR | Azoarcus sp. strain EbN1 | Unknown | Phenylacetate | CAI09182 |
| Rpd1521a | GntR | R. palustris BisB5 | Unknown | Phenylacetate | ABE38758 |
| ORF1a | MarR | T. aromatica K172 | Unknown | 4-Hydroxybenzoate | O33817 |
| PcmQa | MarR | G. metallireducens GS-15 | Unknown | 4-Hydroxybenzoate | ABB32362 |
| BadR | MarR | R. palustris CGA009 | Activator | Benzoate | AAC23923 |
| Rpa1794a | MarR | R. palustris CGA009 | Unknown | p-Coumarate | Q6N8V9 |
| EbA715a | TetR | Azoarcus sp. strain EbN1 | Unknown | 3-Hydroxybenzoate | CAI06487 |
| Gmet1520a | TetR | G. metallireducens AMB-1 | Unknown | Toluene | ABB31754 |
| BadM | Rrf2 | R. palustris CGA009 | Repressor | Benzoate | O07465 |
| Oxygen dependent | |||||
| AadR | FNR/CRP | R. palustris CGA009 | Activator | Benzoate/4-Hydroxybenzoate | B43334 |
| AcpR | FNR/CRP | Azoarcus sp. strain CIB | Activator | Benzoate | AAY81959 |
Predicted.
TCR, two-component regulatory system (histidine kinase/response regulator).
Bacteria thriving in the environment face a range of biological, physical, and chemical signals that need to be processed to achieve a positive or negative physiological response. For instance, bacteria are often confronted with alternative carbon sources, and they need to decide which of them will be preferentially consumed before metabolizing less preferred substrates to optimize their metabolic return. To achieve this goal, bacteria have evolved physiological control mechanisms that govern and adjust the specific regulation of catabolic operons to the physiological and metabolic state of the cells (53, 54, 287, 334). Such overimposed regulation is carried out by global factors that operate on given promoters concomitantly with more specific regulators. The concept of overimposed regulation fits with the idea of connectivity and signal integration in transcriptional regulatory networks; i.e., many regulators in the cell influence the activity of only a few genes, and a few regulators affect many genes (55). Moreover, there are several ways to couple the catabolic promoter to cell physiology and stress responses, which can be regarded as a strategy to facilitate its establishment when transferred to different hosts by having various possibilities for interacting with the physiology of the new recipients (55). Although most of the regulatory proteins involved in the overimposed regulation of the catabolism of aromatic compounds have been described for aerobic pathways (53, 86, 233, 254, 287, 306, 334), a couple of regulators that act in concert with the cognate-specific regulators have been described for some anaerobic catabolic pathways (Table 3).
Effector-Specific Regulation
As it is well known for the aerobic catabolism of aromatic compounds, the individual pathways responsible for the anaerobic catabolism of aromatic compounds generally display rather strict regulation in response to the respective substrate (378). Although the best-studied regulatory circuits are those controlling the expression of the genes responsible for the anaerobic catabolism of benzoate, there are also some reports of specific regulators controlling the anaerobic catabolism of other aromatic acids and hydrocarbons.
Regulation of the central benzoyl-CoA pathway.
The specific regulation of the gene cluster involved in the anaerobic catabolism of benzoate has been studied in two model organisms, the phototroph R. palustris and the denitrifying bacterium Azoarcus sp. strain CIB.
(i) The R. palustris regulatory circuit.
In R. palustris, the benzoate-dependent induction of the bad genes and the corresponding gene products has been confirmed by classical gene expression studies (102, 104, 274, 276) and by more global approaches such as proteomics (267, 362). The current view indicates that the bad genes are transcriptionally organized into five different operons: badDEFGAB, badHI-aliBA-badK, badC, badR, and badM (Fig. 19) (102, 104, 274, 276).
FIG. 19.
Transcriptional organization and regulation of the ali-bad-hba supraoperonic cluster of R. palustris. The ali, bad, and hba genes are represented by yellow, blue, and green boxes, respectively. The enzymes encoded by the ali, bad, and hba genes and the reactions catalyzed by these enzymes are boxed with yellow, blue, and green, respectively. The badR, badM, and hbaR regulatory genes and the corresponding BadR, BadM, and HbaR regulators, respectively, are indicated in red. The AadR regulatory protein, which is encoded outside the clustering, is also shown (violet). The arrows at the top of the genes indicate that these genes constitute an operon (solid arrows) or a putative operon (dashed arrows). The badD, hbaA, and hbaR promoters that were reported to be controlled by the transcriptional regulators are shown (red bent arrows). The − and + symbols indicate transcriptional repression and activation, respectively.
The badDEFG genes, encoding the four-subunit BCRRp, together with badA (encodes the benzoate-CoA ligase) and badB (encodes a ferredoxin) are transcribed as an operon with the transcription start site present 71 nucleotides upstream of the predicted translation initiation codon of the badD gene (104, 276). The badR gene product behaves as a transcriptional activator of the badD promoter (Fig. 19). Although it is likely that BadR binds directly to the badD promoter region to activate gene expression, this notion has yet to be demonstrated (102).
The BadR protein belongs to the MarR family of bacterial regulatory proteins, but it differs from many members of this family in that it activates, rather than represses, gene expression (Table 3). MarR, the best-characterized member of this family, negatively regulates the expression of the antibiotic resistance genes marAB, with 2-hydroxybenzoate (salicylate) representing the inducer molecule (234). In addition to MarR, several other MarR-like regulators, e.g., HpaR and CbaR, are also responding to aromatic compounds (53, 287, 352). In the case of BadR, this activator was suggested to respond to either benzoate or benzoyl-CoA. Of the two compounds, benzoyl-CoA is the more likely effector because (i) benzoyl-CoA accumulated to high levels in the badE::lacZ reporter strain that was used to monitor the activity of the badD promoter; (ii) several compounds, such as 4-HBA, that have been found to induce badE expression are metabolized by R. palustris to form benzoyl-CoA, but not free benzoate, as an intermediate; and (iii) aromatic compounds such as 3-chlorobenzoate that are structurally similar to benzoate but that are not metabolized to benzoyl-CoA do not induce badE expression (102).
In addition to BadR-dependent regulation, the badD promoter is also controlled by the regulatory protein AadR (Table 3) in response to anaerobiosis (overimposed regulation). Together, these two regulators account for the approximately 100-fold induction of badE::lacZ expression that occurs when R. palustris cells grown aerobically on succinate are shifted to anaerobic growth with benzoate: the aadR gene was required for a 20-fold increase in expression that occurred in response to anaerobiosis, and badR was responsible for a further 5-fold increase in expression that occurred in response to benzoate. Accordingly, an R. palustris badR aadR double mutant, in contrast to badR or aadR single mutants, was completely defective in anaerobic growth on benzoate (102).
Although BadR (which senses benzoate or benzoyl-CoA) and AadR (which senses anaerobiosis) appear to account for the full range of benzoate-induced expression of the badDEFG genes (102), a third transcriptional regulator, the BadM protein, was also shown to control the activity of the badD promoter acting as a repressor (Fig. 19) (276). Although it is tempting to speculate that BadM represses benzoate degradation by binding to the badD promoter and that benzoate binds to BadM to induce dissociation from DNA and derepresses gene expression in R. palustris, this assumption has not yet been demonstrated (276). The BadM protein belongs to the Rrf2 family of transcriptional regulators (Table 3), which includes repressors of genes involved in nitrite, nitric oxide, or iron metabolism (130). Although BadM has the Rrf2-type helix-turn-helix (HTH) domain signature presumed to be involved in DNA binding, it does not have the conserved cysteines that are found in the Fe-S cluster of many members of this family (276). Transcriptome analysis revealed that the expression of only a few genes, in addition to the badDEFGAB operon, was also affected by more than fivefold in an R. palustris badM mutant. These genes included a dicarboxylic acid transporter gene (RPA2448), a gene for a conserved hypothetical protein (RPA3401), a possible cytochrome P450 gene (RPA1009), and an operon of conserved hypothetical genes (RPA1209 to RPA1212). However, none of these genes appear to encode Fe-S proteins involved in anaerobic respiration or iron-related metabolism. Thus, the role of BadM as a more general regulatory protein is still under consideration (276).
Despite the studies mentioned above, the understanding of the regulation of anaerobic benzoate degradation in R. palustris is far from complete. For example, several parts of the anaerobic benzoate degradation cluster, as the badHI-aliBA-badK operon (also called chc operon) responsible for the benzoate/cyclohexane carboxylate-inducible modified β-oxidation step after benzoyl-CoA reduction/cyclohexane carboxylate activation (Fig. 19), do not appear to be controlled by the regulators characterized so far, i.e., BadR and BadM (102, 276). How the genes encoding these other benzoate/cyclohexane carboxylate degradation enzymes are regulated is an open question, since based on sequence analysis, the bad gene cluster does not include other genes that seem likely to have a regulatory function. The regulation of the badR, badC, badM, and badL genes should also be addressed to get a global view of the regulation of the benzoate degradation pathway in R. palustris.
(ii) The Azoarcus sp. strain CIB regulatory circuit.
As was shown previously for the bad cluster of R. palustris, the bzd genes responsible for the anaerobic catabolism of benzoate in Azoarcus sp. strain CIB are also specifically induced when the cells are grown in the presence of this aromatic acid (224). The bzdNOPQMSTUVWXYZA catabolic genes constitute a single and large operon that is under the control of the PN promoter (Fig. 20) (224). Upstream of the bzd catabolic operon, and expressed as an independent transcriptional unit, there is a gene, bzdR, whose product controls the expression of the catabolic operon at the level of the PN promoter (Fig. 20), as demonstrated by analyzing the expression of a PN::lacZ translational fusion (14). Thus, it was shown that BzdR negatively regulates the expression of the bzd catabolic operon by repressing the PN promoter when Azoarcus sp. strain CIB cells do not use benzoate as a carbon source. Interestingly, benzoate was not the actual inducer molecule. The results obtained with an engineered E. coli strain expressing the bzdA gene (codes for the anaerobic benzoate-CoA ligase) and, therefore, able to synthesize benzoyl-CoA from the benzoate added to the culture medium revealed that the activation of the PN promoter is triggered by benzoyl-CoA (Fig. 20). In vitro studies confirmed that aromatic acids were not effector molecules, and only benzoyl-CoA, but not some close analogs such as phenylacetyl-CoA, was efficiently recognized by the BzdR protein (14). These results suggest, therefore, that BzdR has evolved to become specifically adapted to control the anaerobic benzoate degradation pathway in Azoarcus by detecting and responding to the presence of the first intermediate of such a pathway, benzoyl-CoA.
FIG. 20.
Transcriptional organization and regulation of the bzd cluster of Azoarcus sp. strain CIB. The genes are grouped into two operons, the bzdR regulatory operon (blue) and the bzdNOPQMSTUVWXYZA catabolic operon (orange), controlled by the PR and PN promoters, respectively. Both promoters are repressed by the BzdR protein (blue), with benzoyl-CoA being the inducer molecule. The activation of the PN promoter under anaerobic conditions is also dependent on the AcpR protein (violet). Some carbon sources, such as organic acids, cause catabolite repression at the PN promoter via BzdR and a still-unknown factor (?). The − and + symbols represent transcriptional repression and activation, respectively.
The transcription initiation site in the catabolic PN promoter was mapped 75 nucleotides upstream of the ATG translation initiation codon of the bzdN gene, showing putative −10 and −35 boxes typical of σ70-dependent promoters. DNase I footprinting experiments revealed that BzdR bound to three different operator regions in the PN promoter: region I (63 bp), spanning positions −32 to −31; region II (21 bp), spanning positions −83 to −63; and region III (21 bp), spanning positions −146 to −126. The three protected regions contain direct repetitions of a sequence, TGCA, which forms part of longer palindromic structures. Whereas the TGCA sequences are separated by 6 nucleotides in regions II and III, the longer region I presents a pair of TGCA sequences separated by one nucleotide and another pair of TGCA sequences separated by 15 nucleotides (14). Other transcriptional regulators that show some similarities to BzdR, such as the SinR regulator from Bacillus subtilis and the Cro and 434 repressors from phages λ and 434, respectively, also bind to short repeated sequences that, in most cases, are located within palindromic regions that span the promoters (58, 196, 232, 372). Moreover, the binding of BzdR induces changes in the DNA structure of PN as revealed by several phosphodiester bonds that become hypersensitive to DNase I cleavage. The fact that BzdR binding region I spans the transcription initiation site as well as the −10 sequence for the recognition of the σ70-RNAP is in agreement with the observed repressor role of BzdR at the PN promoter (14). Nevertheless, the molecular mechanisms of BzdR-mediated repression as well as the benzoyl-CoA-dependent activation of PN are still unknown and should be the subject of future research.
Analysis of the primary structure of the BzdR regulator (298 amino acids) revealed a unique molecular architecture that has not been described so far for any other previously characterized regulatory protein, which makes BzdR the prototype of a new subfamily of transcriptional regulators (14). Thus, BzdR exhibits two distinct domains, an N-terminal domain (residues 1 to 87) and a C-terminal domain (residues 131 to 298), with a recognized folding profile, connected by a putative flexible linker region (43 amino acids). The N-terminal domain (N-BzdR) shows significant sequence similarity with members of the HTH-XRE family of transcriptional regulators, which includes more than 1,300 proteins from eukaryota, archaea, bacteriophages, and bacteria, with an HTH DNA binding motif similar to that of the well-characterized Cro protein of λ phage (316). A three-dimensional model of N-BzdR was generated (Fig. 21) by comparison with the known three-dimensional structure of SinR, a 14-kDa pleiotropic transcriptional regulator from B. subtilis (58) that shares the highest identity with N-BzdR. The presence of a predicted HTH (residues 38 to 76) (Fig. 21), a motif found in numerous DNA binding proteins (165), within N-BzdR provides a reasonable structural basis for suggesting that N-BzdR is the region of BzdR directly interacting with the target DNA. The C-terminal domain of BzdR (C-BzdR) is homologous to shikimate kinases, enzymes that catalyze the conversion of shikimate to shikimate 3-phosphate using ATP as a cosubstrate (366). The template that was used to build a three-dimensional model of C-BzdR (Fig. 21) was shikimate kinase I (AroK) of E. coli (138, 307). Interestingly, the amino acid sequence identity shared between C-BzdR and the selected template (23%) is similar to that observed between the two different isoenzymes of E. coli, shikimate kinases I and II (30% identity) (373). This observation indicates that the protein fold of shikimate kinases is highly versatile, since it can accommodate a wide array of sequences without significant structural departures. According to the three-dimensional model, C-BzdR presents a canonical mononucleotide binding fold found in a number of structurally diverse proteins (198), which is constituted by a five-stranded parallel β-sheet flanked by eight α-helices (Fig. 21). Within this highly conserved fold, a phosphate binding loop (P-loop or Walker A motif) (Fig. 21) and the strictly conserved Gly residue present in the Walker B motif of purine nucleotide binding proteins (315, 367) can be observed (14).
FIG. 21.
Three-dimensional model of the BzdR transcriptional regulator. (A) Ribbon diagram of N-BzdR showing the five-helix bundle. The helices and loop forming the classical HTH motif of XRE-type regulators are in orange. (B) Ribbon diagram of C-BzdR/benzoyl-CoA. The overall fold contains five β-strands (blue arrows) and eight α-helices (salmon ribbons). The phosphate binding site (P loop) is in yellow. Benzoyl-CoA is in red, with the benzoyl and ADP ends being blue and yellow shadowed, respectively. The corresponding N and C termini are labeled as NH2 and COOH, respectively. The figure has been prepared using MOLSCRIPT (197), RASTERED (243), and PyMOL (DeLano Scientific).
The three-dimensional model of C-BzdR allowed a visualization of the complex between C-BzdR and the effector molecule benzoyl-CoA (Fig. 21). The feasibility of this model is accentuated by the striking structural similarity between the ADP and benzoyl moieties of benzoyl-CoA with the two substrates of shikimate kinases, ATP and shikimate, respectively. Manual docking is easily performed, as the C-BzdR fold has a conserved nucleotide binding site where the ADP moiety of benzoyl-CoA fits very well. On the other hand, the pantothenate and β-mercapthoethylamine units of benzoyl-CoA fit into a deep groove whose walls are formed by the loops between residues 163 and 158 and 231 and 271. The groove ends in a cavity, equivalent to the shikimate binding site described for shikimate kinases (138, 198), which perfectly hosts the benzoyl moiety of the effector molecule (Fig. 21) (14). It has been proposed that the enzymatic activity of shikimate kinases proceeds through an induced-fit mechanism, with the loops that constitute the walls of the groove being those suffering important conformational changes upon substrate binding (138). The binding of benzoyl-CoA to BzdR might involve a similar conformational change at C-BzdR that, through the linker region, would then trigger an appropriate conformational change at N-BzdR, leading to the inhibition of the BzdR-mediated repression of the target promoter. The crystallization of BzdR and the BzdR-benzoyl-CoA complex is currently under way, and it will undoubtedly permit an elucidation of the structural basis that determines the biological action of this prototype of a new subfamily of regulatory proteins.
Transcriptional regulators governing the expression of catabolic promoters from aromatic catabolic pathways usually control their own expression (53, 86, 352). Consistent with this, the bzdR gene product also behaves as a repressor of its own expression, and this repression becomes relieved when the cells grow in the presence of benzoate and when benzoyl-CoA is formed (98). The transcription initiation at the promoter of the bzdR gene, termed PR, was located 39 nucleotides upstream of the ATG translation initiation codon of the bzdR gene, showing putative −10 and −35 boxes that significantly match the consensus sequences recognized by the σ70 subunit of RNAP. In vitro transcription experiments have shown that BzdR inhibited PR expression, and the addition of benzoyl-CoA was able to increase the formation of the PR transcript, indicating that this CoA derivative acts as an inducer avoiding the repression effect of BzdR (98). Thus, Azoarcus sp. strain CIB shows a similar pattern of benzoyl-CoA-dependent induction of BzdR-mediated repression at both the PR and PN promoters (Fig. 20). Interestingly, another aromatic CoA derivative, phenylacetyl-CoA, was shown to be the inducer of an aerobic hybrid pathway, the phenylacetic acid degradation pathway, whose intermediates are also CoA-derived compounds (85, 238, 247, 265, 321). Whether transcriptional regulators controlling anaerobic and aerobic hybrid pathways for the catabolism of aromatic acids have evolved to recognize CoA-derived aromatic intermediates rather than the free acids is a hypothesis that requires further confirmation when additional regulatory systems become characterized.
The BzdR operator at the PR promoter was identified by using DNase I footprinting assays, and it spans positions −33 to +25 of the PR promoter. Three direct GCAC repetitions and a short palindromic structure located 2 nucleotides downstream of the +1 site were observed. The fact that the operator of BzdR spans the transcription initiation site as well as the −10 and −35 boxes of PR is in agreement with the role of BzdR as a repressor of the activity of this promoter (98). Since BzdR also overlaps the transcription initiation site as well as the −10 box at the catabolic PN promoter (see above), BzdR appears to act at both the PR and PN promoters using the same repression mechanism. However, the affinity of binding of BzdR to the PR promoter is about 10-fold lower than that observed with the PN promoter, which is also in agreement with the moderate and high levels of repression caused by BzdR at the PR and PN promoters, respectively (14, 98). A comparative analysis revealed that whereas three BzdR operators were identified at the PN promoter, and all of them showed a direct repetition of the TGCA sequence, only one BzdR operator, which contains a sole TGCA sequence, was identified at the PR promoter. Therefore, the lower efficiency of binding of BzdR to PR might reflect the different architectures of the PN and PR promoters (98).
An in silico search in the available databases revealed that the number of identified transcriptional regulators of the BzdR subfamily is continuously increasing, and currently, BzdR orthologs can be identified as being associated to benzoate degradation clusters of over 30 denitrifying proteobacteria from the alpha and beta subgroups. In most cases, these regulatory genes are linked to the hybrid box pathway for the aerobic degradation of benzoate via benzoyl-CoA (83, 124, 125, 238), and therefore, they are likely to be boxR-like genes. So far, only two bzdR-like genes, those from Azoarcus sp. strains CIB and EbN1, are indeed associated with the anaerobic benzoyl-CoA degradation pathway. Despite the finding that the aerobic BoxR and the anaerobic BzdR regulators belong to the same BzdR subfamily, the BoxR regulators cluster together in a branch of the phylogenetic tree separated from that of the BzdR regulators. This finding suggests that although both types of regulators may have evolved from a common ancestor, they have subsequently diverged and adapted to the aerobic (boxR genes) and anaerobic (bzdR genes) gene clusters. Whether the effector molecules and the mechanisms of transcriptional regulation mediated by BoxR and BzdR are similar as well as whether there is cross talk between these two regulators in those bacteria that harbor both the aerobic and anaerobic regulatory circuits are still unknown issues that require further studies. It should be noted, however, that in nondenitrifying bacteria, e.g., R. palustris, G. metallireducens, and S. aciditrophicus, the genes responsible for the anaerobic degradation of benzoate appear to be controlled by a regulatory strategy that differs from that described for the BzdR-dependent systems.
