Abstract
Background
Plasmid encoded blaCTX-M enzymes represent an important sub-group of class A β-lactamases causing the ESBL phenotype which is increasingly found in Enterobacteriaceae including Klebsiella spp. Molecular typing of clinical ESBL-isolates has become more and more important for prevention of the dissemination of ESBL-producers among nosocomial environment.
Methods
Multiple displacement amplified DNA derived from 20 K. pneumoniae and 34 K. oxytoca clinical isolates with an ESBL-phenotype was used in a universal CTX-M PCR amplification assay. Identification and differentiation of blaCTX-M and blaOXY/K1 sequences was obtained by DNA sequencing of M13-sequence-tagged CTX-M PCR-amplicons using a M13-specific sequencing primer.
Results
Nine out of 20 K. pneumoniae clinical isolates had a blaCTX-M genotype. Interestingly, we found that the universal degenerated primers also amplified the chromosomally located K1-gene in all 34 K. oxytoca clinical isolates. Molecular identification and differentiation between blaCTX-M and blaOXY/K1-genes could only been achieved by sequencing of the PCR-amplicons. In silico analysis revealed that the universal degenerated CTX-M primer-pair used here might also amplify the chromosomally located blaOXY and K1-genes in Klebsiella spp. and K1-like genes in other Enterobacteriaceae.
Conclusion
The PCR-based molecular typing method described here enables a rapid and reliable molecular identification of blaCTX-M, and blaOXY/K1-genes. The principles used in this study could also be applied to any situation in which antimicrobial resistance genes would need to be sequenced.
Background
Plasmid encoded blaCTX-M enzymes represent an important sub-group of class-A β-lactamases which hydrolyse broad-spectrum β-lactam antibiotics causing an extended spectrum β-lactamase (ESBL) phenotype, which is increasingly found in enterobacterial species including Klebsiella [1,2]. To date, over 60 different CTX-M-type β-lactamases have been described [3] and divided into five different clusters that reflect similarity at the amino-acid sequence level, namely blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, blaCTX-M-25 [2], respectively. More recently, it has been suggested that blaCTX-M-45 forms a new, separate cluster [4].
Due to constitutive expression of a chromosomal class A β-lactamases [5], Klebsiella oxytoca was shown to have a high level resistance to ceftriaxone and cefotaxime [6]. Originally, this class A β-lactamase was named K1 [7] and later on referred to as KOXY [8] or blaOXY [6]. Sequence diversity of the chromosomally located K. oxytoca K1-gene and the existence of discrete groups of blaOXY-1 and blaOXY-2 genes has been described in detail [5,9].
Numerous PCR-based typing assays for the identification of blaCTX-M genes have been developed. Initially, detection of all members belonging to specific blaCTX-M groups was achieved by combining multiple PCR amplification assays [10,11]. To avoid multiple CTX-M PCR amplification steps, Boyd and co-workers [12] designed a pair of universal, degenerated CTX-M primers, allowing the amplification of hitherto all known blaCTX-M genes. However, identification of a blaCTX-M genotype at the nucleotide level often required cloning of the PCR-amplicons, followed by DNA sequencing. These methods are labour intensive, time-consuming, expensive and moreover, require a battery of amplicon specific sequencing primers.
In this study, we report on the use of a simple, accurate and universal CTX-M PCR amplification and sequencing assay well suited for high-throughput analysis.
Methods
Screening of Klebsiella spp. for cephalosporin resistance during 2001–spring 2007
At the Department of Clinical Microbiology, University Hospital Linköping, susceptibility testing was performed on all Klebsiella pneumoniae and K. oxytoca clinical isolates. Cefadroxil was used for the screening of cephalosporin resistance which was followed up by testing of resistant isolates with cefotaxime and ceftazidime or direct testing with cefotaxime and ceftazidime [14]. A biochemical panel for identification and differentiation of Klebsiella spp. was used. Indole-negative Klebsiella spp. clinical isolates were identified as K. pneumoniae and indole-positive clinical isolates as K. oxytoca.
Phenotypic ESBL-screening
All cefotaxime and/or ceftazidime resistant clinical isolates were phenotypically screened by Etest using ceftazidime and cefotaxime with and without clavulanic acid (bioMerieux Sverige AB, Askim, Sweden). A reduction of MIC by ≥3 twofold dilutions of the cephalosporin in the presence of clavulanic acid, i. e. a MIC ratio of ≥8 or the presence of phantom- or deformation zones was considered indicative of an ESBL-phenotype. Clinical isolates were stored in glycerol containing Nutrient-broth No 2 (Lab M, Bury, UK) at -70°C until analysis.
