Abstract
Self-incompatibility (SI) in the Brassicaceae plant family is controlled by the SRK and SCR genes situated at the S locus. A large number of S haplotypes have been identified, mainly in cultivated species of the Brassica and Raphanus genera, but recently also in wild Arabidopsis species. Here, we used DNA sequences from the SRK and SCR genes of the wild Brassica species Brassica cretica, together with publicly available sequence data from other Brassicaceae species, to investigate the evolutionary relationships among S haplotypes in the Brassicaceae family. The results reveal that wild and cultivated Brassica species have similar levels of SRK diversity, indicating that domestication has had but a minor effect on S-locus diversity in Brassica. Our results also show that a common set of S haplotypes was present in the ancestor of the Brassica and Arabidopsis genera, that only a small number of haplotypes survived in the Brassica lineage after its separation from Arabidopsis, and that diversification within the two Brassica dominance classes occurred after the split between the two lineages. We also found indications that recombination may have occurred between the kinase domain of SRK and the SCR gene in Brassica.
TO avoid self-fertilization, different kinds of self-incompatibility (SI) mechanisms have evolved in many plant families (Richards 1997). In most cases, SI is controlled by a single locus, the S locus, which, even though it often contains several genes, is inherited as a single Mendelian locus (Silva and Goring 2001); for this reason, S alleles are often referred to as S haplotypes (Boyes and Nasrallah 1993). In the Brassicaceae, which has the sporophytic type of SI where the pollen SI phenotype is determined by the S-locus genotype of the diploid parent, two S-locus genes, SRK and SCR, encode the maternal (pistil) and paternal (pollen) specificities, respectively (Stein et al. 1991; Schopfer et al. 1999; Suzuki et al. 1999). The SRK receptor consists of three domains with different functions: an extracellular S domain (encoded by exon 1 in SRK), which is the center for recognition of SCR; a transmembrane domain (exon 2) that passes through the plasma membrane; and an intracellular kinase domain (exons 4–7), which initiates the signaling cascade in the stigma cells (see Takayama and Isogai 2005). The SCR gene product is a small soluble protein molecule present at the pollen surface where it interacts with the S domain of its cognate SRK protein (Takayama et al. 2001). Because of the strong frequency-dependent selection acting on the SI system, the S locus typically maintains a large number of haplotypes (Lawrence 2000), and levels of diversity at both synonymous and nonsynonymous sites are high in the S domain of SRK and in SCR (e.g., Awadalla and Charlesworth 1999; Charlesworth et al. 2003; Takuno et al. 2007). Furthermore, in both Brassica and Arabidopsis, synonymous diversity is elevated in a region encompassing several tens of kilobases beyond the actual targets of selection due to the low level of recombination at the S locus (Kamau and Charlesworth 2005; Castric and Vekemans 2007; Takuno et al. 2007).
There is now firm evidence that the structural elements of SI are homologous among cultivated Brassicaceae species (including Brassica oleracea, Brassica rapa, Brassica nigra, and Raphanus sativus) and the wild Arabidopsis lyrata, indicating that SI has a single origin within the Brassicaceae family (Kusaba et al. 2001; Schierup et al. 2001). Nevertheless, there are characteristic differences in the SI systems between the Brassica and Arabidopsis genera. For example, the Brassica S haplotypes can be divided into two distinct dominance classes with class I haplotypes being dominant over class II haplotypes in pollen but codominant in the stigma (Nasrallah et al. 1991). By contrast, dominance relationships among S haplotypes are more complex in Arabidopsis, where four distinct dominance classes have been identified to date (Prigoda et al. 2005). Furthermore, while data from natural populations support the notion that recombination is effectively abolished at the S locus in Arabidopsis (Charlesworth et al. 2003; Kamau and Charlesworth 2005; Hagenblad et al. 2006), there are indications that some form of recombination or gene conversion has occurred within Brassica (Awadalla and Charlesworth 1999; Takuno et al. 2007).
Two alternative hypotheses on the evolution of the S locus within the Brassicaceae family have been presented (Fobis-Loisy et al. 2004): The first proposes that after the Arabidopsis/Brassica split, diversification of S haplotypes took place independently in the two descendant lineages. The second hypothesis suggests that extant S haplotypes trace their origin back to a common Arabidopsis/Brassica ancestor and that the Brassica lineage lost most of its ancient alleles after its divergence from Arabidopsis. While the time of diversification between class I and class II haplotypes within the Brassica genus has been estimated to be at least 40 million years ago (MYA; Uyenoyama 1995), clearly preceding the split between the Arabidopsis and Brassica lineages, which occurred ∼20 MYA (Yang et al. 1999a; Koch et al. 2001), the historical relationship between extant Arabidopis and Brassica S haplotypes has not been fully resolved (Schierup et al. 2001). Moreover, bottlenecks, which may occur during domestication, can have a significant effect on the number and phylogenetic distribution of S-locus lineages (e.g., Igic et al. 2004). As there have been no S-locus DNA sequence data available from any wild Brassica species, it has not been clear to what extent the domestication of cultivated Brassica species has biased the results of previous studies of this genus.