(iii) Regulation of the lower benzoyl-CoA pathway.
Several genes (pim genes) are involved in the lower pathway responsible for the metabolism of the aliphatic product generated in the central benzoyl-CoA pathway (Fig. 3). In R. palustris, the pimFABCDE genes constitute an operon that is specifically induced when cells grow anaerobically in benzoate (or pimelate) (140, 362). A putative IclR-like regulatory gene and the genes coding for a potential ABC transporter are divergently transcribed from the pim operon (140). In addition to the pim operon, other putative β-oxidation-encoding genes have increased levels of expression when R. palustris cells are grown in benzoate (362). In G. metallireducens, cluster IB contains most of the genes considered to be involved in the lower pathway of benzoate degradation (Fig. 2B), and they are also upregulated in the presence of benzoate (48, 377). However, the only gene of the lower pathway whose expression has been studied to some extent is the gcdH gene, encoding the glutaryl-CoA dehydrogenase activity of Azoarcus sp. strain CIB (Fig. 3) (29). The gcdH gene was inducible when the strain was cultured in the presence of compounds whose degradation generates glutaryl-CoA, such as benzoate or pimelate. RT-PCR experiments revealed that gcdH gene expression is under transcriptional control (29). An analysis of the DNA region harboring the gcdH gene revealed the existence of a divergently transcribed gene, gcdR, which codes for a LysR-type transcriptional activator (Table 3) of the Pg promoter that drives the expression of gcdH. Glutarate and glutaconate were shown to be the specific inducer molecules of GcdR. It is worth noting that the replacement of glutarate and glutaconate by some structural analogs, such as valerate, crotonate, succinate, α-ketoglutarate, glutamate, glutamine, adipate, cis-cis-muconate, or pimelate, did not lead to the activation of the Pg promoter. Thus, the GcdR transcriptional activator shows a highly specific effector profile recognizing only unsubstituted saturated (glutarate) or unsaturated (glutaconate) C5-dicarboxylic acids as inducer molecules (29). Because the CoA derivatives of glutarate and glutaconate (2,3-dihydroglutarate) are the substrates of the two reactions catalyzed by GcdH, it cannot be ruled out that glutaryl-CoA and glutaconyl-CoA can also be inducers of GcdR-mediated Pg activation. The activation of the Pg promoter by glutaryl-CoA/glutaconyl-CoA would facilitate the expression of the gcdH gene when the cells grow in carbon sources, such as aromatic compounds or pimelate, that produce glutaryl-CoA rather than glutaric acid (29).
The transcription start site of the Pg promoter that controls the expression of the gcdH gene was mapped 59 nucleotides upstream of the ATG translation initiation codon. The Pg promoter shows putative −10 and −35 boxes separated by the consensus 17-bp distance typical of σ70-dependent promoters. In the Pg promoter, from positions −71 to −57 with respect to the transcription start site, there is an interrupted inverted repeat, 5′-GTGCGT-N3-ACGCAC-3′, with the consensus LysR-type binding motif T-N11-A (53, 318, 352), that should correspond to the recognition binding site of GcdR (29). A second inverted repeat, 5′-ACGAAATTTTCGT-3′, is located immediately upstream of the −35 box of the Pg promoter, and it might function as the activation binding site of GcdR (29). Interestingly, a nucleotide sequence alignment of putative promoter regions of gcdH genes from different bacteria shows that the more conserved regions are those corresponding to the recognition binding site, activation binding site, and −35 box of Pg, reinforcing the assumption that these are the key regulatory sequences in this promoter (29). Accordingly, a gcdR gene encoding a putative LysR-type transcriptional regulator can be found divergently transcribed from the gcdH genes of many proteobacteria, including those that are able to degrade dicarboxylic acids, but not aromatic compounds, under anaerobic conditions. The fact that the gcdR gene from Azoarcus sp. strain CIB is able to efficiently regulate a gcdH gene from P. putida when the latter was expressed in an Azoarcus sp. strain CIB mutant harboring its own gcdH gene inactivated by disruption revealed the existence of cross talk between GcdR regulators and gcdH promoters coming from members of different phylogenetic subgroups of proteobacteria (29). All these observations suggest that the transcriptional regulation of the gcdH gene has been conserved in many bacteria.
Regulation of the peripheral 4-HBA pathway in R. palustris.
Four different transcriptional units in the hba cluster involved in the peripheral pathway that converts 4-HBA into benzoyl-CoA in R. palustris can be predicted: (i) the hbaR regulatory gene; (ii) the hbaA gene, encoding 4-HBA-CoA ligase; (iii) the hbaBCD genes, encoding 4-hydroxybenzoyl-CoA reductase (dehydroxylating); and (iv) the hbaEFGH genes, encoding a putative ABC transporter (Fig. 19) (104). However, no detailed transcriptional analysis of the hba cluster has been reported so far.
The hbaR gene product is a transcriptional activator that senses 4-HBA as an effector molecule and induces the expression of the hbaA gene, although it does not control the genes responsible for the aerobic degradation of 4-HBA in R. palustris (103). The binding of HbaR to the hbaA promoter has not been demonstrated in vitro with gel mobility shift assays; however, experiments with hbaR expressed in P. aeruginosa cells indicate that HbaR activates the expression of the hbaA promoter (Fig. 19). HbaR is a member of the FNR/CRP superfamily of transcriptional regulators. Sequence comparisons revealed similarities with members of the Dnr group, which comprises proteins that lack the cysteine residues required for iron-sulfur center coordination and oxygen sensing (195). With the exception of HbaR, all the proteins within the Dnr group are global regulators that control the expression of genes involved in denitrification (103). It has also been shown that when expressed in P. aeruginosa cells, HbaR can activate expression from the hbaA promoter in the presence of oxygen. In contrast, hbaA expression was not activated in aerobically grown R. palustris cells, suggesting the participation of a second regulator that responds to anaerobiosis (103). Interestingly, the FNR protein of E. coli influenced the expression of an hbaR-lacZ gene fusion in the absence of oxygen. This finding together with the observation that a putative FNR binding site was centered −42.5 nucleotides from the transcription start site of hbaR strongly suggest that hbaR expression in R. palustris is activated in response to anaerobiosis by an FNR homolog (103). This homolog is, presumably, AadR (see below), since AadR is required for hbaA expression (Fig. 19) (91). Because the expression of an hbaA-lacZ fusion in E. coli was not influenced by the fnr mutation, it is likely that the observed AadR-dependent control of the hbaA gene can be an indirect effect due to its transcriptional control of the hbaR gene, which in turn activates hbaA expression in response to 4-HBA (Fig. 19) (103). The hba gene expression system then involves a regulatory cascade, with one protein (AadR) acting as an oxygen sensor and in turn activating the expression of a substrate-specific regulator (HbaR). The hierarchical expression of two regulators would be an effective strategy to prevent cross talk and provide regulation of relatively specialized target genes, as in the case of 4-HBA degradation, under conditions of oxygen deprivation (103).
To complete our understanding of the regulation of the anaerobic peripheral 4-HBA pathway in R. palustris, control of the expression of the hbaBCD and hbaEFGH genes awaits further studies.
Regulation of the peripheral toluene and ethylbenzene pathways.
The substrate induction of the enzymes involved in toluene catabolism occurs at the transcriptional level (70, 72, 217). In m-xylene-degrading cells, such as in Azoarcus sp. strain T, this hydrocarbon also induces the bss operon (1). The transcription start sites of the bss operon in Azoarcus sp. strain T (1), T. aromatica strain K172 (155), and T. aromatica strain T1 (68) as well as the transcription site of the putative bbs operon in T. aromatica strain K172 (216) (Fig. 16) have been determined. In T. aromatica strain K172 and Azoarcus sp. strain EbN1, the bss genes appear to be transcribed more efficiently than the bbs genes (155, 203). Sequence comparison analyses reveal several conserved sequence motifs upstream of the bssD and bbsA genes from denitrifying bacteria. Two of these motifs are similar to typical −10 and −35 boxes of σ70-dependent promoters. Further conserved motifs located between positions −40 and −50 and around position −65 relative to the mapped (or presumed) transcription start sites might represent the binding sites (operators) of the cognate transcriptional regulator (TdiR proteins) or any other regulatory protein that responds to anaerobiosis and/or toluene availability (202). Nevertheless, in T. aromatica strain T1, an apparent transcription start site has been mapped (68) upstream of the putative operator region identified by sequence comparison analyses (202).
In most denitrifying bacteria, the induction of genes by toluene as a growth substrate appears to be mediated by the tdiSR genes, which are located upstream of the bss genes and code for a putative two-component regulatory system (Fig. 16). These two genes constitute an operon in Azoarcus sp. strain T, and they are divergently transcribed with respect to the bss operon in all Azoarcus strains analyzed so far (Fig. 16) (1). In contrast, in T. aromatica strain K172, the tdiSR genes are transcribed in the same orientation as the bss genes (Fig. 16). The tdiR gene product from T. aromatica strain K172 was shown to bind in vitro to a DNA fragment containing the 5′ region of the bss operon (215). In T. aromatica strain T1, a point mutation in the tutB1 (tdiR) gene was shown to disable anaerobic growth in toluene as well as the expression of the tutE (bssD) gene, which is in agreement with the role of this regulator as an activator of the catabolic promoters (70, 73). Interestingly, T. aromatica strain T1, an organism that is able to grow on toluene aerobically and anaerobically, presents tandem two-component regulatory systems, one (tutCB genes) located immediately upstream and divergently transcribed from the anaerobic tut operon and another (tutC1B1 genes) adjacent to the former and arranged in the opposite orientation (Fig. 16) (215). Sequence comparison analyses revealed that the TutCB proteins show similarity to aerobic two-component regulatory systems (see below), whereas the TutC1B1 proteins are homologous to the anaerobic TdiSR systems, respectively (215).
In the TdiSR regulatory system, the TdiS sensor component consists of two sensory PAS domains and a C-terminal histidine kinase domain that contains the predicted His residue for autophosphorylation, each occupying about one-third of the protein (215). PAS domains are implicated in monitoring light, redox, or hydrocarbon stimuli in diverse sensory proteins (235) and, therefore, should be likely involved in sensing the inducer molecule(s) that interacts with TdiS. TdiS lacks predicted transmembrane regions and is thus likely to be located in the cytoplasm. The TdiR regulator consists of an N-terminal response regulator domain that contains the predicted Asp residue for phosphorylation and a C-terminal HTH motif that would bind to the cognate promoters, classifying it as a member of the FixJ/NarL family of regulators (215). The sensor (TdiS) and regulator (TdiR) components show significant similarity to their aerobic counterparts that are involved in the regulation of the catabolism of toluene, TodS and TodT (214), and styrene, StyS and StyR (360), and all of them constitute a new subfamily of two-component regulatory systems involved in the control of catabolic pathways for the degradation of solvents (47). However, the aerobic histidine kinases are larger and more complex than their anaerobic counterparts, and they contain two supradomains, each containing a PAS and a histidine kinase domain, which are separated by a response regulator receiver domain. The N-terminal PAS domain of TodS from P. putida DOT-T1E binds toluene with high affinity, and this binding increases its basal autophosphorylation rate, leading to the transphosphorylation of TodT and the transcription activation of the cognate promoter by the latter (47, 208). In the anaerobic TdiSR regulatory system, benzylsuccinate (or any other further metabolite in the toluene catabolic pathway) rather than toluene was shown to be the inducer molecule that allows the expression of the tutE and tutFDGH operons in T. aromatica strain T1 despite the fact that this bacterium does not use benzylsuccinate as a carbon source (70). In contrast, such a regulatory scenario appears to be unlikely for Azoarcus sp. strain EbN1, since an upregulation of the expression of bssA or the formation of the Bss and/or Bbs protein was not observed in cells grown with a mixture of pyruvate and benzylsuccinate (203). In strain EbN1, the TdiSR system appears to be specific strictly for toluene since the ethylbenzene analog was unable to induce the expression of the bss and bbs genes (203). In Azoarcus sp. strain T, m-xylene also induces the bss operon (1). Nevertheless, details of the mechanism of action of TdiS and TdiR, controlling the toluene-dependent catabolic promoters in anaerobic pathways, still await further molecular studies.
In contrast to what has been described for denitrifying toluene degraders, orthologs of tdiSR are lacking in G. metallireducens, and therefore, a different regulatory circuit might be present in this iron-reducing bacterium. Consistent with this, XylR-like and TetR-like putative regulators are flanking the bss and bbs clusters, respectively, in G. metallireducens (Fig. 16). Since XylR is the regulatory protein that senses toluene (and some analogs) and controls the σ54-dependent expression of the xyl genes involved in the aerobic catabolism of toluene in several Pseudomonas strains (56, 296), it is tempting to speculate that a σ54-dependent regulatory system is also controlling the expression of anaerobic toluene degradation in G. metallireducens.
For T. aromatica strain K172, a posttranscriptional control of the expression of the bss operon has been suggested (155). Thus, in addition to the bssDCABE transcript originating from the toluene-induced promoter located upstream of bssD, there is a second more abundant mRNA species whose 5′ end is located in front of the bssC gene. The large mRNA containing bssD is very short-lived and generates, likely by RNase processing, the more stable bssCABE transcript. In accordance with this assumption, the RNA sequence surrounding the major 5′ end in front of bssC may form a stem-loop structure resembling some known processing sites of E. coli RNase III (155). This posttranscriptional control would allow that the benzylsuccinate synthase (BssABC) and its activating enzyme (BssD) can be coordinately induced, but the enzyme levels could then be adjusted in the cell to explain the observed 14-fold-lower amounts of the activating enzyme (155). Consistent with this, it might be expected that the activator proteins are needed in smaller amounts than the activated proteins. Interestingly, in T. aromatica strain T1, the different amounts of the activator and benzylsuccinate synthase appear to be controlled transcriptionally by the existence of two different operons, tutE (bssD) and tutFDGE (bssCABE) (68).
In Azoarcus sp. strain EbN1, the tcs1/tcr1 and tcs2/tcr2 genes located between the upper and lower ethylbenzene operons (Fig. 17) encode products that resemble typical two-component regulatory systems controlling the metabolism of aromatic compounds, with Tcs1 and Tcs2 representing the sensors and Tcr1 and Tcr2 representing the response regulators, respectively (292). The predicted Tcs1 histidine kinase was suggested to be implicated in the recognition of the intermediate acetophenone due to its similarity to previously described p-hydroxyacetophenone-sensing systems that regulate the transcription of virulence genes in plant-pathogenic bacteria. The predicted Tcr1 regulator would be involved in controlling the expression of the apc and bal genes, which are responsible for the anaerobic catabolism of acetophenone (292). On the other hand, even though Tcs2 and TdiS are highly similar to each other in the first PAS domain and the histidine kinase domain (41 to 43% identity), their second PAS domains differ markedly (16% identity). This finding suggests that TdiS and Tcs2 may be able to discriminate between toluene and ethylbenzene, respectively, via their second PAS domains (202). Finally, the Tcr2 response regulator may control the expression of the ebd and ped genes (292). Thus, the upper and lower parts of the pathway for anaerobic ethylbenzene degradation can be sequentially induced by their respective substrates, ethylbenzene and acetophenone, which can both be used as growth substrates (202, 290, 292). Such a regulatory scenario is supported by the absence of ebdA expression and EbdCD formation as well as by the concomitant presence of several subunits of acetophenone carboxylase (Apc) and the benzoylacetate-CoA ligase (Bal) in acetophenone-adapted Azoarcus sp. strain EbN1 cells (203). This sequential mode of regulation could provide an economic benefit considering that strain EbN1 probably encounters acetophenone (a plant metabolite) as a substrate in the natural soil environment (203).
The strict specificity shown by the predicted toluene sensor (TdiS) is in contrast to the relaxed specificities observed for the predicted ethylbenzene (Tcs2) and acetophenone (Tcs1) sensors. This relaxed specificity of Tcs1 and Tcs2 could explain the expression of all the genes involved in the anaerobic catabolism of ethylbenzene in Azoarcus sp. strain EbN1 cells grown in toluene, suggesting that toluene acts as a gratuitous inducer for ethylbenzene-specific operons (203). One may speculate that the toluene binding pocket of TdiS cannot accommodate the larger ethylbenzene molecule or that ethylbenzene binding to TdiS does not lead to the autophosphorylation cascade in the sensor protein. In contrast, toluene may access the sensory pockets of Tcs1 and Tcs2 and lead to their productive autophosphorylation. Alternatively, a cross-regulation between TdiSR and the other two systems may occur (203). On the other hand, the simultaneous utilization of toluene and ethylbenzene by Azoarcus sp. strain EbN1 cells resulted in a simultaneous expression of the genes from both degradation pathways, indicating that neither of the two alkylbenzenes is regarded as a preferred substrate. This regulatory behavior of Azoarcus sp. strain EbN1 might reflect an adaptation to low concentrations of the rather insoluble alkylbenzenes, a situation usually encountered in the natural environment (203).
Overimposed Regulation
Two major environmental signals, i.e., oxygen and carbon source availability, have been shown to influence the expression of anaerobic catabolic clusters through regulatory proteins that operate on given catabolic promoters concomitantly with the cognate-specific regulators.
Oxygen-dependent regulation.
A major environmental signal that controls the expression of genes involved in anaerobic aromatic catabolic pathways is oxygen. Thus, the levels of expression of the badDEFG and badA genes of R. palustris, encoding the four subunits of BCRRp and the benzoate-CoA ligase, are decreased under aerobic conditions (104, 276). In T. aromatica, a strong downregulation of the synthesis of BCRTa was found in response to oxygen, since the protein was immunologically detected only in trace amounts in aerobically grown cells (150). The bss genes, encoding the benzylsuccinate synthase involved in toluene degradation in M. magnetotacticum TS-6, were transcribed only in anaerobically toluene-grown cells (335). However, there are some reports showing that genes encoding oxygen-sensitive enzymes such as BCR (bcr genes) from M. magnetotacticum TS-6 (335) and benzylsuccinate synthase (bss genes) from Thauera sp. strain DNT-1 (336) are transcribed not only under anaerobic conditions but also in cells growing aerobically in benzoate and toluene, respectively. Conversely, a low abundance of proteins involved in aerobic benzoate degradation was observed in Azoarcus sp. strain EbN1 cells grown anaerobically with benzoate (378). Therefore, it appears that each organism has evolved a particular regulatory strategy for the oxygen-dependent expression of the genes involved in the anaerobic catabolism of aromatic compounds. The expression of the anaerobic genes under aerobic conditions, or the aerobic genes under anaerobic conditions, can be the result of relaxed regulatory specificities but also could be explained as a mechanism of facultative anaerobes to maintain a basal level of enzymes that facilitate an immediate response in environments subjected to fluctuating oxygen levels. A further adaptation to a range of oxygen concentrations that can induce both the aerobic and anaerobic catabolic pathways is the existence of only one benzoate-CoA ligase that initiates the aerobic and anaerobic catabolism of benzoate in T. aromatica and Magnetospirillum strains (181, 330).
As indicated above, in R. palustris, the badD promoter is controlled by the specific BadR and BadM regulators and, additionally, by the regulatory protein AadR (Table 3) in response to anaerobiosis (Fig. 19) (102). The AadR protein, whose gene lies outside the ali-bad-hba supraoperonic cluster, belongs to the FNR/CRP superfamily of transcriptional regulators, and as such, it contains the conserved features that are important for normal FNR-mediated gene expression in response to oxygen. Since the badD promoter includes an FNR consensus binding sequence (TTGAT-N4-ATCAA) centered at position −39.5 relative to the transcriptional start site, it is likely that the AadR protein responds to anaerobiosis by assuming an active conformation that is proficient in binding to the badD promoter to activate transcription. Nevertheless, the possibility cannot be ruled out that the positive effect of AadR on the activity of the badD promoter can be indirect and occurs through the activation of another regulatory protein (102), as it seems to be the case for the control of the hbaA gene through the AadR-dependent expression of the HbaR activator (see above) (Fig. 19). Since a putative FNR binding box was also detected in the promoter region of the aadR gene, it is reasonable to assume that AadR might bind to this site to autoregulate its synthesis (91).