Susceptibility testing of K. oxytoca with K1-genes and K. pneumoniae with blaCTX-M genes
MIC-values for cefotaxime, ceftazidime and piperacillin/tazobactam were determined with Etest (bioMerieux Sverige AB, Askim, Sweden).
Bacterial type and reference strains
Control strain K. oxytoca 1980K1 was kindly provided by Dr. D. Livermore, Health Protection Agency, Antibiotic Resistance Monitoring and Reference Laboratory, London, UK. Type strains were purchased from the American Type Culture Collection [15] or the Culture Collection University of Gothenburg [16]; K. pneumoniae ATCC 700603, K. pneumoniae CCUG 54718, and K. oxytoca CCUG 15717T.
Multiple displacement amplification of bacterial DNA
To perform multiple genotyping analysis of our growing collection of CTX-M suspected K. pneumoniae and K. oxytoca of clinical origin and omitting tedious bacterial culturing, sufficient amounts of bacterial DNA were produced by means of multiple displacement amplification of bacterial DNA [17]. For that purpose, bacteria from frozen cultures (1 μl) and from the reference strains were added to a GenomiPhi-DNA V2 amplification-kit cocktail as recommended by the manufacturer (GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
Universal blaCTX-M gene PCR amplification
A PCR amplification assay was carried out using 10 pmol of each universal degenerated primer M13-CTX-M.U1.SE (CGTTGTAAAACGACGGCCAGTGAATGTGCAGYACCAGTAARGTKATGGC) and CTX-M.U2.AS (TGGGTRAARTARGTSACCAGAAYCAGCGG) targeting the CTX-M and OXY/K1-enzyme genes [modified from 12] and a HotStarTaq-Master mix (Qiagen GmbH, Hilden, Germany) in a final reaction volume of 25 μl using an Applied Biosystems thermo cycler 2720 (Applied Biosystems, Foster City, USA) and 200 μl thin-walled reaction tubes. This yields an approximately 600 bp PCR amplicon corresponding to 68% of the CTX-M and OXY/K1-enzyme encoding nucleotide sequences.
PCR amplification conditions were as follows: initial denaturation step at 95°C for 15 min; 30 cycles of denaturation at 95°C for 30 s; annealing at 55°C for 30 s; extension at 72°C for 2 min, and a final extension step at 72°C for 10 min. Subsequently, PCR-amplicons were separated electrophoretically on a precast 2% agarose E-gel (Invitrogen, Carlsbad, CA, USA).
DNA sequence analysis
DNA sequence analysis of M13-sequence tagged CTX-M PCR-amplicons was carried out using a M13 uni (-21) primer by a customer DNA sequencing service (Eurofins MWG Operon GmbH, Martinsried, Germany). Prior to DNA sequencing, PCR-amplicons were treated with ExoSAP-IT to inactivate excess of oligonucleotide primers, following the supplier's protocol (USB Europe GmbH, Staufen, Germany). The PCR products were then lyophilised and sequenced. Generated DNA sequences were aligned, edited and compared with blaCTX-M DNA and blaCTX-M-like DNA sequences using the CLC bioinformatics freeware v.3.2.3 [18]. blaCTX-M, blaOXY, K1, and K1-like DNA sequences were retrieved from the NCBI Entrez Nucleotide database [19].