In this study, we have performed phylogenetic analyses of DNA sequence data from the SRK and SCR genes of the wild species B. cretica in combination with publicly available sequence information from cultivated Brassica species and from the wild A. lyrata. We have specifically addressed three questions: (i) Has domestication affected the pattern of S-locus gene genealogies in cultivated Brassica species?, (ii) Have S-locus haplotypes evolved independently in the Arabidopsis and Brassica lineages, or is there shared ancestry across genera?, and (iii) Is there any phylogenetic evidence of historical recombination at the S locus in the wild B. cretica?
MATERIALS AND METHODS
Study species:
B. cretica is a diploid (2n = 18), perennial chasmophyte that belongs to an alliance of Brassica species, including wild B. oleracea, the members of which all carry the Brassica C genome (Harberd 1972; Kianian and Quiros 1992; von Bothmer et al. 1995). B. cretica is found in the Aegean region, mainly on the island of Crete, where it grows in isolated populations in ravines and gorges (Snogerup et al. 1990). As a result of severely restricted gene flow, Cretan populations of B. cretica show an exceptionally high level of neutral genetic differentiation both at molecular marker loci and for quantitative traits (Widén et al. 2002; Edh et al. 2007).
DNA sequencing:
A total of 40 individuals originating from four Cretan B. cretica populations from Crete were included in the study (Edh et al. 2009, accompanying article in this issue). Approximately 100 mg leaf tissue was used for whole-genome DNA extraction with a DNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. Exons 4–7 of the kinase domain of SRK and parts of exons 1 and 2 of SCR were amplified and sequenced as described in Edh et al. (2009). As S-domain-related regions are also found in S-locus genes other than SRK (e.g., SLG; Stein et al. 1991), we chose to work with the SRK kinase domain instead of the S domain to avoid inadvertent PCR amplification of nonhomologous regions. Haplotypes were identified on the basis of total sequence identity, including both introns and exons, in BioEdit version 7.0.5.3 (Hall 1999) after alignment using the Clustal W algorithm (Thompson et al. 1994). For SRK, the 46 haplotypes identified in Edh et al. (2009) were used in the phylogenetic analysis. For SCR, only class II haplotypes could be amplified, and six class II haplotypes were used in this study. DNA sequences from corresponding SRK and SCR regions from A. lyrata, B. oleracea, B. rapa, and R. sativus were obtained from GenBank (supplemental Table 1).
Phylogenetic analyses:
An appropriate substitution model for each data set was found using Modeltest version 3.7 (Posada and Crandall 1998). For SRK, exons 4–7 could be reliably aligned from A. lyrata, B. cretica, B. oleracea, B. rapa, and R. sativus (supplemental Table 1). One 9-bp deletion, which was found in exon 7 in all class II haplotypes and in one class I haplotype (BcrSRK111) from B. cretica, was excluded from the analysis. Two phylogenetic analyses were performed on slightly different versions of the SRK data set. First, a neighbor-joining (NJ) tree based on the TrN substitution model with pairwise deletion of gaps was constructed from a total sequence length of 618 bp using Mega 3.1 software (Kumar et al. 2004). No sequences appropriate for rooting the tree were found and the tree was left unrooted. Statistical support for individual nodes was estimated from 1000 bootstrap resamples. Second, a maximum-likelihood (ML) tree based on the GTR + I + G substitution model, with gamma correction factor G = 1.4874 and the proportion of invariable sites I = 0.2675, was constructed from a subset of 310 bp of SRK sequence (equal length of all sequences) in PAUP*, version 4.0b10 (Swofford 2002). The ML analysis was carried out with a heuristic search approach using the TBR swapping algorithm and the steepest descent option. One thousand bootstrap resamples were used to determine statistical support.
Separate SRK and SCR ML trees were constructed for B. cretica, B. oleracea, and B. rapa class II haplotypes (supplemental Table 1). Here, 485 bp of exons 4–7 from SRK and 174 bp of exon sequence from the SCR gene were aligned separately for the respective genes. The substitution models TVM + G (G = 0.6349) and TVM with equal rates for all sites were used for the SRK class II and SCR class II data sets, respectively. The ML tree was found as described above. For both trees, a class I B. oleracea haplotype (Bol7; supplemental Table 1) was used as the outgroup. Statistical support for individual nodes was estimated from 1000 bootstrap resamples.