The most detailed study of the molecular basis of the oxygen-dependent control of genes involved in the anaerobic catabolism of aromatic compounds was carried out for the bzd cluster from Azoarcus sp. strain CIB. An exhaustive analysis of the PN promoter region revealed a sequence (TTGACTTAGATCAA) centered at position −41.5 from the transcription start point of the bzd catabolic operon that is almost identical to the FNR consensus binding sequence (97). This observation suggested that a protein of the FNR/CRP superfamily could be involved in the regulation of the PN promoter by binding to its cognate sequence in response to oxygen deprivation. In fact, it has been demonstrated that the inactivation of the fnr gene inhibited the anaerobic induction of the PN promoter in E. coli cells and that the FNR* protein, a constitutively active FNR mutant protein that carries a D154A substitution that is able to form a dimer and to bind DNA in the presence of oxygen (185, 388), was able to bind to the PN promoter and protected the FNR binding sequence, thus keeping PN active even under aerobic conditions (97). The key role of FNR in the oxygen-dependent regulation of the bzd catabolic genes was further substantiated by the observation that a recombinant Azoarcus sp. strain CIB mutant harboring the E. coli fnr* gene allowed the expression of the bzd genes when the cells grew aerobically in the presence of benzoate. These results, on the other hand, constitute a nice example of how the expression of an anaerobic pathway for the catabolism of aromatic compounds can be switched to aerobic conditions, which indeed do not allow functional anaerobic catabolism, just by changing a key regulatory protein (97).
The location of the FNR binding site centered at position −41.5 from the transcription start site and overlapping the −35 box fits perfectly with PN being a typical class II FNR-dependent promoter (223, 304). The use of previously characterized E. coli σ70 mutants confirmed that the FNR-σ70-RNAP contact plays a crucial role in the activation of the class II FNR-dependent PN promoter (97). In addition to the FNR-dependent increase in the affinity of RNAP for the PN promoter, in vitro transcription experiments revealed that FNR is essential for the initiation of transcription at some step after RNAP-PN closed-complex formation. As has been shown for other FNR-dependent promoters (376), FNR may activate the transcription of genes controlled by PN by promoting isomerization from the transcriptionally inactive closed complex to the transcriptionally active open complex (97).
AcpR, an FNR homolog, was identified in the genome of Azoarcus sp. strain CIB and other Azoarcus strains such as strain EbN1 (97). As described previously for fnr-like genes in other bacteria, the acpR gene in Azoarcus strains is associated with the hemN gene, which encodes an oxygen-independent coproporphyrinogen III oxidase that is also regulated by the FNR homolog, Anr, in Pseudomonas (195). An Azoarcus sp. strain CIB mutant harboring a disrupted acpR gene was unable to grow anaerobically on aromatic compounds, such as benzoate, phenylacetate, or 4-HBA, that are mineralized through the bzd-encoded central pathway, although aerobic growth on aromatic compounds was not affected in this mutant strain. Moreover, the expression of the PN-lacZ reporter fusion in Azoarcus was shown to be dependent on the presence of a functional acpR gene, which indicates that AcpR is essential for the anaerobic expression of the bzd cluster and that it works as a transcriptional activator required for the activity of the PN promoter when Azoarcus sp. strain CIB is grown with benzoate under anaerobic conditions (Fig. 20) (97). Therefore, the physiological role of AcpR in Azoarcus sp. strain CIB in regulating aromatic compound degradation in response to oxygen appears to be equivalent to that carried out by AadR in R. palustris (see above) (Table 3).
A phylogenetic analysis based on a multiple-sequence alignment of AcpR with other members of the FNR/CRP superfamily revealed that AcpR branches within the Fnr group, which contains the well-characterized E. coli FNR protein (43% amino acid sequence identity) (97). Interestingly, although AcpR and AadR may play similar physiological roles in the cell, they show only 34% amino acid sequence identity, and in fact, AadR has been classified within a different group (FnrN) of the FNR/CRP superfamily (97, 195). In the E. coli FNR protein, four cysteine residues (at positions 20, 23, 29, and 124) contribute to the formation of a [4Fe-4S]2+ cluster that is essential for indirect oxygen sensing, allowing the activation (dimerization) of FNR under anaerobic conditions and inactivation (monomerization) upon exposure to oxygen (137). Whereas these four cysteine residues are perfectly conserved in AcpR from Azoarcus (at positions 18, 21, 27, and 122), in the AadR protein, only two cysteine residues (at positions 20 and 117) are conserved. The AR-1, AR-2, and AR-3 activation regions, which are involved in the interaction of the E. coli FNR protein with the α-C-terminal domain, α-N-terminal domain, and σ70-C-terminal domain subunits of RNAP, respectively (242, 371), are also conserved in AcpR and AadR, although in the latter, there is low amino acid sequence conservation at the AR-1 region (97). A new branch of the FNR/CRP superfamily that has recently emerged includes transcriptional regulators such as CprK (Table 3) that mediate the response to halogenated aromatic compounds in the dehalorespiration of different Desulfitobacterium strains (286). In addition to sensing halogenated compounds through their N-terminal domains, CprK-type regulators contain redox-active thiol groups that also influence the binding to the target DNA through their C-terminal domains (285).
Despite the relevance of the predicted structural similarity between FNR and the AadR and AcpR regulators, the two latter proteins do not play the same regulatory functions as those described for FNR. Thus, the lack of the FNR protein in E. coli has a pleiotropic effect on the expression of a moderate number of genes, including the incapacity of the mutant strain to grow using nitrate or fumarate as a final electron acceptor (355). This pleiotropic character of E. coli FNR was further extended by demonstrating that fnr* is able to efficiently complement the lack of acpR in Azoarcus sp. strain CIB and, therefore, is also able to behave as an aromatic central-pathway regulator (97). In contrast, the lack of AcpR and AadR in Azoarcus sp. strain CIB and R. palustris, respectively, has no obvious effect on the anaerobic growth with nonaromatic carbon sources and other general metabolic features (91, 97). Therefore, both AcpR and AadR appear to be oxygen-dependent regulators that have become specialized to control genes responsible for the anaerobic catabolism of aromatic compounds rather than being global regulatory proteins controlling the expression of different gene programs, as is the case for other members of the Fnr group (195). This specialization could be a general principle in aromatic degraders, such as Azoarcus and R. palustris, that contain a significant array of multiple FNR/CRP regulators, which contrasts with other bacteria, e.g., E. coli, where only one fnr gene is present. Thus, in the genome of Azoarcus sp. strain EbN1, there are seven genes encoding putative transcriptional regulators of the FNR/CRP family, i.e., one FNR-like protein (AcpR), one CRP-like protein, three Dnr-like proteins, and two Nnr-like proteins (293). A multiple-amino-acid sequence alignment of the HTH motifs of these seven regulators revealed significant differences among them, which agrees with the assumption that each individual member evolved to fulfill a particular physiological role (97).
In contrast to what was observed with the PN promoter driving the expression of the bzd catabolic genes, oxygen does not appear to play a major role in the activity of the PR promoter responsible for the transcription of the bzdR regulatory gene in Azoarcus sp. strain CIB. In agreement with this finding, the activity of PR is not subject to the AcpR/oxygen-dependent overimposed regulation that controls the activity of the catabolic PN promoter (97). The aerobic expression of the bzdR gene might contribute to a further decrease in the basal level of expression of the bzd catabolic operon when the cells grow in the presence of oxygen, thus avoiding the unproductive synthesis of some oxygen-sensitive enzymes such as BCR (97).
Catabolite repression control.
Carbon catabolite control is one of the most fundamental environment-sensing mechanisms in bacteria and imparts a competitive advantage by establishing priorities in carbon metabolism. The term catabolite repression describes a number of regulatory processes ensuring that when the cell is exposed to a large amount of a preferred carbon source, the catabolic pathways for other, nonpreferred, substrates are not induced even when the appropriate inducers are present (53, 55, 86, 306). Carbon catabolite repression in the aerobic catabolism of aromatic compounds has been widely reported, and some of the regulatory elements involved have been characterized for different bacteria (53, 66, 233, 254, 264, 287, 306, 334). Catabolite repression control appears to also be a typical feature of organisms under anaerobic growth conditions, and at least a couple of examples of this phenomenon have been described for the anaerobic degradation of aromatic compounds in T. aromatica (150) and Azoarcus sp. strain CIB (224). In the latter organism, catabolite repression of the anaerobic benzoate catabolic pathway is carried out by certain organic acids such as succinate, malate, and acetate. As expected, compounds that cannot be used as sole carbon sources under anaerobic conditions, e.g., citrate, glycerol, fructose, and maltose, did not cause catabolite repression of the anaerobic benzoate pathway (224).
The molecular mechanisms controlling the carbon catabolite repression of aerobic aromatic pathways have been studied for some organisms. Thus, in E. coli, the major component of the catabolite repression of aromatic catabolic pathways is the CRP, which acts primarily as a global transcriptional activator in the presence of cyclic AMP (287). Although glucose is the preferred carbon source for E. coli and many other microorganisms, bacteria of the genus Pseudomonas preferentially metabolize many organic acids or amino acids over sugars (66). Interestingly, not all organic acids can mediate a catabolite repression effect, and even more intriguingly, some of them have a clear effect in some cases but not in others (306). The repression of catabolic pathways frequently results from the integration of several signals rather than just from the presence of a preferred compound in the medium. Although a protein with high similarity to E. coli CRP is encoded in the chromosome of several Pseudomonas strains, this protein does not seem to mediate catabolite repression (249, 306, 348). On the contrary, some other proteins such as Crc (catabolite repression control) (249, 254, 306), PtsN and PtsO (nitrogen phosphotransferase transport system) (233, 281), some o-type terminal oxidases (CyoB) that sense the redox state of the cell (250, 280), and the BphQ response regulator (264) have been reported to be involved in the catabolite repression of several aromatic degradation pathways in nonenteric bacteria.
In contrast to the extensive knowledge about the catabolite repression of aerobic pathways involved in the catabolism of aromatic compounds, the molecular mechanisms underlying carbon catabolite repression in anaerobic pathways are still unknown. So far, the only report that presents a preliminary study of the mechanisms leading to catabolite repression is that dealing with the benzoate degradation pathway in Azoarcus sp. strain CIB (98). In this bacterium, the repressive effect of some carbon sources (e.g., some organic acids such as succinate, malate, and pyruvate, etc.) was carried out at the level of transcription from the PN promoter (Fig. 20) (98, 224). On the contrary, the PR promoter driving the expression of the bzdR regulatory gene is not subject to catabolite repression by organic acids in Azoarcus sp. strain CIB (98). There are several reports showing that the downregulation of promoters from catabolic pathways when the cells grow in the presence of the particular substrate and a preferred carbon source is mediated by shifting the levels of the cognate transcriptional regulators. Thus, the catabolite repression of the alkane degradation pathway (alk genes) encoded in the OCT plasmid from P. putida strain GPo1 parallels a decrease in the level of expression of the AlkS activator (51). Similarly, the catabolite repression of phenol degradation (phl genes) in P. putida strain H occurs by interfering with the activating function of the PhlR transcriptional regulator (257). However, the catabolite repression system operating at the PN promoter is so far unique among those that control the degradation of aromatic compounds, because the expression of the specific transcriptional regulator is not subject to such physiological control (98). The existence of additional factors that might account for the catabolite repression of the anaerobic benzoate degradation pathway in Azoarcus sp. strain CIB has been suggested. A possible scenario is that an additional regulatory factor binds to the PN promoter in response to some organic acids such as succinate, thus preventing the basal levels of bzd readthrough that allow the formation of the effector molecule benzoyl-CoA (Fig. 20). Consistent with this, orthologs of some of the carbon repression mediators in proteobacteria described so far have been found in the genome of Azoarcus strains. Thus, Azoarcus sp. strain EbN1 has orthologs of Crc (ebA3323), CRP (ebA7043), PtsN/PtsO (ebA2793/ebA2794), and BphQ (ebA125) as well as three orthologs of the CyoB protein (ebA4228, ebA156, and ebA4554). The involvement of some of these proteins in carbon catabolite repression needs to be explored further to complete the current view on the overimposed regulation of the bzd gene cluster, which is so far the best-studied regulatory system in the anaerobic catabolism of aromatic compounds.
Although the overimposed regulation of the anaerobic catabolism of toluene has not been studied in detail so far, the catabolite repression control of the bss and bbs genes responsible for anaerobic toluene degradation in Azoarcus sp. strain CIB was observed when the cells were cultivated simultaneously in toluene and nonaromatic organic acids such as succinate and pyruvate (B. Blázquez, unpublished data). A similar catabolite repression control by substrates like succinate was reported previously for aerobic toluene degradation pathways (95, 309). Nevertheless, the molecular basis underlying catabolite repression control of the anaerobic toluene degradation pathway in Azoarcus sp. strain CIB and other toluene degraders is still unknown and remains to be characterized.
GENETIC DIVERSITY AND ECOPHYSIOLOGY OF ANAEROBIC BIODEGRADERS OF AROMATIC COMPOUNDS
Since less than 1% of the bacteria present in environmental samples can be cultured in the laboratory, molecular methods (often based on 16S rRNA genes) have been used to provide a more explicit accounting of the genetic diversity and the ecophysiology of the anaerobic biodegraders of aromatic compounds within the microbial population. A more complete understanding of the community structure and activity can also be relevant for a better prediction and control of environmentally relevant processes such as the natural attenuation of aromatic pollutants (114, 184, 193). Since there is an enormous amount of work published about the ecophysiology/diversity of the anaerobic catabolism of aromatic compounds, here, we present mainly some examples of the tools and principles described for anaerobic BTEX degradation.
16S rRNA Gene-Based Studies
The structure of the microbial communities and its dynamics in environmental samples enriched with aromatic compounds have been analyzed using whole-cell hybridization with fluorescently labeled 16S rRNA-targeted oligonucleotide probes (156, 275). A 16S rRNA biomarker was also developed to estimate the concentration of putative benzene degraders under strongly anaerobic (sulfate-reducing and methanogenic) conditions by real-time quantitative PCR. Since natural attenuation is often the most cost-effective approach to manage groundwater contamination by benzene, the biomarker has potential applicability for forensic analysis of the natural attenuation of strongly anaerobic benzene plumes (79). DNA microarrays have also been constructed to assess the diversity of 16S rRNA genes in natural environmental samples. For example, mesophilic toluene-degrading and ethylbenzene-degrading sulfate-reducing consortia were first characterized by denaturing gradient gel electrophoresis (DGGE) fingerprinting of PCR-amplified 16S rRNA gene fragments, followed by sequencing. The sequences of the major bands were affiliated with the family Desulfobacteriaceae. Oligonucleotide probes specific for the 16S rRNA genes of target organisms were designed, and hybridization conditions were optimized for two analytical formats, membrane and DNA microarray hybridization. Both formats were used to characterize the toluene- and ethylbenzene-degrading consortia, and the results of both were consistent with results of the DGGE analyses (193).
The use of stable-isotope-probing (SIP) methods has allowed the tracking of the actual players that influence aromatic compound removal within microbial populations (114, 184). Thus, a microbial population that was capable of degrading [13C]benzene in gasoline-contaminated groundwater under nitrate-reducing conditions was identified by RNA-based SIP coupled to DGGE of RT-PCR-amplified 16S 13C rRNA gene fragments isolated from the community. Sequence analysis of the major 16S rRNA gene bands was performed, and a phylotype affiliated with the genus Azoarcus was observed. To isolate the Azoarcus strains, the groundwater sample was streaked onto agar plates containing nonselective medium, and DGGE analysis was used to screen colonies formed on the plates. Among 60 colonies obtained, five bacterial isolates corresponded to the SIP-identified Azoarcus phylotype. However, only two strains, strains DN11 and AN9, were shown to degrade benzene under denitrifying conditions. Therefore, these data show that RNA-SIP identification coupled to the phylogenetic screening of nonselective isolates facilitates the isolation of microorganisms with a specific function. Moreover, these data indicate that functional heterogeneity exists among strains within the Azoarcus phylotype, and therefore, specific molecular markers will be necessary for discriminating benzene-degrading Azoarcus populations from other closely related Azoarcus populations in the gasoline-contaminated aquifer (180). The degradative capabilities and bioaugmentation potential of Azoarcus sp. strain DN11 was further evaluated. Strain DN11 could grow on benzene, toluene, m-xylene, and benzoate as the sole carbon and energy sources under nitrate-reducing conditions, and o- and p-xylenes were transformed in the presence of toluene. The addition of strain DN11 to benzene-contaminated groundwater and further anaerobic incubation in laboratory bottles supplemented with nitrate showed significant benzene consumption, suggesting that Azoarcus sp. strain DN11 is potentially useful for the bioremediation of benzene that contaminates underground aquifers at relatively low concentrations (179).
Another study using the DNA-SIP technology reported a detailed functional and phylogenetic characterization of an iron-reducing enrichment culture maintained with benzene as the sole carbon and energy source. The most active microbes in the assimilation of [13C]benzene were shown to be uncultured bacteria within the gram-positive family Peptococcaceae (clostridia), which are closely related to environmental clones retrieved from contaminated aquifers worldwide and only distantly related to cultured representatives of the genus Thermincola (205). It has been suggested that benzene degradation involves an unusual syntrophy, where members of the Clostridia oxidize primarily benzene and where the electrons from the contaminant are both directly transferred to ferric iron by the primary oxidizers and also partially shared with the Desulfobulbaceae (deltaproteobacteria) as syntrophic partners. Alternatively, electrons may also be quantitatively transferred to the syntrophic partners, which then reduce the ferric iron. These results provide evidence for the importance of a novel clade of gram-positive iron reducers in anaerobic benzene degradation and the major role played by the syntrophic interactions in this process (205).
Catabolic Marker Genes
Due to the large phylogenetic diversity of bacteria that are able to anaerobically degrade some aromatic compounds, e.g., toluene, a selective characterization of such microorganisms in the environment based on rRNA marker genes is not possible. In this case, it is helpful to target a gene encoding a conserved key enzyme of the pathway, a so-called functional marker gene. The bssA gene, encoding the α-subunit of benzylsuccinate synthase (see above), has been used as a functional marker gene, which allows the specific detection of both known and unknown anaerobic toluene (and m-xylene) degraders that use fumarate-adding key reactions for the initial activation of the aromatic substrate (17, 78, 375). Degenerate bssA primers were even used to identify a couple of gene clusters (ass1 and ass2) encoding new glycyl radical enzymes that are likely to be involved in anaerobic n-alkane metabolism in sulfate-reducing strain AK-01 (50). The use of different degenerate bssA-targeted primer sets with pure cultures of anaerobic toluene degraders allowed an expansion of the phylogenetic framework of bssA reference sequences, especially within the deltaproteobacterium subgroup, e.g., Desulfobacula toluolica, D. cetonicum, the sulfate reducer TRM1 (Desulfocapsa related), Geobacter grbiciae, and Geobacter sp. strain TMJ1, and probably also with nonproteobacteria such as Desulfotomaculum spp. and Sedimentibacter spp. within the clostridial fermenters (375). The bssA gene has also been PCR amplified in a toluene-degrading methanogenic consortium (369). The phylogenetic comparison of ribosomal and bssA marker genes for different anaerobic toluene degraders suggests the existence of several events of lateral gene transfer for this catabolic marker. The bssA-based studies shed new light on the considerable environmental diversity of anaerobic hydrocarbon degraders, namely, within sulfate reducers, which is fundamental for the understanding of biodegradation processes in polluted sites (375). The combination of terminal restriction fragment length polymorphism fingerprinting and quantitative PCR of bacterial 16S rRNA genes and bssA genes allowed a high-resolution depth profile of a tar-oil-impacted aquifer to quantify the distribution of total microbiota and specific anaerobic toluene degraders, respectively. This study revealed that a highly specialized degrader community of microbes related to the iron and sulfate reducers of the deltaproteobacteria as well as clostridial fermenters resides within the biogeochemical gradient zone underneath the highly contaminated plume core. These findings support the presumed “plume fringe concept,” which postulates that the biodegradation of groundwater contaminants occurs mostly within the biogeochemical gradients surrounding contaminant plumes, and they show that the distribution of specific aquifer microbiota as well as degradation processes in contaminated aquifers are tightly coupled (375).