In silico DNA sequence analysis
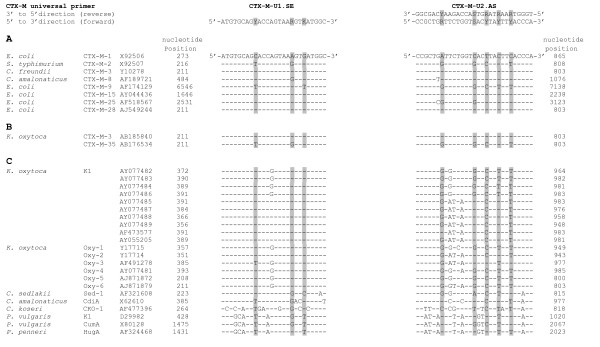
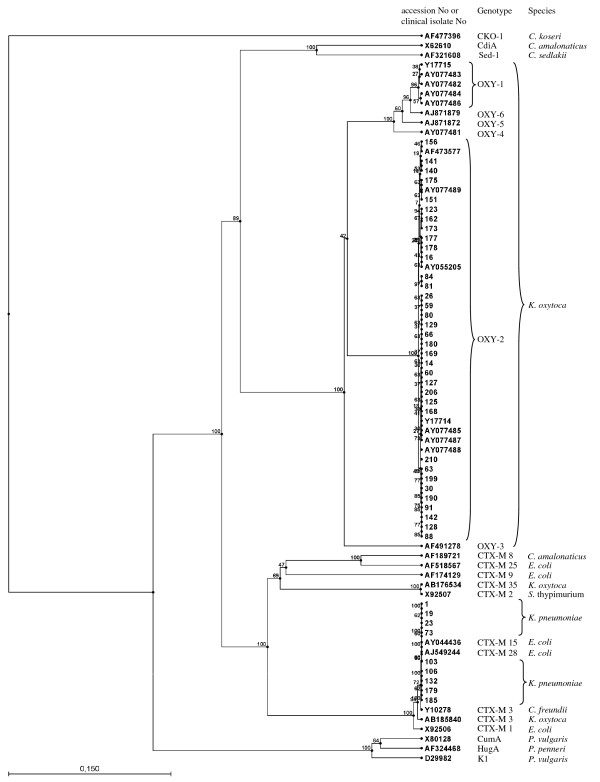
blaCTX-M type strains E. coli blaCTX-M-1 [GenBank:X92506] (blaCTX-M Group 1), E. coli blaCTX-M-9 [GenBank:AF189721] (blaCTX-M Group 9), E. coli blaCTX-M-15 [GenBank:AY044436] (blaCTX-M Group 1), E. coli blaCTX-M-25 [GenBank:AF518567] (blaCTX-M Group 25), E. coli blaCTX-M-28 [GenBank:AJ549244] (blaCTX-M Group 25), Salmonella Typhimurium blaCTX-M-2 [GenBank:X925079] (blaCTX-M Group 2), Citrobacter freundii blaCTX-M-3 [GenBank:Y10278] (blaCTX-M-Group 1), Citrobacter amalonaticus blaCTX-M-8 [GenBank:AF189721] (blaCTX-M Group 8), K. oxytoca K1-genes [GenBank:AY077482–AY077489, AF473577, AY055205], K. oxytoca blaOXY-1 [GenBank:Y17715], blaOXY-2 [GenBank:Y17714], blaOXY-3 [GenBank:AF491278], blaOXY-4 [GenBank:AY077481], blaOXY-5 [GenBank:AJ871872] and blaOXY-6 [GenBank:AJ871879], K. oxytoca blaCTX-M-3 [GenBank:AB185840] (blaCTX-M Group 1) and blaCTX-M-35 [GenBank:AB176534] (blaCTX-M Group 1), chromosomally encoded β-lactamase genes from Citrobacter sedlakii Sed-1 [GenBank:AF321608], Citrobacter amalonaticus CdiA [GenBank:X62610], Citrobacter koseri CKO [GenBank:AF477396], Proteus vulgaris K1 [GenBank:D29982], Proteus vulgaris CumA [GenBank:X80128], Proteus penneri HugA [GenBank:AF324468] and the universal degenerated primers CTX-M.U1.SE and CTX-M.U2.AS, respectively (Fig. 1), were aligned using ClustalW [20]. A dendrogram consisting of all partial DNA sequences used for primer comparison in figure 1 and DNA sequences derived from the clinical isolates was constructed using the CLC bioinformatics freeware v.3.2.3 [18] and UPGMA clustering (Fig. 2). DNA sequences were edited to comprise the relevant DNA sequences between the two universal degenerated CTX-M primers (Fig. 1).
Figure 1.
