Abstract
Binding of infected erythrocytes to brain venules is a central pathogenic event in the lethal malaria disease complication, cerebral malaria. The only parasite adhesion trait linked to cerebral sequestration is binding to intercellular adhesion molecule-1 (ICAM-1). In this report, we show that Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) binds ICAM-1. We have cloned and expressed PfEMP1 recombinant proteins from the A4tres parasite. Using heterologous expression in mammalian cells, the minimal ICAM-1 binding domain was a complex domain consisting of the second Duffy binding-like (DBL) domain and the C2 domain. Constructs that contained either domain alone did not bind ICAM-1. Based on phylogenetic criteria, there are five distinct PfEMP1 DBL types designated α, β, γ, δ, and ɛ. The DBL domain from the A4tres that binds ICAM-1 is DBLβ type. A PfEMP1 cloned from a distinct ICAM-1 binding variant, the A4 parasite, contains a DBLβ domain and a C2 domain in tandem arrangement similar to the A4tres PfEMP1. Anti-PfEMP1 antisera implicate the DBLβ domain from A4var PfEMP1 in ICAM-1 adhesion. The identification of a P. falciparum ICAM-1 binding domain may clarify mechanisms responsible for the pathogenesis of cerebral malaria and lead to interventions or vaccines that reduce malarial disease.
The parasitic protozoan, Plasmodium falciparum, is a major cause of morbidity and mortality in sub-Saharan Africa (1). A significant proportion of malarial deaths are the result of cerebral malaria. The molecular mechanisms responsible for cerebral malaria are complex and incompletely understood (2). However, a prominent pathological feature noted in postmortem analyses of individuals who died from cerebral malaria is the increased number of infected erythrocytes present in brain venules relative to those who died from other malaria complications (3, 4). These observations have spurred interest in defining molecules involved in infected erythrocyte cerebral sequestration.
Infected erythrocytes have a diverse and varied binding potential, and a number of host receptors have been shown to support parasite adherence, including CD36 (5) and intercellular adhesion molecule-1 (ICAM-1) (6). Studies correlating the binding attributes of field isolates to disease outcome and postmortem histopathological analyses of individuals who died from cerebral malaria have implicated ICAM-1 as a potential host receptor involved in cerebral malaria (7, 8). In addition, a dimorphism in human ICAM-1 alters the risk of cerebral malaria in Kenya (9). However, direct evidence for this role is lacking. We believe that it is important to identify parasite ICAM-1 binding protein(s) because they are potential vaccine candidates that may reduce morbidity and mortality from cerebral malaria.
A leading candidate is P. falciparum erythrocyte membrane protein 1 (PfEMP1), encoded by the large multigene family var (10–12). Members of the PfEMP1 protein family are parasite adhesion ligands that are exported to the surface of infected erythrocytes (10). Each parasite clone appears to express a single PfEMP1 (13) that can switch at the next cycle of erythrocytic invasion (14). Two distinct binding domains have been identified in PfEMP1: the Duffy binding-like (DBL) domain, which was originally described as an adhesive region in other Plasmodium proteins involved in erythrocyte invasion (15–19), and the cysteine-rich interdomain region (CIDR), which binds CD36 (20, 21). PfEMP1 DBL domains have a diverse binding potential that depends on their primary sequence. DBL1 domains from two distinct parasite variants that form rosettes have been found to bind complement receptor 1 (22) and heparan sulfate (23), respectively, on erythrocytes. In addition, DBL domains have been implicated both by anti-PfEMP1 antisera (24) and in direct binding experiments to adhere to chondroitin sulfate A (CSA) (25).
PfEMP1 molecules contain between two and seven DBL domains and one and two CIDR domains. By phylogenetic criteria, PfEMP1 DBL domains group as five distinct types: α, β, γ, δ, and ɛ (J.D.S., unpublished observations). Because the domain architecture of PfEMP1 is variable, we identify PfEMP1 DBL domains first by position in the protein and second by type. For example, the amino-terminal (first DBL) domain of all known PfEMP1 is DBLα type. Thus, it is referred to as DBL1α. The DBL1α domain is always followed by a CIDR1 domain, and this tandem arrangement of domains has been proposed to form a conserved head structure for PfEMP1 molecules (12). In contrast, beginning with the second DBL domain, the order and number of DBL domains is not conserved between PfEMP1.