The hypothesis of the absence of historical recombination at the S locus was tested by investigating the topological congruency between the SRK class II and SCR class II phylogenetic trees. The difference in log likelihood (Δl) between the ML SRK tree and an SRK tree having the same topology as the ML SCR tree, and vice versa, was calculated. For each gene, alternative trees having significantly different likelihoods were regarded as topologically different. The statistical significance of the difference in log likelihood was assessed on the basis of the standard error (SE) in log likelihood for each tree, as described by Kishino and Hasegawa (1989).
RESULTS
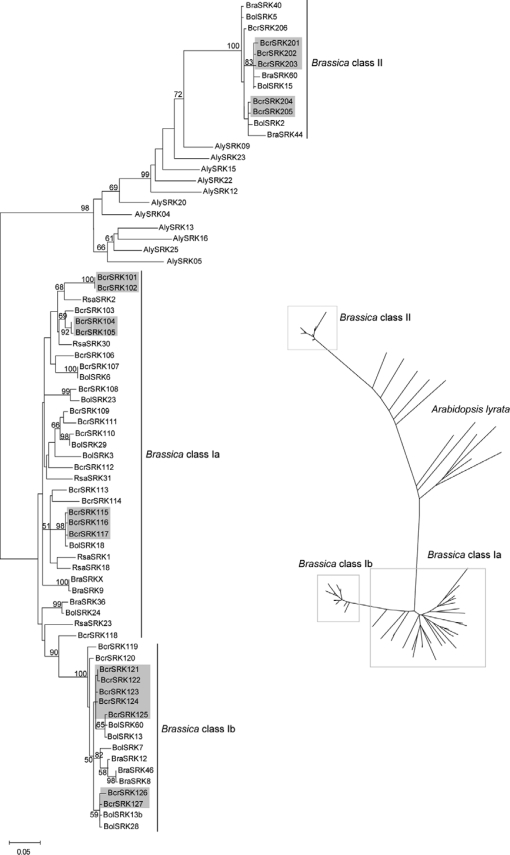
The ML tree of the SRK kinase domain displayed two distinct clusters corresponding to class I and class II haplotypes from Brassica and Raphanus species (Figure 1). A subset of the B. cretica, B. oleracea, and B. rapa class I haplotypes formed a separate, well-supported clade referred to here as class Ib, while the remaining class I haplotypes are referred to as class Ia. All three major groups of haplotypes (classes Ia, Ib, and II) included haplotypes originating from B. cretica, B. oleracea, and B. rapa, whereas R. sativus haplotypes were found only among the class Ia haplotypes. While all A. lyrata SRK haplotypes were well separated from the Brassica haplotypes, the Brassica class I and class II haplotypes did not form a monophyletic group (Figure 1). Overall, A. lyrata haplotypes displayed longer branches than Brassica or Raphanus haplotypes. The SRK NJ tree (not shown) was similar to the SRK ML tree.
Figure 1.—
Unrooted maximum-likelihood tree for SRK kinase domain sequences (exons 4–7) from B. cretica (Bcr), B. oleracea (Bol), B. rapa (Bra), R. sativus (Rsa), and A. lyrata (Aly). (Left) Bootstrap support values (from 1000 replicates) in tree are indicated when >50%. B. cretica haplotypes enclosed in shaded boxes belong to the same putative functional S haplotypes (Edh et al. 2009). (Right) Tree for improved visualization of relationships among major phylogenetic groups.