Partial DNA sequence alignment of (A) CTX-M type sequences E. coli CTX-M 1 (Group 1), E. coli CTX-M-9 (Group 9), E. coli CTX-M-15 (Group 1), E. coli CTX-M-25 (Group 25), E. coli CTX-M-28 (Group 2), S. typhimurium CTX-M-2 (Group 2), C. freundii CTX-M-3 (Group 1), C. amalonaticus CTX-M-8 (Group 8); (B), K. oxytoca CTX-M-3 (Group 1) and K. oxytoca CTX-M-35 (Group 2), (C) chromosomally CTX-M-like sequences K. oxytoca K1, K. oxytoca OXY-1 to OXY-6, C. sedlakii Sed-1, C. amalonaticus CdiA, C. koseri CKO, P. vulgaris K1, K. vulgaris CumA, and P. penneri HugA genes and the universal degenerated CTX-M-U1.SE and CTX-M-U2. AS primers. Dashes indicate sequence homologies. The degenerated nucleotide sequence positions in the primers and its corresponding nucleotides in the aligned genes are indicated in grey. For clarity, sense and antisense DNA sequences of the universal degenerated CTX-M-U2.AS (reverse) primer are given.
Figure 2.
Phylogenetic tree from partial Enterobacteriaceae blaCTX-M and K1-like DNA sequences. K. pneumoniae blaCTX-M-15/28 and K. oxytoca K1/OXY-2 form distinct cluster groups, distinguishable from the K1/OXY-1 cluster, respectively. P. penneri HugA, P. vulgaris CumA and K1, C. sedlakii and C. amalonaticus form separate clades.
Results
Distribution of Klebsiella spp
The number of K. pneumoniae and K. oxytoca clinical isolates of each year was in the range 457 to 864 (K. pneumoniae) and 270 to 455 (K. oxytoca), respectively, comprising 99% of the genus Klebsiella collected and analysed at the Department of Clinical Microbiology, University Hospital, Linköping, Sweden.
Screening of cephalosporin resistance and ESBL-phenotype
The cefadroxil resistance rate was in the range of 1.5% to 3.2% for K. pneumoniae and 3.7% to 6.4% for K. oxytoca. Twenty K. pneumoniae and 34 K. oxytoca clinical isolates were selected from a positive ESBL-phenotypic screening during 2001 to spring 2007. All K. oxytoca isolates (n = 34) revealed an ESBL-phenotype by screening with Etest using cefotaxime with and without clavulanic acid. All isolates were negative in the corresponding test with ceftazidime. Similarly, a majority of K. pneumoniae isolates (16 of 20) revealed an ESBL-phenotype in both ESBL Etests. Two of the K. pneumoniae isolates revealed an ESBL-phenotype only with cefotaxime and two isolates only with ceftazidime, respectively.
blaCTX-M PCR amplification and partial DNA sequence analysis
DNA sequencing of M13-sequence-tagged universal CTX-M PCR-amplicons of MDA-DNA derived from K. pneumoniae and K. oxytoca of clinical origin revealed the presence of blaCTX-M genes in 9 out of 20 K. pneumoniae and the presence of the K1-gene in all 34 K. oxytoca clinical isolates. According to the phylogenetic tree constructed from partial blaCTX-M, blaOXY and K1-DNA sequences located between the two universal degenerated primers (Fig. 1), nine K. pneumoniae clinical isolates formed a unique cluster with E. coli blaCTX-M-15, [GenBank:AY044436] and E. coli blaCTX-M-28 [GenBank:AJ549244] which itself is closely related to the C. freundii CTX-M-3 [GenBank:Y10278], K. oxytoca blaCTX-M-3 [GenBank:AB185840], and E. coli blaCTX-M-1 [GenBank:X92506] cluster, respectively (Fig. 2). Similarly, the K. oxytoca K1 clinical isolates form a unique K1/blaOXY-2 cluster together with K. oxytoca K1 [GenBank:AF473577, AY077489, AY055205, AY077487, AY077485, and AY077488] and K. oxytoca blaOXY-2 [GenBank:Y17714], respectively (Fig 2). None of the K. oxytoca K1 clinical isolates clustered within the K1/blaOXY-1 cluster. Clearly, partial DNA sequence analysis of the CTX-M PCR-amplicons did not allow an unequivocal discrimination of the blaCTX-M genes. However, our data indicate the presence of a blaCTX-M-15/28 genotype in the K. pneumoniae clinical isolates.
Susceptibility testing of K. oxytoca with K1-genes and K. pneumoniae with blaCTX-M genes
The MIC-values for cefotaxime and ceftazidime for the K. oxytoca isolates were in the range of 0.5 to 8 mg/l and 0.125 to 4 mg/l, respectively. Corresponding MIC-values for the K. pneumoniae isolates with CTX-M genotypes were in the range of 64 to 256 mg/l and 16 to 256 mg/l, respectively. The susceptibility for piperacillin/tazobactam was lower in the K. oxytoca isolates with MIC-values ≥128 mg/l compared to MIC-values between 4 to 64 mg/l for the K. pneumoniae isolates.