The genes encoding BCR, the key enzyme in the anaerobic degradation of many aromatic compounds (see above), have been successfully used as functional markers to determine the distribution and the capability for anaerobic degradation of aromatic compounds in bacterial isolates and in the environment (163, 343). Degenerate oligonucleotide primer pairs were developed for the genes encoding the α-subunit of BCRs from T. aromatica (bcrA), R. palustris (badF), and A. evansii (bzdQ). PCR amplification using such primers detected BCR genes in 21 denitrifying alpha-, beta-, or gammaproteobacterial isolates, i.e., strains from the genera Bradyrhizobium, Paracoccus, Ensifer, Acidovorax, Azoarcus, Thauera, and Pseudomonas, as well as in sediment samples capable of the anaerobic degradation of aromatic compounds. Phylogenetic analyses, sequence similarity comparisons, and conserved indel determinations grouped the PCR-amplified sequences into either the bcr type (similar to those of T. aromatica, R. palustris, and Magnetospirillum strains) or the bzd type (similar to that of Azoarcus strains). These two types of genes appear to have been transferred horizontally across generic lines in different bacteria. Thus, all Thauera strains; isolates from the genera Acidovorax, Bradyrhizobium, Paracoccus, and Ensifer; and some Pseudomonas strains had bcr-type BCRs with amino acid sequence similarities of more than 97%. The genes detected from Azoarcus strains were assigned to the bzd type. However, BCR genes from strains of the genera Ochrobactrum and Mesorhizobium and other Pseudomonas strains that are able to anaerobically degrade benzoate could not be amplified with any of the primers designed. Therefore, either such genes must be highly divergent from the bcr- and bzd-type genes or these denitrifying strains contain BCRs that are significantly different from the classical ATP-dependent BCR, as has been shown to be the case for the ATP-independent BCRs from strict anaerobes (277, 343, 377). The BCR genes detected from the environmental samples are much more diverse than those found in pure cultures. Among 50 environmental clones, 28 clones were considered to harbor bcr- or bzd-type BCRs, with the 22 remaining clones being derived from denitrifying bacteria whose BCR genes did not cluster closely with those from bacterial isolates. These results suggest that the genes involved in anaerobic benzoate degradation are more diverse than previously thought and, more importantly, that the capability for the anaerobic degradation of aromatic compounds is widespread among cultivated isolates and commonly present in natural environments (343).
Nevertheless, the use of gene probes to detect BCRs has some drawbacks such as the possibility of false-positive results due to the similarity among BCRs and the 2-hydroxyglutaryl-CoA dehydratases present in anaerobic bacteria. The construction of gene probes based on conserved regions of the dienoyl-CoA hydratase from the central benzoyl-CoA pathway (Fig. 2) has also been proposed as an alternative to the BCR probes for detecting both facultative and obligate anaerobic aromatic degraders (279). However, the similarities at the nucleotide level were not sufficient to use the dienoyl-CoA hydratase gene as a target for PCR amplification (206). Recently, a degenerate oligonucleotide primer pair was deduced from conserved regions of the genes (bamA, oah, or bzdY) encoding the ring-opening enzyme in the central benzoyl-CoA pathway (Fig. 2) and used to amplify the expected DNA fragment of hydrolase genes from all known facultative and obligate anaerobes that use aromatic growth substrates by PCR. The only exception was an R. palustris strain, which uniquely uses a different modified β-oxidation for benzoyl-CoA degradation (see above) (Fig. 2). Using oligonucleotide primers, the expected DNA fragment was also amplified in D. multivorans, a sulfate-reducing strain that degrades aromatic compounds, as well as in two sulfate-reducing enrichment cultures that degrade toluene and m-xylene, thus demonstrating its potential use in less defined bacterial communities (206). The combination of general probes for most anaerobic aromatic degraders, such as the bamA gene probe for the central benzoyl-CoA pathway, with probes for specific processes, such as the bssA probe for toluene degradation, will allow us to obtain a more thorough picture of the composition of microbial communities and the metabolic pathways involved in the bioremediation of aromatic compounds (206).
Detection of catabolic genes has also been achieved using functional gene arrays. For instance, a 50-mer-based oligonucleotide microarray based on most of the known genes and pathways involved in biodegradation and metal resistance has been designed (303). The array included 100 probes that target anaerobic degradation genes of monoaromatic compounds. Under the experimental conditions used, the microarray-differentiated sequences had <88% similarity. A linear relationship was found between signal intensities and DNA concentrations of pure cultures and mixed DNA templates. However, the microarray failed to detect less abundant functional genes, as it was only sensitive enough to detect dominant catabolic genes (303). A novel comprehensive microarray, termed GeoChip, has been developed. This microarray contains 24,243 oligonucleotide (50-mer) probes and covers more than 10,000 genes belonging to more than 150 functional groups involved in nitrogen, carbon, sulfur, and phosphorus cycling; metal reduction and resistance; and organic contaminant degradation, e.g., genes responsible for the anaerobic degradation of aromatic pollutants such as phenol, toluene, and ethylbenzene, etc. (146).
Metabolic Biomarkers
Specific intermediates of metabolic pathways can serve as biomarkers in the environment, where their detection is indicative of the presence of an active microbial population utilizing the metabolic substrate concerned. Monitoring of the disappearance of the substrate concomitant with the appearance of the biomarker over time is a reliable indication of in situ biodegradation (59, 383). The fact that the fumarate addition (benzylsuccinate formation) pathway for anaerobic aromatic hydrocarbon degradation is widely distributed in the environment (see above) prompted the successful detection of benzylsuccinate-like metabolites as indicators of in situ anaerobic bioremediation of hydrocarbon-contaminated sites (2, 16, 383).
An alternative strategy for monitoring the in situ biodegradation of a particular compound is to monitor the changes in stable isotope composition of the molecule of interest. This methodology is based on the fact that the relative abundances of the stable (nonradioactive) isotopes are effectively constant for each element, but microbial processes make small, although significant, changes to isotopic compositions by preferentially utilizing the lighter isotopes. Isotopic fractionation studies to monitor in situ anaerobic bioremediation of BTEX-contaminated habitats have been successfully implemented (59, 113, 231, 252, 300). In some cases, the carbon isotopic ratio of the dissolved inorganic carbon in oil-contaminated seawater samples was used as a rapid method to directly assess the level of activity of microbes on the oil components (241). Isotope-labeling studies were also used to identify the specific microbial subpopulations involved in the anaerobic degradation of toluene in hydrocarbon-contaminated aquifer sediments by tracing the incorporation of [13C]toluene into the polar lipid-derived fatty acids of a sulfate-reducing bacterial community (275).
Metagenomic Approaches
Metagenomics provides a wonderful source of information on new gene clusters and their distribution in the environment. In contrast to more classical genetic methods, e.g., PCR amplification of one type of degradative gene, the metagenomic approaches allow the simultaneous retrieval of different types of genes from environmental samples, facilitating the reconstruction of complete degradative pathways in some cases. A metagenome library for a Black Sea shelf microbial mat was constructed, and the nucleotide sequences of several clones were determined (201). One 79-kb contig, named FC1, harbored all genes required for complete anaerobic benzoate degradation as well as genes similar to those involved in the initial reactions of anaerobic phenol degradation and orthologs of the genes encoding 4-hydroxybenzoyl-CoA reductases in denitrifying organisms such as Azoarcus and Thauera strains (Fig. 2 and 7). The clustering of all these catabolic genes suggests that the metagenomic sequence represents an aromatic catabolic island of an organism that uses phenol, benzoate, and 4-HBA as carbon sources. The contig displayed no sequence deviations, indicating that it was derived from only one genotype, even though its location on the chromosome or on a plasmid could not be determined (201). No established taxonomic marker like 16S rRNA genes was present in the metagenomic sequence. Since the GC content (64.6%) of the FC1 contig differs significantly from that (43%) determined for an archaeal metagenomic sequence from the Black Sea, it can be assumed that it does not belong to a strain belonging to the Archaea. On the other hand, the fact that the genes encoding the putative BCR show strong similarities to those of facultative anaerobes, e.g., Azoarcus strains, suggests that the metagenomic sequence cannot be derived from an obligate anaerobe, e.g., a sulfate-reducing organism of the Black Sea mat, whose BCR enzyme does not show sequence similarities with the classical ATP-dependent BCR. These findings indicate that the metabolic diversity of the Black Sea mat is wider than currently known and that probably bacteria other than those of the methane-oxidizing consortia, for example, nitrate reducers that produce nitrite for anaerobic ammonia oxidation, contribute to the degradation of aromatic compounds in this anoxic habitat (201).
CONCLUSIONS AND PERSPECTIVES
The genetic and the more recent genomic and proteomic approaches that have been undertaken to study the anaerobic catabolism of aromatic compounds have contributed significantly to accelerating and completing our understanding of different aspects of the physiology, ecology, biochemistry, and regulatory mechanisms underlying a secondary metabolism that allows the use of this highly abundant carbon source by some anaerobic bacteria. These studies revealed that the anaerobic catabolism of aromatic compounds is more diverse than previously thought and, more importantly, that the capability for the anaerobic degradation of aromatic compounds is widespread in natural environments.
Comparative genetics and genomics suggest that the overall organization of anaerobic catabolic clusters is conserved across a wide variety of microorganisms. However, genus- and species-specific variations account for differences in gene arrangements, substrate specificities, and regulatory elements. A more detailed reconstruction of the evolutionary history of the catabolic clusters will benefit from the continued effort to sequence environmentally relevant strains. The arrangement of several catabolic clusters that are functionally related (catabolon) in a supraoperonic clustering, or even in a catabolic island, as well as the frequent location of mobile genetic elements next to the catabolic genes (48, 290) provide a molecular basis to explain the spreading of some catabolic genes among several members of the bacterial community, pointing to lateral gene transfer as a major mechanism for the adaptation and evolution of novel catabolic abilities for aromatic compounds in the anaerobic ecosystem.
However, there are still many issues regarding the anaerobic catabolism of aromatic compounds that remain unknown or poorly studied and that therefore require future attention. Thus, although much is currently known regarding the enzymes that catalyze the anaerobic dearomatization of benzoyl-CoA in facultative anaerobes, there is still a gap of knowledge regarding the dearomatization activities in strict anaerobes (30, 31). Moreover, the gene clusters encoding aromatic central pathways other than the benzoyl-CoA pathway are also poorly studied, and further efforts should be devoted to the characterization of such gene clusters. The anaerobic catabolism of some toxic aromatic hydrocarbons such as benzene and PAHs (including heterocyclic compounds) should also be further investigated to clarify both the biochemistry of these processes and the genes involved. For instance, the anaerobic dearomatization of benzene, which is the compound with the highest C-H bond dissociation energy known in nature, may involve a completely novel enzyme (204).
Although the number of complete genomes of anaerobic biodegraders available is still very limited, the first genomic analyses have already pointed to the existence of many paralogous genes that are likely to be involved in the anaerobic degradation of aromatic compounds, which, in turn, raises the question of the physiological role of this genetic redundancy (48, 213, 290). Whether such paralogous genes may operate under different ecological conditions (ecoparalogs) in anaerobes that inhabit multiple niches or whether they are standard paralogs and account for the catabolism of yet-unknown aromatic compounds are still open questions that need to be answered.
In contrast to the extensive knowledge generated about the regulation of the genes responsible for the aerobic degradation of aromatic compounds in many different bacteria, the mechanisms that control the gene expression of catabolic clusters have not been deciphered in most anaerobes. Increasing evidence suggests that a wide diversity of regulatory circuits also controls the expression of aromatic catabolic clusters in many anaerobes, and some of these regulatory networks in R. palustris and Azoarcus strains are being characterized (53). Integrating the effector-specific regulatory circuits into the global regulatory network of the cell will allow a better understanding and redesign of the expression of the anaerobic catabolic clusters and therefore will contribute to the better prediction and control of the behavior of anaerobic biodegraders.
Another important aspect that should be stressed in the near future is the transmembrane trafficking of aromatic substrates and metabolites. Different transport systems have been predicted to be involved in the uptake and/or efflux of aromatic compounds in anaerobic bacteria, but none has yet been characterized. Intracellular and intercellular transports of metabolic intermediates are also of great relevance, especially in those situations that involve syntrophic interactions between two or more different partners for the complete mineralization of aromatic substrates (205). Moreover, the ability of some anaerobes to sense and swim toward aromatic compounds has so far not been studied at the molecular level. Chemotaxis to aromatic compounds under anaerobic conditions is an important issue that deserves future investigation and that may be of great interest to fully understand the behavioral responses of anaerobes to the presence of such compounds (269).
The recent transcriptomic and proteomic approaches carried out with some anaerobic biodegraders shed some light on additional (and unexpected) genes and proteins that are not present in the cognate catabolic clusters but that are also involved, either directly or indirectly, in the anaerobic degradation of aromatic compounds. Some of these auxiliary functions correspond to proteins involved in the central metabolism of the cell, and they play a major role in basic processes such as the regeneration of particular cofactors and the transport of metabolites, etc. (48, 362, 378). The characterization of many auxiliary genes whose functions are still unknown should be the subject of future research, and it will allow a broadening of our current view of the constellation of genes and/or proteins that are essential for the anaerobic biodegradation of aromatic compounds in different host bacteria.
Many aromatic compounds, e.g., hydrocarbons and phenolic compounds, simultaneously serve as potential nutrients to be metabolized by bacteria and also as stressors since they are membrane-damaging compounds, and their metabolism can also generate stress signals within the cell. In aerobic biodegraders, the adaptative solution to the biodegradation-versus-stress dilemma appears to be temporally subordinating the expression of biodegradative genes (metabolic program) to adaptation to physicochemical stresses (stress program) (361). In solvent-shocked anaerobic cells, such as in Azoarcus sp. strain EbN1 cells, several stress responses, including oxidative stress, have been detected by proteomic studies (351). Nevertheless, further research should be carried out to better define the anaerobic stress program and to extend these analyses to strict anaerobes.
The increased use of high-throughput techniques such as genomics, metagenomics, proteomics, metabolomics, as well as systems biology approaches for addressing biological complexity from a holistic perspective and the application of the network theory to biology will certainly contribute significantly to unraveling the intricate regulatory and metabolic networks that govern the anaerobic biodegradation of aromatic compounds as well as their distribution and ecophysiological relevance to carbon flux in the environment. Knowledge of the full range of metabolic capabilities of the microorganisms influencing bioremediation as well as monitoring of microbial capacities and their distributions at contaminated sites will allow predictions of how these organisms are likely to respond to changes in environmental conditions that will take place during the course of natural attenuation.
Finally, all the knowledge generated so far should accelerate the development of anaerobic bioremediation technologies, e.g., bioaugmentation. For instance, energy recovery from oil fields in the form of methane, based on accelerating natural methanogenic biodegradation, may offer a route to the economic production of difficult-to-recover energy from oil fields (175). Genetically engineered anaerobes efficiently expressing multiple aromatic biodegradative pathways and the sequential use of anaerobic and aerobic recombinants will provide useful means to address the removal of toxic pollutants. Anaerobic biotransformation processes based on unprecedented enzymes and pathways with novel metabolic capabilities as well as the design of novel regulatory circuits and catabolic networks of great biotechnological potential in synthetic biology are now feasible to approach, and they should also be accomplished in the near future.
Acknowledgments
Work in our laboratory was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (BIO2003-01482, VEM2003-20075-C02-02, GEN2006-27750-C5-3-E, BIO2006-05957, and CSD2007-00005) and the Comunidad Autónoma de Madrid (P-AMB-259-0505).