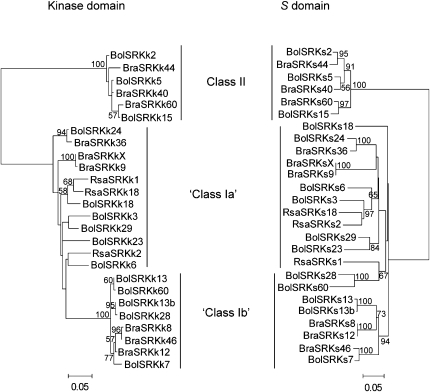
To further investigate the apparent subdivision of Brassica class I haplotypes into class Ia and class Ib, we used publicly available SRK sequence data from a total of 26 B. oleracea, B. rapa, and R. sativus haplotypes (supplemental Table 1) to construct separate phylogenetic trees for the kinase and S domains. ML trees were constructed using 330 bp from exons 4–7 of the kinase domain and 1147 bp from exon 1 of the S domain. The GTR + G (G = 0.7881) and the TrN + I + G (I = 0.1519, G = 0.8863) substitution models were used for the kinase and S domains, respectively. The ML trees were found as described above and bootstrap support values were derived from 1000 resamples. In the kinase domain tree, the three groups of SRK haplotypes (classes Ia, Ib, and II) were clearly evident (Figure 2). By contrast, the separation of class I haplotypes into class Ia and class Ib was not, however, obvious in the SRK S-domain ML tree (Figure 2). Instead, a well-supported clade was found that included those SRK haplotypes classified as class Ib in the kinase domain tree, except for the B. oleracea BolSRK28 and BolSRK60 haplotypes (Figure 2). These haplotypes were instead grouped with the putative class Ia haplotypes. Topological incongruence between the two phylogenies was further indicated by the statistically significant differences in log likelihood, both between the ML SRK kinase domain tree and a kinase domain tree with the SRK S-domain topology (Δl ± SE = −244 ± 35.4) and between the ML SRK S-domain tree and an S-domain tree with the SRK kinase domain topology (Δl ± SE = −1026 ± 63.5).
Figure 2.—
Maximum-likelihood trees of the kinase domain (left) and the S domain (right) of the SRK gene based on publicly available sequence data from B. oleracea (Bol), B. rapa (Bra), and R. sativus (Rsa) (supplemental Table 1). Both trees are unrooted and bootstrap support values are given when >50%. The division of class Ia and class Ib sequences is based on the kinase domain phylogeny.
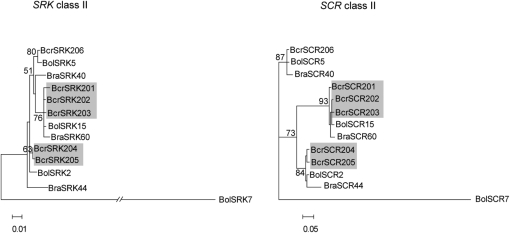
The ML trees of the class II haplotypes from the SRK and SCR genes displayed similar topologies even though the SCR tree was better resolved (Figure 3). In the SCR tree, and to a lesser extent in the SRK class II tree, three strongly supported clades were found, each of which included one of the three known B. oleracea/B. rapa class II haplotype pairs (BolSCR2 and BraSCR44, BolSCR5 and BraSCR40, and BolSCR15 and BraSCR60; Sato et al. 2003; Takuno et al. 2007). Each of the three clades also included either a single or two or three closely related B. cretica haplotypes. There was no significant likelihood difference between the SRK ML tree and an SRK tree with the SCR ML topology (Δl ± SE = −14.8 ± 8.4), whereas the ML SCR tree had a slightly but significantly higher likelihood than an SCR tree with the SRK ML topology (Δl ± SE = −44.3 ± 11.8).
Figure 3.—
Maximum-likelihood trees for B. cretica (Bcr), B. oleracea (Bol), and B. rapa (Bra) class II sequences from SRK (left) and SCR (right). The trees were rooted with the B. oleracea class I haplotypes BolSRK7 and BolSCR7, respectively. Bootstrap support values are indicated when >50%. B. cretica haplotypes enclosed in shaded boxes belong to the same putative functional S haplotype (Edh et al. 2009).
DISCUSSION
Brassica S-locus haplotypes:
Phylogenetic analysis of SRK kinase domain sequences shows tight clustering of all Brassica sequences according to dominance class (Figure 1). The class II cluster includes the three B. oleracea and three B. rapa haplotypes previously known to be pollen recessive. Since the B. cretica haplotypes form the same dominance class-specific clusters as the other Brassica species, we suggest that the dominance pattern is the same in wild B. cretica as in cultivated Brassica, with dominant class I and recessive class II haplotypes. In the absence of crossing data, we previously assigned B. cretica SRK haplotypes into putative groups of haplotypes with identical SI specificity on the basis of DNA sequence similarity (Edh et al. 2009). As revealed by the ML SRK kinase domain tree, clusters of similar haplotypes assigned to the same functional group are, in all cases except one, monophyletic (Figure 1). The only exception is the BcrSRK125 haplotype, which is not monophyletic with the BcrSRK121-124 haplotypes assigned to the same functional group. Thus, the BcrSRK125 is either a functionally distinct haplotype or belongs, together with the BcrSRK121-124 haplotypes, to an interspecific pair of functionally identical haplotypes, the other member of which would be either of the very similar B. oleracea haplotypes BolSRK13 and BolSRK60. A number of such interspecific haplotype pairs involving B. oleracea and B. rapa have been detected (Kimura et al. 2002; Sato et al. 2003; Takuno et al. 2007). Indeed, in the class II SCR tree, the clustering of interspecific triplets consisting of one haplotype from each of B. oleracea and B. rapa, together with one putatively functional B. cretica haplotype, is particularly evident (Figure 3). In fact, the entire reservoir of class II haplotypes in cultivated B. oleracea can be found within a single B. cretica population (Edh et al. 2009) and, thus, we apparently have not found any functional class II haplotypes not previously known from Brassica in the wild B. cretica. Several closely related pairs of B. cretica/B. oleracea SRK haplotypes may represent additional examples of such interspecific pairs, a finding that would be expected, given that the divergence between B. cretica and B. oleracea dates back <1 million years (Edh et al. 2007), compared to nearly 4 million years for the B. oleracea–B. rapa divergence (Inaba and Nishio 2002).