In silico DNA sequence comparison
The finding that the universal degenerated CTX-M primer-pair amplified the chromosomally located K1-enzyme in K. oxytoca prompted us to perform a DNA sequence alignment of the universal CTX-M primer-pair with Enterobacteriaceae blaCTX-M, blaOXY, K1, and K1-like genes retrieved from the Entrez Nucleotide database (Methods). As illustrated in figure 1, the universal degenerated CTX-M primers revealed a high degree of DNA sequence similarity between the target sequences present in the E. coli, S. Typhimurium, C. freundii and C. amalonaticus blaCTX-M type-gene; K. oxytoca blaCTX-M-3 and blaCTX-M-35 genes, K. oxytoca K1 and blaOXY-1 to blaOXY-6 genes; the chromosomally encoded C. sedlakii Sed-1 and C. amalonaticus CdiA showed a lower degree of sequence similarity compared to C. koseri CKO, P. vulgaris K1, P. vulgaris CumA, and P. penneri HugA genes, respectively. With the exception of C. koseri CKO, P. vulgaris K1, P. vulgaris CumA and P. penneri HugA DNA sequences, most of the nucleotide variations are observed at 5'-Y, R, K-3' positions in primer CTX-M-U1.SE and 5'-R, R, R, S, Y-3' positions (were R stands for purine, Y stands for pyrimidine, S stands for G or C, and K stands for G or T) in primer CTX-M-U2.AS (Fig. 1). The GC-rich 3'-ends of the primers are highly conserved within the corresponding blaCTX-M, blaOXY, and K1 target DNA sequences. This may explain why the universal degenerated CTX-M primer-pair amplified blaCTX-M and K1 sequences.
Discussion
The increased prevalence of Enterobacteriaceae that produce blaCTX-M enzymes makes new demands on clinical routine microbiology laboratories to perform blaCTX-M typing. Due to the growing number of blaCTX-M enzymes, the traditional iso-electrofocusing appears not to be the method of choice for establishing an enterobacterial blaCTX-M genotype any longer. Molecular techniques for identification and classification of blaCTX-M genes in clinical isolates on a large scale have been described. Recently, a multiplex CTX-M PCR (MP-PCR) amplification assay was described which allows differentiation between different blaCTX-M subtype groups [21]. However, using this particular MP-PCR assay, we often observed non-specific PCR amplification in K. oxytoca isolates. Subsequent cloning and DNA sequencing analysis revealed that the unspecific PCR-amplicons represented K1-enzyme gene sequences. Thus, misinterpretation of strains as active blaCTX-M producers based on false positive PCR amplification might cause false reporting of blaCTX-M genes in K. oxytoca.
A different approach has been used by Galas and co-workers [22]. These authors described the use of a CTX-M-consensus primer-pair to establish a blaCTX-M-genotype in Enterobacteriaceae, including K. oxytoca [22]. However, in silico DNA sequence analysis reveals that this consensus CTX-M primer-pair [23] also targets blaOXY/K1-genes such as K. oxytoca blaOXY-2 [Genbank:Y17714] at positions 348–367 (MA1 primer) and 872–391 (MA2 primer), and K1 gene [GenBank:AY077482] at positions 369–378 (MA1 Primer) and 893–911 (MA2 primer), respectively. Thus, based on PCR amplification alone, it seems to be far from clear whether these K. oxytoca isolates would have a blaCTX-M, or a blaOXY/K1-genotype, respectively. This question can only be settled by DNA-sequencing of the PCR amplicons.
The use of M13-sequence tagged PCR-amplicons in combination with M13-specific sequencing primers was originally described for sequencing of Staphylococcus aureus protein A (Spa-typing) PCR-amplicons [24]. Our results employing the same technique for sequencing of β-lactamase PCR amplicons convincingly demonstrate that that the use of a M13-sequence tagged CTX-M.U1.SE primer allowed for an unequivocal discrimination of blaCTX-M and K1-genes. Moreover, our results support our previous findings indicating that blaCTX-M and K1-enzyme genes might have some degree of sequence similarity [13]. Extended in silico analysis furthermore revealed a high degree of sequence similarities between Enterobacteriaceae blaCTX-M, blaOXY 1–6, and K1 related sequences (Fig. 1), respectively. Thus, based on PCR amplification alone using universal degenerated CTX-M primer-pairs, it would be difficult, not to say impossible to distinguish between an Enterobacteriaceae blaCTX-M or K1/OXY genotype if sequencing had not been performed.