REFERENCES
- 1.Achong, G. R., A. M. Rodríguez, and A. M. Spormann. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 1836763-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitken, C. M., D. M. Jones, and S. R. Larter. 2004. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 431291-294. [DOI] [PubMed] [Google Scholar]
- 3.Alder, E. 1977. Lignin chemistry past, present and future. Wood Sci. Technol. 11169-218. [Google Scholar]
- 4.Alhapel, A., D. J. Darley, N. Wagener, E. Eckel, N. Elsner, and A. J. Pierik. 2006. Molecular and funcional analysis of nicotinate catabolism in Eubacterium barkeri. Proc. Natl. Acad. Sci. USA 10312341-12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altenschmidt, U., B. Oswald, and G. Fuchs. 1991. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligase from a denitrifying Pseudomonas sp. J. Bacteriol. 1735494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreesen, J. R., and S. Fetzner. 2002. The molybdenum-containing hydroxylases of nicotinate, isonicotinate, and nicotine. Met. Ions Biol. Syst. 39405-430. [PubMed] [Google Scholar]
- 7.Annweiler, E., A. Materna, M. Safinowski, A. Kappler, H. H. Richnow, W. Michaelis, and R. U. Meckenstock. 2000. Anaerobic degradation of 2-methylnaphthalene by a sulfate-reducing enrichment culture. Appl. Environ. Microbiol. 665329-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Annweiler, E., W. Michaelis, and R. U. Meckenstock. 2002. Identical ring cleavage products during anaerobic degradation of naphthalene, 2-methylnaphthalene, and tetralin indicate a new metabolic pathway. Appl. Environ. Microbiol. 68852-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auburger, G., and J. Winter. 1992. Purification and characterization of benzoyl-CoA ligase from a syntrophic, benzoate-degrading, anaerobic mixed culture. Appl. Microbiol. Biotechnol. 37789-795. [DOI] [PubMed] [Google Scholar]
- 10.Auburger, G., and J. Winter. 1996. Activation and degradation of benzoate, 3-phenylpropionate and crotonate by Syntrophus buswellii strain GA. Evidence for electron-transport phosphorylation during crotonate respiration. Appl. Microbiol. Biotechnol. 44807-815. [DOI] [PubMed] [Google Scholar]
- 11.Baas, D., and J. Rétey. 1999. Cloning, sequencing and heterologous expression of pyrogallol-phloroglucinol transhydroxylase from Pelobacter acidigallici. Eur. J. Biochem. 265896-901. [DOI] [PubMed] [Google Scholar]
- 12.Ball, H. A., H. A. Johnson, M. Reinhard, and A. M. Spormann. 1996. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J. Bacteriol. 1785755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker, H. A. 1981. Amino acid degradation by anaerobic bacteria. Annu. Rev. Biochem. 5023-40. [DOI] [PubMed] [Google Scholar]
- 14.Barragán, M. J. L., B. Blázquez, M. T. Zamarro, J. M. Mancheño, J. L. García, E. Díaz, and M. Carmona. 2005. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 28010683-10694. [DOI] [PubMed] [Google Scholar]
- 15.Beatrix, B., O. Zelder, D. Linder, and W. Buckel. 1994. Cloning, sequencing and expression of the gene encoding the coenzyme B12-dependent 2-methyleneglutarate mutase from Clostridium barkeri in Escherichia coli. Eur. J. Biochem. 221101-109. [DOI] [PubMed] [Google Scholar]
- 16.Beller, H. R., W. H. Ding, and M. Reinhard. 1995. Byproducts of anaerobic alkylbenzene metabolism useful as indicators of in situ bioremediation. Environ. Sci. Technol. 292864-2869. [DOI] [PubMed] [Google Scholar]
- 17.Beller, H. R., S. R. Kane, T. C. Legler, and P. J. J. Alvarez. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 363977-3984. [DOI] [PubMed] [Google Scholar]
- 18.Beller, H. R., and A. M. Spormann. 1998. Analysis of the novel benzylsuccinate synthase reaction for anaerobic toluene activation based on structural studies of the product. J. Bacteriol. 1805454-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beller, H. R., and A. M. Spormann. 1999. Substrate range of benzylsuccinate synthase from Azoarcus sp. strain T. FEMS Microbiol. Lett. 178147-153. [DOI] [PubMed] [Google Scholar]
- 20.Berman-Frank, I., P. Lundgren, and P. Falkowski. 2003. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 154157-164. [DOI] [PubMed] [Google Scholar]
- 21.Berry, D. F., A. J. Francis, and J. M. Bollag. 1987. Microbial metabolism of homocyclic and heterocyclic aromatic compounds under anaerobic conditions. Microbiol. Rev. 5143-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beuerle, T., and E. Pichersky. 2002. Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal. Biochem. 302305-312. [DOI] [PubMed] [Google Scholar]
- 23.Bhandare, R., M. Calabro, and P. Coschigano. 2006. Site-directed mutagenesis of the Thauera aromatica strain T1 tutE tutFDGH gene cluster. Biochem. Biophys. Res. Commun. 346992-998. [DOI] [PubMed] [Google Scholar]
- 24.Biegert, T., U. Altenschmidt, C. Eckerskorn, and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoate-CoA ligase from a denitrifying Pseudomonas species. Eur. J. Biochem. 213555-561. [DOI] [PubMed] [Google Scholar]
- 25.Biegert, T., U. Altenschmidt, C. Eckerskorn, and G. Fuchs. 1995. Purification and properties of benzyl alcohol dehydrogenase from a denitrifying Thauera sp. Arch. Microbiol. 163418-423. [DOI] [PubMed] [Google Scholar]
- 26.Biegert, T., G. Fuchs, and J. Heider. 1996. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur. J. Biochem. 238661-668. [DOI] [PubMed] [Google Scholar]
- 27.Blake, C. K., and G. D. Hegeman. 1987. Plasmid pCBI carries genes for anaerobic benzoate catabolism in Alcaligenes xylosoxidans subsp. denitrificans PN-1. J. Bacteriol. 1694878-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blakley, E. R. 1978. The microbial degradation of cyclohexanecarboxylic acid by a β-oxidation pathway with simultaneous induction to the utilisation of benzoate. Can. J. Microbiol. 24847-855. [DOI] [PubMed] [Google Scholar]
- 29.Blázquez, B., M. Carmona, J. L. García, and E. Díaz. 2008. Identification and analysis of a glutaryl-CoA dehydrogenase-encoding gene and its cognate transcriptional regulator from Azoarcus sp. CIB. Environ. Microbiol. 10474-482. [DOI] [PubMed] [Google Scholar]
- 30.Boll, M. 2005. Dearomatizing benzene ring reductases. J. Mol. Microbiol. Biotechnol. 10132-142. [DOI] [PubMed] [Google Scholar]
- 31.Boll, M. 2005. Key enzymes in the anaerobic aromatic metabolism catalysing Birch-like reductions. Biochim. Biophys. Acta 170734-50. [DOI] [PubMed] [Google Scholar]
- 32.Boll, M., and G. Fuchs. 1998. Identification and characterization of the natural electron donor ferredoxin and of FAD as a possible prosthetic group of benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. Eur. J. Biochem. 251946-954. [DOI] [PubMed] [Google Scholar]
- 33.Boll, M., G. Fuchs, and J. Heider. 2002. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Chem. Biol. 6604-611. [DOI] [PubMed] [Google Scholar]
- 34.Boll, M., G. Fuchs, C. Meier, A. Trautwein, A. El Kasmi, S. W. Ragsdale, G. Buchanan, and D. J. Lowe. 2001. Redox centers of 4-hydroxybenzoyl-CoA reductase, a member of the xanthine oxidase family of molybdenum-containing enzymes. J. Biol. Chem. 27647853-47862. [DOI] [PubMed] [Google Scholar]
- 35.Boll, M., D. Laempe, W. Eisenreich, A. Bacher, T. Mittelberger, J. Heinze, and G. Fuchs. 2000. Nonaromatic products from anoxic conversion of benzoyl-CoA with benzoyl-CoA reductase and cyclohexa-1,5-diene-1-carbonyl-CoA hydratase. J. Biol. Chem. 27521889-21895. [DOI] [PubMed] [Google Scholar]
- 36.Bonin, I., B. M. Martins, V. Purvanov, S. Fetzner, R. Huber, and H. Dobbek. 2004. Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase. Structure 121425-1435. [DOI] [PubMed] [Google Scholar]
- 37.Bonting, C. F., and G. Fuchs. 1996. Anaerobic metabolism of 2-hydroxybenzoic acid (salicylic acid) by a denitrifying bacterium. Arch. Microbiol. 165402-408. [DOI] [PubMed] [Google Scholar]
- 38.Bossert, I. D., G. Whited, D. T. Gibson, and L. Y. Young. 1989. Anaerobic oxidation of p-cresol mediated by a partially purified methylhydroxylase from a denitrifying bacterium. J. Bacteriol. 1712956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breese, K., M. Boll, J. Alt-Mörbe, H. Schägger, and G. Fuchs. 1998. Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium Thauera aromatica. Eur. J. Biochem. 256148-154. [DOI] [PubMed] [Google Scholar]
- 40.Breese, K., and G. Fuchs. 1998. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 251916-923. [DOI] [PubMed] [Google Scholar]
- 41.Breining, S., E. Schiltz, and G. Fuchs. 2000. Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica. J. Bacteriol. 1825849-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brune, A., E. Miambi, and J. A. Breznak. 1995. Roles of oxygen and the intestinal microflora in the metabolism of lignin-derived phenylpropanoids and other monoaromatic compounds by termites. Appl. Environ. Microbiol. 612688-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brune, A., S. Schnell, and B. Schink. 1992. Sequential transhydroxylations converting hydroxyhydroquinone to phloroglucinol in the strictly anaerobic, fermentative bacterium Pelobacter massiliensis. Appl. Environ. Microbiol. 581861-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buckel, W., and R. Semmler. 1983. Purification, characterisation and reconstitution of glutaconyl-CoA decarboxylase, a biotin-dependent sodium pump from anaerobic bacteria. Eur. J. Biochem. 136427-434. [DOI] [PubMed] [Google Scholar]
- 45.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421357-360. [DOI] [PubMed] [Google Scholar]
- 46.Burland, S. M., and E. A. Edwards. 1999. Anaerobic benzene biodegradation linked to nitrate reduction. Appl. Environ. Microbiol. 65529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busch, A., J. Lacal, A. Martos, J. L. Ramos, and T. Krell. 2007. Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proc. Natl. Acad. Sci. USA 10413774-13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler, J. E., Q. He, K. P. Nevin, Z. He, J. Zhou, and D. R. Lovley. 2007. Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site. BMC Genomics 8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caldwell, M. E., and J. M. Suflita. 2000. Detection of phenol and benzoate as intermediates of anaerobic benzene biodegradation under different terminal electron-accepting conditions. Environ. Sci. Technol. 341216-1220. [Google Scholar]
- 50.Callaghan, A. V., B. Wawrik, S. M. N. Chadhain, L. Y. Young, and G. J. Zylstra. 2008. Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes. Biochem. Biophys. Res. Commun. 366142-148. [DOI] [PubMed] [Google Scholar]
- 51.Canosa, I., J. M. Sánchez-Romero, L. Yuste, and F. Rojo. 2000. A positive feedback mechanism controls expression of AlkS, the transcriptional regulator of the Pseudomonas oleovorans alkane degradation pathway. Mol. Microbiol. 35791-799. [DOI] [PubMed] [Google Scholar]
- 52.Carmona, M., and E. Díaz. 2005. Iron-reducing bacteria unravel novel strategies for the anaerobic catabolism of aromatic compounds. Mol. Microbiol. 581210-1215. [DOI] [PubMed] [Google Scholar]
- 53.Carmona, M., M. A. Prieto, B. Galán, J. L. García, and E. Díaz. 2008. Signaling networks and design of pollutants biosensors, p. 97-143. In E. Díaz (ed.), Microbial biodegradation: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 54.Cases, I., and V. de Lorenzo. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cases, I., and V. de Lorenzo. 2005. Promoters in the environment: transcriptional regulation in its natural context. Nat. Rev. Microbiol. 3105-118. [DOI] [PubMed] [Google Scholar]
- 56.Cases, I., D. W. Ussery, and V. de Lorenzo. 2003. The sigma54 regulon (sigmulon) of Pseudomonas putida. Environ. Microbiol. 51281-1293. [DOI] [PubMed] [Google Scholar]
- 57.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3351-368. [Google Scholar]
- 58.Cervin, M. A., R. J. Lewis, J. A. Brannigan, and G. B. Spiegelman. 1998. The Bacillus subtilis regulator SinR inhibits spoIIG promoter transcription in vitro without displacing RNA polymerase. Nucleic Acids Res. 263806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakraborty, R., and J. D. Coates. 2004. Anaerobic degradation of monoaromatic hydrocarbons. Appl. Microbiol. Biotechnol. 64437-446. [DOI] [PubMed] [Google Scholar]
- 60.Chakraborty, R., and J. D. Coates. 2005. Hydroxylation and carboxylation—two crucial steps of anaerobic benzene degradation by Dechloromonas strain RCB. Appl. Environ. Microbiol. 715427-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakraborty, R., S. M. O'Connor, E. Chan, and J. D. Coates. 2005. Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene compounds by Dechloromonas strain RCB. Appl. Environ. Microbiol. 718649-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chalmers, R. M., J. N. Keen, and C. A. Fewson. 1991. Comparison of benzyl alcohol dehydrogenases and benzaldehyde dehydrogenases from the benzyl alcohol and mandelate pathways in Acinetobacter calcoaceticus and from the TOL-plasmid-encoded toluene pathway in Pseudomonas putida. N-terminal amino acid sequences, amino acid compositions and immunological cross-reactions. Biochem. J. 27399-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Champion, K. M., K. Zengler, and R. Rabus. 1999. Anaerobic degradation of ethylbenzene and toluene in denitrifying strain EbN1 proceeds via independent substrate-induced pathways. J. Mol. Microbiol. Biotechnol. 1157-164. [PubMed] [Google Scholar]
- 64.Coates, J. D., R. Chakraborty, J. G. Lack, S. M. O'Connor, K. A. Cole, K. S. Bender, and L. A. Achenbach. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 4111039-1043. [DOI] [PubMed] [Google Scholar]
- 65.Coates, J. D., R. Chakraborty, and M. J. McInerney. 2002. Anaerobic benzene biodegradation—a new era. Res. Microbiol. 153621-628. [DOI] [PubMed] [Google Scholar]
- 66.Collier, D. N., P. W. Hager, and P. V. Phibbs, Jr. 1996. Catabolite repression control in the pseudomonads. Res. Microbiol. 147551-561. [DOI] [PubMed] [Google Scholar]
- 67.Collier, L. S., N. N. Nichols, and E. L. Neidle. 1997. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J. Bacteriol. 1795943-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coschigano, P. W. 2000. Transcriptional analysis of the tutE tutFDGH gene cluster from Thauera aromatica strain T1. Appl. Environ. Microbiol. 661147-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coschigano, P. W. 2002. Construction and characterization of insertion/deletion mutations of the tutF, tutD, and tutG genes of Thauera aromatica strain T1. FEMS Microbiol. Lett. 21737-42. [DOI] [PubMed] [Google Scholar]
- 70.Coschigano, P. W., and B. J. Bishop. 2004. Role of benzylsuccinate in the induction of the tutE tutFDGH gene complex of T. aromatica strain T1. FEMS Microbiol. Lett. 16261-266. [DOI] [PubMed] [Google Scholar]
- 71.Coschigano, P. W., M. M. Häggblom, and L. Y. Young. 1994. Metabolism of both 4-chlorobenzoate and toluene under denitrifying conditions by a constructed bacterial strain. Appl. Environ. Microbiol. 60989-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coschigano, P. W., T. S. Wehrman, and L. Y. Young. 1998. Identification and analysis of genes involved in anaerobic toluene metabolic by strain T1: putative role of a glycine free radical. Appl. Environ. Microbiol. 641650-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coschigano, P. W., and L. Y. Young. 1997. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl. Environ. Microbiol. 63652-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crawford, J. J., G. K. Sims, R. L. Mulvaney, and M. Radosevich. 1998. Biodegradation of atrazine under denitrifying conditions. Appl. Microbiol. Biotechnol. 49618-623. [DOI] [PubMed] [Google Scholar]
- 75.Cunane, L. M., Z. W. Chen, N. Shamala, F. S. Mathews, C. N. Cronin, and W. S. McIntire. 2000. Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism. J. Mol. Biol. 295357-374. [DOI] [PubMed] [Google Scholar]
- 76.Dagley, S. 1978. Determinants of biodegradability. Q. Rev. Biophys. 11577-602. [DOI] [PubMed] [Google Scholar]
- 77.Darley, P. I., J. A. Hellstern, J. I. Medina-Bellver, S. Marqués, B. Schink, and B. Philipp. 2007. Heterologous expression and identification of the genes involved in anaerobic degradation of 1,3-dihydroxybenzene (resorcinol) in Azoarcus anaerobius. J. Bacteriol. 1893824-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Da Silva, M. L. B., and P. J. J. Alvarez. 2004. Enhanced anaerobic biodegradation of benzene-toluene-ethylbenzene-xylene-ethanol mixtures in bioaugmented aquifer columns. Appl. Environ. Microbiol. 704720-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Da Silva, M. L. B., and P. J. J. Alvarez. 2007. Assessment of anaerobic benzene degradation potential using 16S rRNA gene-targeted real-time PCR. Environ. Microbiol. 972-80. [DOI] [PubMed] [Google Scholar]
- 80.Dayan, F. E., S. B. Watson, and N. P. Nanayakkara. 2007. Biosynthesis of lipid resorcinols and benzoquinones in isolated secretory plant root hairs. J. Exp. Bot. 583263-3272. [DOI] [PubMed] [Google Scholar]
- 81.de Lorenzo, V., and J. Pérez-Martín. 1996. Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol. Microbiol. 191177-1184. [DOI] [PubMed] [Google Scholar]
- 82.del Peso-Santos, T., D. Bartolomé-Martín, C. Fernández, S. Alonso, J. L. García, E. Díaz, V. Shingler, and J. Perera. 2006. Coregulation by phenylacetyl-coenzyme A-responsive PaaX integrates control of the upper and lower pathways for catabolism of styrene by Pseudomonas sp. strain Y2. J. Bacteriol. 1884812-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Denef, V. J., J. A. Klappenbach, M. A. Patrauchan, C. Florizone, J. L. Rodrigues, T. V. Tsoi, W. Verstraete, L. D. Eltis, and J. M. Tiedje. 2006. Genetic and genomic insights into the role of benzoate-catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 72585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Díaz, E. 2004. Bacterial degradation of aromatic pollutants: a paradigm of metabolic versatility. Int. Microbiol. 7173-180. [PubMed] [Google Scholar]
- 85.Díaz, E., A. Ferrández, M. A. Prieto, and J. L. García. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Díaz, E., and M. A. Prieto. 2000. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11467-475. [DOI] [PubMed] [Google Scholar]
- 87.Dickert, S., A. J. Pierik, D. Linder, and W. Buckel. 2000. The involvement of coenzyme A esters in the dehydration of (R)-phenyllactate to (E)-cinnamate by Clostridium sporogenes. Eur. J. Biochem. 2673874-3884. [DOI] [PubMed] [Google Scholar]
- 88.Dilling, S., F. Imkamp, S. Schmidt, and V. Müller. 2007. Regulation of caffeate respiration in the acetogenic bacterium Acetobacterium woodii. Appl. Environ. Microbiol. 733630-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dimroth, P., and B. Schink. 1998. Energy conservation in the decarboxylation of dicarboxylic acids by fermenting bacteria. Arch. Microbiol. 17069-77. [DOI] [PubMed] [Google Scholar]
- 90.Ding, B., S. Schmeling, and G. Fuchs. 2008. Anaerobic metabolism of catechol by the denitrifying bacterium Thauera aromatica—a result of promiscuous enzymes and regulators? J. Bacteriol. 1901620-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dispensa, M., C. T. Thomas, M. K. Kim, J. A. Perrotta, J. Gibson, and C. S. Harwood. 1992. Anaerobic growth of Rhodopseudomonas palustris on 4-hydroxybenzoate is dependent on AadR, a member of the cyclic AMP receptor protein family of transcriptional regulators. J. Bacteriol. 1745803-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2414-424. [DOI] [PubMed] [Google Scholar]
- 93.Domínguez-Cuevas, P., J. E. González-Pastor, S. Marqués, J. L. Ramos, and V. de Lorenzo. 2006. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J. Biol. Chem. 28111981-11991. [DOI] [PubMed] [Google Scholar]
- 94.Dörner, E., and M. Boll. 2002. Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J. Bacteriol. 1843975-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duetz, W. A., S. Marqués, B. Wind, J. L. Ramos, and J. G. van Andel. 1996. Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWW0 under various conditions of nutrient limitation in chemostat culture. Appl. Environ. Microbiol. 62601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Duldhardt, I., I. Nijenhuis, F. Schauer, and H. J. Heipieper. 2007. Anaerobically grown Thauera aromatica, Desulfococcus multivorans, Geobacter sulfurreducens are more sensitive towards organic solvents than aerobic bacteria. Appl. Microbiol. Biotechnol. 77:705-711. [DOI] [PubMed] [Google Scholar]
- 97.Durante-Rodríguez, G., M. T. Zamarro, J. L. García, E. Díaz, and M. Carmona. 2006. Oxygen-dependent regulation of the central pathway for the anaerobic catabolism of aromatic compounds in Azoarcus sp. strain CIB. J. Bacteriol. 1882343-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Durante-Rodríguez, G., M. T. Zamarro, J. L. García, E. Díaz, and M. Carmona. 2008. New insights into the BzdR-mediated transcriptional regulation of the anaerobic catabolism of benzoate in Azoarcus sp. CIB. Microbiology 154306-316. [DOI] [PubMed] [Google Scholar]