Several independent lines of evidence suggest that PfEMP1 is a parasite ICAM-1 binding protein. First, ICAM-1 can affinity purify PfEMP1 proteins from detergent extracts of infected erythrocytes (26). Second, antigenically variant clonal lines are differentially susceptible to proteases in their binding to ICAM-1 (27). Third, in a well-characterized ICAM-1-binding parasite clonal line, expression of a particular PfEMP1 protein is linked to ICAM-1 adhesion (11, 21). We have cloned var genes from two antigenically distinct ICAM-1 binding parasites. In this paper, we report that a complex PfEMP1 domain of DBLβ and C2 is responsible for adhesion to ICAM-1 and that antisera raised to the DBLβ domain block this interaction.
Materials and Methods
Parasite Selection and Cultivation.
Parasites were grown in tissue culture flasks with daily changes of medium as described by Gardner et al. (27). The A4 clone is derived from P. falciparum line IT 4/25/5 by micromanipulation (28). A4tres was derived from A4 by selection on ICAM-1 after trypsinization at 1 mg/ml trypsin for 5 min at room temperature by using a methodology described by Gardner et al. (27).
Cell Culture of Cos-7.
Cos-7 cells, obtained from the American Type Culture Collection, were used for transient expression of PfEMP1 expression constructs. Cos-7 cells were cultured in DMEM (Biofluids, Rockville, MD) containing 10% heat-inactivated FCS (Life Technologies, Gaithersburg, MD).
Cloning of the A4tres PfEMP1.
The gene coding for the major var gene expressed by A4tres parasites was identified and sequenced by using standard techniques. Briefly, reverse transcription (RT)-PCR using universal primers to the DBL1 domain (S. K., unpublished observations) was carried out at the trophozoite stage, and the products were cloned and sequenced. Nine of sixteen clones were identical in sequence. The majority sequence was extended by carrying out PCR with unique primers in the sequenced region and a series of vectorette libraries (HindIII, DraI, BclI, and EcoRI) (29). At each walk, the unique PCR product was cloned into PTZ, and at least three independent colonies were sequenced, using the vector sequencing primers followed by primer walking. The 5′ end was completed by PCR, using a unique primer in DBL1 and a primer to the relatively conserved 5′UTR of var genes (5′-GATATATACATCCACCATGC).
Expression of DBL Domains in Escherichia coli for Production of Domain-Specific Antibody.
Regions representing the five DBL domains from A4var and the second DBL domain of A4tres were amplified from cDNA by using the following primers (forward and reverse): A4var1α (atgaatatcatact and atattccgtatgagaand), A4var2β (acgaaccaatattcc and attttttgcatgtag), A4var3δ (accaagttggatgtg and agaagaataaccttt), A4var4γ (ggtaaggttataaac and atattgatctttcca), A4var5β (tctattttagacagt and tgtcctatcctgtgt), and A4tres2β (cgtggtaatggcggtggacct and ccaccattagcggcagcagt). PCR products were cloned into the EcoRI site of pGEX4T-1 (Amersham Pharmacia). Glutathione S-transferase (GST) fusion proteins were expressed in E. coli BL21. Fusion proteins were purified on glutathione-agarose (Sigma). Antisera to the expressed protein were prepared in rabbits. IgG was purified from the antisera by using protein A Sepharose and then exhaustively absorbed with the same normal red cell population used to grow parasites prior to flow cytometric analysis of unfixed infected cells or use in cytoadherence reversal assays.
Construction of Recombinant Plasmids for Surface Expression in Mammalian Cell Lines.
A4tres and A4var expression constructs were amplified from genomic DNA by PCR and cloned into NotI and EcoRI restriction sites of the pSRα5 vector (21). Amino acid boundaries of constructs are shown in Fig. 1. For transfection, fresh monolayers of Cos-7 cells were seeded onto coverslips and grown overnight. The next day, 2.5 μg of plasmid DNA was introduced into cells by using Superfect reagent (Qiagen, Valencia, CA) according to methods supplied by the manufacturer. Cells were analyzed for immunofluorescence and binding to CD36 or ICAM-1 48–60 h after transfection.
Figure 1.
Schematic of A4tres and A4var PfEMP1 domain organization and expression constructs. The entire extracellular region of PfEMP1, including a putative transmembrane domain, is encoded in exon 1. The intron and exon 2 (partially sequenced for each PfEMP1) are labeled. Recombinant PfEMP1 proteins expressed in Cos-7 cells are shown beneath the protein schematics with domain boundaries listed. The A4var DBLβC2 was synthesized and tested for ICAM-1 binding in this paper. Previously, other A4var PfEMP1 recombinant proteins that covered the whole extracellular domain were tested for ICAM-1 binding (21).
Binding CD36- or ICAM-1-Coated Dynal Beads to Cos-7 Cell Lines Transfected with PfEMP1 Constructs and Reversal with Antibody.