The presence of haplotypes from all three Brassica species in both class I and class II provides further evidence that previous observations of trans-specific polymorphisms at the S locus in cultivated Brassica species (Kusaba et al. 1997; Shiba et al. 2002; Takuno et al. 2007) extend to the wild B. cretica. Moreover, SRK lineages from the cultivated B. oleracea and B. rapa are widely dispersed among lineages from the wild B. cretica. Together with the observation of almost equal levels of SRK sequence polymorphism in wild and cultivated Brassica species (Edh et al. 2009), this suggests that domestication has not entailed any severe bottleneck, with subsequent reduction in the number and diversity of lineages, at the Brassica S locus (cf. Igic et al. 2004). The domestication history of B. oleracea is not known in detail although multiple domestication events as well as gene flow between wild and cultivated forms have been suggested (von Bothmer et al. 1995), situations that may have failed to create a significant bottleneck effect in cultivated Brassica.
Evolution of the Brassicaceae S locus:
Whereas there is good evidence that the origin of SI is monophyletic in the Brassicaceae, the phylogenetic relationship among S haplotypes in the different genera is less clear (Schierup et al. 2001; Fobis-Loisy et al. 2004). Previous observations of an apparently independent phylogenetic clustering of SRK haplotypes from A. lyrata and cultivated Brassica species (Schierup et al. 2001) have also seemed to favor the notion of separate diversification in the Arabidopsis and Brassica lineages. On the other hand, Schierup et al. (2001) identified a “deviant” A. lyrata SRK haplotype (Aly13-9/AlSRK09), which was more closely related to Brassica class II SRK haplotypes than to other A. lyrata haplotypes. In a recent study, Paetsch et al. (2006) found that putative SRK haplotypes from the self-incompatible Capsella grandiflora, which is more closely related to Arabidopsis than to Brassica (Yang et al. 1999b), clustered with A. lyrata haplotypes but separately from the Brassica haplotypes. From this observation, Paetsch et al. (2006) concluded that the S locus had evolved differently in the Brassica/Raphanus and Arabidopsis/Capsella lineages. However, their phylogenetic analysis included neither the A. lyrata AlSRK09 haplotype nor any Brassica class II SRK haplotypes. With additional SRK sequence data from B. cretica, especially from class II haplotypes, our study lends more support to a scenario in which there is sharing of ancient SRK haplotypes between the Arabidopsis and Brassica lineages. We found that all SRK sequences from the Brassica lineage (including the wild B. cretica) cluster in well-supported clades on the basis of dominance class, whereas the A. lyrata SRK sequences form a more diverse group with AlySRK09 most closely related, of all A. lyrata haplotypes, to the Brassica class II haplotypes (Figure 1). In particular, we find that, even though the phylogenetic tree is unrooted, irrespective of where the root would be placed, SRK haplotypes are apparently not monophyletic within Brassica. Thus, there are not only trans-specific, but also trans-generic polymorphisms at the SRK gene involving the Arabidopsis and Brassica lineages. Since the age of the last common ancestor of Arabidopsis and Brassica is estimated to be ∼20 million years (Yang et al. 1999a; Koch et al. 2001), our results are consistent with the estimated time of at least 40 million years since the split between the class I and class II lineages of the Brassica S locus (Uyenoyama 1995). Our data further show that extant SRK haplotypes from both wild and cultivated Brassica species have descended from only two of the lineages presumably present in the Arabidopsis–Brassica ancestor and that diversification of the Brassica haplotypes took place after the separation of the two genera.
