Abstract
Many aspects of plant biology depend on the ubiquitin proteasome system for degradation of regulatory proteins. Ubiquitin E3 ligases confer substrate specificity in this pathway, and SCF-type ligases comprise a major class of E3s. SCF ligases have four subunits: SKP1, CUL1, RBX1, and an F-box protein for substrate recognition. The Aux/IAAs are a well-characterized family of SCF substrates in plants. Here, we report characterization of a mutant isolated from a genetic screen in Arabidopsis thaliana designed to identify plants defective in degradation of an Aux/IAA fusion protein, Aux/IAA1-luciferase (IAA1-LUC). This mutant exhibited fourfold slower IAA1-LUC degradation compared with the progenitor line, and seedlings displayed altered auxin responses. Experiments identified the mutant as an allele of CUL1, named cul1-7. The cul1-7 mutation affects the C terminus of the protein, results in reduced cul1-7 levels, and interferes with RBX1 interaction. cul1-7 seedlings are defective in degradation of an endogenous SCF substrate, Repressor of ga1-3 (RGA), and have altered responses to gibberellins. cul1-7 seedlings exhibit slower degradation of the light-labile red/far-red photoreceptor phytochrome A and are photomorphogenic in the dark. This mutation represents the first reported allele of CUL1 to directly affect subunit interactions at the CUL1 C terminus.
THE ubiquitin pathway catalyzes the post-translational modification of substrate proteins with the small protein ubiquitin, and it includes the enzymes catabolizing ubiquitylated proteins and additional proteins regulating the process. Conjugation of ubiquitin to proteins in the cytosol and nucleus of eukaryotes has diverse biological consequences and is involved in almost every aspect of eukaryotic biology (Weissman 2001; Dreher and Callis 2007; Schwechheimer et al. 2009). Ubiquitination is catalyzed by a series of enzymes consisting of an E1 (ubiquitin activating enzyme), an E2 (ubiquitin conjugating enzyme), and an E3 (ubiquitin ligase). Ubiquitin is ultimately transferred to a lysyl residue within a substrate protein or to one or more lysyl residues of a previously attached ubiquitin, forming a polyubiquitin chain. One fate of polyubiquitylated proteins is hydrolysis by the 26S proteasome, a megadalton barrel protease complex (reviewed in Smalle and Vierstra 2004). The specificity and regulation of ubiquitylation remains a major focus of research.
Ubiquitin ligases constitute the largest enzyme class in the ubiquitin pathway and in most cases confer substrate specificity. They are responsible for interacting with both the substrate and the E2 carrying activated ubiquitin. One major class of E3s is the multisubunit SCF type. Each member of the SCF family contains a scaffolding CULLIN1 (CUL1) subunit that binds substrate-recognizing subunits at its N terminus and the RING-finger protein RBX1 (Ring-box1) at its C terminus (Cardozo and Pagano 2004; Bosu and Kipreos 2008; Hotton and Callis 2008). The RING domain of RBX1 recruits the ubiquitin-charged E2 and brings it into proximity of the substrate. The substrate recognition subunits of the SCF are the adaptor SKP1-like protein (ASK1 in plants) and a substrate binding F-box protein (reviewed in Cardozo and Pagano 2004). The F-box protein is the variable subunit of the complex and interacts directly with the substrate. Over 700 different F-box proteins have been postulated in Arabidopsis and rice (Gagne et al. 2002; Jain et al. 2007). Substrate specificity appears to be determined in large part by the nature of the F-box protein present. Thus, the assembly and regulation of SCF activity is of considerable interest.
SCF activity in animals, fission yeast, and plants is maximally active after the CUL1 subunit is covalently modified by a ubiquitin-like protein called Neural-Precursor-Cell-Expressed, Developmentally Downregulated 8 (NEDD8) in metazoa and fission yeast and Related to Ubiquitin (RUB) in plants and budding yeast (Lammer et al. 1998; Del Pozo and Estelle 1999; Ohh et al. 2002; Dharmasiri et al. 2003; Bostick et al. 2004). This attachment follows an enzymatic cascade similar to that of ubiquitin, beginning with RUB1/2 activation by a heterodimeric E1, then transfer to the RUB1/2 E2, and finally transfer to CUL1 using RBX1 as part of the RUB1/2 E3 (Del Pozo et al. 1998, 2002; Kamura et al. 1999; Gray et al. 2002; Bostick et al. 2004; Larsen and Cancel 2004; Woodward et al. 2007). One of the original auxin resistant mutants, axr1, is an allele of the N-terminal half of the RUB1/2 E1 heterodimer (Leyser et al. 1993; Del Pozo et al. 1998).
Just as RUB conjugation to CUL1 is required for proper function, so is its removal by the protease activity of the COP9 signalosome (reviewed in Cope and Deshaies 2003; Wei and Deng 2003). This cycle of conjugation–deconjugation is, in part, regulated by CAND1 (Cullin-Associated-Neddylation-Disassociated 1) (Liu et al. 2002; Zheng et al. 2002a; Oshikawa et al. 2003; Goldenberg et al. 2004; Min et al. 2005; Bornstein et al. 2006; Lo and Hannink 2006; Chew and Hagen 2007). Many outstanding questions remain regarding regulation of SCF ligases. The identification of Defective-In-Cullin-Neddylation 1 (DCN1) as a scaffold-type E3 for NEDD8, as opposed to RBX1 alone, has added a new dimension to this complexity (Kurz et al. 2005, 2008; Biswas et al. 2007; Yang et al. 2007). Work is also needed to sort out how RBX1 discriminates between the RUB E2 and the UBQ E2.
Much evidence has implicated SCF ligases in plant hormonal signaling mechanisms (Thomann et al. 2005). The first discoveries linked auxin signaling to a specific SCF, SCFTIR1 (Gray et al. 1999) and subsequently to a small family of related F-box proteins, the AFBs (Dharmasiri et al. 2005b). These proteins are also auxin receptors, binding both auxin and Auxin/Indole-3-Acetic Acid (Aux/IAA) proteins (Dharmasiri et al. 2005a,b; Kepinski and Leyser 2005). Auxin binds at the base of a pocket in a region of the TIR1 leucine-rich-repeat domain, facilitating binding of the core Aux/IAA sequences in the same pocket (Tan et al. 2007). Aux/IAA proteins are the substrates of SCFTIR1 and they function in auxin signaling as short-lived transcriptional regulators (reviewed in Quint and Gray 2006). High levels of exogenous auxin stimulate rapid ubiquitin-mediated degradation of some Aux/IAA proteins (Gray et al. 2001; Tiwari et al. 2001; Zenser et al. 2001). Auxin-responsive degradation of the Aux/IAAs requires a core sequence of GWPPL/V/I within a region conserved in many Aux/IAAs called domain II, identified by using Aux/IAA-luciferase (IAA-LUC) fusion proteins, and degradation is slowed when these residues are substituted (Ramos et al. 2001).
In addition, forward genetic screens searching for Arabidopsis thaliana plants with altered responses to auxin as well as for developmental responses not necessarily directly linked to auxin-identified semidominant mutations with substitutions in the Aux/IAA core sequence (above), suggest defects in degradation. In many cases, slowed degradation of the mutant protein was experimentally verified (Timpte et al. 1994; Leyser et al. 1996; Rouse et al. 1998; Nagpal et al. 2000; Tian et al. 2002; Tatematsu et al. 2004; Yang et al. 2004). Other auxin response mutants include plants with defective subunits of the SCF ubiquitin E3 ligase or mutations that cause misregulation of SCF activity/assembly (Ruegger et al. 1998; Gray et al. 2003; Hellmann et al. 2003; Cheng et al. 2004; Chuang et al. 2004; Feng et al. 2004; Dharmasiri et al. 2005b; Quint et al. 2005; Alonso-Peral et al. 2006; Walsh et al. 2006; Moon et al. 2007; Woodward et al. 2007).
SCF ubiquitin ligases are also required for other plant hormonal signaling pathways, including jasmonic acid (JA), ethylene, and gibberellic acid (GA) (reviewed in Thomann et al. 2005). SCFCOI1 functions to target for degradation a group of transcriptional repressors called Jasmonate ZIM-Domain (JAZ) proteins, and remarkably has been identified as a receptor for jasmonic acid and its derivatives (Chini et al. 2007; Thines et al. 2007; Katsir et al. 2008). Ethylene signaling requires stabilization of EIN3, which is constitutively targeted for degradation by SCFEBF1/2 (Gagne et al. 2004) and may require phosphorylation prior to ubiquitylation (Yoo et al. 2008). DELLA proteins, which are negative regulators of the GA signaling pathway, are rapidly degraded in response to the hormone (Fleet and Sun 2005). In Arabidopsis, the DELLA, Repressor of ga1-3 (RGA) degradation requires the SCFSLY1 E3 ubiquitin ligase (McGinnis et al. 2003; Dill et al. 2004).
SCF ligases have a defined role in cell cycle control in various eukaryotes (reviewed in Petroski and Deshaies 2005). Recent progress in this area has also demonstrated the importance of the F-box protein SKP2A in regulating cell division in Arabidopsis by contributing to KRP1 degradation, and SKP2A degradation itself is regulated by auxin (Jurado et al. 2008; Ren et al. 2008). Aside from their roles in hormonal signaling and cell cycle control, SCF ubiquitin ligases are important for flower development, circadian rhythms, phosphate starvation, and myriad other processes, as suggested by the abundance of F-box proteins in Arabidopsis (Lechner et al. 2006).
Homozygous null mutations in CUL1 are embryonic lethal and exhibit various defects when heterozygous due to haplo-insufficiency (Shen et al. 2002; Hellmann et al. 2003). Missense CUL1 mutants have also been characterized. Two semidominant alleles of CUL1, namely axr6-1 and axr6-2 cause substitutions at the same N-terminal residue. Corresponding mutant proteins, consequently, are affected in their interaction with ASK1 (Hellmann et al. 2003). axr6-3, a recessive allele, has a mutation at the N terminus of the protein as well and affects ASK1 binding, but is a temperature-sensitive allele (Quint et al. 2005). The substitution in a fourth missense mutation, which is also recessive, cul1-6, is only four amino acids away from the substitutions in axr6-1 and axr6-2 and affects CAND1 binding, but not ASK1 interaction (Moon et al. 2007).
Here, we report the identification and characterization of a novel, missense, recessive allele of CUL1 (called cul1-7) identified from a screen designed to isolate mutants defective in the degradation of an IAA1-LUC fusion protein. cul1-7 is the only recessive allele to affect function at the C terminus of CUL1. Because of its unique biochemical phenotype and its strong photomorphogenic phenotype at 28°, cul1-7 will be a useful tool in understanding regulation of SCF function and in sorting out the role of SCF-ligases in photomorphogenesis.
MATERIALS AND METHODS
Plant materials and growth conditions:
All plants were A. thaliana, ecotype Columbia (Col-0). Phenotypic studies of homozygous cul1-7 used in this study were F3 plants from the third backcross to CUL1 with the exception of cul1-7 axr1-30, which was created by crossing pollen from a cul1-7 M3 plant onto an axr1-30 pistil. Transgenic complementation lines were created using the floral dip method (Clough and Bent 1998), and all lines were either homozygous T3 or T4 generation. Growth media (GM) consisted of 4.3 g/liter Murashige and Skoog (MS) basal salts (Sigma-Aldrich), 1% sucrose, 2.5 mm MES, 1× B vitamins (0.5 μg/ml nicotinic acid, 1.0 μg/ml thiamine·HCl, 0.5 μg/ml pyroxidine·Cl, 0.1 μg/ml myo-inositol), 8 g/liter BactoAgar (Becton, Dickinson and Company), pH 5.7 unless otherwise noted. Plants grown on soil were first started on GM then transferred to soil and grown as described. All seeds were surface sterilized prior to use in any experiment.
Genetic screen:
T4 seeds of a plant line harboring a UBQ10:IAA1-LUC expression cassette (referred to here as CUL1), (Worley et al. 2000) were mutagenized with 0.2% ethyl methanesulfonate (EMS) for 24 hr and prepared for planting on soil using standard protocols. Seeds were collected from 7475 M1 plants and initially screened for high LUC activity by plating 200 seeds on a GM plate, growing for 1 week, adding 1 ml 1mm luciferin and incubating in the dark for 1 hr. Plates were then visualized using a CCD camera (Princeton Instruments model NTE/CCD-TKD D12990) using WinView/32 version 2.4 software (Roper Scientific, Trenton, NJ). Seedlings with higher LUC activity were transferred to soil, and M3 seeds collected for IAA1-LUC degradation analysis. The IAA1-LUC coding region was sequenced from higher light emitting plants and found to be identical to that introduced into the progenitor line.