Magnetic 450 beads coated with sheep antibodies directed against murine IgG were purchased from Dynal (Lake Success, NY). CD36 or ICAM-1 were coated on beads by using previously described methods (21). In brief, a soluble CD36 recombinant protein (sCD36 provided by Affymax Research Institute, Santa Clara, CA) was coated on beads by using M1 anti-Flag mAb (Sigma) at a concentration of 1.0 μg mAb/107 beads. To coat beads with ICAM-1, sheep anti-mouse antibody-coated dynal beads were reacted with mouse anti-human IgG Fc antisera (Jackson ImmunoResearch) at a concentration of 1.0 μg antibody/107 beads. Washed anti-Fc-coated beads were used to bind a chimeric ICAM-1-Fc protein at 1.0 μg/107 beads. The ICAM-1/Fc had previously been shown to support infected erythrocyte adhesion in vitro (30).
To assess binding of coated beads, transfected Cos-7 cells grown on coverslips were transferred to fresh wells of a 6-well plate (Falcon) and overlaid with 40 μl of RPMI 1640 binding medium containing 1.5–2.0 × 106 beads. RPMI 1640 washing and binding medium (Life Technologies) included 25 mM Hepes (Biofluids) and 0.5% BSA (ICN). Binding of CD36-coated and ICAM-1-coated beads was tested at pH 6.8 and 7.0, respectively. Binding and washing conditions have been described previously (21). For reversal of binding, ICAM-1/Fc-coated beads were incubated with cells grown on coverslip and then washed by inverting the coverslips in binding medium. Washed coverslips were transferred to a new 6-well plate and covered with RPMI-binding medium containing 10 μg/ml ICAM-1 mAb. After a 30-min incubation, coverslips were washed again by inversion and fixed with paraformaldehyde for immunofluorescence.
Reversal Assays Using Polyclonal Antibodies Derived from DBL Regions.
Antibodies to specific DBL regions were raised in rabbits and purified on protein A Sepharose, as described for ICAM-1-Fc production (30), omitting the pH 5 elution step. These were tested for their ability to label live cells (by FACS analysis) and to reverse adhesion of ITO4-A4 and A4Tres to ICAM-1 coated on plastic.
The appropriate parasite line was allowed to adhere to ICAM-1 at low haematocrit (1%) and low parasitaemia (1%). These conditions were used to minimize the effect of agglutination during antibody treatment of the dishes. After washing, to remove the unbound cells, the plates were incubated in 1.25 ml of binding medium (RPMI 1640/1% BSA) supplemented with 5 mg/ml polyclonal antibody, dialyzed against PBS, and preadsorbed on uninfected erythrocytes. After 60 min at 37°C, the dishes were washed again, fixed in glutaraldehyde, and stained with Giemsa. The number of bound cells was counted by microscopy.
Sequence Analysis.
Sequence analysis was performed on DBLβC2 domains from eight PfEMP1 deposited in the GenBank database. Sequences are identified by gi accession number, gene name, and amino acid boundaries of the DBLβC2 domains numbering from the initial methionine: 3540145, A4var (aa 810-1243); 886375, Dd2var1 (aa 926-1365); 2944095, varph17 (aa 901-1368); 886377, FCR2var3 (aa 816-1241); 886378, FCR3var2 (aa 825-1298); 1517814, ItGvar (aa 820-1285); and 1809295, FCR3varT11-1 (aa 820-1286). DBL domain boundaries were set by using previously described criteria (12). Multiple alignments were performed with the bcm search launcher available at http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html. Conserved amino acid features were identified by using the consensus program at http://www.bork.embl-heidelberg.de/Alignment/consensus.html, and protein secondary structure was predicted by using predator at the web site, http://www.embl-heidelberg.de/cgi/predator_serv.pl.
Results
Selection of A4tres Parasites on ICAM-1.
Previously, we reported on the A4var PfEMP1, which was cloned from the A4 parasite that binds ICAM-1. Although we were unable to identify an ICAM-1 binding domain in A4var by heterologous expression, we used molecular biological techniques and antisera to directly correlate expression of the A4var message and protein to parasite adhesion to ICAM-1 (11, 21).