Interestingly, the A. lyrata haplotypes AlySRK04 and AlySRK09, which are most closely related to the Brassica class I and II haplotypes, respectively (Figure 1; see also Schierup et al. 2001), belong to the same Arabidopsis dominance class (Prigoda et al. 2005), implying that dominance in Brassica arose after the split from Arabidopsis. The divergence of the Brassica class I haplotypes is believed to have begun ∼24 MYA (Uyenoyama 1995) perhaps coinciding with, or directly following, the separation of the Arabidopsis and Brassica lineages. Kusaba et al. (2001) found that the S locus occupies different chromosomal locations in Arabidopsis and Brassica, presumably as a result of a translocation. A translocation event involving a single S haplotype would have imposed a severe bottleneck on the S-locus region in the derived lineage. And, if the translocation happened at the time of the Arabidopsis–Brassica split, as proposed by Kusaba et al. (2001), this could explain the significant reduction in the number of ancient S haplotypes in the Brassica lineage (Fobis-Loisy et al. 2004), given that the chromosomal position of the S locus in Brassica represents the derived state with respect to the translocation.
An interesting observation in the SRK kinase domain tree is the presence of two phylogenetically separate subgroups among the Brassica class I haplotypes (Figure 1). In particular, the class Ib haplotypes formed a strongly supported, monophyletic clade and included haplotypes from all three Brassica species (B. cretica, B. oleracea, and B. rapa). In contrast, the class Ia group of haplotypes was not monophyletic (Figure 1). Using complete SRK haplotype sequence data from B. oleracea and B. rapa, the class Ib group was again apparent in the kinase domain tree (Figure 2). In the S-domain tree, two monophyletic subgroups of class I were found; however, they did not fully correspond to the class Ia and class Ib groups in the kinase domain tree because the B. oleracea BolSRK28 and BolSRK60 haplotypes clustered with the class Ia haplotypes in the S-domain tree (Figure 2). As the S domain is the center of recognition of SCR, it is possible that the two groups represent distinct dominance classes. We currently have no crossing data to support this hypothesis; however, Ockendon (1982) reported that the B. oleracea S6 haplotype (class Ia) was dominant to the B. oleracea S13 haplotype (class Ib) in the stigma but not in the pollen. On the other hand, Takuno et al. (2007) did not find any obvious phylogenetic separation of class Ia and class Ib haplotypes using a data set of 52 S-domain haplotypes from B. oleracea and B. rapa. Thus, from a phenotypic point of view, the status of the class Ia and Ib haplotypes remains obscure.
Recombination at the S locus:
Due to the requirement of matching pistil and pollen specificities, recombination within the S locus has been presumed to be very rare or entirely absent (e.g., Uyenoyama and Newbigin 2000). Data from Arabidopsis appear to support this notion: Population-based studies have found no traces of recombination at the S locus (Charlesworth et al. 2003; Kamau and Charlesworth 2005; Hagenblad et al. 2006). In cultivated Brassica, on the other hand, a small number of studies have reported evidence of historical recombination at the S locus. For example, Awadalla and Charlesworth (1999) found a statistically significant decline of LD with distance at the SLG gene, an S-locus gene not directly involved in SI specificity determination (Cabrillac et al. 1999; Kusaba et al. 2001; Silva et al. 2001). In a recent study of four B. oleracea/B. rapa haplotype pairs, Takuno et al. (2007) showed that whereas the SRK S domain and the SCR gene (termed SP11 by Takuno et al. 2007) had identical genealogies, the topologies of S-locus gene genealogies became increasingly different the farther from the SRK/SCR “core region” the genes were located. In particular, from differences in topology between phylogenetic trees inferred from the SRK kinase and S-domain sequence data, Takuno et al. (2007) showed that the SRK kinase domain had undergone recombination relative to the SRK S domain and SCR. A similar result was obtained here from a smaller SRK kinase and S-domain data set (Figure 2). Furthermore, as shown by the comparison between the ML class II SRK and SCR phylogenetic trees (Figure 3), the SCR tree was topologically different from the SRK tree, whereas the opposite was true for the reciprocal comparison. This apparently conflicting result could be due to the relatively low resolution of the class II SRK kinase domain data set; i.e., there might be several topologies, of which one is represented by the ML class II SCR tree, with likelihoods not significantly lower than the ML class II SRK tree. These results, although ambiguous, tentatively indicate that recombination might have happened between the SRK kinase domain and SCR in the wild B. cretica. Also, the presence of a 9-bp deletion in exon 7 of all SRK class II haplotypes and in only one of the class I haplotypes (BcrSRK111) could be the result of recombination; however, as the parts of the BcrSRK111 haplotype flanking the deletion were very dissimilar to any class II haplotype, we cannot exclude multiple deletion events as an explanation for the phylogenetic distribution of this particular feature. Moreover, in a population study of SRK and SCR in B. cretica, Edh et al. (2009) detected configurations of pairs of polymorphic sites (the four-gamete test; Hudson and Kaplan 1985) within the kinase domain of the SRK gene that could be explained only by recombination or gene conversion. The specific arrangement of genes at the S locus varies among some Brassica haplotypes (e.g., Fobis-Loisy et al. 2004), so that the SCR gene in some cases is closer to the kinase domain than to the S domain. Thus, recombination between the SCR gene and the kinase domain of SRK might break up the association between the pollen and pistil specificities in certain haplotypes. Nevertheless, taken together, available evidence suggests that recombination, or gene conversion, has occurred at the Brassica S locus but outside the actual pollen and pistil specificity determinants.