- 99.Ebenau-Jehle, C., M. Boll, and G. Fuchs. 2003. 2-Oxoglutarate:NADP+ oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J. Bacteriol. 1856119-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Egland, P. G., J. Gibson, and C. S. Harwood. 1995. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J. Bacteriol. 1776545-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Egland, P. G., J. Gibson, and C. S. Harwood. 2001. Reductive, coenzyme A-mediated pathway for 3-chlorobenzoate degradation in the phototrophic bacterium Rhodopseudomonas palustris. Appl. Environ. Microbiol. 671396-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Egland, P. G., and C. S. Harwood. 1999. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an FNR family member. J. Bacteriol. 1812102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Egland, P. G., and C. S. Harwood. 2000. HbaR, a 4-hydroxybenzoate sensor and FNR-CRP superfamily member, regulates anaerobic 4-hydroxybenzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 182100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 946484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.El Fantroussi, S., H. Naveau, and S. N. Agathos. 1998. Anaerobic dechlorinating bacteria. Biotechnol. Prog. 14167-188. [DOI] [PubMed] [Google Scholar]
- 106.Elshahed, M. S., V. K. Bhupathiraju, N. Q. Wofford, M. A. Nanny, and M. J. McInerney. 2001. Metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by “Syntrophus aciditrophicus” strain SB in syntrophic association with H2-using microorganisms. Appl. Environ. Microbiol. 671728-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Elshahed, M. S., and M. J. McInerney. 2001. Benzoate fermentation by the anaerobic bacterium Syntrophus aciditrophicus in the absence of hydrogen-using microorganisms. Appl. Environ. Microbiol. 675520-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Esteve-Núñez, A., A. Caballero, and J. L. Ramos. 2001. Biological degradation of 2,4,6-trinitrotoluene. Microbiol. Mol. Biol. Rev. 65335-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Esteve-Nuñez, A., G. Lucchesi, B. Philipp, B. Schink, and J. L. Ramos. 2000. Respiration of 2,4,6-trinitrotoluene by Pseudomonas sp. strain JLR11. J. Bacteriol. 1821352-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Evans, W. C., and G. Fuchs. 1988. Anaerobic degradation of aromatic compounds. Annu. Rev. Microbiol. 42289-317. [DOI] [PubMed] [Google Scholar]
- 111.Fetzner, S. 1998. Bacterial degradation of pyridine, indole, quinoline, and their derivatives under different redox conditions. Appl. Microbiol. Biotechnol. 49237-250. [Google Scholar]
- 112.Fetzner, S. 2002. Enzymes involved in the aerobic bacterial degradation of N-heteroaromatic compounds: molybdenum hydroxylases and ring-opening 2,4-dioxygenases. Naturwissenschaften 8759-69. [DOI] [PubMed] [Google Scholar]
- 113.Fischer, A., I. Herklotz, S. Herrmann, M. Thullner, S. A. Weelink, A. J. Stams, M. Schlömann, H. H. Richnow, and C. Vogt. 2008. Combined carbon and hydrogen isotope fractionation investigations for elucidating benzene biodegradation pathways. Environ. Sci. Technol. 424356-4363. [DOI] [PubMed] [Google Scholar]
- 114.Friedrich, M. W. 2006. Stable-isotope probing of DNA: insights into the function of uncultivated microorganisms from isotopically labeled metagenomes. Curr. Opin. Biotechnol. 1759-66. [DOI] [PubMed] [Google Scholar]
- 115.Fu, Z., M. Wang, R. Paschke, S. Rao, F. E. Frerman, and J. J. P. Kim. 2004. Crystal structures of human glutaryl-CoA dehydrogenases with and without an alternate substrate: structural bases of dehydrogenation and decarboxylation reactions. Biochemistry 439674-9684. [DOI] [PubMed] [Google Scholar]
- 116.Fuchs, G. 2008. Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 112582-99. [DOI] [PubMed] [Google Scholar]
- 117.Gallegos, M. T., A. J. Molina-Henares, X. Zhang, W. Terán, P. Bernal, Y. Alguel, M. E. Guazzaroni, T. Krell, A. Segura, and J. L. Ramos. 2008. Genomic insights into solvent tolerance and pumps that extrude toxic chemicals, p. 189-217. In E. Díaz (ed.), Microbial biodegradation: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 118.Gallus, C., and B. Schink. 1994. Anaerobic degradation of pimelate by newly isolated denitrifying bacteria. Microbiology 140409-416. [DOI] [PubMed] [Google Scholar]
- 119.Gallus, C., and B. Schink. 1998. Anaerobic degradation of α-resorcylate by Thauera aromatica strain AR-1 proceeds via oxidation and decarboxylation to hydroxyhydroquinone. Arch. Microbiol. 169333-338. [DOI] [PubMed] [Google Scholar]
- 120.Galushko, A., D. Minz, B. Schink, and F. Widdel. 1999. Anaerobic degradation of naphthalene by a pure culture of a novel type of marine sulphate-reducing bacterium. Environ. Microbiol. 1415-420. [DOI] [PubMed] [Google Scholar]
- 121.Galushko, A. S., and B. Schink. 2000. Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch. Microbiol. 174314-321. [DOI] [PubMed] [Google Scholar]
- 122.Gasson, M. J., Y. Kitamura, W. R. McLauchlan, A. Narbad, A. J. Parr, E. L. Parsons, J. Payne, M. J. Rhodes, and N. J. Walton. 1998. Metabolism of ferulic acid to vanillin. A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester. J. Biol. Chem. 2734163-4170. [DOI] [PubMed] [Google Scholar]
- 123.Geissler, J. F., C. S. Harwood, and J. Gibson. 1988. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J. Bacteriol. 1701709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gescher, J., W. Ismail, E. Olgeschläger, W. Eisenreich, J. Wörth, and G. Fuchs. 2006. Aerobic benzoyl-coenzyme A (CoA) catabolic pathway in Azoarcus evansii: conversion of ring cleavage product by 3,4-dehydroadipyl-CoA semialdehyde dehydrogenase. J. Bacteriol. 1882919-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gescher, J., A. Zaar, M. Mohamed, H. Schägger, and G. Fuchs. 2002. Genes coding for a new pathway of aerobic benzoate metabolism in Azoarcus evansii. J. Bacteriol. 1846301-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gibson, J., M. Dispensa, G. C. Fogg, D. T. Evans, and C. S. Harwood. 1994. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J. Bacteriol. 176634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gibson, J., M. Dispensa, and C. S. Harwood. 1997. 4-Hydroxybenzoyl coenzyme A reductase (dehydroxylating) is required for anaerobic degradation of 4-hydroxybenzoate by Rhodopseudomonas palustris and shares features with molybdenum-containing hydroxylases. J. Bacteriol. 179634-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56345-369. [DOI] [PubMed] [Google Scholar]
- 129.Gibson, K. J., and J. Gibson. 1992. Potential early intermediates in anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl. Environ. Microbiol. 58696-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Giel, J. L., D. Rodionov, M. Liu, F. R. Blattner, and P. J. Kiley. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 601058-1075. [DOI] [PubMed] [Google Scholar]
- 131.Gillooly, D. J., A. G. Robertson, and C. A. Fewson. 1998. Molecular characterization of benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II of Acinetobacter calcoaceticus. Biochem. J. 3301375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gladyshev, V. N., S. V. Khangulov, and T. C. Stadtman. 1996. Properties of the selenium- and molybdenum-containing nicotinic acid hydroxylase from Clostridium barkeri. Biochemistry 35212-223. [DOI] [PubMed] [Google Scholar]
- 133.Glöckler, R., A. Tschech, and G. Fuchs. 1989. Reductive dehydroxylation of 4-hydroxybenzoyl-CoA to benzoyl-CoA in a denitrifying, phenol-degrading Pseudomonas species. FEBS Lett. 251237-240. [DOI] [PubMed] [Google Scholar]
- 134.Gomes, B., G. Fendrich, and R. H. Abeles. 1981. Mechanism of action of glutaryl-CoA and butyryl-CoA dehydrogenases. Purification of glutaryl-CoA dehydrogenase. Biochemistry 201481-1490. [DOI] [PubMed] [Google Scholar]
- 135.Gorny, N., and B. Schink. 1994. Anaerobic degradation of catechol by Desulfobacterium sp. strain Cat2 proceeds via carboxylation to protocatechuate. Appl. Environ. Microbiol. 603396-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gorny, N., G. Wahl, A. Brune, and B. Schink. 1992. A strictly anaerobic nitrate-reducing bacterium growing with resorcinol and other aromatic compounds. Arch. Microbiol. 15848-53. [DOI] [PubMed] [Google Scholar]
- 137.Green, J., B. Bennett, P. Jordan, E. T. Ralph, A. J. Thomson, and J. R. Guest. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gu, Y., L. Reshetnikova, Y. Li, Y. Wu, H. Yan, S. Singh, and X. Ji. 2002. Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis. J. Mol. Biol. 319779-789. [DOI] [PubMed] [Google Scholar]
- 139.Gülensoy, N., and P. J. Alvarez. 1999. Diversity and correlation of specific aromatic hydrocarbon biodegradation capabilities. Biodegradation 10331-340. [DOI] [PubMed] [Google Scholar]
- 140.Harrison, F. H., and C. S. Harwood. 2005. The pimFABCDE operon from Rhodopseudomonas palustris mediates dicarboxylic acid degradation and participates in anaerobic benzoate degradation. Microbiology 151727-736. [DOI] [PubMed] [Google Scholar]
- 141.Härtel, U., E. Eckel, J. Koch, G. Fuchs, D. Linder, and W. Buckel. 1993. Purification of glutaryl-CoA dehydrogenase from Pseudomonas sp., an enzyme involved in the anaerobic degradation of benzoate. Arch. Microbiol. 159174-181. [DOI] [PubMed] [Google Scholar]
- 142.Harwood, C. S., G. Burchhardt, H. Herrmann, and G. Fuchs. 1999. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22439-458. [Google Scholar]
- 143.Harwood, C. S., and J. Gibson. 1988. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl. Environ. Microbiol. 54712-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Harwood, C. S., and J. Gibson. 1997. Shedding light on anaerobic benzene ring degradation: a process unique to prokaryotes? J. Bacteriol. 179301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Harwood, C. S., and R. E. Parales. 1996. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50553-590. [DOI] [PubMed] [Google Scholar]
- 146.He, Z., T. J. Gentry, C. W. Schadt, L. Wu, J. Liebich, S. C. Chong, Z. Huang, W. Wu, B. Gu, P. Jardine, C. Criddle, and J. Zhou. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 167-77. [DOI] [PubMed] [Google Scholar]
- 147.Head, I. M., D. M. Jones, and S. R. Larter. 2003. Biological activity in the deep subsurface and the origin of heavy oil. Nature 426344-352. [DOI] [PubMed] [Google Scholar]
- 148.Hearn, E. M., D. R. Patel, and B. van den Berg. 2008. Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proc. Natl. Acad. Sci. USA 1058601-8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Heider, J. 2007. Adding handles to unhandy substrates: anaerobic hydrocarbon activation mechanisms. Curr. Opin. Chem. Biol. 11188-194. [DOI] [PubMed] [Google Scholar]
- 150.Heider, J., M. Boll, K. Breese, S. Breinig, C. Ebenau-Jehle, U. Feil, N. Gad'on, D. Laempe, B. Leuthner, M. El-Said Mohamed, S. Schneider, G. Burchhardt, and G. Fuchs. 1998. Differential induction of enzymes involved in anaerobic metabolism of aromatic compounds in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 170120-131. [DOI] [PubMed] [Google Scholar]
- 151.Heider, J., and G. Fuchs. 1997. Anaerobic metabolism of aromatic compounds. Eur. J. Biochem. 243577-596. [DOI] [PubMed] [Google Scholar]
- 152.Heider, J., and R. Rabus. 2008. Genomic insights in the anaerobic biodegradation of organic pollutants, p. 25-54. In E. Díaz (ed.), Microbial biodegradation: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 153.Heider, J., A. M. Spormann, H. R. Beller, and F. Widdel. 1999. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev. 22459-473. [Google Scholar]
- 154.Heipieper, H. J., G. Neumann, S. Cornelissen, and F. Meinhardt. 2007. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 74961-973. [DOI] [PubMed] [Google Scholar]
- 155.Hermuth, K., B. Leuthner, and J. Heider. 2002. Operon structure and expression of the genes for benzylsuccinate synthase in Thauera aromatica strain K172. Arch. Microbiol. 177132-138. [DOI] [PubMed] [Google Scholar]
- 156.Hess, A., B. Zarda, D. Hahn, A. Häner, D. Stax, P. Höhener, and J. Zeyer. 1997. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl. Environ. Microbiol. 632136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hirsch, W., H. Schägger, and G. Fuchs. 1998. Phenylglyoxylate:NAD+ oxidoreductase (CoA benzoylating), a new enzyme of anaerobic phenylalanine metabolism in the denitrifying bacterium Azoarcus evansii. Eur. J. Biochem. 251907-915. [DOI] [PubMed] [Google Scholar]
- 158.Höffken, H. W., M. Duong, T. Friedrich, M. Breuer, B. Hauer, R. Reinhardt, R. Rabus, and J. Heider. 2006. Crystal structure and enzyme kinetics of the (S)-specific 1-phenylethanol dehydrogenase of the denitrifying bacterium strain EbN1. Biochemistry 4582-93. [DOI] [PubMed] [Google Scholar]
- 159.Holcenberg, J. S., and L. Tsai. 1969. Nicotinic acid metabolism. IV. Ferredoxin-dependent reduction of 6-hydroxynicotinic acid to 6-oxo-1,4,5,6-tetrahydronicotinic acid. J. Biol. Chem. 2441204-1211. [PubMed] [Google Scholar]
- 160.Holliger, C., G. Wohlfarth, and G. Dieckert. 1999. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22383-398. [Google Scholar]
- 161.Holm, L., and C. Sander. 1997. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins 2872-82. [PubMed] [Google Scholar]
- 162.Hopper, D. J., I. D. Bossert, and M. E. Rhodes-Roberts. 1991. p-Cresol methylhydroxylase from a denitrifying bacterium involved in anaerobic degradation of p-cresol. J. Bacteriol. 1731298-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hosoda, A., Y. Kasai, N. Hamamura, Y. Takahata, and K. Watanabe. 2005. Development of a PCR method for the detection and quantification of benzoyl-CoA reductase genes and its application to monitored natural attenuation. Biodegradation 16591-601. [DOI] [PubMed] [Google Scholar]
- 164.Huang, J., Z. He, and J. Wiegel. 1999. Cloning, characterization, and expression of a novel gene encoding a reversible 4-hydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. J. Bacteriol. 1815119-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huffman, J. L., and R. G. Brennan. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 1298-106. [DOI] [PubMed] [Google Scholar]
- 166.Hughes, M. N. 1999. Relationships between nitric oxide, nitroxyl ion, nitrosonium cation and peroxynitrite. Biochim. Biophys. Acta 1411263-272. [DOI] [PubMed] [Google Scholar]
- 167.Ismail, W., M. El-Said Mohamed, B. L. Wanner, K. A. Datsenko, W. Eisenreich, F. Rohdich, A. Bacher, and G. Fuchs. 2003. Functional genomics by NMR spectroscopy. Phenylacetate catabolism in Escherichia coli. Eur. J. Biochem. 2703047-3054. [DOI] [PubMed] [Google Scholar]
- 168.Jackson, B. E., and M. J. McInerney. 2002. Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415454-456. [DOI] [PubMed] [Google Scholar]
- 169.Jannsen, D. B., J. E. Oppentocht, and G. J. Poelarends. 2001. Microbial dehalogenation. Curr. Opin. Biotechnol. 12254-258. [DOI] [PubMed] [Google Scholar]
- 170.Jiménez, J. I., B. Miñambres, J. L. García, and E. Díaz. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4824-841. [DOI] [PubMed] [Google Scholar]
- 171.Johannes, J., A. Bluschke, N. Jehmlich, M. von Bergen, and M. Boll. 2008. Purification and characterization of active-site components of the putative p-cresol methylhydroxylase membrane complex from Geobacter metallireducens. J. Bacteriol. 1906493-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Johannes, J., M. C. Unciuleac, T. Friedrich, E. Warkentin, U. Ermler, and M. Boll. 2008. Inhibitors of the molybdenum cofactor containing 4-hydroxybenzoyl-CoA reductase. Biochemistry 474964-4972. [DOI] [PubMed] [Google Scholar]
- 173.Johnson, H. A., D. A. Pelletier, and A. M. Spormann. 2001. Isolation and characterization of anaerobic ethylbenzene dehydrogenase, a novel Mo-Fe-S enzyme. J. Bacteriol. 1834536-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Johnson, H. A., and A. M. Spormann. 1999. In vitro studies on the initial reactions of anaerobic ethylbenzene mineralization. J. Bacteriol. 1815662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jones, D. M., I. M. Head, N. D. Gray, J. J. Adams, A. K. Rowan, C. M. Aitken, B. Bennett, H. Huang, A. Brown, B. F. J. Bowler, T. Oldenburg, M. Erdmann, and S. R. Larter. 2007. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451176-180. [DOI] [PubMed] [Google Scholar]
- 176.Kaiser, J. P., Y. Feng, and J. M. Bollag. 1996. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol. Rev. 60483-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kane, S. R., H. R. Beller, T. C. Legler, and R. T. Anderson. 2002. Biochemical and genetic evidence of benzylsuccinate synthase in toluene-degrading, ferric iron-reducing Geobacter metallireducens. Biodegradation 13149-154. [DOI] [PubMed] [Google Scholar]
- 178.Karns, J. S. 1999. Gene sequence and properties of an s-triazine ring-cleavage enzyme from Pseudomonas sp. strain NRRLB-12227. Appl. Environ. Microbiol. 653512-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kasai, Y., Y. Kodama, Y. Takahata, T. Hoaki, and K. Watanabe. 2007. Degradative capacities and bioaugmentation potential of an anaerobic benzene-degrading bacterium strain DN11. Environ. Sci. Technol. 416222-6227. [DOI] [PubMed] [Google Scholar]
- 180.Kasai, Y., Y. Takahata, M. Manefield, and K. Watanabe. 2006. RNA-based stable isotope probing and isolation of anaerobic benzene-degrading bacteria from gasoline-contaminated groundwater. Appl. Environ. Microbiol. 723586-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kawaguchi, K., Y. Shinoda, H. Yurimoto, Y. Sakai, and N. Kato. 2006. Purification and characterization of benzoate-CoA ligase from Magnetospirillum sp. strain TS-6 capable of aerobic and anaerobic degradation of aromatic compounds. FEMS Microbiol. Lett. 257208-213. [DOI] [PubMed] [Google Scholar]
- 182.Kazumi, J., M. E. Caldwell, J. M. Suflita, D. R. Lovley, and L. Y. Young. 1997. Anaerobic degradation of benzene in diverse anoxic environments. Environ. Sci. Technol. 31813-818. [Google Scholar]
- 183.Keat, M. J., and D. J. Hopper. 1978. p-Cresol and 3,5-xylenol methylhydroxylases in Pseudomonas putida N.C.I.B. 9896. Biochem. J. 175649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kerkhof, L. J., and M. M. Häggblom. 2008. Detecting active bacteria involved in biodegradation in the environment, p. 55-70. In E. Díaz (ed.), Microbial biodegradation: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 185.Kiley, P. J., and W. S. Reznikoff. 1991. Fnr mutants that activate gene expression in the presence of oxygen. J. Bacteriol. 17316-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kim, T. S., H. Y. Kim, and B. H. Kim. 1990. Petroleum desulfurization by Desulfovibrio desulfuricans M6 using electrochemically supplied reducing equivalent. Biotechnol. Lett. 12757-761. [Google Scholar]
- 187.Kitts, C. L., J. P. Lapointe, V. T. Lam, and R. A. Ludwig. 1992. Elucidation of the complete Azorhizobium nicotinate catabolism pathway. J. Bacteriol. 1747791-7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kloer, D. P., C. Hagel, J. Heider, and G. E. Schulz. 2006. Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure 141377-1388. [DOI] [PubMed] [Google Scholar]
- 189.Kluge, C., A. Tschech, and G. Fuchs. 1990. Anaerobic metabolism of resorcyclic acids (m-dihydroxybenzoic acids) and resorcinol (1,3-benzenediol) in a fermenting and in a denitrifying bacterium. Arch. Microbiol. 15568-74. [Google Scholar]
- 190.Kniemeyer, O., T. Fischer, H. Wilkes, F. O. Glöckner, and F. Widdel. 2003. Anaerobic degradation of ethylbenzene by a new type of marine sulfate-reducing bacterium. Appl. Environ. Microbiol. 69760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kniemeyer, O., and J. Heider. 2001. (S)-1-Phenylethanol dehydrogenase of Azoarcus sp. strain EbN1, an enzyme of anaerobic ethylbenzene catabolism. Arch. Microbiol. 176129-135. [DOI] [PubMed] [Google Scholar]
- 192.Kniemeyer, O., and J. Heider. 2001. Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidizing molybdenum/iron-sulfur/heme enzyme. J. Biol. Chem. 27621381-21386. [DOI] [PubMed] [Google Scholar]
- 193.Koizumi, Y., J. J. Nelly, T. Nakagawa, H. Urakawa, S. El-Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa, and D. A. Stahl. 2002. Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl. Environ. Microbiol. 683215-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kollmann-Koch, A., and H. Eggerer. 1984. Nicotinic acid metabolism. Dimethylmaleate hydratase. Hoppe Syler's Z. Physiol. Chem. 365847-857. [DOI] [PubMed] [Google Scholar]
- 195.Körner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27559-592. [DOI] [PubMed] [Google Scholar]