Because of the high rate of clonal antigenic variation, cloned parasites rapidly become a mixture of different antigenic types (28). Within the A4 clonal line there are parasites that bind ICAM-1 in a trypsin-independent fashion (27). To enrich for this parasite variant(s), an A4 parasite culture was treated with trypsin and selected on ICAM-1. Two distinct ICAM-1 binding parasites variants, A4tres and A4trpi, were selected with this approach. A4tres used in the present study and A4trpi described in Gardner et al. (27) are phenotypically similar in that they bind ICAM-1 in a trypsin-resistant fashion, but are antigenically distinct and express PfEMP1 of different molecular weight. Both A4tres and A4trpi parasites show greatly enhanced binding to ICAM-1 after treatment of infected cells with trypsin, and in the case of A4tres, the expressed PfEMP-1 molecule is totally resistant to degradation by this enzyme in intact cells (data not shown).
Cloning of the A4tres Extracellular Region.
The gene coding for the predominant var gene expressed by A4tres parasites was identified by RT-PCR with universal primers to the DBL1 domain. To test whether the majority DBL1 sequence identified by RT-PCR was expressed in a pattern that correlated with A4tres parasite adhesion to ICAM-1, its expression was analyzed in A4tres parasites maintained in the presence or absence of ICAM-1 selection. For this comparison, the DBL1 sequence was hybridized to a Northern blot of RNA from A4tres parasites that had been very recently selected on ICAM-1 following trypsinization or to RNA from parasites several cycles removed from selection. In the former case, both a probe derived from the majority DBL1 sequence and a probe to the conserved exon 2 of var genes detected a single major product of 8 kb in A4tres parasites (data not shown). In contrast, two var gene products were detected by the exon 2 probe in the A4tres parasite line maintained without ICAM-1 selection, but only a single 8-kb band by the majority DBL1 sequence probe (data not shown). Thus, the majority DBL1 sequence belonged to a predominant PfEMP1 variant expressed by A4tres lines maintained under ICAM-1 selection.
The majority DBL1 sequence, designated the A4tres var, was then extended in the 3′ direction by vectorette cloning (see Materials and Methods). To confirm that the correct gene had been sequenced, A4tres RNA was hybridized with an exon 2 probe or multiple different probes to exon 1. In each case, a single major product of approximately 8 kb was detected (data not shown). In addition, RT-PCR was carried out with A4tres RNA by using sets of primers spanning the entire coding region. In all cases, bands of the predicted size were amplified (data not shown). The A4tres PfEMP1 contains three DBL domains and one CIDR domain. Based on phylogenetic criteria, the A4tres DBL domains 1, 2, and 3 are α, β, and γ, respectively (Fig. 1).
Both DBL2β and C2 Domains of A4tres PfEMP1 Are Required to Bind ICAM-1.
To identify the ICAM-1 binding domain in the A4tres PfEMP1, DBL and CIDR domain recombinant proteins encompassing the whole extracellular region were expressed at the surface of Cos-7 cells (Fig. 1). For binding assays, ICAM-1/Fc was immobilized onto Dynal 450 magnetic beads by using an anti-human IgG Fc antisera and incubated with transfected cells. After washing, specific binding was quantified by counting the number of transfected cells covered with ICAM-1-coated beads (Fig. 2).
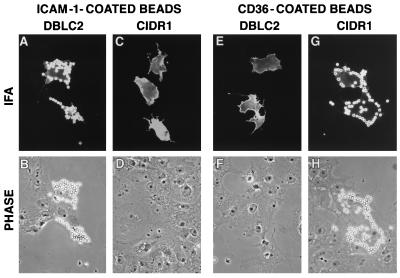
Figure 2.
A4tres PfEMP1 binding domains for ICAM-1 and CD36. ICAM-1-coated beads specifically bind DBLβC2-transfected cells (A and B) and do not bind CIDR1 transfectants (C and D). In contrast, CD36-coated beads bind CIDR1-transfected cells (G and H) but do not adhere to DBLβC2 transfectants (E and F). (Top; A, C, E, and G) Immunofluorescent images from the staining of transfected cells with an antibody against an epitope tag in the recombinant protein. (Bottom; B, D, F, and H) Phase images.
The minimal recombinant protein from the A4tres PfEMP1 that bound ICAM-1 was contained in the DBL2βC2 recombinant protein (Fig. 3). A larger DBL2βC2DBL3γ recombinant protein also bound ICAM-1 (data not shown). In contrast, a DBL2β recombinant protein that extended approximately one-third into the C2 domain did not bind ICAM-1 nor did the C2DBL3γC3 recombinant protein (Fig. 3). Thus, a complex domain consisting of PfEMP1 DBLβ and C2 domains was required for ICAM-1 binding in this mammalian expression system.
Figure 3.