Genetic mapping:
To create a mapping population, the mutant was crossed to Ler, and a total of 99 F2 individuals displaying the mutant phenotype were obtained. Bulked segregant analysis linked the mutation to ciw5 (http://www.arabidopsis.org), an SSLP marker on chromosome IV. Genotyping F2 mapping individuals for ciw5, GA1.1, and nga8, placed the mutation between ciw5 and GA1.1 (http://www.arabidopsis.org). CUL1 was a good candidate gene within this interval; therefore, 825 bp upstream of the translational start to 224 bp downstream of the translational stop in genomic DNA prepared from the mutant was sequenced.
Auxin and GA sensitivity tests:
For auxin sensitivity experiments, seedlings were grown on GM for 5 days, transferred to vertical plates containing the indicated concentration of 2,4-D (Sigma) or the corresponding amount of 0.1 m KOH solvent. The length of the primary root was marked. Plants were grown vertically for 7 additional days, and primary root length was determined. The percentage of inhibition for a given concentration is the average of that calculated from three independent experiments for 5–12 roots for each plate. Percentage of root growth inhibition was calculated by the following formula: percent inhibition = (1 − growth on 2,4-D/growth on KOH) × 100. For GA sensitivity experiments, seeds were cold stratified overnight in water, and plated directly on GM plates with 10 μm paclobutrazol (PAC) (Riedel-de Haën) containing 1, 10, or 100 μm GA3 (Sigma). Plates were wrapped in foil, and grown for 5 days at 20° or 28°. Hypocotyl length was measured from the point of cotyledon attachment to the point of root hair emergence. National Institutes of Health Image J 1.36b was used to measure roots and hypocotyls.
Protein degradation experiments:
For single-seedling degradation assays, seeds were plated in individual wells of white polystyrene flat-bottom 96-well plates (Whatman, Clifton, NJ) that contained 100 μl of GM. The plates were sealed to prevent desiccation with microplate adhesive film (USA Scientific), cold stratified overnight, and placed at 22° under constant light for 6 days. On day 7, 50 μl of 1 mm D-luciferin potassium salt (Gold Bio Technology) dissolved in liquid GM was added to the seedlings in the plates, and the plates were incubated in the dark for 1 hr. Cycloheximide (Sigma-Aldrich) dissolved in GM was then added to a final concentration of 200 μg/ml. Light emission from each seedling was then monitored every 15 min over a 60-min time course using a MicroLumat LB 96 P luminometer (EG&G Berthold Instruments). For data analysis, the relative light unit (RLU) initial readings from seedlings of the same genotype were averaged, and then the readings from the time course, including the initial for each individual, were normalized (divided) by that average. To linearize the data, the natural log of the normalized RLU for each seedling at each time point was calculated. The average ln(normalized RLU) for each time point with its corresponding standard deviation was plotted on the y-axis against time on the x-axis. To calculate the half-life, ln(0.5) was divided by the slope of degradation line. Pooled-seedling IAA1-LUC degradation experiments were performed as described in Dreher et al. (2006).
For RGA degradation experiments, CUL1 and cul1-7 seeds imbibed in water in the dark at 4° for 3 days. Seeds were then plated on 1× MS, 1.2% agar in 100 × 15 mm Petri dishes, and incubated under constant light at 22°. On day 5, 3 ml of a 100 μm PAC solution in GM was added to the plates and the plates were incubated for 4 more days. The remaining PAC solution was then removed, and an 8-ml GM solution, containing 200 μg/ml cycloheximide, 100 μm PAC ± 20 nM GA4, was added to the plates. Samples collected at each time point were immediately frozen in liquid nitrogen. Protein extraction and immunoblot analysis using affinity-purified RGA polyclonal antibodies were performed as described previously (Silverstone et al. 2001). Half-life estimates were based on densitometry and comparison to the values for dilutions of the “0” time point.
For phytochrome A degradation experiments, seeds (∼2 mg in 1 ml liquid GM) were aliquoted into 60 × 15 mm plates. Plates were sealed, wrapped in foil, and cold stratified at 4° for 2 days. Seedlings were dark grown for 7 days at 22°. On the day of the experiment, plates were opened under dim green light; “dark” samples were immediately collected, and frozen in liquid nitrogen. The solution was removed from the remaining plates and replaced with 1 ml of 200 μg/ml cycloheximide dissolved in GM and then placed under 20 μmol m−2 s−1 red light for the indicated time, then flash frozen in liquid nitrogen. Protein extracts were prepared as described below.
Genotyping:
Because the mutation in cul1-7 does not create a CAPS marker, we used dCAPS (Michaels and Amasino 1998; Neff et al. 1998) to follow the allele. Using dCAPS Finder 2.0 (http://helix.wustl.edu/dcaps/dcaps.html), we designed a forward primer (5′-GATTGACTTGACCGTCACTGTTGATA-3′) that has two mismatches, which in combination with the mutation in cul1-7, creates an EcoRV site in the PCR product; the site is missing in the PCR product produced from CUL1 with the same primer. With the reverse primer (5′-CTGTGTTTCGTTTTCGTTTCA-3′) the full-length product is 220 bp and becomes 194 bp after EcoRV digestion. Fragments were resolved on 4% agarose gels. To genotype the CUL1-FLAG cul1-7 complementation lines, a 1.3-kb genomic fragment specific to the endogenous locus using the forward primer 5′-GTGACAGGTGACGGATTTGA-3′ and the reverse primer 5′- CATTAAGGCCATTTCTCCATCT-3′ were used as a template for the dCAPS PCR.
A similar approach using dCAPS was used for genotyping axr6-3. The forward primer (5′-GTTCTTCTGTCAGGTTGATCTA-3′) was designed with two mismatches, resulting in an XbaI site in the fragment produced from CUL1, which is not present in axr6-3. The full-length fragment produced with the reverse primer (5′-CACGAGTCATGCCTTCAACA-3′) is 229 bp, 210 bp after digestion with XbaI.
Western blots and antibodies:
All plant extracts for Western blot analysis were prepared by grinding frozen samples in 150–200 μl extraction buffer [50 mm Tris-HCl pH 8.0, 150 mm NaCl, 20 mm EDTA pH 8.0, 1% glycerol, 0.15% NP-40, 1 mm PMSF, 1 tablet/10 ml Complete Mini protease inhibitor pill (Roche)] unless noted otherwise. Samples were cleared by centrifugation at 16,000 × g for 20 min. Protein concentration was determined by Bradford assay (Bio-Rad). Protein was transferred to Immobilon-P membrane (Millipore) and prepared for probing with antibodies using standard techniques. Protein on membrane was visualized using an Amersham ECL Plus detection kit (GE Healthcare). All blocking and incubation of antibodies with membrane was done in Blotto (1× TBS, 0.1% Tween-20, 5% powdered nonfat dry milk). Rabbit primary antibodies against PhyA, CUL1, and RGA have been previously described (Elich and Lagarias 1987; Gray et al. 1999; Silverstone et al. 2001). Other antibodies were goat anti-rabbit IgG-horseradish peroxidase secondary antibodies (Jackson ImmunoResearch), mouse anti-FLAG primary antibodies (Sigma), rabbit anti-GST polyclonal antibodies (Santa Cruz Biotechnology, catalog no. sc-459), goat anti-mouse IgG-horseradish peroxidase secondary antibodies (Kirkegaard and Perry Laboratories), and a rabbit anti-ROC1 polyclonal antibody (AHO0402, Invitrogen) previously demonstrated to be immunoreactive with AtRBX1 (Xu et al. 2002).
Protein expression and immunoprecipitations:
A cDNA of AtRBX1 was cloned into pDest15 using Gateway Technology (Invitrogen) for expression of GST-RBX1 in Escherichia coli as described in Stone et al. (2005). This construct was transformed into BL21 arabinose-inducible E. coli (Invitrogen). Cultures were grown to an OD600 of 0.6 and induced with 0.2% arabinose at 37° for 3 hr. Cells were harvested by centrifugation at 16,000 × g for 10 min and lysed using standard techniques in GST lysis buffer (25 mm Tris-HCl pH 7.5, 500 mm NaCl, 0.1% Triton X-100). Lysate was cleared by centrifugation at 16,000 × g for 20 min; GST-RBX1 was affinity purified from the supernatant using 1 ml of glutathione-sepharose beads (GE Healthcare). Beads were washed three times for 20 min in GST wash buffer (25 mm Tris-HCl pH 7.5, 300 mm NaCl, 0.01% Triton X-100), then mixed with 1 ml GST elution buffer (25 mm Tris-HCl pH 7.5, 150 mm NaCl, 0.01% Triton X-100, 50 mm reduced glutathione) overnight to elute GST-RBX1. Eluted GST-RBX1 was buffer exchanged at a 1:3840 dilution into 25 mm Tris-HCl pH 7.5, 25 mm NaCl using a 10,000 NMWL Amicon Ultra-4 centrifugal filter device (Millipore). GST-RBX1 was then concentrated to ∼250 ng/μl using an Ultrafree-0.5 centrifugal filter device (Millipore). GST was expressed from the pGEX2T construct (GE Healthcare) in BL21 pLysS E. coli by inducing 1 liter of cells (OD600) with 1.0 mm IPTG for 2 hr at 37°. GST was purified and concentrated as described above for GST-RBX1.
The CUL1 coding sequence in the pUNI51 (Yamada et al. 2003) backbone (clone U09998, The Arabidopsis Resource Center) was changed to the cul1-7 sequence by site-directed mutagenesis using the forward primer 5′-GACTTGACCGTCACTGTTCTTATCACTGGTTTCTGGCC-3′ and its reverse complement as the reverse primer. The CUL1 and cul1-7 coding sequences were then moved into pDONR201 (Invitrogen) using Gateway Technology, sequence verified, and subsequently recombined into pEXP1-DEST (Invitrogen). The resulting HIS6x-EXP-CUL1 and HIS6x-EXP-cull-7 expression vectors were added to a TnT T7 Coupled Reticulocyte Lysate System (Promega). The translation mixes included 14 ng plasmid template per μl translation and 0.1 μCi/μl 3H-leucine (PerkinElmer) and were incubated for 2 hr at 30°.
For in vitro pull-down experiments, 100-μl translation reactions for HIS6x-EXP-CUL1 and HIS6x-EXP-cull-7 were supplemented with ∼500 ng of purified GST-RBX1 or GST (prepared as described above) and incubated for 2 hr at 30°. Input fractions were removed and the remaining reactions were diluted with 1 ml GST wash buffer. Glutathione-sepharose beads (20 μl) were then added and incubated with mixing overnight at 4°. Beads were collected by centrifugation and washed three times for 30 min in 1 ml GST wash buffer. After the last wash, beads were boiled in 40 μl of 5× Laemmeli sample buffer (LSB). Proteins were separated by SDS–PAGE, and 3H labeled HIS6x-EXP-CUL1 and HIS6x-EXP-cul-7 were visualized by autoradiography and GST-RBX by Western blot.
For RBX1-CUL1 co-immunoprecipitation experiments, 1-week-old seedlings grown in liquid GM were flash frozen and ground in 1 μl/mg fresh weight in IP buffer [50 mm Tris pH 7.5, 150 mm NaCl, 0.5% NP-40, 1 mm PMSF, 1 tablet/10 ml Complete Mini protease inhibitor pill (Roche), 50 μm MG132 (Peptides International)]. Extracts were cleared by centrifugation at 16,000 × g for 20 min at 4°. Protein concentration was determined by Bradford assay, and immunoprecipitations were performed using 5–10 mg total protein from each genotype. Extracts were precleared for 2 hr at 4° with 50 μl 1:1 v:v Protein A agarose slurry (Sigma, P-1406) per 5 mg total protein. To immunoprecipitate RBX1, 4 μg of anti-ROC1 antibody (Invitrogen) per 1 mg total protein were added to the extract. After 2 hr of rocking at 4°, 10 μl of Protein A agarose slurry per 1 mg total protein were added and samples allowed to incubate overnight with gentle rocking. Immunocomplexes were collected by centrifugation at 2,000 × g and washed four times in 1ml IP buffer for 20 min. Remaining wash buffer was aspirated and protein was eluted from the Protein A agarose into 40 μl of 100 mm glycine pH 1.9 by vortexing gently for 1 hr at room temperature. Eluant was neutralized with 1 m NaOH, SDS sample buffer added, and boiled for 5 min. Equal volumes were loaded onto 8 and 15% polyacrylamide gels for subsequent anti-CUL1 and anti-RBX1 Western blots, respectively.