- 196.Koudelka, G. B., and C. Y. Lam. 1993. Differential recognition of OR1 and OR3 by bacteriophage 434 repressor and Cro. J. Biol. Chem. 26823812-23817. [PubMed] [Google Scholar]
- 197.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24946-950. [Google Scholar]
- 198.Krell, T., J. R. Coggins, and A. J. Lapthorn. 1998. The three-dimensional structure of shikimate kinase. J. Mol. Biol. 278983-987. [DOI] [PubMed] [Google Scholar]
- 199.Krieger, C. J., H. R. Beller, M. Reinhard, and A. M. Spormann. 1999. Initial reactions in anaerobic oxidation of m-xylene by the denitrifying bacterium Azoarcus sp. strain T. J. Bacteriol. 1816403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Krieger, C. J., W. Roseboom, S. P. Albracht, and A. M. Spormann. 2001. A stable organic free radical in anaerobic benzylsuccinate synthase of Azoarcus sp. strain T. J. Biol. Chem. 27612924-12927. [DOI] [PubMed] [Google Scholar]
- 201.Kube, M., A. Beck, A. Meyerdierks, R. Amann, R. Reinhardt, and R. Rabus. 2005. A catabolic gene cluster for anaerobic benzoate degradation in methanotrophic microbial Black Sea mats. Syst. Appl. Microbiol. 28287-294. [DOI] [PubMed] [Google Scholar]
- 202.Kube, M., J. Heider, J. Amann, P. Hufnagel, S. Kühner, A. Beck, R. Reinhardt, and R. Rabus. 2004. Genes involved in the anaerobic degradation of toluene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 181182-194. [DOI] [PubMed] [Google Scholar]
- 203.Kühner, S., L. Wöhlbrand, I. Fritz, W. Wruck, C. Hultschig, P. Hufnagel, M. Kube, R. Reinhardt, and R. Rabus. 2005. Substrate-dependent regulation of anaerobic degradation pathways for toluene and ethylbenzene in a denitrifying bacterium, strain EbN1. J. Bacteriol. 1871493-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kunapuli, U., C. Griebler, H. R. Beller, and R. U. Meckenstock. 2008. Identification of intermediates formed during anaerobic benzene degradation by an iron-reducing enrichment culture. Environ. Microbiol. 101703-1712. [DOI] [PubMed] [Google Scholar]
- 205.Kunapuli, U., T. Lueders, and R. U. Meckenstock. 2007. The use of stable isotope probing to identify key iron-reducing microorganisms involved in anaerobic benzene degradation. ISME J. 1643-653. [DOI] [PubMed] [Google Scholar]
- 206.Kuntze, K., Y. Shinoda, H. Moutakki, M. J. McInerney, C. Vogt, H. H. Richnow, and M. Boll. 2008. 6-Oxocyclohex-1-ene-1-carbonyl-coenzyme A hydrolases from obligately anaerobic bacteria: characterization and identification of its gene as a functional marker for aromatic compounds degrading anaerobes. Environ. Microbiol. 101547-1556. [DOI] [PubMed] [Google Scholar]
- 207.Küver, J., Y. Xu, and J. Gibson. 1995. Metabolism of cyclohexane carboxylic acid by the photosynthetic bacterium Rhodopseudomonas palustris. Arch. Microbiol. 164337-345. [DOI] [PubMed] [Google Scholar]
- 208.Lacal, J., A. Busch, M. E. Guazzaroni, T. Krell, and J. L. Ramos. 2006. The TodS-TodT two-component regulatory system recognizes a wide range of effectors and works with DNA-bending proteins. Proc. Natl. Acad. Sci. USA 1038191-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lack, A., and G. Fuchs. 1992. Carboxylation of phenylphosphate by phenol carboxylase, an enzyme system of anaerobic phenol metabolism. J. Bacteriol. 1743629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Laempe, D., W. Eisenreich, A. Bacher, and G. Fuchs. 1998. Cyclohexa-1,5-diene-1-carbonyl-CoA hydratase, an enzyme involved in anaerobic metabolism of benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 255618-627. [DOI] [PubMed] [Google Scholar]
- 211.Laempe, D., M. Jahn, K. Breese, H. Schägger, and G. Fuchs. 2001. Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica. J. Bacteriol. 183968-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Laempe, D., M. Jahn, and G. Fuchs. 1999. 6-Hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 263420-429. [DOI] [PubMed] [Google Scholar]
- 213.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 2255-61. [DOI] [PubMed] [Google Scholar]
- 214.Lau, P. C., Y. Wang, A. Patel, D. Labbé, H. Bergeron, R. Brousseau, Y. Konishi, and M. Rawlings. 1997. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc. Natl. Acad. Sci. USA 181453-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Leuthner, B., and J. Heider. 1998. A two-component system involved in regulation of anaerobic toluene metabolism in Thauera aromatica. FEMS Microbiol. Lett. 16635-41. [DOI] [PubMed] [Google Scholar]
- 216.Leuthner, B., and J. Heider. 2000. Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of β-oxidation of the intermediate benzylsuccinate. J. Bacteriol. 182272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Leuthner, B., C. Leutwein, H. Schulz, P. Hörth, W. Haehnel, E. Schiltz, H. Schägger, and J. Heider. 1998. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol. Microbiol. 28615-628. [DOI] [PubMed] [Google Scholar]
- 218.Leutwein, C., and J. Heider. 2001. Succinyl-CoA:(R)-benzylsuccinate CoA-transferase: an enzyme of the anaerobic toluene catabolic pathway in denitrifying bacteria. J. Bacteriol. 1834288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Leutwein, C., and J. Heider. 2002. (R)-Benzylsuccinyl-CoA dehydrogenase of Thauera aromatica, an enzyme of the anaerobic toluene catabolic pathway. Arch. Microbiol. 178517-524. [DOI] [PubMed] [Google Scholar]
- 220.Lochmeyer, C., J. Koch, and G. Fuchs. 1992. Anaerobic degradation of 2-aminobenzoic acid (anthranilic acid) via benzoyl-coenzyme A (CoA) and cyclohex-1-enecarboxyl-CoA in a denitrifying bacterium. J. Bacteriol. 1743621-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.London, J., and M. Knight. 1966. Concentrations of nicotinamide nucleotide coenzymes in microorganisms. J. Gen. Microbiol. 44241-254. [DOI] [PubMed] [Google Scholar]
- 222.Londry, K. L., J. M. Suflita, and R. S. Tanner. 1999. Cresol metabolism by the sulfate-reducing bacterium Desulfotomaculum sp. strain Groll. Can. J. Microbiol. 45458-463. [PubMed] [Google Scholar]
- 223.Lonetto, M. A., V. Rhodius, K. Lamberg, P. Kiley, S. Busby, and C. Gross. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J. Mol. Biol. 2841353-1365. [DOI] [PubMed] [Google Scholar]
- 224.López-Barragán, M. J., M. Carmona, M. T. Zamarro, B. Thiele, M. Boll, G. Fuchs, J. L. García, and E. Díaz. 2004. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 1865762-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.López-Barragán, M. J., E. Díaz, J. L. García, and M. Carmona. 2004. Genetic clues on the evolution of anaerobic catabolism of aromatic compounds. Microbiology 1502018-2021. [DOI] [PubMed] [Google Scholar]
- 226.Lovley, D. R. 2001. Bioremediation. Anaerobes to the rescue. Science 2931444-1446. [DOI] [PubMed] [Google Scholar]
- 227.Lovley, D. R. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 135-44. [DOI] [PubMed] [Google Scholar]
- 228.Lovley, D. R., J. D. Coates, J. C. Woodward, and E. Phillips. 1995. Benzene oxidation coupled to sulfate reduction. Appl. Environ. Microbiol. 61953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1996. Rapid anaerobic benzene oxidation with a variety of chelated Fe(III) forms. Appl. Environ. Microbiol. 62288-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lupa, B., D. Lyon, M. D. Gibbs, R. A. Reeves, and J. Wiegel. 2005. Distribution of genes encoding the microbial non-oxidative reversible hydroxyarylic acid decarboxylases/phenol carboxylases. Genomics 86342-351. [DOI] [PubMed] [Google Scholar]
- 231.Mancini, S. A., A. C. Ulrich, G. Lacrampe-Couloume, B. Sleep, E. A. Edwards, and B. S. Lollar. 2003. Carbon and hydrogen isotopic fractionation during anaerobic biodegradation of benzene. Appl. Environ. Microbiol. 69191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Mandic-Mulec, I., L. Doukhan, and I. Smith. 1995. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J. Bacteriol. 1774619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Marqués, S., I. Aranda-Olmedo, and J. L. Ramos. 2006. Controlling bacterial physiology for optimal expression of gene reporter constructs. Curr. Opin. Biotechnol. 1750-56. [DOI] [PubMed] [Google Scholar]
- 234.Martin, R. G., and J. L. Rosner. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 925456-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Matsunaga, T., Y. Okamura, Y. Fukuda, A. T. Wahyudi, Y. Murase, and H. Takeyama. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 12157-166. [DOI] [PubMed] [Google Scholar]
- 237.McInerney, M., L. Rohlin, H. Mouttaki, U. Kim, R. S. Krupp, L. Rios-Hernández, J. Sieber, C. G. Struchtemeyer, A. Bhattacharyya, J. W. Campbell, and R. P. Gunsalus. 2007. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc. Natl. Acad. Sci. USA 1047600-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.McLeod, M. P., and L. D. Eltis. 2008. Genomic insights into the aerobic pathways for degradation of organic pollutants, p. 1-23. In E. Díaz (ed.), Microbial biodegradation: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 239.Mechichi, T., E. Stackebrandt, N. Gad'on, and G. Fuchs. 2002. Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch. Microbiol. 17826-35. [DOI] [PubMed] [Google Scholar]
- 240.Meckenstock, R. U., M. Safinowski, and C. Griebler. 2004. Anaerobic degradation of polycyclic aromatic hydrocarbons: a review. FEMS Microbiol. Ecol. 4927-36. [DOI] [PubMed] [Google Scholar]
- 241.Medina-Bellver, J. I., P. Marín, A. Delgado, A. Rodríguez-Sánchez, E. Reyes, J. L. Ramos, and S. Marqués. 2005. Evidence for in situ crude oil biodegradation after the Prestige oil spill. Environ. Microbiol. 7773-779. [DOI] [PubMed] [Google Scholar]
- 242.Melville, S. B., and R. P. Gunsalus. 1990. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J. Biol. Chem. 26518733-18736. [PubMed] [Google Scholar]
- 243.Merritt, E. A., and M. E. Murphy. 1994. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1869-873. [DOI] [PubMed] [Google Scholar]
- 244.Messerschmidt, A., H. Niessen, D. Abt, O. Einsle, B. Schink, and P. M. Kroneck. 2004. Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. Proc. Natl. Acad. Sci. USA 10111571-11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Möbitz, H., and M. Boll. 2002. A Birch-like mechanism in enzymatic benzoyl-CoA reduction: a kinetic study of substrate analogues combined with an ab initio model. Biochemistry 411752-1758. [DOI] [PubMed] [Google Scholar]
- 246.Mohamed, M. E. 2000. Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J. Bacteriol. 182286-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mohamed, M. E., and G. Fuchs. 1993. Purification and characterization of phenylacetate-coenzyme A ligase from a denitrifying Pseudomonas sp., an enzyme involved in the anaerobic degradation of phenylacetate. Arch. Microbiol. 159554-562. [DOI] [PubMed] [Google Scholar]
- 248.Mohamed, M. E., B. Seyfried, A. Tschech, and G. Fuchs. 1993. Anaerobic oxidation of phenylacetate and 4-hydroxyphenylacetate to benzoyl-coenzyme A and CO2 in denitrifying Pseudomonas sp. Arch. Microbiol. 159563-573. [DOI] [PubMed] [Google Scholar]
- 249.Morales, G., J. F. Linares, A. Beloso, J. P. Albar, J. L. Martínez, and F. Rojo. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 1861337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Morales, G., A. Ugidos, and F. Rojo. 2006. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 81764-1774. [DOI] [PubMed] [Google Scholar]
- 251.Morasch, B., and R. U. Meckenstock. 2005. Anaerobic degradation of p-xylene by a sulfate-reducing enrichment culture. Curr. Microbiol. 51127-130. [DOI] [PubMed] [Google Scholar]
- 252.Morasch, B., H. H. Richnow, A. Vieth, B. Schink, and R. U. Meckenstock. 2004. Stable isotope fractionation caused by glycyl radical enzymes during bacterial degradation of aromatic compounds. Appl. Environ. Microbiol. 702935-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Morasch, B., B. Schink, C. C. Tebbe, and R. U. Meckenstock. 2004. Degradation of o-xylene and m-xylene by a novel sulphate-reducer belonging to the genus Desulfatomaculum. Arch. Microbiol. 181407-417. [DOI] [PubMed] [Google Scholar]
- 254.Moreno, R., and F. Rojo. 2008. The target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the BenR transcriptional regulator. J. Bacteriol. 1901539-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mosqueda, G., M. I. Ramos-González, and J. L. Ramos. 1999. Toluene metabolism by the solvent-tolerant Pseudomonas putida DOT-T1 strain, and its role in solvent impermeabilization. Gene 23269-76. [DOI] [PubMed] [Google Scholar]
- 256.Mouttaki, H., M. A. Nanny, and M. J. McInerney. 2007. Cyclohexane carboxylate and benzoate formation from crotonate in Syntrophus aciditrophicus. Appl. Environ. Microbiol. 73930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Müller, C., L. Petruschka, H. Cuypers, G. Burchhardt, and H. Herrmann. 1996. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J. Bacteriol. 1782030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Müller, J. A., A. S. Galushko, A. Kappler, and B. Schink. 2001. Initiation of anaerobic degradation of p-cresol by formation of 4-hydroxybenzylsuccinate in Desulfobacterium cetonicum. J. Bacteriol. 183752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Müller, J. A., and B. Schink. 2000. Initial steps in the fermentation of 3-hydroxybenzoate by Sporotomaculum hydroxybenzoicum. Arch. Microbiol. 173288-295. [DOI] [PubMed] [Google Scholar]
- 260.Musat, F., A. Galushko, J. Jacob, F. Widdel, M. Kube, R. Reinhardt, H. Wilkes, B. Schink, and R. Rabus. 22 September 2008. Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria. Environ. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1462-2920.2008.01756.x [DOI] [PubMed]
- 261.Narasingarao, P., and M. M. Häggblom. 2006. Sedimenticola selenatireducens, gen. nov., sp. nov., an anaerobic selenate-respiring bacterium isolated from estuarine sediment. Syst. Appl. Microbiol. 29382-388. [DOI] [PubMed] [Google Scholar]
- 262.Narmandakh, A., N. Gad'on, F. Drepper, B. Knapp, W. Haehnel, and G. Fuchs. 2006. Phosphorylation of phenol by phenylphosphate synthase: role of histidine phosphate in catalysis. J. Bacteriol. 1887815-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Oda, Y., W. G. Meijer, J. L. Gibson, J. C. Gottschal, and L. J. Forney. 2004. Analysis of diversity among 3-chlorobenzoate-degrading strains of Rhodopseudomonas palustris. Microb. Ecol. 4768-79. [DOI] [PubMed] [Google Scholar]
- 264.Ohtsubo, Y., H. Goto, Y. Nagata, T. Kudo, and M. Tsuda. 2006. Identification of a response regulator gene for catabolite control from a PCB-degrading beta-proteobacteria, Acidovorax sp. KKS102. Mol. Microbiol. 601563-1575. [DOI] [PubMed] [Google Scholar]
- 265.Olivera, E. R., B. Miñambres, B. García, C. Muñiz, M. A. Moreno, A. Ferrández, E. Díaz, J. L. García, and J. M. Luengo. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. USA 956419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ostermann, A., C. Gallus, and B. Schink. 1997. Decarboxylation of 2,3-dihydroxybenzoate to catechol supports growth of fermenting bacteria. Curr. Microbiol. 35270-273. [Google Scholar]
- 267.Pan, C., Y. Oda, P. K. Lankford, B. Zhang, N. F. Samatova, D. A. Pelletier, C. S. Harwood, and R. L. Hettich. 2008. Characterization of anaerobic catabolism of p-coumarate in Rhodopseudomonas palustris by integrating transcriptomics and quantitative proteomics. Mol. Cell. Proteomics 7938-948. [DOI] [PubMed] [Google Scholar]
- 268.Parales, R. E., and C. S. Harwood. 2002. Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr. Opin. Microbiol. 5266-273. [DOI] [PubMed] [Google Scholar]
- 269.Parales, R. E., K. S. Ju, J. B. Rollefson, and J. L. Ditty. 2008. Bioavailability, chemotaxis, and transport of organic pollutants, p. 145-187. In E. Díaz (ed.), Microbial biodegradation: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 270.Parales, R. E., and S. M. Resnick. 2006. Aromatic ring hydroxylating dioxygenases, p. 287-340. In J. L. Ramos and R. C. Levesque (ed.), Pseudomonas, vol. 4. Molecular biology of emerging issues. Springer, New York, NY. [Google Scholar]
- 271.Parke, D., M. A. García, and L. N. Ornston. 2001. Cloning and genetic characterization of dca genes required for β-oxidation of straight-chain dicarboxylic acids in Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 674817-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Parke, D., and L. N. Ornston. 2004. Toxicity caused by hydroxycinnamoyl-coenzyme A thioester accumulation in mutants of Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 702974-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Pelletier, D. A., and C. S. Harwood. 1998. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 1802330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Pelletier, D. A., and C. S. Harwood. 2000. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J. Bacteriol. 1822753-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Pelz, O., A. Chatzinotas, N. Andersen, S. M. Bernasconi, C. Hesse, W. R. Abraham, and J. Zeyer. 2001. Use of isotopic and molecular techniques to link toluene degradation in denitrifying aquifers microcosms to specific microbial populations. Arch. Microbiol. 175270-281. [DOI] [PubMed] [Google Scholar]
- 276.Peres, C., and C. S. Harwood. 2006. BadM is a transcriptional regulator and one of three regulators that control benzoyl coenzyme A reductase gene expression in Rhodopseudomonas palustris. J. Bacteriol. 1888662-8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Peters, F., D. Heintz, J. Johannes, A. V. Dorsselaer, and M. Boll. 2007. Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens. J. Bacteriol. 1894729-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Peters, F., M. Rother, and M. Boll. 2004. Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J. Bacteriol. 1862156-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Peters, F., Y. Shinoda, M. J. McInerney, and M. Boll. 2007. Cyclohexa-1,5-diene-1-carbonyl-coenzyme A (CoA) hydratases of Geobacter metallireducens and Syntrophus aciditrophicus: evidence for a common benzoyl-CoA degradation pathway in facultative and strict anaerobes. J. Bacteriol. 1891055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Petruschka, L., G. Burchhardt, C. Müller, C. Weihe, and H. Herrmann. 2001. The cyo operon of Pseudomonas putida is involved in carbon catabolite repression of phenol degradation. Mol. Genet. Genomics 266199-206. [DOI] [PubMed] [Google Scholar]
- 281.Pflüger, K., and V. de Lorenzo. 2008. Evidence of in vivo cross talk between the nitrogen-related and fructose-related branches of the carbohydrate phosphotransferase system of Pseudomonas putida. J. Bacteriol. 1903374-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Philipp, B., D. Kemmler, J. Hellstern, N. Gorny, A. Caballero, and B. Schink. 2002. Anaerobic degradation of protocatechuate (3,4-dihydroxybenzoate) by Thauera aromatica strain AR-1. FEMS Microbiol. Lett. 212139-143. [DOI] [PubMed] [Google Scholar]
- 283.Philipp, B., and B. Schink. 1998. Evidence of two oxidative reaction steps initiating anaerobic degradation of resorcinol (1,3-dihydroxybenzene) by the denitrifying bacterium Azoarcus anaerobius. J. Bacteriol. 1803644-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Pieper, D. H., V. A. P. Martins dos Santos, and P. N. Golyshin. 2004. Genomic and mechanistic insights into the biodegradation of organic pollutants. Curr. Opin. Biotechnol. 15215-224. [DOI] [PubMed] [Google Scholar]
- 285.Pop, S. M., N. Gupta, A. S. Raza, and S. W. Ragsdale. 2006. Transcriptional activation of dehalorespiration. Identification of redox-active cysteines regulating dimerization and DNA binding. J. Biol. Chem. 28126382-26390. [DOI] [PubMed] [Google Scholar]
- 286.Pop, S. M., R. J. Kolarik, and S. W. Ragsdale. 2004. Regulation of anaerobic dehalorespiration by the transcriptional activator CprK. J. Biol. Chem. 2649910-49918. [DOI] [PubMed] [Google Scholar]
- 287.Prieto, M., B. Galán, B. Torres, A. Ferrández, C. Fernández, B. Miñambres, J. L. García, and E. Díaz. 2004. Aromatic metabolism versus carbon availability: the regulatory network that controls catabolism of less-preferred carbon sources in Escherichia coli. FEMS Microbiol. Rev. 28503-518. [DOI] [PubMed] [Google Scholar]
- 288.Qiu, Y. L., Y. Sekiguchi, S. Hanada. H. Imachi, I. C. Tseng, S. S. Cheng, A. Ohashi, H. Harada, and Y. Kamagata. 2006. Pelotomaculum terephtalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch. Microbiol. 185172-182. [DOI] [PubMed] [Google Scholar]
- 289.Qiu, Y. L., Y. Sekiguchi, H. Imachi, Y. Kamagata, I. C. Tseng, S. S. Cheng, A. Ohashi, and H. Harada. 2003. Sporotomaculum syntrophicum sp. nov., a novel anaerobic, syntrophic benzoate-degrading bacterium isolated from methanogenic sludge treating wastewater from terephthalate manufacturing. Arch. Microbiol. 179242-249. [DOI] [PubMed] [Google Scholar]