Binding of PfEMP1 recombinant proteins to ICAM-1 and CD36. PfEMP1 recombinant proteins were expressed at the surface of Cos-7 cells and tested for binding to ICAM-1/Fc or CD36-coated dynal magnetic beads. The percentage of binding for each PfEMP1 recombinant proteins was calculated by counting 100 positive transfectants and scoring Cos-7 cells with 5 or more beads as positive. (A and B) Binding to ICAM-1/Fc. (C) Binding to CD36. Results in B and C are the mean of binding plus standard deviation of experiments performed in duplicate.
DBL2βC2 Recombinant Proteins Bind ICAM-1 with Similar Specificity to A4tres-Infected Erythrocytes.
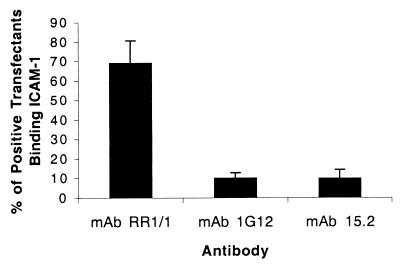
ICAM-1 contains five immunoglobulin domains. To test the specificity of A4tres DBLβC2 adhesion to ICAM-1, ICAM-1 antibodies were used that recognize the first domain of ICAM-1 and differ in their ability to block A4tres-infected erythrocyte adhesion to ICAM-1. ICAM-1 mAbs 1G12 and 15.2 blocked A4tres parasite adhesion (data not shown) and significantly reversed binding of ICAM-1-coated beads to DBLβC2-transfected Cos-7 approximately 90% (Fig. 4). In contrast, mAb RR1/1 antibody, which maps to the first ICAM-1 domain but does not block A4tres binding (data not shown), was only marginally more active in reversal assays than polyclonal normal mouse sera (Fig. 4). Thus, domain 1-specific ICAM-1 antibodies have identical activities on the A4tres DBLβC2 recombinant protein and parasites, strengthening the conclusion that the DBLβC2 is the A4tres ICAM-1 binding domain.
Figure 4.
Reversal of A4tres DBLβC2 recombinant protein binding to ICAM-1 with ICAM-1 antibodies. A4tres DBLβC2-transfected Cos-7 cells were incubated with ICAM-1/Fc coated beads, washed by inversion, and then incubated in the presence or absence of 10 μg/ml domain 1-specific ICAM-1 antibodies (1G12, 15.2, or RR1/1) or normal mouse IgG. Binding was quantified by inspecting 100 positive transfectants and scoring Cos-7 cells with 5 or more beads as positive. The percentage binding has been normalized to binding observed in the presence of normal mouse IgG. Results are the mean of six experiments plus standard error.
The DBLβ Domain from the A4var PfEMP1 Is Implicated in Parasite Adhesion to ICAM-1.
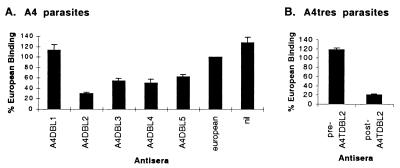
A tandem arrangement of a DBLβ and C2 domains is also present in the A4var PfEMP1 (Fig. 1 and 5). Previously, we had expressed recombinant proteins from the entire A4var extracellular region but had not tested the DBLβC2 domain combination (21). Despite the homology with the A4tres ICAM-1 binding domain, an A4var DBLβC2 recombinant protein did not bind ICAM-1, although it was well expressed at the cell surface (Fig. 3).
Figure 5.
Alignments of A4tres and A4var PfEMP1 DBLβC2 domains. The start of DBLβ and C2 domains is labeled above the alignment. Dashes (-) indicate gaps introduced to maintain alignment. A 100% amino acid consensus for eight DBLβC2 domains, including A4tres and A4var, is shown below the multiple alignment. Invariant residues are identified in bold with capital letters of the single amino acid code. Residues with a similar amino acid character are shown in bold: c (charge), h (hydrophobic), p (polar), s (small), u (tiny), b (big). Shown below the consensus is a secondary structural prediction of folding for the A4tres DBLβC2. H stands for α helical; β strands were not predicted. The structural prediction is highly representative of the seven other sequences in the alignment; helical boundaries were identical or varied by only a few amino acids (data not shown).
As a different approach, antisera were prepared against each of the five DBL domains of A4var and the DBLβ domain of A4tres. Purified antibody from these sera were tested for an ability to reverse binding of infected erythrocytes to ICAM-1. Reversal assays were necessary because all of the antisera agglutinated infected erythrocytes to different extents, leading to indirect inhibition in direct binding assays.