CUL1-FLAG was immunoprecipitated from 12-day-old light-grown complementation lines with anti-FLAG M2 agarose beads (Sigma, A2220). Extracts were prepared in IP buffer without MG132, and 4 mg total protein from each genotype were mixed with 30 μl anti-FLAG beads overnight at 4°. Beads were collected by centrifugation, washed three times for 20 min in 1 ml IP buffer, boiled in 20 μl LSB, and separated by SDS–PAGE.
RESULTS
A screen for mutants defective in IAA1-LUC degradation identifies a new allele of CUL1:
To identify genes important for regulating Aux/IAA protein degradation, a genetic screen based on an increase in LUC activity from plants expressing an Aux/IAA-LUC fusion protein from a transgene in A. thaliana (Col) was performed. In vivo LUC activity from individual 7- to 10-day-old M2 seedlings expressing full-length IAA1-LUC (Ramos et al. 2001; Zenser et al. 2001, 2003; Dreher et al. 2006) was measured. The substrate luciferin was added to intact seedlings, and light emission, a product of LUC activity, from each seedling was measured. Seedlings emitting >50% more light than the progenitor line were propagated. To determine if increases in light emission result from slowed IAA-LUC degradation, an assay to measure protein degradation directly in intact single seedlings was designed. Single M4 or control seeds were sown directly into individual wells of a 96-well plate and after 7 days, luciferin and cycloheximide, a protein synthesis inhibitor, were added. The amount of light emitted was monitored over a 60-min time course.
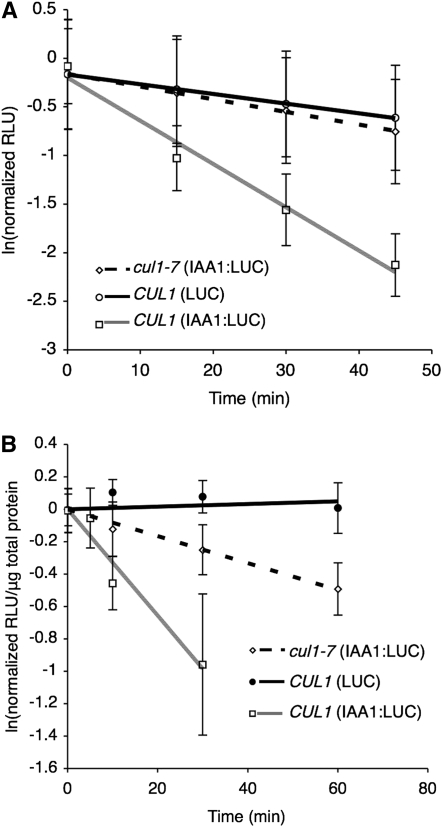
Using this screen, we identified one line that exhibited slower IAA1-LUC degradation (Figure 1A, dotted line). The half-life of IAA1-LUC in the mutant seedlings (designated cul1-7, see below) was ∼50 min, ∼3.5 times slower than the ∼15-min half-life for IAA1-LUC in the nonmutagenized seedlings. (Figure 1A, shaded line, designated CUL1). The half-life of IAA1-LUC in cul1-7 was similar to that of LUC (∼70 min), which lacks the IAA1 degron (Figure 1A, solid line). This measurable loss of activity from plants expressing LUC alone has been observed previously and ascribed to increased degradation when luciferin is added to intact cells (N. Zenser and J. Callis, unpublished data). The increased rate of loss of LUC alone from in vivo addition of its substrate prevents using this specific in vivo degradation assay to measure the half-lives of LUC fusion proteins if the fusion protein half-life is slower than that observed for LUC alone, which is ∼70 min.
Figure 1.—
Degradation of IAA1-LUC in cul1-7. (A) Single-seedling degradation assay. Experiment performed on 7-day-old seedlings. Zero represents the initial luciferase activity of seedlings in the initial plate reading. Values represent averages ± 1 SD from a total of at least 56 seedlings from two independent experiments. T1/2 (IAA1-LUC) = 15 and 53 min, respectively, for CUL1 and cul1-7. T1/2 (LUC) = 70 min in CUL1. (B) Pooled-seedling degradation assay. Values represent averages ± SD from a total of 9 replicates, from three independent experiments. T1/2 (IAA1-LUC) = 21 and 83 min, respectively, for CUL1 and cul1-7. Loss of LUC in CUL1 is not detected.
To confirm the half-life differences observed using the screening method and single-seedling degradation assay and to more accurately measure the IAA1-LUC degradation rate in cul1-7, we determined degradation rates in these same lines using our traditional pooled-seedling degradation assay (Worley et al. 2000; Dreher et al. 2006) (Figure 1B). In this case, only cycloheximide is added to the intact seedlings and LUC activity is determined in extracts prepared at various times after addition. In these assays, LUC alone shows no loss of activity in the time course (Figure 1B, solid line), consistent with previous results (Worley et al. 2000; Ramos et al. 2001; Dreher et al. 2006). The half-life of IAA1-LUC was ∼80 and 21 min in mutant and wild-type seedlings, respectively, confirming that the mutant shows altered rates of IAA1-LUC degradation (Figure 1B, dashed and shaded lines, respectively). This ∼80-min half-life was consistent between generations and in homozygous seedlings after several backcrosses to the nonmutagenized transgenic line (data not shown).
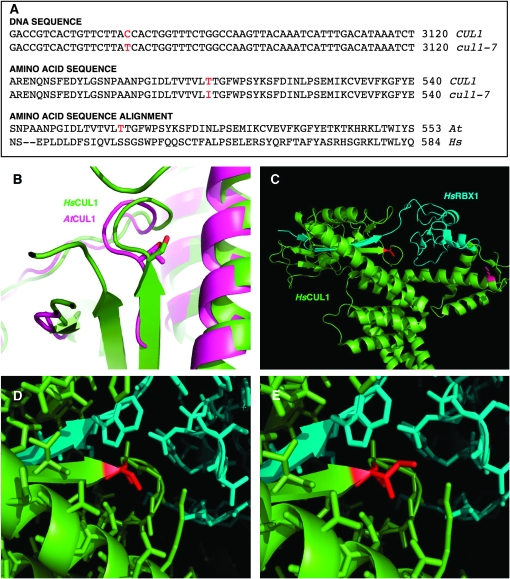
Bulked segregant analysis (Michelmore et al. 1991; Lukowitz et al. 2000) placed the mutation on the short arm of chromosome IV. Using a series of SSLP and CAPS markers spanning the short arm of chromosome IV, the mutation was located within a genetic interval that included the CULLIN1 (CUL1) gene, which encodes the cullin subunit of SCF-type ubiquitin ligases (Gray et al. 1999). We sequenced the CUL1 coding region from the mutant line and found one difference from wild type—a C-to-T transition in exon 16 of CUL1 resulting in a T510I substitution (Figure 2A). We called this allele cul1-7. The threonine residue in wild-type CUL1 is conserved among other cullin family members; AtCUL1, AtCUL2, AtCUL3a, AtCUL3b, and AtCUL4. Additionally, amino acid sequence alignment revealed that Thr510 of AtCUL1 aligns with Ser541 of HsCUL1, suggesting a functional conservation of this residue between the species (Figure 2A).
Figure 2.—
Identification and molecular modeling of cul1-7 allele. (A) DNA and amino acid sequence comparison of cul1-7 and CUL1, and amino acid sequence alignment of AtCUL1 with HsCUL1. The positions affected by the cul1-7 are highlighted in red. Sequence alignments were performed using standard parameters of ClustalW (http://www.ebi.ac.uk/clustalw/). (B) Structural overlay of AtCUL1 (pink) and HsCUL1 (green). The structure of AtCUL1 was predicted using SWISS-MODEL protein modeling server (http://swissmodel.expasy.org), and the overlay was generated using Coot software. (C) Crystal structure of HsCUL1 (green) with HsRBX1 (blue). The PDB file for this structure (Zheng et al. 2002b) was downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) and manipulated using PyMol molecular graphics system (http://www.pymol.org). Ser541 of HsCUL1, which aligns with Thr510 of AtCUL1, is highlighted in red. Lys720, site of RUB modification, is highlighted in purple. (D and E) Modeling of cul1-7 in HsCUL1. (D) Close-up view of HsCUL1 (green), Ser541 with HsRBX1 (blue). (E) HsCUL1 Ser541Ile with HsRBX1. Residue 541 is highlighted in red.
We modeled the sequence of AtCUL1 with the known crystal structure of human CUL1 (HsCUL1) in complex with HsRBX1 (Zheng et al. 2002b), and Thr510 of AtCUL1 overlapped with Ser541 of HsCUL1 as suggested by the primary sequence alignment (Figure 2B). Ser541 is at the end of an HsCUL1 β-strand near the beginning of a loop in HsCUL1. This HsCUL1 β-strand interacts with a β-strand of HsRBX1 (Figure 2, C and D). While the hydroxyl group of HsCUL1 Ser541 does not participate in hydrogen bonding with any residues of HsRBX1, it is within hydrogen bonding distance of Asp510 of HsCUL1, which is conserved as Asp477 in AtCUL1. Moreover, the backbone nitrogen of Leu540 is within hydrogen bonding distance of the backbone carbonyl of Ala31 of HsRBX1, and these residues are conserved in the corresponding Arabidopsis homologs. There is insufficient room for the side group of isoleucine (the amino acid in cul1-7) in the crystal structure when substituted for Ser541 in silico (Figure 2E), and such a substitution could potentially affect the described interaction with RBX1 in this region.
cul1-7 is recessive and plants display pleiotropic developmental defects similar to other CUL1 alleles:
To assess the recessivity of the cul1-7 allele, we performed single-seedling degradation assays (as described in Figure 1A) on a segregating F2 population derived from the self of a backcross of cul1-7 with the progenitor transgenic line. The defect in IAA1-LUC degradation segregated 3:1 (χ2 = 0.68, P = 0.410, d.f. = 1, n = 49), indicating that the trait was recessive. The cul1-7 allele cosegregated with the mutant phenotype after three backcrosses (data not shown), suggesting that the mutation in cul1-7 is responsible for the observed phenotypic differences.
cul1-7 plants display pleiotropic phenotypes at almost all stages of development (Figures 3–5). Adult cul1-7 plants are dwarfed, exhibit a reduction in apical dominance, and have numerous curly leaves (Figure 3, A and B). We more directly determined that the lesion in cul1-7 was responsible for the observed phenotypes by performing an allelism test with axr6-3, a recessive, temperature-sensitive allele of CUL1 that contains a missense mutation near the N terminus (Quint et al. 2005). We used a dCAPS-based method to distinguish the mutant alleles from wild type (Michaels and Amasino 1998; Neff et al. 1998) and to verify the genotypes of individuals from crosses (see materials and methods and supplemental Figure 1). The phenotypes of the cul1-7/axr6-3 heteroallelic F1 plants are equivalent to cul1-7 homozygotes (Figure 3C), indicating that the lesion in cul1-7 is likely responsible for the observed phenotypes.