- 290.Rabus, R. 2005. Functional genomics of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Appl. Microbiol. Biotechnol. 68580-587. [DOI] [PubMed] [Google Scholar]
- 291.Rabus, R., and J. Heider. 1998. Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria. Arch. Microbiol. 170377-384. [Google Scholar]
- 292.Rabus, R., M. Kube, A. Beck, F. Widdel, and R. Reinhardt. 2002. Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 178506-516. [DOI] [PubMed] [Google Scholar]
- 293.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 18327-36. [DOI] [PubMed] [Google Scholar]
- 294.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 16396-103. [DOI] [PubMed] [Google Scholar]
- 295.Rabus, R., H. Wilkes, A. Behrends, A. Armstroff, T. Fischer, A. J. Pierik, and F. Widdel. 2001. Anaerobic initial reaction of n-alkanes in a denitrifying bacterium: evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in n-hexane metabolism. Appl. Environ. Microbiol. 1831707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ramos, J. L., S. Marqués, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 297.Reams, A. B., and E. L. Neidle. 2004. Selection for gene clustering by tandem duplication. Annu. Rev. Microbiol. 58119-142. [DOI] [PubMed] [Google Scholar]
- 298.Reeve, C. D., M. A. Carver, and D. J. Hopper. 1989. The purification and characterization of 4-ethylphenol methylenehydroxylase, a flavocytochrome from Pseudomonas putida JD1. Biochem. J. 263431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Reichenbecher, W., B. Philipp, M. J. Suter, and B. Schink. 2000. Hydroxyhydroquinone reductase, the initial enzyme involved in the degradation of hydroxyhydroquinone (1,2,4-trihydroxybenzene) by Desulfovibrio inopinatus. Arch. Microbiol. 173206-212. [DOI] [PubMed] [Google Scholar]
- 300.Reusser, D. E., J. D. Istok, H. R. Beller, and J. A. Field. 2002. In situ transformation of deuterated toluene and xylene to benzylsuccinic acid analogues in BTEX-contaminated aquifers. Environ. Sci. Technol. 364127-4134. [DOI] [PubMed] [Google Scholar]
- 301.Rhee, S. K., and G. Fuchs. 1999. Phenylacetyl-CoA:acceptor oxidoreductase, a membrane-bound molybdenum-iron-sulfur enzyme involved in anaerobic metabolism of phenylalanine in the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 262507-515. [DOI] [PubMed] [Google Scholar]
- 302.Rhee, S. K., G. M. Lee, J. H. Yoon, Y. H. Park, H. S. Bae, and S. T. Lee. 1997. Anaerobic and aerobic degradation of pyridine by a newly isolated denitrifying bacterium. Appl. Environ. Microbiol. 632578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Rhee, S. K., X. Liu, L. Wu. S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformations in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 704303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Rhodius, V. A., and S. J. W. Busby. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1151-159. [DOI] [PubMed] [Google Scholar]
- 305.Rieger, P. G., H. M. Meier, M. Gerle, U. Vogt, T. Groth, and H. J. Knackmuss. 2002. Xenobiotics in the environment: present and future strategies to obviate the problem of biological persistence. J. Biotechnol. 94101-123. [DOI] [PubMed] [Google Scholar]
- 306.Rojo, F., and A. Dinamarca. 2004. Catabolite repression and physiological control, p. 365-387. In J. L. Ramos (ed.), Pseudomonas, vol. 2. Virulence and gene regulation. Kluwer Academic Press, New York, NY. [Google Scholar]
- 307.Romanowski, M. J., and S. K. Burley. 2002. Crystal structure of the Escherichia coli shikimate kinase I (AroK) that confers sensitivity to mecillinam. Proteins 47558-562. [DOI] [PubMed] [Google Scholar]
- 308.Rudolphi, A., A. Tschech, and G. Fuchs. 1991. Anaerobic degradation of cresols by denitrifying bacteria. Arch. Microbiol. 155238-248. [DOI] [PubMed] [Google Scholar]
- 309.Ruíz, R., M. I. Aranda-Olmedo, P. Domínguez-Cuevas, M. I. Ramos-González, and S. Marqués. 2004. Transcriptional regulation of the toluene catabolic parthways, p. 509-537. In J. L. Ramos (ed.), Pseudomonas, vol. 2. Virulence and gene regulation. Kluwer Academic Press, New York, NY. [Google Scholar]
- 310.Safinowski, M., C. Griebler, and R. U. Meckenstock. 2006. Anaerobic cometabolic transformation of polycyclic and heterocyclic aromatic hydrocarbons: evidence from laboratory and field studies. Environ. Sci. Technol. 404165-4173. [DOI] [PubMed] [Google Scholar]
- 311.Safinowski, M., and R. U. Meckenstock. 2006. Methylation is the initial reaction in anaerobic naphthalene degradation by a sulfate-reducing enrichment culture. Environ. Microbiol. 8347-352. [DOI] [PubMed] [Google Scholar]
- 312.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Samanta, S. K., and C. S. Harwood. 2005. Use of the Rhodopseudomonas palustris genome sequence to identify a single amino acid that contributes to the activity of coenzyme A ligase with chlorinated substrates. Mol. Microbiol. 551151-1159. [DOI] [PubMed] [Google Scholar]
- 314.Sánchez-Pérez, G., A. Mira, G. Nyiro, L. Pasić, and F. Rodríguez-Valera. 2008. Adapting to environmental changes using specialized paralogs. Trends Genet. 24154-158. [DOI] [PubMed] [Google Scholar]
- 315.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1543-44. [DOI] [PubMed] [Google Scholar]
- 316.Sauer, R. T., R. R. Yocum, R. F. Doolittle, M. Lewis, and C. O. Pabo. 1982. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature 298447-451. [DOI] [PubMed] [Google Scholar]
- 317.Schaefer, A. L., E. P. Greenberg, C. M. Oliver, Y. Oda, J. J. Huang, G. Bittan-Banin, C. M. Peres, S. Schmidt, K. Juhaszova, J. R. Sufrin, and C. S. Harwood. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454595-599. [DOI] [PubMed] [Google Scholar]
- 318.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47597-626. [DOI] [PubMed] [Google Scholar]
- 319.Schennen, U., K. Braun, and H. J. Knasckmuss. 1985. Anaerobic degradation of 2-fluorobenzoate by benzoate-degrading denitrifying bacteria. J. Bacteriol. 161321-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Schink, B., B. Phillips, and J. Müller. 2000. Anaerobic degradation of phenolic compounds. Naturwissenschaften 8712-23. [DOI] [PubMed] [Google Scholar]
- 321.Schleissner, C., E. R. Olivera, M. Fernández-Valverde, and J. M. Luengo. 1994. Aerobic catabolism of phenylacetic acid in Pseudomonas putida U: biochemical characterization of a specific phenylacetic acid transport system and formal demonstration that phenylacetyl-coenzyme A is a catabolic intermediate. J. Bacteriol. 1767667-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Schmeling, S., A. Narmandakh, O. Schmitt, N. Gad'on, K. Schühle, and G. Fuchs. 2004. Phenylphosphate synthase: a new phosphotransferase catalyzing the first step in aerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 1868044-8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Schneider, S., and G. Fuchs. 1998. Phenylacetyl-CoA:acceptor oxidoreductase, a new alpha-oxidizing enzyme that produces phenylglyoxylate. Assay, membrane localization, and differential production in Thauera aromatica. Arch. Microbiol. 169509-516. [DOI] [PubMed] [Google Scholar]
- 324.Schneider, S., M. E. S. Mohamed, and G. Fuchs. 1997. Anaerobic metabolism of L-phenylalanine via benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 168310-320. [DOI] [PubMed] [Google Scholar]
- 325.Schnell, S., F. Bak, and N. Pfennig. 1989. Anaerobic degradation of aniline and dihydroxybenzenes by newly isolated sulfate-reducing bacteria and description of Desulfobacterium anilini. Arch. Microbiol. 152556-563. [DOI] [PubMed] [Google Scholar]
- 326.Schnell, S., and B. Schink. 1991. Anaerobic aniline degradation via reductive deamination of 4-aminobenzoyl-CoA in Desulfobacterium anilini. Arch. Microbiol. 155183-190. [Google Scholar]
- 327.Schöcke, L., and B. Schink. 1998. Membrane-bound proton-translocating pyrophosphatase of Syntrophus gentianae, a syntrophically benzoate-degrading fermenting bacterium. Eur. J. Biochem. 15589-594. [DOI] [PubMed] [Google Scholar]
- 328.Schöcke, L., and B. Schink. 1999. Energetics and biochemistry of fermentative benzoate degradation by Syntrophus gentianae. Arch. Microbiol. 171331-337. [DOI] [PubMed] [Google Scholar]
- 329.Schühle, K., and G. Fuchs. 2004. Phenylphosphate carboxylase: a new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 1864556-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Schühle, K., J. Gescher, U. Feil, M. Paul, M. Jahn, H. Schägger, and G. Fuchs. 2003. Benzoate-coenzyme A ligase from Thauera aromatica: an enzyme acting in anaerobic and aerobic pathways. J. Bacteriol. 1854920-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Schühle, K., M. Jahn, S. Ghisla, and G. Fuchs. 2001. Two similar gene clusters coding for enzymes of a new type of aerobic 2-aminobenzoate (anthranilate) metabolism in the bacterium Azoarcus evansii. J. Bacteriol. 1835268-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Selmer, T., A. J. Pierik, and J. Heider. 2005. New glycyl radical enzymes catalysing key metabolic steps in anaerobic bacteria. Biol. Chem. 386981-988. [DOI] [PubMed] [Google Scholar]
- 333.Seyfried, B., A. Tschech, and G. Fuchs. 1991. Anaerobic degradation of phenylacetate and 4-hydroxyphenylacetate by denitrifying bacteria. Arch. Microbiol. 155249-255. [DOI] [PubMed] [Google Scholar]
- 334.Shingler, V. 2003. Integrated regulation in response to aromatic compounds: from signal sensing to attractive behaviour. Environ. Microbiol. 51226-1241. [DOI] [PubMed] [Google Scholar]
- 335.Shinoda, Y., J. Akagi, Y. Uchihashi, A. Hiraishi, H. Yukawa, H. Yurimoto, Y. Sakai, and N. Kato. 2005. Anaerobic degradation of aromatic compounds by Magnetospirillum strains: isolation and degradation genes. Biosci. Biotechnol. Biochem. 691483-1491. [DOI] [PubMed] [Google Scholar]
- 336.Shinoda, Y., Y. Sakai, H. Uenishi, Y. Uchihashi, A. Hiraishi, H. Yukawa, H. Yurimoto, and N. Kato. 2004. Aerobic and anaerobic toluene degradation by a new isolated denitrifying bacterium, Thauera sp. strain DNT-1. Appl. Environ. Microbiol. 701385-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Sikkema, J., J. A. de Bont, and B. Poolman. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59201-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 339.Song, B., L. J. Kerkhof, and M. M. Häggblom. 2002. Characterization of bacterial consortia capable of degrading 4-chlorobenzoate and 4-bromobenzoate under denitrifying conditions. FEMS Microbiol. Lett. 213183-188. [DOI] [PubMed] [Google Scholar]
- 340.Song, B., N. J. Palleroni, and M. M. Häggblom. 2000. Isolation and characterization of diverse halobenzoate-degrading denitrifying bacteria from soils and sediments. Appl. Environ. Microbiol. 663446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Song, B., N. J. Palleroni, L. J. Kerkhof, and M. M. Häggblom. 2001. Characterization of halobenzoate-degrading, denitrifying Azoarcus and Thauera isolates and description of Thauera chlorobenzoica sp. nov. Int. J. Syst. Evol. Microbiol. 51589-602. [DOI] [PubMed] [Google Scholar]
- 342.Song, B., and B. B. Ward. 2003. Nitrate reductase genes in halobenzoate degrading denitrifying bacteria. FEMS Microbiol. Ecol. 43349-357. [DOI] [PubMed] [Google Scholar]
- 343.Song, B., and B. B. Ward. 2005. Genetic diversity of benzoyl coenzyme A reductase genes detected in denitrifying isolates and estuarine sediment communities. Appl. Environ. Microbiol. 712036-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Song, F., Z. Zhuang, L. Finci, D. Dunaway-Mariano, R. Kniewel, J. A. Buglino, V. Solorzano, J. Wu, and C. D. Lima. 2006. Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J. Biol. Chem. 28111028-11038. [DOI] [PubMed] [Google Scholar]
- 345.Spormann, A. M., and F. Widdel. 2000. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 1185-105. [DOI] [PubMed] [Google Scholar]
- 346.Springer, N., W. Ludwig, B. Philipp, and B. Schink. 1998. Azoarcus anaerobius sp. nov., a resorcinol-degrading, strictly anaerobic, denitrifying bacterium. Int. J. Syst. Bacteriol. 48953-956. [DOI] [PubMed] [Google Scholar]
- 347.Stadtman, E. R., T. C. Stadtman, I. Pastan, and L. D. Smith. 1972. Clostridium barkeri sp. n. J. Bacteriol. 110758-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Suh, S. J., L. J. Runyen-Janecky, T. C. Maleniak, P. Hager, C. H. MacGregor, N. A. Zielinski-Mozny, P. V. Phibbs, Jr., and S. E. West. 2002. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology 148:1561-1569. [DOI] [PubMed] [Google Scholar]
- 349.Thiele, B., O. Rieder, N. Jehmlich, M. von Bergen, M. Müller, and M. Boll. 2008. Aromatizing cyclohexa-1,5-diene-1-carbonyl-coenzyme A oxidase: characterization and its role in anaerobic aromatic metabolism. J. Biol. Chem. 28320713-20721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Touati, D. 2000. Sensing and protecting against superoxide stress in Escherichia coli—how many ways are there to trigger soxRS response? Redox Res. 5287-293. [DOI] [PubMed] [Google Scholar]
- 351.Trautwein, K., S. Kühner, L. Wöhlbrand, T. Halder, K. Kuchta, A. Steinbüchel, and R. Rabus. 2008. Solvent stress response of the denitrifying bacterium “Aromatoleum aromaticum” strain EbN1. Appl. Environ. Microbiol. 742267-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Tropel, D., and J. R. van der Meer. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68474-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Tschech, A., and B. Schink. 1985. Fermentative degradation of resorcinol and resorcylic acids. Arch. Microbiol. 14352-59. [Google Scholar]
- 354.Unciuleac, M., E. Warkentin, C. C. Page, M. Boll, and U. Ermler. 2004. Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow. Structure 122249-2256. [DOI] [PubMed] [Google Scholar]
- 355.Unden, G., S. Becker, J. Bongaerts, G. Hollighaus, J. Schirawsky, and S. Six. 1995. O2-sensing and O2-dependent gene regulation in facultatively anaerobic bacteria. Arch. Microbiol. 16481-90. [PubMed] [Google Scholar]
- 356.Vaillancourt, F. H., J. T. Bolin, and L. D. Eltis. 2006. The ins and outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 41241-267. [DOI] [PubMed] [Google Scholar]
- 357.Vallenet, D., P. Nordmann, V. Barbe, L. Poirel, S. Mangenot, E. Bataille, C. Dossat, S. Gas, A. Kreimeyer, P. Lenoble, S. Oztas, J. Poulain, B. Segurens, C. Robert, C. Abergel, J. M. Claverie, D. Raoult, C. Médigue, J. Weissenbach, and S. Cruveiller. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS ONE 3e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.van der Meer, J. R. 2008. A genomic view on the evolution of catabolic pathways and bacterial adaptations to xenobiotics compounds, p. 219-267. In E. Díaz (ed.), Microbial biodegradation: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 359.van Hamme, J. D., A. Singh, and O. P. Ward. 2003. Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 67503-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Velasco, A., S. Alonso, J. L. García, J. Perera, and E. Díaz. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 1801063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Velázquez, F., V. de Lorenzo, and M. Valls. 2006. The m-xylene biodegradation capacity of Pseudomonas putida mt-2 is submitted to adaptation to abiotic stresses: evidence from expression profiling of xyl genes. Environ. Microbiol. 8:591-602. [DOI] [PubMed] [Google Scholar]
- 362.verBerkmoes, N. C., M. B. Shah, P. K. Lankford, D. A. Pelletier, M. B. Strader, D. L. Tabb, W. H. McDonald, J. W. Barton, G. B. Hurst, L. Hauser, B. H. Davison, J. T. Beatty, C. S. Harwood, F. R. Tabita, R. L. Hettich, and F. W. Larimer. 2006. Determination and comparison of the baseline proteomes of the versatile microbe Rhodopseudomonas palustris under its major metabolic states. J. Proteome Res. 5287-298. [DOI] [PubMed] [Google Scholar]
- 363.Verfürth, K., A. J. Pierik, C. Leutwein, S. Zorn, and J. Heider. 2004. Substrate specificities and electron paramagnetic resonance properties of benzylsuccinate synthases in anaerobic toluene and m-xylene metabolism. Arch. Microbiol. 181155-162. [DOI] [PubMed] [Google Scholar]
- 364.Villemur, R. 1995. Coenzyme A ligases involved in anaerobic biodegradation of aromatic compounds. Can. J. Microbiol. 41855-861. [DOI] [PubMed] [Google Scholar]
- 365.Vogel, T. M., and D. Grbìc-Galìc. 1986. Incorporation of oxygen from water into toluene and benzene during anaerobic fermentative transformation. Appl. Environ. Microbiol. 52200-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Vogels, G. D., and C. van der Drift. 1976. Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 40403-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Wang, Y., M. Rawlings, D. T. Gibson, D. Labbé, H. Bergeron, R. Brousseau, and P. C. Lau. 1995. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol. Gen. Genet. 246570-579. [DOI] [PubMed] [Google Scholar]
- 369.Washer, C. E., and E. A. Edwards. 2007. Identification and expression of benzylsuccinate synthase genes in a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 731367-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Weber, F. J., and J. A. de Bont. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286225-245. [DOI] [PubMed] [Google Scholar]
- 371.Weber, K. D., O. D. Vincent, and P. J. Kiley. 2005. Additional determinants within Escherichia coli FNR activating region 1 and RNA polymerase α subunit required for transcription activation. J. Bacteriol. 1871724-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Weickert, M. J., and S. Adhya. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 26715869-15874. [PubMed] [Google Scholar]
- 373.Whipp, M. J., and A. J. Pittard. 1995. A reassessment of the relationship between aroK- and aroL-encoded shikimate kinase enzymes of Escherichia coli. J. Bacteriol. 1771627-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12259-276. [DOI] [PubMed] [Google Scholar]
- 375.Winderl, C., S. Schaefer, and T. Lueders. 2007. Detection of anaerobic toluene and hydrocarbon degraders in contaminated aquifers using benzylsuccinate synthase (bssA) genes as a functional marker. Environ. Microbiol. 91035-1046. [DOI] [PubMed] [Google Scholar]
- 376.Wing, H. J., J. Green, J. R. Guest, and S. J. Busby. 2000. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoters. J. Biol. Chem. 27529061-29065. [DOI] [PubMed] [Google Scholar]
- 377.Wischgoll, S., D. Heintz, F. Peters, A. Erxleben, E. Sarnighausen, R. Reski, A. V. Dorssalaer, and M. Boll. 2005. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 581238-1252. [DOI] [PubMed] [Google Scholar]
- 378.Wöhlbrand, L., B. Kallerhoff, D. Lange, P. Hufnagel, J. Thiermann, R. Reinhardt, and R. Rabus. 2007. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics 72222-2239. [DOI] [PubMed] [Google Scholar]
- 379.Wöhlbrand, L., H. Wilkes, T. Halder, and R. Rabus. 2008. Anaerobic degradation of p-ethylphenol by “Aromatoleum aromaticum” strain EbN1: pathway, regulation, and involved proteins. J. Bacteriol. 1905699-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Xiang, L., and B. S. Moore. 2003. Characterization of benzoyl coenzyme A biosynthesis genes in the enterocin-producing bacterium “Streptomyces maritimus.” J. Bacteriol. 185399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Yoshida, T., and T. Nagasawa. 2000. Enzymatic functionalization of aromatic N-heterocycles: hydroxylation and carboxylation. J. Biosci. Bioeng. 89111-118. [DOI] [PubMed] [Google Scholar]
- 382.Young, D. M., D. Parke, and L. N. Ornston. 2005. Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu. Rev. Microbiol. 59519-551. [DOI] [PubMed] [Google Scholar]
- 383.Young, L. Y., and C. D. Phelps. 2005. Metabolic biomarkers for monitoring in situ anaerobic hydrocarbon degradation. Environ. Health Perspect. 11362-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Yu, L., M. Blaser, P. I. Andrei, A. J. Pierik, and T. Selmer. 2006. 4-Hydroxyphenylacetate decarboxylases: properties of a novel subclass of glycyl radical enzyme systems. Biochemistry 459584-9592. [DOI] [PubMed] [Google Scholar]
- 385.Zengler, K., J. Heider, R. Rossello-Mora, and F. Widdel. 1999. Phototrophic utilization of toluene under anoxic conditions by a new strain of Blastochloris sulfoviridis. Arch. Microbiol. 172204-212. [DOI] [PubMed] [Google Scholar]
- 386.Zhang, C., and G. N. Bennett. 2005. Biodegradation of xenobiotics by anaerobic bacteria. Appl. Microbiol. Biotechnol. 67600-618. [DOI] [PubMed] [Google Scholar]
- 387.Zhang, X., and L. Y. Young. 1997. Carboxylation as an initial reaction in the anaerobic metabolism of naphthalene and phenanthrene by sulfidogenic consortia. Appl. Environ. Microbiol. 634759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Ziegelhoffer, E. C., and P. J. Kiley. 1995. In vitro analysis of a constitutively active mutant form of the Escherichia coli global transcription factor FNR. J. Mol. Biol. 245351-361. [DOI] [PubMed] [Google Scholar]