Acknowledgments
We thank Maarit Jaarola for assisting in the phylogenetic analyses and Bengt Jacobsson for taking care of the plant material. We also thank two anonymous reviewers for constructive and helpful comments. This work was financially supported by a grant from the Crafoord Foundation to A.C.
References
- Awadalla, P., and D. Charlesworth, 1999. Recombination and selection at Brassica self-incompatibility loci. Genetics 152 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D. C., and J. B. Nasrallah, 1993. Physical linkage of the SLG and SRK genes at the self-incompatibility locus Brassica oleracea. Mol. Gen. Genet. 236 369–373. [DOI] [PubMed] [Google Scholar]
- Cabrillac, D., V. Delorme, J. Garin, V. Ruffio-Châble, J. L. Giranton et al., 1999. The S-15 self-incompatibility haplotype in Brassica oleracea includes three S gene family members expressed in stigmas. Plant Cell 11 971–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric, V., and X. Vekemans, 2007. Evolution under strong balancing selection: How many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae? BMC Evol. Biol. 7 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., C. Bartolomé, M. H. Schierup and B. K. Mable, 2003. Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol. Biol. Evol. 20 1741–1753. [DOI] [PubMed] [Google Scholar]
- Edh, K., B. Widén and A. Ceplitis, 2007. Nuclear and chloroplast microsatellites reveal extreme population differentiation and limited gene flow in the Aegean endemic Brassica cretica (Brassicaceae). Mol. Ecol. 16 4972–4983. [DOI] [PubMed] [Google Scholar]
- Edh, K., B. Widén and A. Ceplitis, 2009. Molecular population genetics of the SRK and SCR self-incompatibility genes in the wild plant species Brassica cretica (Brassicaceae). Genetics 181 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobis-Loisy, I., C. Miege and T. Gaude, 2004. Molecular evolution of the S locus controlling mating in the Brassicaceae. Plant Biol. 6 109–118. [DOI] [PubMed] [Google Scholar]
- Hagenblad, J., J. Bechsgaard and D. Charlesworth, 2006. Linkage disequilibrium between incompatibility locus region genes in the plant Arabidopsis lyrata. Genetics 173 1057–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Harberd, D. J., 1972. A contribution to the cyto-taxonomy of Brassica (Cruciferae) and its allies. Bot. J. Linn. Soc. 65 1–23. [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic, B., L. Bohs and J. R. Kohn, 2004. Historical inferences from the self-incompatibility locus. New Phytol. 161 97–105. [Google Scholar]
- Inaba, R., and T. Nishio, 2002. Phylogenetic analysis of Brassicaceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theor. Appl. Genet. 105 1159–1165. [DOI] [PubMed] [Google Scholar]
- Kamau, E., and D. Charlesworth, 2005. Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr. Biol. 15 1773–1778. [DOI] [PubMed] [Google Scholar]
- Kianian, S. F., and C. F. Quiros, 1992. Trait inheritance, fertility, and genomic relationships of some n = 9 Brassica species. Genet. Res. Crop Evol. 39 165–175. [Google Scholar]
- Kimura, R., K. Sato, R. Fujimoto and T. Nishio, 2002. Recognition specificity of self-incompatibility maintained after the divergence of Brassica oleracea and Brassica rapa. Plant J. 29 215–223. [DOI] [PubMed] [Google Scholar]
- Kishino, H., and M. Hasegawa, 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29 170–179. [DOI] [PubMed] [Google Scholar]
- Koch, M., B. Haubold and T. Mitchell-Olds, 2001. Molecular systematics of the Brassicaeae: evidence from coding plastidic matK and nuclear Chs sequences. Am. J. Bot. 88 534–544. [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5 150–163. [DOI] [PubMed] [Google Scholar]
- Kusaba, M., T. Nishio, Y. Satta, K. Hinata and D. Ockendon, 1997. Striking sequence similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: implications for the evolution and recognition mechanism. Proc. Natl. Acad. Sci. USA 94 7673–7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba, M., K. G. Dwyer, J. Hendershot, J. Vrebalov, J. B. Nasrallah et al., 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13 627–643. [PMC free article] [PubMed] [Google Scholar]
- Lawrence, M. J., 2000. Population genetics of the homomorphic self-incompatibility polymorphisms in flowering plants. Ann. Bot. 85 221–226. [Google Scholar]
- Nasrallah, J. B., T. Nishio and M. E. Nasrallah, 1991. The self-incompatibility genes of Brassica: expression and use in genetic ablation of floral tissues. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42 393–422. [Google Scholar]
- Ockendon, D. J., 1982. An S-allele survey of cabbage (Brassica oleracea var. capitata). Euphytica 31 325–331. [Google Scholar]
- Paetsch, M., S. Mayland-Quellhorst and B. Neuffer, 2006. Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity 97 283–290. [DOI] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14 817–818. [DOI] [PubMed] [Google Scholar]
- Prigoda, N. L, A. Nassuth and B. K. Mable, 2005. Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22 1609–1620. [DOI] [PubMed] [Google Scholar]
- Richards, A. J., 1997. Plant Breeding Systems. Chapman & Hall, London.