Figure 3.—
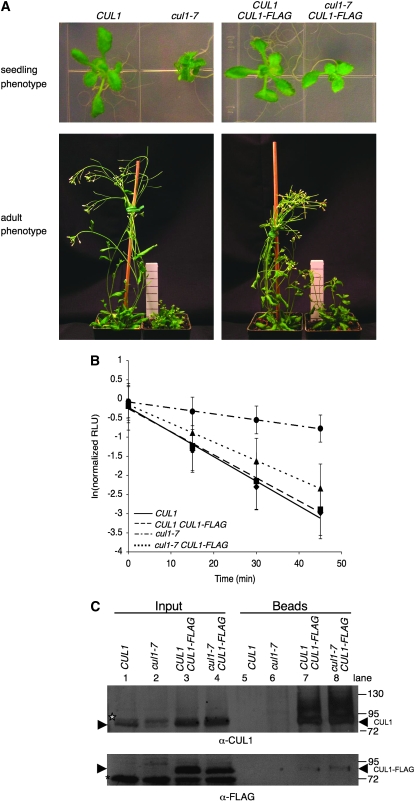
Morphological phenotypes of cul1-7 in comparison to wild type and axr6-3. (A and B) Aerial phenotype of cul1-7. (A) One week-old cul1-7 (left) and the progenitor line CUL1 (right) seedlings grown on GM were transferred to soil and grown 4 weeks more under a 16-hr photoperiod. (B) Close up of cul1-7 phenotype in A. Bars, 1 cm. (C) Allelism test of cul1-7 with axr6-3. cul1-7 was crossed to axr6-3, and the resulting F1 progeny were grown 2 weeks at 22° under constant light on GM plates, genotyped, then transferred to soil for an additional 6 weeks.
Figure 4.—
Genetic complementation of cul1-7 with CUL1-FLAG. (A) Seedling and adult phenotypes of CUL1, cul1-7, CUL1 CUL1-FLAG, and cul1-7 CUL1-FLAG lines. Complementation lines were homozygous T3 generation and expressing CUL1-FLAG from the same genetic locus. (B) Degradation of IAA1-LUC in cul1-7 complementation lines. Single-seedling degradation assays, as described in Figure 1A, were performed on 2-week-old seedlings. Values represent averages ± 1 SD from a total of at least 50 seedlings from six individual 96-well plates. T1/2 for IAA1-LUC = 11 min for CUL1 and CUL1 CUL1-FLAG, 14 min for cul1-7 CUL1-FLAG, and 44 min for cul1-7. (C) CUL1-FLAG expression in complementation lines. CUL1-FLAG was immunoprecipitated using anti-FLAG agarose from 4 mg total protein from the genotypes described in A and B. Plants used were 12-day-old light-grown seedlings. For inputs, 40 μg and 100 μg total protein were loaded for anti-CUL1 and anti-FLAG blots, respectively. For the immunoprecipitation, 90% of the volume was loaded for anti-CUL1 blot and 10% loaded for the anti-FLAG blot. Numbers on the left represent migration of molecular weight markers, in kilodaltons (kDa). Arrowheads represent the location of CUL1 and CUL1-FLAG bands. The CUL1RUB band is starred, and an asterisk denotes a nonspecific anti-FLAG immunoreactive species.
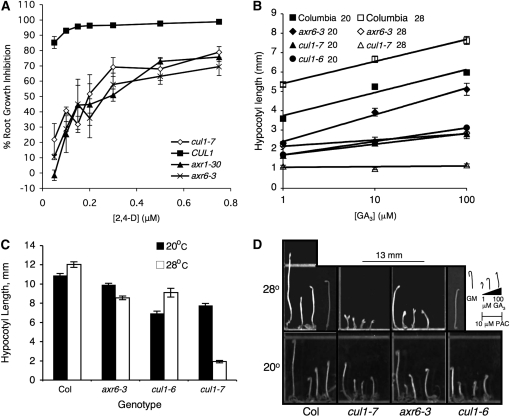
Figure 5.—
cul1-7 confers reduced auxin and GA responses. (A) Root growth inhibition on 2,4-D. Values represent the average ± SE for three independent experiments, each of which represented the percentage of inhibition from an average of at least six seedlings. (B) Hypocotyl elongation on GA3. Seeds were germinated on 10 μm PAC media with various concentrations of GA3 and grown for 5 days in the dark at either 20° or 28°. Values represent the average ± SE for at least 17 seedlings. The equations for linear regression at 20° are y = 0.5164ln(x) + 3.7477, R2 = 0.9549; y = 0.2403ln(x) + 1.7205, R2 = 0.9925; y = 0.5999ln(x) + 2.4086, R2 = 0.9929; y = 0.302ln(x) + 1.722, R2 = 0.9986 for Columbia, cul1-7, axr6-3, and cul1-6, respectively. The equations for linear regression at 28° are y = 0.4934ln(x) + 5.4062, R2 = 0.9928; y = 0.0146ln(x) + 1.0826, R2 = 0.1331; y = 0.139ln(x) + 2.1705, R2 = 0.9643 for Columbia, cul1-7, and axr6-3, respectively. (C) Quantification of hypocotyl lengths from 5-day-old etiolated Columbia (Col), cul1-7, axr6-3, and cul1-6 seedlings grown at two temperatures. Seedlings were grown 5 days in the dark on GM plates without GA or PAC at either 20° or 28°, then transferred to a plastic sheet protector and imaged using a flat bed scanner. Values represent averages from at least 19 seedlings ± SE. (D) Etiolated phenotypes of Col, cul1-7, axr6-3, and cul1-6 seedlings from experiments in B and C. Grids represent 13 × 13 mm. Seedlings of each genotype were arranged from left to right—GM, 1, 10, 100 μm GA3.
cul1-7 can be partially complemented by expression of CUL1-FLAG:
We attempted to complement the cul1-7 phenotype by expressing epitope-tagged CUL1 under its own 5′ flanking region. We used an expression cassette containing a 5221-bp genomic fragment from the CUL1 locus that includes 1103 bp of sequence upstream of the translational start site and the entire coding region with the addition of in-frame codons for the FLAG epitope before a stop codon (Ren et al. 2005). CUL1-FLAG expression can be detected in transgenic Columbia plants containing this construct, and CUL1-FLAG was additionally shown to interact with COI1, ASK1, and RBX1 in co-immunoprecipitation experiments (Ren et al. 2005).
Because cul1-7 plants are difficult to transform, we transformed CUL1/cul1-7 heterozygotes and obtained 98 T1 individuals, of which 25 genotyped as CUL1/cul1-7 (data not shown). Seventeen of the 25 independent T1 lines were carried forward to the T2 generation. None of the T2 plants that genotyped as homozygous cul1-7 looked wild type, but had phenotypes resembling partial complementation. Six homozygous cul1-7 T2 lines were carried to the T3 generation, and individuals homozygous for CUL1-FLAG expressing transgene were identified (called cul1-7 CUL1-FLAG). We were also able to obtain the corresponding homozygous wild-type CUL1 CUL1-FLAG siblings for two of these lines from the segregating T2 family. The seedling and adult phenotypes for one of these pairs (T3 generation) are shown in Figure 4A. Expressing CUL1-FLAG in CUL1 does not cause any observable phenotypic difference from Columbia wild type in either of the two lines (Figure 4A and data not shown), consistent with a previous report (Ren et al. 2005).
Expression of CUL1-FLAG in cul1-7 partially complements the mutant aerial phenotype. Compared with cul1-7, cul1-7 CUL1-FLAG seedlings have larger true leaves and are less dwarfed (Figure 4A, top). As adults, some of the apical dominance is restored in cul1-7 CUL1-FLAG as evidenced by a reduction in the number of inflorescences and reduced dwarfism compared with cul1-7 plants (Figure 4A, bottom). Surprisingly, the IAA1-LUC half-life in cul1-7 CUL1-FLAG was threefold different from that in cul1-7, ∼14 and ∼44 min, respectively. The half-lives in CUL1 and CUL1 CUL1-FLAG were equivalent, indicating that expression of CUL1-FLAG does not change the IAA1-LUC degradation rate (Figure 4B).
CUL1-FLAG can be detected in both cul1-7 CUL1-FLAG and CUL CUL1-FLAG plants and results in increased total CUL1 levels. Immunoblot analysis using anti-FLAG antibodies on total protein extracts from these two genotypes shows a FLAG immunoreactive species (Figure 4C, bottom, lanes 3 and 4) that is missing in CUL1 and cul1-7 extracts (Figure 4C, bottom, lanes 1 and 2), indicating that these plants are expressing CUL1-FLAG. Immunoblot analysis of the same extracts using anti-CUL1 antibodies revealed that the level of CUL1 protein is much higher in both CUL1-FLAG lines with both CUL1 and cul1-7, presumably because of the presence of CUL1-FLAG migrating at the same size as the endogenous CUL1 in these gels (Figure 4C, top, compare lanes 1 and 2 with 3 and 4). This was confirmed by performing a FLAG-IP using anti-FLAG beads with protein extracts prepared from the same four genotypes and immunoblotting with anti-CUL1 or anti-FLAG antibodies. Only CUL1-FLAG expressing plants enriched for a CUL1 immunoreactive protein (Figure 4C, compare lanes 5 and 6 with 7 and 8). Protein pulled down by anti-FLAG beads from cul1-7 CUL1-FLAG and CUL1 CUL1-FLAG extracts comigrates with endogenous CUL1 from CUL1 and cul1-7 extracts (Figure 4C, top, compare lanes 1 and 2 with 7 and 8). In addition, slower migrating anti-CUL1 reactive forms were also visualized in the anti-FLAG pull-down. Thus, CUL1-FLAG is present in both cul1-7 and CUL1 backgrounds and CUL1 levels (composed of CUL1 + CUL1-FLAG) in the cul1-7 CUL1-FLAG and CUL1 CUL1-FLAG lines are greater than seen in CUL1 and cul1-7 plants.
Auxin and GA responses are altered and skotomorphogenesis is affected in cul1-7:
We hypothesized that the phenotypic differences between cul1-7 and CUL1 plants result from altered degradation of endogenous SCF substrates. Due to its extremely low abundance, we were not able to detect endogenous IAA3 protein from total cul1-7 protein extracts using IAA3 anti-serum (Colon-Carmona et al. 2000). Instead, we used another indicator of defective degradation of endogenous Aux/IAA proteolysis. Plants with defects in Aux/IAA degradation have altered auxin responses. Mutations that result in substitutions in the Aux/IAA core degron sequence, or mutations that affect SCF function have been shown to confer auxin-resistant phenotypes (Rouse et al. 1998; Ruegger et al. 1998; Tian and Reed 1999; Hobbie et al. 2000; Nagpal et al. 2000; Schwechheimer et al. 2001; Tiwari et al. 2001; Gray et al. 2002, 2003; Park et al. 2002; Hellmann et al. 2003; Bostick et al. 2004; Chuang et al. 2004; Feng et al. 2004; Tatematsu et al. 2004; Yang et al. 2004; Dharmasiri et al. 2005b; Quint et al. 2005; Alonso-Peral et al. 2006; Walsh et al. 2006; Moon et al. 2007; Woodward et al. 2007).
We measured inhibition of primary root elongation conferred by exogenous auxin in cul1-7 as a measure of auxin response (Figure 5A). CUL1 was very sensitive to 2,4-D, a synthetic auxin, reaching a nearly complete inhibition on concentrations >0.2 μm, consistent with Columbia wild-type response previously reported (Estelle and Somerville 1987). The dose response for cul1-7 was very similar to that of axr1-30 and axr6-3, two previously characterized auxin-resistant mutants. axr1-30 is a T-DNA insertional allele of AXR1 characterized in our laboratory. This allele is phenotypically identical to published axr1 null alleles (M. Bostick, unpublished data). All three mutants appear ∼25% less sensitive than CUL1 on high concentrations of 2,4-D. These results are consistent with longer Aux/IAA half-lives in cul1-7 compared with CUL1 (Figure 1).
As CUL1 is a core subunit of all SCF complexes, proteolysis of other SCF substrates should be affected in cul1-7. Similar to auxin, GA signaling depends on the degradation of negative regulators called DELLA proteins, and in Arabidopsis, SCFSLY1 is required for their ubiquitylation (Silverstone et al. 1998, 2001; Dill et al. 2001, 2004; Wen and Chang 2002; McGinnis et al. 2003). Like the Aux/IAAs, DELLA proteins are rapidly reduced in abundance in response to GA (Fleet and Sun 2005). We therefore hypothesized that cul1-7 plants should have increased longevity of DELLA proteins and consequently, be less sensitive to GA.