Complementing direct binding results, the A4tres DBLβ antisera reversed binding of A4tres-infected erythrocytes to ICAM-1 greater than 80% compared with preimmune sera (Fig. 6). By comparison, many of the A4var antisera possessed a low level of reversal activity, but the A4var DBLβ sera was the most active (Fig. 6). These results do not directly identify the A4var PfEMP1 binding domain. However, coupled with the observation that A4 and A4tres parasites have similar ICAM-1 binding specificity, they are consistent with the conclusion that the A4var DBL2β binds ICAM-1, perhaps in cooperation with the C2 domain.
Figure 6.
Reversal of A4 or A4tres parasite binding to ICAM-1 with PfEMP1 antisera. Infected erythrocytes were bound to ICAM-1 coated onto plastic, and the binding was reversed by using PfEMP1 antisera raised against different DBL domains. (A) A4-infected erythrocyte treated with A4var PfEMP1 antisera, nonimmune European sera, or no antibody (nil). (B) A4tres-infected erythrocytes treated with preimmune sera or anti-DBLβ sera (postimmune). (A and B) The binding is presented as a percentage of that observed using nonimmune European sera for reversal. Results in A are the mean of four experiments and in B the mean of two experiments plus standard error.
The CIDR Domain from the A4tres PfEMP1 Binds CD36.
A4tres-infected erythrocytes bind both ICAM-1 and CD36. These two proteins have been shown to synergize in mediating infected erythrocyte adhesion to cell lines that express both of these molecules (31). A CD36 binding domain has previously been shown to reside in the CIDR domain of PfEMP1 that bind CD36 (20, 21). We tested each of the DBL and CIDR domains in A4tres by using CD36-coated beads and showed that only the CIDR bound CD36 (Figs. 2 and 3). PfEMP1 proteins possess multiple DBL and one to two CIDR domains that may expand the adherence characteristics of a clone. The identification of ICAM-1 and CD36 binding domains in A4tres makes it the first PfEMP1 in which multiple adhesion domains have been defined at a molecular level.
Discussion
Of the known P. falciparum adhesion traits, ICAM-1 binding is to date most closely associated with cerebral sequestration (7, 8). We have been interested in defining the parasite protein(s) responsible for ICAM-1 adhesion. Previously, we demonstrated by both serological and molecular criteria that the A4var PfEMP1 molecule was expressed in a pattern that correlated with A4 parasite adhesion to ICAM-1 (11, 21). These results suggested that PfEMP1 molecules, which are known to participate in infected erythrocyte cytoadherent events, might be responsible for ICAM-1 binding.
In this report, we have cloned a distinct PfEMP1 molecule from another ICAM-1 binding line, A4tres, and have expressed recombinant proteins to identify the binding domain. We demonstrate that a complex A4tres PfEMP1 domain of DBLβ and C2 binds ICAM-1. Recombinant proteins that contained the DBLβ domain alone or the C2 domain without the DBLβ domain did not bind ICAM-1. ICAM-1 mAbs were used to demonstrate that the A4tres DBLβC2 recombinant protein bound ICAM-1 with similar specificity to A4tres parasites, strengthening the conclusion that this is the parasite ICAM-1 binding domain.
Like A4tres, the A4var PfEMP1 that we previously studied contains a DBLβC2 complex domain. However, neither an A4var DBLβC2 recombinant protein (Fig. 3) nor other DBL or CIDR recombinant proteins from A4var that collectively encompass the entire extracellular domain (21) bind ICAM-1 in any of the assays that we have tested. The explanation for the lack of binding is unclear. As a different approach to identify the ICAM-1 binding domain, antisera were prepared to each of the DBL domains in the A4var PfEMP1. Antibodies to the DBL2β domain were the most active in reversing A4 parasite adhesion to ICAM-1 (Fig. 6). These results do not prove that the A4varDBL2β domain binds ICAM-1 but are consistent with this interpretation. In addition, our findings raise the possibility that the ICAM-1 binding domain in other PfEMP1 molecules may reside in a DBLβ domain type.
The PfEMP1 C2 domain was previously uncharacterized. To explore the structural relationship of DBLβ and C2 domains in PfEMP1 molecules, a sequence analysis was performed on 18 PfEMP1 in the GenBank database. In 8 of 18 PfEMP1, a C2 domain followed a DBLβ domain and the C2 domain never followed other DBL types (data not shown). The tandem arrangement of DBLβC2 is universally present when a DBLβ domain is at the second DBL position (Fig. 1, and data not shown). The A4var DBL5β domain (Fig. 1) is one of three exceptions in which the DBLβ is not associated with C2. The exceptions are distinguished from other DBLβ family members in that they have diverged in sequence, do not follow a CIDR domain, and are not present at the second DBL position (J.D.S., unpublished observations).