Fournier and co-workers [8] have described the existence of two discrete groups of K. oxytoca blaOXY-1 and blaOXY-2 enzymes. Later on, it was shown that blaOXY-1 and blaOXY-2 genes are expressed in two genetic K. oxytoca groups, namely K. oxytoca strain SG266 and SG271, respectively [25]. So far, six groups of OXY β-lactamases have been identified and characterised in K. oxytoca [26]. In our study we have found that all K. oxytoca clinical isolates form a distinct K. oxytoca K1/OXY-2 cluster group distinguishable from the K1/OXY-1 and the K. pneumoniae CTX-M15/28 cluster group, respectively. Moreover, the phylogenetic tree that was established implies the existence of a chromosomally located β-lactamase super-gene family in Enterobacteriaceae (Fig. 2). This is in agreement with previous reports describing that chromosomally encoded class A β-lactamases found in Klebsiella species are highly conserved at the amino-acid level compared to class A β-lactamases found in other Enterobacteriaceae [27-31]. The Citrobacter spp. DNA sequences included in the phylogenetic tree form separate clades. This is in agreement with earlier reports showing that C. koseri CKO-1 and C. amalonaticus CdiA isolates carry highly divergent β-lactamase genes despite the fact that they show a highly similar biochemical profile and 16S rDNA sequence similarity [32]. Biochemical methods may not always be adequate to identify Klebsiella spp. and their phylogenetic groups in clinical microbiology laboratories because several species share similar biochemical profiles [33,34]. Therefore, molecular techniques as applied in the present study may help to accomplish bacterial genotyping at reasonable costs and time.
MIC-value determination of piperacillin/tazobactam in this study also distinguished between K. oxytoca with K1 β-lactamase from CTX-M producing K. pneumoniae, showing higher MIC-values. Furthermore, the MIC-values for cefotaxime and ceftazidime for K. oxytoca with K1-genes was lower than for CTX-M producing K. pneumoniae. Similar results have been reported by Potz and co-workers [35].
Conclusion
The PCR-based molecular typing method described here enables a rapid and reliable identification of CTX-M and OXY/K1-genes. The principles used in the present study can be applied to any situation in where antimicrobial resistance genes are to be sequenced. This is desirable because only sequencing of full-length reading frames will allow for an unequivocal discrimination between various subtypes of antimicrobial resistance genes such as blaCTX-M, blaSHV and blaTEM gene-families, respectively. Moreover, the use of M13-sequence tagged primers in PCR amplification assays facilitates amplicon sequencing since only one single sequencing primer (M13) is required.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
HJM, MT and LEN participated in the conception, design, drafting of the manuscript, and final approval of the version to be published. HJM and MT were responsible for the acquisition, analysis and interpretation of the molecular biology based data. MT and LEN were responsible for the clinical strain collection and phenotypic screening, analysis and interpretation of phenotypic data.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Hans-Jürg Monstein, Email: hanmo@ibk.liu.se.
Maria Tärnberg, Email: maria.tarnberg@imk.liu.se.
Lennart E Nilsson, Email: len@imk.liu.se.
Acknowledgements
This study was financially supported in part by a County Medical Centre Östergötland (LMC) Grant (21403-ESBL) the Research Council in the South East of Sweden (FORSS-3971), and the Molecular Biology Program, Clinical Microbiology-LMC, Linköping, Sweden. We greatly acknowledge the technical assistance of M. V. Nilsson during the initial phase of this study and the critical reading and commenting on the manuscript by Dr. Jon Jonasson.