As a test of GA sensitivity, the response of dark-grown hypocotyls to exogenous GA3 was measured in plants carrying the two previously published recessive alleles of CUL1, axr6-3 (Quint et al. 2005) and cul1-6 (Moon et al. 2007) and in cul1-7 (this report) and Columbia (Col) under the same growth conditions. It is well established that GA promotes stem elongation, and GA signaling mutants have a dampened response (reviewed in Sun and Gubler 2004). Because auxin signaling directly influences GA levels (Wolbang and Ross 2001; O'neill and Ross 2002; Wolbang et al. 2004; Frigerio et al. 2006), and since all cul1 plants have altered auxin responses (Figure 5A), 10 μm paclobutrazol (PAC), a GA biosynthesis inhibitor, was added to eliminate differences in the endogenous GA content between Col and the cul1 alleles. We conducted the experiment at 20° and 28° to compare the responses to axr6-3, which has a temperature-sensitive phenotype (Quint et al. 2005). We were unable to perform the GA experiment with cul1-6 at 28°, because the seeds had very poor germination. Results are quantified in Figure 5B and representative seedlings are shown in Figure 5D. For Col, the slopes of the response curve at these two temperatures are equivalent, indicating that temperature does not affect GA sensitivity (Figure 5B). axr6-3 responds similarly to Col at 20°, but is ∼3.5-fold less sensitive to GA3 at 28°. At 20°, both cul1-6 and cul1-7 are approximately twofold less sensitive to GA than Columbia. Surprisingly, cul1-7 was completely insensitive to GA at 28°, demonstrating a more extreme temperature sensitivity than axr6-3 in this response.
To further explore the effects of cul1 on development, we compared the dark-grown phenotype of cul1 mutants. Additionally, because of the temperature-sensitive GA response of axr6-3 and cul1-7 (Figure 5B), we compared dark-grown seedlings at the same two temperatures. At 20°, etiolated axr6-3, cul1-6, and cul1-7 seedlings have an apical hook and closed cotyledons only differing from wild type in hypocotyl length (Figure 5, C and D). At 28°, all genotypes, including wild type, are unhooked; however, cul1-7 is strongly photomorphogenic with open cotyledons and a dramatically shortened hypocotyl (Figure 5D). All three cul1 alleles are statistically shorter than Col at both temperatures and each genotype is statistically different from itself at the two temperatures as determined by a Student's t-test with a Bonferroni correction for testing multiple hypotheses (α = 0.005, P ≤ 0.001). At 28°, both Col and cul1-6 have longer hypocotyls than at 20°, while axr6-3 and cul1-7 have shorter hypocotyls, suggesting that both of these alleles are temperature sensitive (Figure 5C).
Degradation of a negative regulator of GA responses is slowed in cul1-7:
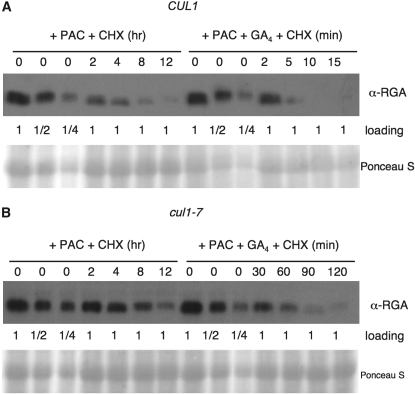
We tested directly whether the degradation rate of the DELLA protein RGA is affected in cul1-7. Similar to the physiological test, to minimize the effect of endogenous GA and the effect that reduced auxin responsiveness has on endogenous GA levels, CUL1 and cul1-7 seedlings were preincubated with PAC for 4 days and then treated with cycloheximide in the presence or absence of 20 nm GA4. Immunoblot analysis indicates that RGA half-life in cul1-7 is longer than in CUL1 (Figure 6). Without GA application (in the presence of PAC), RGA half-life is ∼2 hr in CUL1 (Figure 6A) and ∼8 hr in cul1-7 (Figure 6B). In the presence of 20 nm GA4, RGA has a much shorter half-life, ∼2 min in CUL1. RGA degradation in cul1-7 is also faster in the presence of GA, but its half-life of ∼30 min is still longer than in CUL1. These changes in RGA degradation rate are consistent with the physiological data in Figure 5B and represent the first examination of RGA half-life in plants that consider the effect of endogenous gibberellins.
Figure 6.—
SCF-mediated RGA protein degradation is impaired in cul1-7. Nine-day-old wild type (A) and cul1-7 (B) seedlings that were preincubated with 100 μm PAC for 4 days were treated with 200 μg/ml cycloheximide ± 20 nm GA4. At various time points (as indicated), total proteins were extracted and analyzed by immunoblot analysis using affinity purified anti-RGA antibodies. All lanes had equal loading of total protein (120 μg in A, 80 μg in B) except that zero time points also had loadings of one-half and one-quarter amounts. Images of Ponceau S stained blots were included to show equal loading.
Phytochrome A degradation requires CUL1:
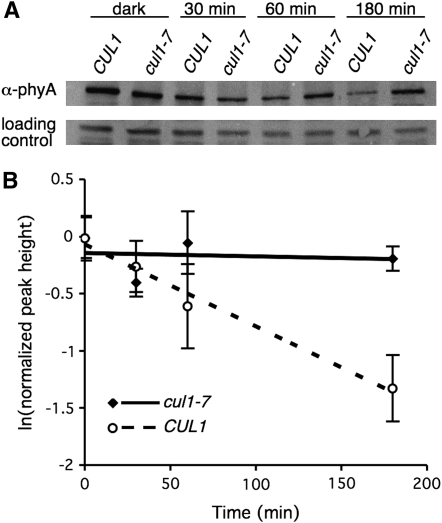
Phytochrome A degradation has been shown to be essential for proper photomorphogenesis. A long-lived red light-absorbing form of PhyA, termed Pr, exists in the cytoplasm of dark-grown cells. Upon illumination with red light, Pr undergoes a conformational change to the active far-red light-absorbing form (Pfr), translocates to the nucleus, and is subsequently degraded by the ubiquitin-proteasome system (Shanklin et al. 1987). COP1 is thought to be the major E3 ligase contributing to PhyA ubiquitylation (Seo et al. 2004); however, evidence is emerging that SCF-type ligases are also required. An F-box protein, EID1, was identified that specifically acts in the PhyA signaling pathway (Dieterle et al. 2001). Second, the two reported recessive alleles of CUL1 have altered PhyA degradation kinetics (Quint et al. 2005; Moon et al. 2007); therefore, we measured PhyA degradation rate in cul1-7 to see if PhyA degradation was similarly affected (Figure 7). Etiolated seedlings were treated with cycloheximide and then transferred from dark to 20 μmol m−2 s−1 red light for the indicated time. Total protein was extracted, separated by SDS–PAGE, and analyzed for PhyA protein by Western blot with anti-PhyA anti-serum (Elich and Lagarias 1987). Over the 2-hr time course, PhyA protein rapidly disappears in CUL1, but is not appreciably lost in a cul1-7 background (Figure 7A). Densitometry of PhyA reveals the t1/2 of PhyA in CUL1 to be ∼100 min (Figure 7B); however, in cul1-7 PhyA does not appear to change abundance during this time course. Thus, a degradation rate cannot be calculated in cul1-7.
Figure 7.—
cul1-7 stabilizes phytochrome A protein. (A) PhyA degradation in cul1-7. Protein extracts were made, 40 μg total protein were separated by SDS–PAGE, and PhyA levels determined by immunoblotting with an anti-PhyA antibody. A cross-reactive band was included as a loading control. (B) Quantification of PhyA degradation in cul1-7. ImageQuant 1.0 software (Molecular Dynamics) was used to quantify relative PhyA levels from the Western blot analyses described in A. Values represent averages ± SD from a total of at least three Western blots from three independent experiments. T1/2 = 96 min in CUL1. PhyA is not degraded appreciably in cul1-7.
cul1-7 disrupts SCF regulation at the C terminus of CUL1:
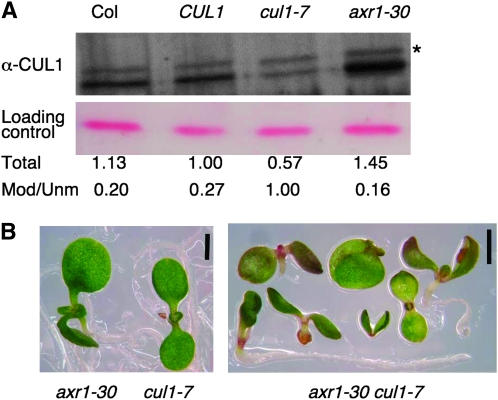
A modification that is important for full SCF activity is the attachment of the ubiquitin-like protein, RUB, to a lysyl residue of CUL1. To determine whether the mutation in cul1-7 affects the ability of the protein to be RUB modified, we performed a Western blot analysis on total protein prepared from 9-day-old seedlings grown on GM plates. The amount of RUB-modified CUL1 appears unaffected by the mutation, as cul1-7 has the same amount of modified protein as axr1-30 [as previously observed in axr1-12 (Dharmasiri et al. 2003)], the Columbia ecotype control and the transgenic IAA1-LUC progenitor line, CUL1 (Figure 8A). Surprisingly, the amount of unmodified protein in cul1-7 is drastically reduced compared with wild type, accounting for a 43% reduction of total CUL1 protein in the mutant. This reduction increases the ratio of modified-to-unmodified CUL1 from ∼0.2–0.27 in wild type to ∼1.0 in cul1-7. In contrast, in axr1-30, where the RUB-conjugation pathway is compromised, the total amount of CUL1 increases ∼45% from wild-type levels. Thus, the modified-to-unmodified ratio is reduced further than wild type, strikingly different from cul1-7 (Figure 8A).
Figure 8.—
Unmodified CUL1 levels are reduced in cul1-7. (A) CUL1 immunoblot analysis. Western blot analysis using anti-CUL1 antisera (Gray et al. 1999) was done on total protein extracts separated by SDS–PAGE. Each lane represents 12.5 μg total protein from 9-day-old light-grown seedlings. Levels were quantified as in Figure 7, using local background correction. Total levels were normalized to CUL1, while the modified-to-unmodified ratio (Mod/Unm) represents the amount of CUL1RUB(*) to CUL1 within a given genotype. (B) One-week-old cul1-7, axr1-30, and cul1-7 axr1-30 seedlings grown on GM. cul1-7 was crossed to axr1-30 and the resulting F3 double homozygotes were obtained from both CUL1/cul1-7 axr1-30 and cul1-7 AXR1/axr1-30 F2 parents. Bars, 1 mm.
A previously described cul1 allele, axr6-3, demonstrated strong genetic interaction with axr1 (Quint et al. 2005). To determine whether cul1-7 shows a genetic interaction with axr1, we crossed cul1-7 with axr1-30. Genotypes were confirmed by PCR analysis (data not shown). Double axr1-30 cul1-7 mutants were consistently much more strongly affected than either single mutant (Figure 8B). axr1-30 cul1-7 seedlings accumulated anthocyanin at the cotyledon margin and no seedling developed more than two to six leaves. Some failed to develop hypocotyls or roots, producing only one or two cotyledons, while others lacked hypocotyls and roots but did go on to produce two true leaves (Figure 8B). Less severe double mutants developed normal cotyledons and a hypocotyl but no roots. The most severe double-mutant phenotype resembles that of homozygous axr6-1 and axr6-2, semidominant alleles of CUL1 that disrupt ASK1 interaction (Hobbie et al. 2000; Hellmann et al. 2003). These double-mutant phenotypes are also consistent with double-mutant phenotypes of axr1-12 axr6-3 plants (Quint et al. 2005). This strong genetic interaction with loss of AXR1 is consistent with the mutation in cul1-7 being responsible for the observed phenotypes.