Fig. 5 aligns the DBL2β and C2 domains of A4tres and A4var and illustrates the consensus features of the DBLβ and C2 domain types based on a multiple alignment of eight sequences. Despite considerable variation of sequence, DBLβ-type domains share many similar features, including blocks of strong amino acid conservation that are flanked by regions of more extensive antigenic polymorphism. Many of the regions of DBLβ conservation are predicted to possess α helical structure (Fig. 5). C2 domains are predicted to possess globular features and regions of α helical secondary structure and have several conserved residues, including four invariant cysteines.
The tight linkage of DBLβ and C2 domains in PfEMP1 proteins raises the possibility that this tandem domain combination may form an extended domain that folds differently than DBL domains alone. This might explain why both domains were required for A4tres ICAM-1 binding activity. However, our results do not exclude the possibility that the C2 domain acts as a spacer to present the DBLβ binding domain above the glycocalyx of the cell. Formal proof of the involvement of both domains in ICAM-1 adhesion will require further structural studies and site-directed mutagenesis to identify the critical amino acids involved in binding.
The identification of parasite ICAM-1 binding domains is a first step in developing interventions that may protect against malarial disease. It may be possible to immunologically target PfEMP1 binding variants that are responsible for disease if these PfEMP1 contain common structural or antigenic features that are a requirement for adhesion activity. As an example, pregnant women develop strain-independent anti-CSA adhesion antibodies that correlate with protection against placental malaria (32). Although the epitope(s) recognized by antiadhesion antibodies are not known, CSA-binding PfEMP1 (24, 25) may possess conserved features that are targets of strain-independent protection. In terms of the general population, it has been reported (33) that variants isolated from individuals with severe disease are more commonly recognized by serum from children in the community than those from cases of mild disease. It may be that these variants are more common in the parasite population, or that there exists some degree of crossreactivity between them, explaining the apparent rapid acquisition of immunity to severe malaria (34). Based on these considerations, the DBLβC2 domains of ICAM-1 binding PfEMP1 variants, because of their potential involvement in cerebral sequestration, are candidates for a vaccine against cerebral malaria.
Acknowledgments
We thank Sarah Lee and Zoe Christodolou for technical assistance, G. Subramanian for advice and support, and E. Whithorn and Affymax Research Institute for providing the pSRα5 vector, mAb179, and soluble CD36. This work was supported by The Wellcome Trust (U.K.) and the Human Frontiers Science Program (Strassburg, France).
Abbreviations
- ICAM-1
intercellular adhesion molecule 1
- PfEMP1
Plasmodium falciparum erythrocyte membrane protein 1
- DBL
Duffy binding-like
- CSA
chondroitin sulfate A
- CIDR
cysteine-rich interdomain region
- RT
reverse transcription
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF193424).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040545897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040545897
References
- 1.Warrell D A, Molyneux M E, Beales P F. Trans R Soc Trop Med Hyg. 1990;84:1–65. [Google Scholar]
- 2.Miller L H, Good M F, Milon G. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 3.MacPherson G G, Warrell M J, White N J, Looareesuwan S, Warrell D A. Am J Pathol. 1985;119:385–401. [PMC free article] [PubMed] [Google Scholar]
- 4.Pongponratn E, Riganti M, Punpoowong B, Aikawa M. Am J Trop Med Hyg. 1991;44:168–175. doi: 10.4269/ajtmh.1991.44.168. [DOI] [PubMed] [Google Scholar]
- 5.Barnwell J W, Asch A S, Nachman R L, Yamaya M, Aikawa M, Ingravallo P. J Clin Invest. 1989;84:765–772. doi: 10.1172/JCI114234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berendt A R, Simmons D L, Tansey J, Newbold C I, Marsh K. Nature (London) 1989;341:57–59. doi: 10.1038/341057a0. [DOI] [PubMed] [Google Scholar]
- 7.Newbold C, Warn P, Black G, Berendt A, Craig A, Snow B, Msobo M, Peshu N, Marsh K. Am J Trop Med Hyg. 1997;57:389–398. doi: 10.4269/ajtmh.1997.57.389. [DOI] [PubMed] [Google Scholar]