- Sato, Y., R. Fujimoto, K. Toriyama and T. Nishio, 2003. Commonality of self-recognition specificity of S haplotypes between Brassica oleracea and Brassica rapa. Plant Mol. Biol. 52 617–626. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., B. K. Mable, P. Awadalla and D. Charlesworth, 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer, C. R., M. E. Nasrallah and J. B. Nasrallah, 1999. The male determinant of self-incompatibility in Brassica. Science 286 1697–1700. [DOI] [PubMed] [Google Scholar]
- Shiba, H., M. Iwano, T. Entani, K. Ishimoto, H. Shimosato et al., 2002. The dominanance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, N. F., and D. R. Goring, 2001. Mechanisms of self-incompatibility in flowering plants. Cell. Mol. Life Sci. 58 1988–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, N. F., S. L. Stone, L. N: Christie, W. Sulaman, K. A. P. Nazarian et al., 2001. Expression of the S receptor kinase in self-incompatible Brassica napus cv. Westar leads to the allele specific rejection of self-incompatible Brassica napus pollen. Mol. Genet. Genomics 265 552–559. [DOI] [PubMed] [Google Scholar]
- Snogerup, S., M. Gustavsson and R. von Bothmer, 1990. Brassica sect. Brassica (Brassicaceae) I. Taxonomy and variation. Willdenowia 19 271–365. [Google Scholar]
- Stein, J. C., B. Howlett, D. Boyes, M. E. Nasrallah and J. B. Nasrallah, 1991. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88 8816–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, G., N. Kai, T. Hirose, K. Fukui, T. Nishio et al., 1999. Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA.
- Takayama, S., and A. Isogai, 2005. Self-incompatibility in plants. Annu. Rev. Plant Biol. 56 467–489. [DOI] [PubMed] [Google Scholar]
- Takayama, S., H. Shimosato, H. Shiba, M. Funato, F.-S. Che et al., 2001. Direct ligand-receptor complex interactions control Brassica self-incompatibility. Nature 413 534–538. [DOI] [PubMed] [Google Scholar]
- Takuno, S., R. Fujimoto, T. Sugimura, K. Sato, S. Okamoto et al., 2007. Effects of recombination on hitchhiking diversity in the Brassica self-incompatibility locus complex. Genetics 177 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama, M. K., 1995. A generalized least-squares estimate for the origin of sporophytic self-incompatibility. Genetics 139 975–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama, M. K., and E. Newbigin, 2000. Evolutionary dynamics of dual-specificity self-incompatibility alleles. Plant Cell 12 310–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bothmer, R., M. Gustavsson and S. Snogerup, 1995. Brassica sect. Brassica (Brassicaceae) II. Inter- and intraspecific crosses with cultivars of B. oleracea. Genet. Res. Crop Evol. 42 165–178. [Google Scholar]
- Widén, B., S. Andersson, G-Y. Rao and M. Widén, 2002. Population divergence of genetic (co)variance matrices in a subdivided plant species, Brassica cretica. J. Evol. Biol. 15 961–970. [Google Scholar]
- Yang, Y. W., K. N. Lai, P. Y. Tai and W. H. Li, 1999. a Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48 597–604. [DOI] [PubMed] [Google Scholar]
- Yang, Y.-W., K.-N. Lai, P.-Y. Tai, D.-P. Ma and W.-H. Li, 1999. b Molecular phylogenetic studies of Brassica, Rorippa, Arabidopsis and allied genera based on the internal transcribed spacer region of 18S–25S rDNA. Mol. Phylogenet. Evol. 13 455–462. [DOI] [PubMed] [Google Scholar]