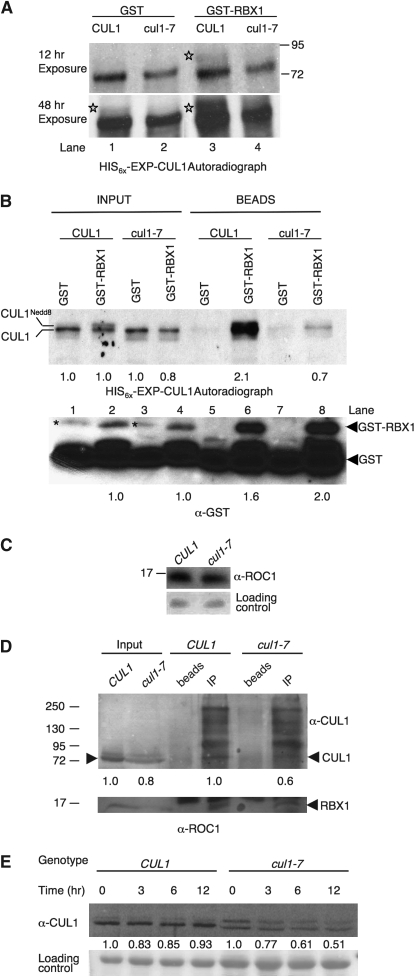
On the basis of the location of the amino acid change in cul1-7 (Figure 2), cul1-7 could have impaired interaction with RBX1. To determine whether cul1-7 is affected in RBX1 binding, we synthesized epitope-tagged versions of wild-type and mutant CUL1, HIS6x-EXP-CUL1, and HIS6x-EXP-cul1-7, respectively, in a rabbit reticulocyte lysate system, which has the ability to conjugate NEDD8 to CUL1 (Furukawa et al. 2000). In plants overexpressing RBX1, the majority of CUL1 is in the RUB-modified form (Gray et al. 2002), suggesting that RBX1 interaction is limiting CUL1 modification. We hypothesized that the addition of recombinant RBX1 (here as GST-RBX1) to the in vitro translation reaction could increase production of Nedd8-modified CUL1, but not equivalently NEDD8-modified cul1-7 if the substitution in cul1-7 impairs RBX1 interaction. GST-RBX1 and GST were added at the initiation of translation so that the proteins are translated in the presence of RBX1, which gives maximal neddylation. Addition of GST-RBX1 resulted in increased levels of Nedd8-modified CUL1, more easily visualized in a long exposure (bottom), compared with addition of GST alone (Figure 9A, lanes 1 and 3). Addition of GST-RBX1 to the translation reaction synthesizing cul1-7 did not promote equivalent neddylation (Figure 9A, lane 4). These results are consistent with cul1-7 having reduced RBX1 interaction.
Figure 9.—
RBX1 interaction with cul1-7 is impaired and cul-7 destabilizes CUL1 protein. (A) In vitro translations of HIS6x-EXP-CUL1 and HIS6x-EXP-cul1-7 in the presence of GST-RBX1. Proteins were translated in vitro and radiolabeled with 3H −Leu. Reactions were supplemented with ∼125 ng either GST or GST-RBX1. Stars were placed to the left of CUL1Nedd8 bands. (B) Pull-down of in vitro translated HIS6x-EXP-CUL1 and HIS6x-EXP-cul1-7 with GST-RBX1. HIS6x-EXP-CUL1 and HIS6x-EXP-cul1-7 proteins were translated in reactions supplemented with ∼500 ng of GST or GST-RBX1. Translations were incubated with glutathione-sepharose beads to collect GST-RBX1 complexes. Input represents 1% of the total for the autoradiogram and 4% for the anti-GST blot. Beads represents 75% of the total pull-down for the autoradiogram and 25% for the anti-GST blot. Inputs were normalized to either the amount of HIS6x-EXP-CUL1 translated with GST for the autoradiograph or the amount of GST-RBX1 in HIS6x-EXP-CUL1 translation for the anti-GST blot, and the amount in the pull downs were normalized to their respective inputs. The asterisk represents a nonspecific, cross-reactive band. A GST cleavage product that copurified with GST-RBX1 is also detectable in the GST-RBX1 lanes. (C) RBX1 immunoblot analysis. Western blot analysis using anti-ROC1 antisera was performed on total protein extracts. Each lane represents 120 μg total protein from week-old light-grown seedlings. (D) Co-immunoprecipitation of CUL1 and RBX1 from CUL1 and cul1-7 plant extracts. RBX1 was immunoprecipitated using 40 μg anti-ROC1 antibody from 5 and 10 mg total protein from CUL1 and cul1-7, respectively, to allow for more equal CUL1 input. Immunocomplexes were eluted from Protein A agarose and equal amounts were resolved by SDS–PAGE for Western blotting. Input represents 1% and 2% of the total for the anti-CUL1 and anti-ROC1 blots, respectively. cul1-7 input was normalized to CUL1 input, and the amount of co-immunoprecipitated cul1-7 was normalized to the amount of co-immunoprecipitated CUL1 as denoted by numbers at the bottom of the blot. (E) CUL1 and cul1-7 degradation. CUL1 protein degradation was examined in the7-day-old progenitor and cul1-7 lines. The zero time point represents a mock cycloheximide sample. Each lane represents 20 μg and 40 μg total protein for CUL1 and cul1-7, respectively. CUL1 levels and image quantification were determined as in Figure 7. Quantification denotes the amount of total CUL1 relative to 0 time point for the given genotype. Migration of molecular weight markers (sizes in kDa) is marked in A and C.
To demonstrate more directly a difference in RBX1 interaction between CUL1 and cul1-7, we pulled down GST-RBX1 or GST from in vitro translation reactions with glutathione sepharose and determined the amount of the HIS6x-EXP-CUL1 or HIS6x-EXP-cul1-7 present in the pull-down fraction (Figure 9B). GST and GST-RBX1 translation master mixes were prepared and added to HIS6x-EXP-CUL1 and HIS6x-EXP-cul1-7 DNA templates. The amounts of CUL1 and cul1-7 produced in these reactions are nearly identical (Figure 9B, INPUT lanes and quantified below). As observed before, CUL1 translation reactions with added GST-RBX1 had increased levels of a slower migrating band, CUL1Nedd8 compared with GST-containing reactions. After the pull-down with glutathione beads from the GST-RBX1-containing reactions, much more CUL1 was present than cul1-7, approximately threefold after normalization to respective input (Figure 9B, compare lane 6 with lane 8). The enhancement of NEDD8 modification by RBX1, together with the reduced recovery of cul1-7 in RBX1 pull-down assays, indicate that cul1-7 has impaired interaction with RBX1.
To determine whether interaction with RBX1 is affected in vivo, we immunoprecipitated RBX1 from total protein extracts and immunoblotted for CUL1. CUL1 and cul1-7 plants have roughly equivalent amounts of RBX1 (Figure 9C), indicating that the mutation in cul1-7 does not indirectly affect RBX1 levels. When RBX1 was immunoprecipitated from equal amounts of total protein from wild-type and mutant extracts, CUL1 could be visualized from wild-type extracts, but no cul1-7 protein was detected from mutant extracts (data not shown). Because extracts from cul1-7 have less cul1-7 protein compared with wild-type extracts on a total protein basis (Figure 8A), we performed the experiment controlling for CUL1 input by immunoprecipitating from twice as much total protein from cul1-7 extracts than from wild-type extracts. The CUL1 input is roughly equivalent, with slightly more CUL1 than cul1-7 according to densitometry measurements (Figure 9D and data not shown). Under these conditions, cul1-7 is visible in the RBX1 immunoprecipitation; however, there is less (∼40%) than in IPs from CUL1 plants. This difference cannot simply be due to input (as there is only ∼20% less cul1-7 input), indicating that cul1-7 interaction with RBX1 is also impaired in vivo.
RBX1 levels appear equal in the two genotypes (Figure 9C), indicating that the differences in CUL1 levels are not due to reduced RBX1 levels. Several anti-CUL1 immunoreactive species with molecular weights greater than CUL1 appeared to co-IP with RBX1, from both mutant and wild-type extracts. The identity of these species is not known, and they were not immunoreactive with anti-ubiquitin antibodies (data not shown). The species that appeared just above 95 kDa, also appeared in the anti-FLAG IP from the complementation lines (Figure 4C), adding to the validity that species could represent modified species of CUL1.
On the basis of the results of Figure 8A and evidence that RBX1 abundance affects total CUL1 protein levels (Gray et al. 2002; Lechner et al. 2002), we hypothesized that unmodified cul1-7 is less stable in vivo than CUL1. We performed a cycloheximide degradation assay over a 12-hr time course for CUL1 and cul1-7 and determined CUL1 levels as described above. Because there is approximately twice as much CUL1 in wild type as in cul1-7, twice the amount of total protein was loaded in cul1-7 lanes. The amount of total CUL1 does not significantly change over this time course; however, half of the total cul1-7 protein is degraded in 12 hr (Figure 9E). We did not analyze the stability of either CUL1 or CUL1RUB singly in these experiments, but rather total CUL1, because CUL1 could enter the CUL1RUB pool and vice versa during the course of the experiment thereby confounding the interpretation.
DISCUSSION
SCF ligases are of special significance in plants. The large number of potential F-box proteins (∼700 in Arabidopsis and ∼700 in rice) (Gagne et al. 2002; Jain et al. 2007) suggests broad roles for SCFs throughout the life of the organism. Genetic screens have revealed that mutations in specific F-box protein-encoding genes play critical roles in multiple hormone signaling pathways, in cell division, and in developmental processes not yet linked to a specific hormonal pathway. For example, branching, flowering, and pathogen response are dependent on the F-box proteins MAX2, UFO, and SON1, respectively (Samach et al. 1999; Zhao et al. 1999; Kim and Delaney 2002; Stirnberg et al. 2002). Significantly, hormone perception and proteolysis in several cases, such as in auxin and JA signaling pathways, are directly linked, in that hormone binding to the F-box protein regulates substrate ubiquitylation (Napier 2005; Chini et al. 2007; Thines et al. 2007). Finally, mutations in proteins that regulate SCF function have pleiotropic phenotypes that severely affect plant development, such as the csn mutants, which are defective in the ability to remove RUB/Nedd8 from CUL1 and are seedling lethal (Hotton and Callis 2008), again demonstrating the importance of SCF function.
Mutations in various SCF components and factors regulating SCF activity have phenotypic differences that have not been reconciled, suggesting that we do not understand completely the regulation of SCF activity in plants. Thus, the isolation and characterization of multiple viable, partial loss-of-function alleles affecting SCF function will greatly facilitate studies on determining the scope of processes that regulate SCF ligases.
Toward this goal of understanding SCF function, we report here a new recessive allele of CUL1 that causes misregulation of SCF complexes at the CUL1 C terminus. The lesion in cul1-7 results in a T510I change near the C terminus, and T510 is predicted to be in a region of CUL1 that binds RBX1. An allelism test indicates that this mutation is responsible for the observed phenotypes, and partial complementation with CUL1-FLAG also supports this conclusion.
Several alleles of CUL1 in Arabidopsis have been reported previously, and in contrast to cul1-7, their changes map to the N-terminal region: two point mutations with semidominant phenotypes, axr6-1 and axr6-2, that disrupt interaction with ASK1 and are homozygous lethal (Hellmann et al. 2003); a recessive, viable temperature-sensitive allele, axr6-3, that also disrupts ASK1 binding (Quint et al. 2005); cul1-6, a recessive viable allele that affects CAND1 interaction (Moon et al. 2007); and cul1-1 to cul1-4, four T-DNA insertional alleles that are homozygous embryo lethal and display phenotypes as heterozygotes (Shen et al. 2002; Hellmann et al. 2003).
Responses to several hormones known to require SCF function were assayed in all three viable cul1 mutant backgrounds to directly compare their responses. Like the other alleles, cul1-7 has altered responses to auxin and slowed Aux/IAA degradation. Aux/IAA degradation rates are similarly affected in cul1-7 and axr6-3 (Quint et al. 2005), with four- to fivefold longer half-lives. This difference is also comparable to that seen in axr1 mutant backgrounds, where the RUB conjugation pathway is compromised (Zenser et al. 2003). In axr6-1/+ and axr6-2/+ seedlings, Aux/IAA degradation rates were more mildly affected (Hellmann et al. 2003). In contrast to auxin responses, GA responses were previously reported to be unaffected in axr6-3 (Quint et al. 2005) and not determined for cul1-6 (Moon et al. 2007). Here, we compare the GA response for all three cul1 alleles and show that all have reduced responses to GA. As expected for a temperature-sensitive allele, axr6-3 is similar to wild type at 20°, but less sensitive to GA at 28°. Surprisingly, cul1-7 also shows a strong temperature dependency, and in contrast to the other cul1 alleles, GA had no effect on hypocotyl length at 28°.