References
- Tzouvelekis LS, Tzelepi E, Tassios PT, Legakis NJ. CTX-M-type β-lactamases: An emerging group of extended-spectrum enzymes. Int J Antimicrob Agents. 2000;14:137–142. doi: 10.1016/S0924-8579(99)00165-X. [DOI] [PubMed] [Google Scholar]
- Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amino Acid Sequences for TEM, SHV and OXA Extended-Spectrum and Inhibitor Resistant β-Lactamases. http://www.lahey.org/studies
- Rossolini GM, Andrea MMD, Mugnaioli C. The spread of CTX-M-type extended-spectrum β-lactamase. Clin Microbiol Infect Dis. 2008;14:33–41. doi: 10.1111/j.1469-0691.2007.01867.x. [DOI] [PubMed] [Google Scholar]
- Arakawa Y, Ohta M, Kido N, Mori M, Ito H, Komatsu T, Fujii Y, Kato N. Chromosomal β-lactamase of Klebsiella oxytoca, a new class A enzyme that hydrolyzes broad-spectrum β-lactam antibiotics. Antimicrob Agents Chemother. 1989;33:63–70. doi: 10.1128/aac.33.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B, Arlet G, Lagrange PH, Philippon. Klebsiella oxytoca: Resistance to aztreonam by overproduction of the chromosomally encoded beta-lactamase. FEMS Microbiol Lett. 1994;116(1):31–36. doi: 10.1111/j.1574-6968.1994.tb06671.x. [DOI] [PubMed] [Google Scholar]
- Emanuel EL, Gagnon J, Waley G. Structural and kinetic studies on β-lactamase K1 from Klebsiella aerogenes. Biochem J. 1986;234:343–347. doi: 10.1042/bj2340343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM, Yuan M. Antibiotic resistance and production of extended-spectrum β-lactamases amongst Klebsiella spp. from intensive care units in Europe. J Antimicrob Chemother. 1996;38:409–424. doi: 10.1093/jac/38.3.409. [DOI] [PubMed] [Google Scholar]
- Fournier B, Roy PH, Lagrange PH, Philippon A. Chromosomal β-lactamase of Klebsiella oxytoca are divided into two main groups, blaOXY-1 and bla OXY-2. Antimicrob Agents Chemother. 1996;40:454–459. doi: 10.1128/aac.40.2.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout JDD, Hossain A, Hanson ND. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J Clin Microbiol. 2004;42:5715–5721. doi: 10.1128/JCM.42.12.5715-5721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soge O, Queenan, Ojo KK, Adeniyi BA, Roberts MC. CTX-M-15 extended-spectrum β-lactamase from Nigerian Klebsiella pneumoniae. J Antimicrob Chemother. 2006;57:24–30. doi: 10.1093/jac/dki429. [DOI] [PubMed] [Google Scholar]
- Boyd D, Tyler AS, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. the Canadian nosocomal infection surveillance program, Health Care. Complete nucleotide sequence of a 92-Kliobase plasmid harbouring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother. 2004;48:3758–64. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monstein H-J, Östholm-Balkhed Å, Nilsson MV, Nilsson M, Dornbusch K, Nilsson LE. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and bla CTX-M genes in Enterobacteriaceae. APMIS. 2007;115:1400–1408. doi: 10.1111/j.1600-0463.2007.00722.x. [DOI] [PubMed] [Google Scholar]
- The Swedish Reference Group for Antibiotics. http://www.srga.org
- The American Type Culture Collection. http://www.lgcstandards-atcc.org
- The Culture Collection, University of Gothenburg. http://www.ccug.se
- Monstein H-J, Olsson C, Nilsson I, Grahn N, Benoni C, Ahrné S. Multiple displacement amplification of DNA from human colon and rectum biopsies. Bacterial profiling and identification of Helicobacter pylori- DNA by means of 16s rDNA-based TTGE and pyrosequencing analysis. J Microbiol Meth. 2005;63:239–247. doi: 10.1016/j.mimet.2005.03.012. [DOI] [PubMed] [Google Scholar]
- CLC bio. http://www.clcbio.com
- NCBI Entrez Nucleotide. http://www.ncbi.nlm.nih.gov/nucleotide
- EMBL-EBI ClustalW2. http://www.ebi.ac.uk/Tools/clustalw2
- Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for the rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2006;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- Galas M, Decousser J-W, Breton N, Godard T, Allouch PY, Pina P. the College de Bacteriologie virology Hygiène (ColBVH) study group. Nationwide study of the prevalence, characteristics, and molecular epidemiology of extended-spectrum β-lactamase-producing Enterobacteriaceae in France. Antimicrob Agents Chemother. 2008;52:786–789. doi: 10.1128/AAC.00906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladin M, Cao VTB, Lambert T, Donay J-L, Herrmann J-L, Ould-Hocine Z, Verdet C, Delisle F, Philippon A, Arlet G. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol Lett. 2002;209:161–168. doi: 10.1111/j.1574-6968.2002.tb11126.x. [DOI] [PubMed] [Google Scholar]
- Frénay HMEM, Bunschoten AE, Schouls LM, van Leeuwen WJ, Vandenbroucke-Grauls CMJE, Verhof J, Mooi RF. Molecular typing of Methicillin-resistant Staphylococcus aureus on the basis of Protein A gene polymorphism. Eur J Clinical Microbiol Infect Dis. 1996;15:60–64. doi: 10.1007/BF01586186. [DOI] [PubMed] [Google Scholar]
- Granier SA, Leflon-Guibout V, Goldstein FW, Nicolas-Chanoine M-H. New Klebsiella oxytoca β-lactamase genes blaOXY-3 and blaOXY-4 and a third group of K. oxytoca based on blaOXY-3. Antimicrob Agents Chemother. 2003;47:2922–2928. doi: 10.1128/AAC.47.9.2922-2928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevre C, Jbel M, Passet V, Weill FX, Grimont PAD, Brisse S. Six groups of OXY β-lactamase evolved over millions of years in Klebsiella oxytoca. Antimicrob Agents Chemother. 2005;49:3453–3462. doi: 10.1128/AAC.49.8.3453-3462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli M, Franceschini N, Segatore B, Amicosante G, Orator A, Duez C, Joris B, Frère JM. Cloning and nucleotide sequencing of the gene encoding the β-lactamase from Citrobacter diversus. FEMS Microbiol Lett. 1991;67:79–84. doi: 10.1016/0378-1097(91)90448-j. [DOI] [PubMed] [Google Scholar]
- Péduzzi J, Farzaneh S, Reynaud A, Barthélémy M, Labia R. Characterization and amino acid sequence analysis of a new oxyimino cephalosporin-hydrolyzing class A β-lactamase from Serratia fonticola. Biochim Biophys Acta. 1997;1341:58–70. doi: 10.1016/s0167-4838(97)00020-4. [DOI] [PubMed] [Google Scholar]
- Nukaga M, Mayama K, Crichlow GV, Knox JR. Structure of the extended-spectrum class A β-lactamase from Proteus vulgaris K1. J Mol Biol. 2002;317:109–117. doi: 10.1006/jmbi.2002.5420. [DOI] [PubMed] [Google Scholar]
- Petrella S, Clermont D, Casin I, Jarlier V, Sougakoff W. Novel class A β-lactamase Sed-1 from Citrobacter sedlakii: Genetic diversity of β-lactamases within the Citrobacter genus. Antimicrob Agents Chemother. 2001;45:2287–2298. doi: 10.1128/AAC.45.8.2287-2298.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella S, Renard M, Ziental-Gelus N, Clermont D, Jarlier V, Sougakoff W. Characterization of the chromosomal class A β-lactamase CKO from Citrobacter koseri. FEMS Microbiol Lett. 2006;254:285–292. doi: 10.1111/j.1574-6968.2005.00028.x. [DOI] [PubMed] [Google Scholar]
- Underwood S, Avison MB. Citrobacter koseri and Citrobacter amalonaticus isolates carry highly divergent β-lactamase genes despite the having high levels of biochemical similarity and 16S rRNA sequence homology. J Antimicrob Chemother. 2004;53:1076–1080. doi: 10.1093/jac/dkh235. [DOI] [PubMed] [Google Scholar]
- Hansen DS, Aucken HM, Abiola T, Podschun R. Recommended test panel for differentiation of Klebsiella species on the basis of a trilateral interlaboratory evaluation of 18 biochemical tests. J Clin Microbiol. 2004;42:3665–3669. doi: 10.1128/JCM.42.8.3665-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves MS, Dias de Castro AC, Riley LW, Moreira BM. Identification of clinical isolates of Indole-positive and Indole-negative Klebsiella spp. J Clin Microbiol. 2006;44:3640–3646. doi: 10.1128/JCM.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potz NA, Colman M, Warner M, Reynolds R, Livermore DM. False-positive extended-spectrum beta-lactamase tests for Klebsiella oxytoca hyperproducing K1 beta-lactamase. J Antimicrob Chemother. 2004;53:545–547. doi: 10.1093/jac/dkh112. [DOI] [PubMed] [Google Scholar]