- 8.Turner G D, Morrison H, Jones M, Davis T M, Looareesuwan S, Buley I D, Gatter K C, Newbold C I, Pukritayakamee S, Nagachinta B, et al. Am J Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Reyes D, Craig A G, Kyes S A, Peshu N, Snow R W, Berendt A R, Marsh K, Newbold C I. Hum Mol Genet. 1997;6:1357–1360. doi: 10.1093/hmg/6.8.1357. [DOI] [PubMed] [Google Scholar]
- 10.Baruch D I, Pasloske B L, Singh H B, Bi X, Ma X C, Feldman M, Taraschi T F, Howard R J. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 11.Smith J D, Chitnis C E, Craig A G, Roberts D J, Hudson-Taylor D E, Peterson D S, Pinches R, Newbold C I, Miller L H. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su X Z, Heatwole V M, Wertheimer S P, Guinet F, Herrfeldt J A, Peterson D S, Ravetch J A, Wellems T E. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, Fernandez V, Sundstrom A, Schlichtherle M, Datta S, Hagblom P, Wahlgren M. Nature (London) 1998;394:392–395. doi: 10.1038/28660. [DOI] [PubMed] [Google Scholar]
- 14.Biggs B A, Gooze L, Wycherley K, Wollish W, Southwell B, Leech J H, Brown G V. Proc Natl Acad Sci USA. 1991;88:9171–9174. doi: 10.1073/pnas.88.20.9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams J H, Hudson D E, Torii M, Ward G E, Wellems T E, Aikawa M, Miller L H. Cell. 1990;63:141–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- 16.Adams J H, Sim B K, Dolan S A, Fang X, Kaslow D C, Miller L H. Proc Natl Acad Sci USA. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chitnis C E, Miller L H. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sim B K, Chitnis C E, Wasniowska K, Hadley T J, Miller L H. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 19.Peterson D S, Miller L H, Wellems T. Proc Natl Acad Sci USA. 1995;92:7100–7104. doi: 10.1073/pnas.92.15.7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baruch D I, Ma X C, Singh H B, Bi X, Pasloske B L, Howard R J. Blood. 1997;90:3766–3775. [PubMed] [Google Scholar]
- 21.Smith J D, Kyes S, Craig A G, Fagan T, Hudson-Taylor D, Miller L H, Baruch D I, Newbold C I. Mol Biochem Parasitol. 1998;97:133–148. doi: 10.1016/s0166-6851(98)00145-5. [DOI] [PubMed] [Google Scholar]
- 22.Rowe J A, Moulds J M, Newbold C I, Miller L H. Nature (London) 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- 23.Chen Q, Barragan A, Fernandez V, Sundstrom A, Schlichtherle M, Sahlen A, Carlson J, Datta S, Wahlgren M. J Exp Med. 1998;187:15–23. doi: 10.1084/jem.187.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeder J C, Cowman A F, Davern K M, Beeson J G, Thompson J K, Rogerson S J, Brown G V. Proc Natl Acad Sci USA. 1999;96:5198–5202. doi: 10.1073/pnas.96.9.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buffet P, Gamain B, Scheidig C, Baruch D, Smith J, Hernandez-Rivas R, Pouvelle B, Oishi S, Fujii N, Fusai T, et al. Proc Natl Acad Sci USA. 1999;96:12743–12748. doi: 10.1073/pnas.96.22.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baruch D I, Gormely J A, Ma C, Howard R J, Pasloske B L. Proc Natl Acad Sci USA. 1996;93:3497–3502. doi: 10.1073/pnas.93.8.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner J P, Pinches R A, Roberts D J, Newbold C I. Proc Natl Acad Sci USA. 1996;93:3503–3508. doi: 10.1073/pnas.93.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts D J, Craig A G, Berendt A R, Pinches R, Nash G, Marsh K, Newbold C I. Nature (London) 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold C, Hodgson I J. PCR Methods Appl. 1991;1:39–42. doi: 10.1101/gr.1.1.39. [DOI] [PubMed] [Google Scholar]
- 30.Craig A G, Pinches R, Khan S, Roberts D J, Turner G D, Newbold C I, Berendt A R. Infect Immun. 1997;65:4580–4585. doi: 10.1128/iai.65.11.4580-4585.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormick C J, Craig A, Roberts D, Newbold C I, Berendt A R. J Clin Invest. 1997;100:2521–2529. doi: 10.1172/JCI119794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fried M, Nosten F, Brockman A, Brabin B J, Duffy P E. Nature (London) 1998;395:851–852. doi: 10.1038/27570. [DOI] [PubMed] [Google Scholar]
- 33.Bull P C, Lowe B S, Kortok M, Marsh K. Infect Immun. 1999;67:733–739. doi: 10.1128/iai.67.2.733-739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta S, Snow R W, Donnelly C A, Marsh K, Newbold C. Nat Med. 1999;5:340–343. doi: 10.1038/6560. [DOI] [PubMed] [Google Scholar]