While cul1-7 was isolated because of slower degradation of an IAA1-LUC fusion protein expressed from a transgene, we confirmed the prediction that the degradation of endogenous SCF substrates distinct from Aux/IAA proteins would also be affected, given that the CUL1 subunit is shared among SCF ligases. The degradation rate of an endogenous SCF substrate, RGA, a DELLA protein, was slowed in cul1-7 seedlings. In wild-type seedlings, RGA is short lived and has a much shorter half-life in the presence of exogenous GA (Willige et al. 2007). To understand how RGA is degraded in the cul1-7 mutant and to avoid the consequences of auxin's influence on GA metabolism, which would be altered in cul1-7, we examined RGA degradation in seedlings that had been pretreated with paclobutrazol (PAC), a GA biosynthesis inhibitor. In wild-type seedlings, RGA degradation still occurs, but is much slower in the presence of PAC. Whether RGA is degraded in the absence of GA is not known. The slow, but measurable, degradation observed here could be the result of PAC's inability to completely eliminate endogenous GA and/or result from a GA-independent degradation mechanism. When exogenous GA is added to PAC-treated CUL1 seedlings, the degradation rate of RGA increases dramatically. In cul1-7, RGA degradation rate is also affected by GA application (in the presence of PAC), but with or without added GA it is still slower than wild type. This demonstrates that degradation of a known endogenous substrate of an SCF E3 ligase is slowed in cul1-7.
In addition, we demonstrate that red light-induced degradation of PhyA depends on CUL1. COP1 has been shown to ubiquitylate PhyA in vitro, and in cop1-6 and cop1-4, PhyA protein is stabilized (Seo et al. 2004). However, in axr6-3 and cul1-6 PhyA degradation is slowed, so we asked whether cul1-7 similarly affects PhyA degradation and sought to quantitatively measure PhyA degradation rate. In cul1-7, PhyA was not detectably degraded over a 180-min time course compared to an ∼100-min half-life in CUL1. This result is comparable to the reported 130-min half-life of oat PhyA protein when seedlings are given a pulse of red light at the beginning of the time course before transfer to darkness (Shanklin et al. 1987), but contrary to the reported half-life of 30 min for Arabidopsis PhyA when seedlings are kept in continuous red light up to the time of sample collection (Hennig et al. 1999). Two F-box proteins, EID1 and AFR, have been shown to be required for far-red light signaling, suggesting that SCF ligases are important mediators of far-red response (Dieterle et al. 2001; Harmon and Kay 2003; Marrocco et al. 2006), although the substrates of these ligases are not known. While our results and those of others demonstrate a strong dependency of PhyA degradation on CUL1, they do not establish a direct connection. Thus, it is still uncertain if there are ligases in addition to COP1 that catalyze PhyA ubiquitylation in vivo.
cul1-7 and the other cul1 alleles differ dramatically in their dark-grown seedling phenotype and response to added GA. When grown at 28° in the dark, cul1-7 seedlings are photomorphogenic with a greatly reduced hypocotyl length compared with 20°, are unhooked with open cotyledons, and addition of GA has no effect on hypocotyl elongation. In contrast, axr6-3 hypocotyls are only slightly shorter at 28° than at 20°, and GA treatment is effective in further increasing hypocotyl length. cul1-6 is longer at 28° than at 20° like wild type. Several recent reports have shown that GA represses photomorphogenesis in the dark and that this response depends on degradation of DELLA proteins (Alabadi et al. 2004, 2008; Achard et al. 2007). Our data also support this model. The cul1-7 dark-grown phenotype suggests that hyperaccumulation of DELLA proteins in the dark promotes photomorphogenesis, and exogenous GA is unable to accelerate sufficient degradation to block the photomorphogenic program promoted by the DELLA proteins. Thus, the cul1-7 allele specifically is a valuable allele for dissecting the role of SCF in light signaling because of its unique phenotype in the dark.
To begin to understand the molecular mechanism responsible for reduced SCF function in cul1-7, we determined cul1-7 protein levels, degradation rate, extent of RUB modification, and ability to interact with RBX1. All reported alleles seem to affect the abundance of total CUL1 protein. Despite all mutant backgrounds slowing degradation of tested substrates, axr6-1 and axr6-2 have an increase in abundance of both unmodified and RUB-modified CUL1 (Hellmann et al. 2003), cul1-6 has an increase in unmodified cul1-6, but not modified (Moon et al. 2007), while axr6-3 and cul1-7 have reduced levels of only unmodified cul1 (Quint et al. 2005 and this work). The semidominant phenotype of the insertional alleles, cul1-3 and cul1-4, suggests haplo-insufficiency at the CUL1 locus (Shen et al. 2002; Dharmasiri et al. 2003), and thus the reduction of CUL1 levels in axr6-3 and cul1-7 could contribute to the overall defect in these mutants. However, together these studies indicate that there is no simple relationship in vivo between steady state CUL1 levels, extent of RUB modification, and SCF activity.
We explored how the mutation in cul1-7 affects cul1-7 abundance and found that cul1-7 was significantly less stable than wild type, accounting for its lower accumulation than wild type. We also demonstrated that cul1-7 is defective in RBX1 binding. Previous studies have linked RBX1 and CUL1 levels. In RBX1 dsRNA lines (Lechner et al. 2002), CUL1 levels are severely reduced. In 35S:RBX1 overexpressing lines, CUL1 levels increased with nearly all in the RUB-modified form (Gray et al. 2002). Taken together, these results suggest a model that RBX1-CUL1 interaction plays a role in regulating CUL1 abundance and that CUL1-RBX1 is a stable subcomplex of the SCF.
Multiple alleles of CUL1 are useful in dissecting SCF function in vivo. The cul1-7 allele is a valuable addition to the collection of cul1 alleles by being the only one reported to directly affect subunit interactions at the CUL1 C terminus. Thus, the molecular consequences in the cul1-7 background may be different than in the other viable cul1 mutants, axr6-3 and cul1-6, which are different from each other. In support of this idea, cul1-7 plants have phenotypic differences from previously described cul1 mutants. The diversity of phenotypes from the few reported cul1 alleles demonstrates the complexity of SCF function in plants and indicates that analyses of SCF substrates should include multiple cul1 alleles to approach a more accurate estimate of SCF contribution. Further analyses of SCF function in plants could likely benefit from isolation and characterization of even more mutant cul1 alleles than are currently available.
Acknowledgments
The authors thank members of the Callis laboratory for helpful discussion and the University of California-Davis Controlled Environment Facility for assistance with the propagation of transgenic plants. We thank Bayer Corporation, Berkeley, CA, for their equipment donation of a luminometer that made this work possible. We thank Raymond Tam, Mai Dao, Stephanie Lochhead, and Linh Tran for assistance with plant propagation. We also thank Clark Lagarias for anti-PhyA antibodies and helpful discussions, Mark Estelle and William Gray for anti-CUL1 antibodies, and Steffan Abel for anti-IAA3 antibodies. We thank William Gray and Yunde Zhao for seed and Daoxin Xie for CUL1-FLAG DNA. We thank Mark Wogulis for assistance with modeling and Ellen Martin-Tryon for mapping primers and excellent advice. The authors gratefully acknowledge the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (contract DE-FG02-03ER15416) for funding the characterization of cul1-7 and a grant from the National Science Foundation (IBN 0212659) for development of the genetic screen and the Paul K. and Ruth R. Stumpf Endowed Professorship in Plant Biochemistry (to J.C.). This work was also supported in part by the National Science Foundation (IBN-0348814) (to T.-p.S.).
References
- Achard, P., L. Liao, C. Jiang, T. Desnos, J. Bartlett et al., 2007. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadi, D., J. Gil, M. A. Blazquez and J. L. Garcia-Martinez, 2004. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadi, D., J. Gallego-Bartolome, L. Orlando, L. Garcia-Carcel, V. Rubio et al., 2008. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53 324–335. [DOI] [PubMed] [Google Scholar]
- Alonso-Peral, M. M., H. Candela, J. C. del Pozo, A. Martinez-Laborda, M. R. Ponce et al., 2006. The HVE/CAND1 gene is required for the early patterning of leaf venation in Arabidopsis. Development 133 3755–3766. [DOI] [PubMed] [Google Scholar]
- Biswas, K. K., C. Ooura, K. Higuchi, Y. Miyazaki, V. Van Nguyen et al., 2007. Genetic characterization of mutants resistant to the antiauxin p-chlorophenoxyisobutyric acid reveals that AAR3, a gene encoding a DCN1-like protein, regulates responses to the synthetic auxin 2,4-dichlorophenoxyacetic acid in Arabidopsis roots. Plant Physiol. 145 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein, G., D. Ganoth and A. Hershko, 2006. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc. Natl. Acad. Sci. USA 103 11515–11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick, M., S. R. Lochhead, A. Honda, S. Palmer and J. Callis, 2004. RELATED TO UBIQUITIN1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell 16 2418–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu, D. R., and E. T. Kipreos, 2008. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 3 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo, T., and M. Pagano, 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5 739–751. [DOI] [PubMed] [Google Scholar]
- Cheng, Y., X. Dai and Y. Zhao, 2004. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol. 135 1020–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew, E.-H., and T. Hagen, 2007. Substrate-mediated regulation of cullin neddylation. J. Biol. Chem. 282 17032–17040. [DOI] [PubMed] [Google Scholar]
- Chini, A., S. Fonseca, G. Fernandez, B. Adie, J. M. Chico et al., 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666–673. [DOI] [PubMed] [Google Scholar]
- Chuang, H. W., W. Zhang and W. M. Gray, 2004. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCFTIR1 ubiquitin ligase. Plant Cell 16 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona, A., D. L. Chen, K. C. Yeh and S. Abel, 2000. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope, G. A., and R. J. Deshaies, 2003. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114 663–671. [DOI] [PubMed] [Google Scholar]
- del Pozo, J. C., and M. Estelle, 1999. The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J. C., C. Timpte, S. Tan, J. Callis and M. Estelle, 1998. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280 1760–1763. [DOI] [PubMed] [Google Scholar]
- del Pozo, J. C., S. Dharmasiri, H. Hellmann, L. Walker, W. M. Gray et al., 2002. AXR1–ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri, N., S. Dharmasiri and M. Estelle, 2005. a The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., S. Dharmasiri, D. Weijers, E. Lechner, M. Yamada et al., 2005. b Plant development is regulated by a family of auxin receptor F-box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, S., N. Dharmasiri, H. Hellmann and M. Estelle, 2003. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 22 1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle, M., Y. C. Zhou, E. Schafer, M. Funk and T. Kretsch, 2001. EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 15 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., H. S. Jung and T. P. Sun, 2001. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., S. G. Thomas, J. Hu, C. M. Steber and T. P. Sun, 2004. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher, K. A., and J. Callis, 2007. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 99 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher, K. A., R. Saw, J. Brown and J. Callis, 2006. Diversity in the degradation and auxin responsiveness of Arabidopsis Aux/IAA proteins. Plant Cell 18 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elich, T. D., and J. C. Lagarias, 1987. Phytochrome chromophore biosynthesis: both 5-aminolevulinic acid and biliverdin overcome inhibition by gabaculine in etiolated Avena sativa L. seedlings. Plant Physiol. 84 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle, M., and C. Somerville, 1987. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206 200–206. [Google Scholar]
- Feng, S., Y. Shen, J. A. Sullivan, V. Rubio, Y. Xiong et al., 2004. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16 1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet, C. M., and T.-p. Sun, 2005. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85. [DOI] [PubMed] [Google Scholar]
- Frigerio, M., D. Alabadi, J. Perez-Gomez, L. Garcia-Carcel, A. L. Phillips et al., 2006. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, M., Y. Zhang, J. McCarville, T. Ohta and Y. Xiong, 2000. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20 8185–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J. M., B. P. Downes, S. H. Shiu, A. M. Durski and R. D. Vierstra, 2002. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J. M., J. Smalle, D. J. Gingerich, J. M. Walker, S. D. Yoo et al., 2004. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg, S. J., T. C. Cascio, S. D. Shumway, K. C. Garbutt, J. Liu et al., 2004. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119 517–528. [DOI] [PubMed] [Google Scholar]
- Gray, W. M., J. C. del Pozo, L. Walker, L. Hobbie, E. Risseeuw et al., 1999. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W. M., S. Kepinski, D. Rouse, O. Leyser and M. Estelle, 2001. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414 271–276. [DOI] [PubMed] [Google Scholar]
- Gray, W. M., H. Hellmann, S. Dharmasiri and M. Estelle, 2002. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W. M., P. R. Muskett, H. W. Chuang and J. E. Parker, 2003. Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon, F. G., and S. A. Kay, 2003. The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr. Biol. 13 2091–2096. [DOI] [PubMed] [Google Scholar]
- Hellmann, H., L. Hobbie, A. Chapman, S. Dharmasiri, N. Dharmasiri et al., 2003. Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligase in auxin regulation of embryogenesis. EMBO J. 22 3314–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig, L., C. Buche, K. Eichenberg and E. Schafer, 1999. Dynamic properties of endogenous phytochrome A in Arabidopsis seedlings. Plant Physiol. 121 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, L., M. McGovern, L. R. Hurwitz, A. Pierro, N. Y. Liu et al., 2000. The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127 23–32. [DOI] [PubMed] [Google Scholar]
- Hotton, S., and J. Callis, 2008. Regulation of Cullin RING ligases. Annu. Rev. Plant Biol. 59 467–489. [DOI] [PubMed] [Google Scholar]
- Jain, M., A. Nijhawan, R. Arora, P. Agarwal, S. Ray et al., 2007. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado, S., S. Diaz-Trivino, Z. Abraham, C. Manzano, C. Gutierrez et al., 2008. SKP2A, an F-box protein that regulates cell division, is degraded via the ubiquitin pathway. Plant J. 53 828–841. [DOI] [PubMed] [Google Scholar]
- Kamura, T., M. N. Conrad, Q. Yan, R. C. Conaway and J. W. Conaway, 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir, L., A. L. Schilmiller, P. E. Staswick, S. Y. He and G. A. Howe, 2008. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski, S., and O. Leyser, 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Kim, H. S., and T. P. Delaney, 2002. Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz, T., Y. C. Chou, A. R. Willems, N. Meyer-Schaller, M. L. Hecht et al., 2008. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell 29 23–35. [DOI] [PubMed] [Google Scholar]
- Kurz, T., N. Ozlu, F. Rudolf, S. M. O'Rourke, B. Luke et al., 2005. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435 1257–1261. [DOI] [PubMed] [Google Scholar]
- Lammer, D., N. Mathias, J. M. Laplaza, W. Jiang, Y. Lui et al., 1998. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 12 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, P. B., and J. D. Cancel, 2004. A recessive mutation in the RUB1-conjugating enzyme, RCE1, reveals a requirement for RUB modification for control of ethylene biosynthesis and proper induction of basic chitinase and PDF1.2 in Arabidopsis. Plant J. 38 626–638. [DOI] [PubMed] [Google Scholar]
- Lechner, E., D. Xie, S. Grava, E. Pigaglio, S. Planchais et al., 2002. The AtRBX1 protein is part of plant SCF complexes, and its down-regulation causes severe growth and developmental defects. J. Biol. Chem. 277 50069–50080. [DOI] [PubMed] [Google Scholar]
- Lechner, E., P. Achard, A. Vansiri, T. Potuschak and P. Genschik, 2006. F-box proteins everywhere. Curr. Opin. Plant Biol. 9 631–638. [DOI] [PubMed] [Google Scholar]
- Leyser, H. M. O., C. A. Lincoln, C. Timpte, D. Lammer, J. Turner et al., 1993. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364 161–164. [DOI] [PubMed] [Google Scholar]
- Leyser, O. H., F. B. Pickett, S. Dharmaseri and M. Estelle, 1996. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10 403–413. [DOI] [PubMed] [Google Scholar]
- Liu, J., M. Furukawa, T. Matsumoto and Y. Xiong, 2002. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1–SKP1 binding and SCF ligases. Mol. Cell 10 1511–1518. [DOI] [PubMed] [Google Scholar]
- Lo, S. C., and M. Hannink, 2006. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol. Cell. Biol. 26 1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., C. S. Gillmor and W. R. Scheible, 2000. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco, K., Y. Zhou, E. Bury, M. Dieterle, M. Funk et al., 2006. Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J. 45 423–438. [DOI] [PubMed] [Google Scholar]
- McGinnis, K. M., S. G. Thomas, J. D. Soule, L. C. Strader, J. M. Zale et al., 2003. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 1998. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14 381–385. [DOI] [PubMed] [Google Scholar]
- Michelmore, R., I. Paran and R. Kesseli, 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, K. W., M. J. Kwon, H. S. Park, Y. Park, S. K. Yoon et al., 2005. CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem. Biophys. Res. Commun. 334 867–874. [DOI] [PubMed] [Google Scholar]
- Moon, J., Y. Zhao, X. Dai, W. Zhang, W. M. Gray et al., 2007. A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol. 143 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, P., L. M. Walker, J. C. Young, A. Sonawala, C. Timpte et al., 2000. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier, R. M., 2005. TIRs of joy: new receptors for auxin. BioEssays 27 1213–1217. [DOI] [PubMed] [Google Scholar]
- Neff, M., J. Neff, J. Chory and A. Pepper, 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14 387–392. [DOI] [PubMed] [Google Scholar]
- O'Neill, D. P., and J. J. Ross, 2002. Auxin regulation of the gibberellin pathway in pea. Plant Physiol. 130 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh, M., W. Y. Kim, J. J. Moslehi, Y. Z. Chen, V. Chau et al., 2002. An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 3 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa, K., M. Matsumoto, M. Yada, T. Kamura, S. Hatakeyama et al., 2003. Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem. Biophys. Res. Commun. 303 1209–1216. [DOI] [PubMed] [Google Scholar]
- Park, J.-Y., H.-J. Kim and J. A. G. Kim, 2002. Mutation in domain II of IAA1 confers diverse auxin-relatd phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J. 32 669–683. [DOI] [PubMed] [Google Scholar]
- Petroski, M. D., and R. J. Deshaies, 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6 9–20. [DOI] [PubMed] [Google Scholar]
- Quint, M., and W. M. Gray, 2006. Auxin sigalling. Curr. Opin. Plant Biol. 9 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint, M., H. Ito, W. Zhang and W. M. Gray, 2005. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of the SCF ubiquitin ligase. Plant J. 43 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, J., N. Zenser, O. Leyser and J. Callis, 2001. Rapid degradation of Auxin/Indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, C., J. Pan, W. Peng, P. Genschik, L. Hobbie et al., 2005. Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 42 514–524. [DOI] [PubMed] [Google Scholar]
- Ren, H., A. Santner, J. C. del Pozo, J. A. Murray and M. Estelle, 2008. Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant J. 53 705–716. [DOI] [PubMed] [Google Scholar]
- Rouse, D., P. Mackay, P. Stirnberg, M. Estelle and O. Leyser, 1998. Changes in auxin response from mutations in an AUX/IAA gene. Science 279 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., E. Dewey, W. Gray, L. Hobbie, J. Turner et al., 1998. The TIR protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., J. E. Klenz, S. E. Kohalmi, E. Risseeuw, G. W. Haughn et al., 1999. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20 433–445. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., G. Serino, J. Callis, W. L. Crosby, S. Lyapina et al., 2001. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292 1379–1382. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., B. C. Willige, M. Zourelidou and E. M. Dohmann, 2009. Examining protein stability and its relevance for plant growth and development. Methods Mol. Biol. 479 1–25. [DOI] [PubMed] [Google Scholar]
- Seo, H. S., E. Watanabe, S. Tokutomi, A. Nagatani and N. H. Chua, 2004. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin, J., M. Jabben and R. D. Vierstra, 1987. Red light-induced formation of ubiquitin-phytochrome conjugates: identification of possible intermediates of phytochrome degradation. Proc. Natl. Acad. Sci. USA 84 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W. H., Y. Parmentier, H. Hellmann, E. Lechner, A. W. Dong et al., 2002. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A. L., C. N. Ciampaglio and T. P. Sun, 1998. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A. L., H. S. Jung, A. Dill, H. Kawaide, Y. Kamiya et al., 2001. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and R. D. Vierstra, 2004. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Stirnberg, P., K. van De Sande and H. Leyser, 2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129 1131–1141. [DOI] [PubMed] [Google Scholar]
- Stone, S. L., H. Hauksdottir, A. Troy, J. Herschleb, E. Kraft et al., 2005. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T. P., and F. Gubler, 2004. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 197–223. [DOI] [PubMed] [Google Scholar]
- Tan, X., L. I. Calderon-Villalobos, M. Sharon, C. Zheng, C. V. Robinson et al., 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645. [DOI] [PubMed] [Google Scholar]
- Tatematsu, K., S. Kumagai, H. Muto, A. Sato, M. K. Watahiki et al., 2004. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, B., L. Katsir, M. Melotto, Y. Niu, A. Mandaokar et al., 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448 661–665. [DOI] [PubMed] [Google Scholar]
- Thomann, A., M. Dieterle and P. Genschik, 2005. Plant CULLIN-based E3s: phytohormones come first. FEBS Lett. 579 3239–3245. [DOI] [PubMed] [Google Scholar]
- Tian, Q., and J. Reed, 1999. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126 711–721. [DOI] [PubMed] [Google Scholar]
- Tian, Q., N. J. Uhlir and J. W. Reed, 2002. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte, C., A. K. Wilson and M. Estelle, 1994. The Axr2–1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S., G. Hagen and T. Guilfoyle, 2001. Aux/IAA proteins are active repressors and their stability and activity are modulated by auxin. Plant Cell 13 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, T. A., R. Neal, A. O. Merlo, M. Honma, G. R. Hicks et al., 2006. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 142 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and X. W. Deng, 2003. The COP9 signalosome. Annu. Rev. Cell. Dev. Biol. 19 261–286. [DOI] [PubMed] [Google Scholar]
- Weissman, A. M., 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2 169–178. [DOI] [PubMed] [Google Scholar]
- Wen, C. K., and C. Chang, 2002. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige, B. C., S. Ghosh, C. Nill, M. Zourelidou, E. M. Dohmann et al., 2007. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang, C. M., P. M. Chandler, J. J. Smith and J. J. Ross, 2004. Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiol. 134 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang, C. M., and J. J. Ross, 2001. Auxin promotes gibberellin biosynthesis in decaptitated tobacco plants. Planta 214 153–157. [DOI] [PubMed] [Google Scholar]
- Woodward, A. W., S. E. Ratzel, E. E. Woodward, Y. Shamoo and B. Bartel, 2007. Mutation of E1-CONJUGATING ENZYME-RELATED1 decreases RELATED TO UBIQUITIN conjugation and alters auxin response and development. Plant Physiol. 144 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley, C. K., N. Zenser, J. Ramos, D. Rouse, O. Leyser et al., 2000. Degradation of Aux/IAA proteins is essential for normal auxin signaling. Plant J. 21 553–562. [DOI] [PubMed] [Google Scholar]
- Xu, L., F. Liu, E. Lechner, P. Genschik, W. L. Crosby et al., 2002. The SCFCOI ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., J. Lim, J. M. Dale, H. Chen, P. Shinn et al., 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846. [DOI] [PubMed] [Google Scholar]
- Yang, X., S. Lee, J. H. So, S. Dharmasiri, N. Dharmasiri et al., 2004. The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J. 40 772–782. [DOI] [PubMed] [Google Scholar]
- Yang, X., J. Zhou, L. Sun, Z. Wei, J. Gao et al., 2007. Structural basis for DCN-1's function in protein neddylation. J. Biol. Chem. 282 24490–24494. [DOI] [PubMed] [Google Scholar]
- Yoo, S. D., Y. H. Cho, G. Tena, Y. Xiong and J. Sheen, 2008. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser, N., K. A. Dreher, S. R. Edwards and J. Callis, 2003. Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J. 35 285–294. [DOI] [PubMed] [Google Scholar]
- Zenser, N., A. Ellsmore, C. Leasure and J. Callis, 2001. Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D. H., M. Yang, J. Solava and H. Ma, 1999. The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev. Genet. 25 209–223. [DOI] [PubMed] [Google Scholar]
- Zheng, J., X. Yang, J. M. Harrell, S. Ryzhikov, E. H. Shim et al., 2002. a CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 10 1519–1526. [DOI] [PubMed] [Google Scholar]
- Zheng, N., B. A. Schulman, L. Z. Song, J. J. Miller, P. D. Jeffrey et al., 2002. b Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416 703–709. [DOI] [PubMed] [Google Scholar]