Abstract
An increasing number of cancer types have been found to be linked to specific mutations in the mitochondrial DNA, which result in specific structural changes of the respiratory enzyme complexes. In this study, we have investigated the effect of 2 such mutations identified in colon cancer patients, leading to the amino acid substitutions Ser458Pro and Gly125Asp in subunit I of cytochrome c oxidase (complex IV) [Greaves et al. (2006) Proc Natl Acad Sci USA 103:714–719]. We introduced these mutations in Rhodobacter sphaeroides, which carries an oxidase that serves as a model of the mitochondrial counterpart. The lack of expression of the former variant indicates that the amino acid substitution results in severely altered overall structure of the enzyme. The latter mutation (Gly171Asp in the bacterial oxidase) resulted in a structurally intact enzyme, but with reduced activity (approximately 30%), mainly due to slowed reduction of the redox site heme a. Furthermore, even though the Gly171Asp CytcO pumps protons, an intrinsic proton leak was identified, which would lead to a decreased overall energy-conversion efficiency of the respiratory chain, and would also perturb transport processes such as protein, ion, and metabolite trafficking. Furthermore, the specific leak may act to alter the balance between the electrical and chemical components of the proton electrochemical gradient.
Keywords: cytochrome aa3, electrochemical, electron transfer, pump, respiratory oxidase
The mammalian mitochondrial DNA (mtDNA) is a double-stranded circular molecule of 16.6 kb, which encodes 13 of the polypeptides of the respiratory chain complexes. In recent years, an increasing number of diseases have been found to be associated with mutations in the mtDNA (1–7). Furthermore, a number of different cancer types have been found to be linked to such mutations, and in many cases, these mutations result in structural modifications of enzymes of the electron-transport chain (8–11). A possible factor contributing to the development of the disease is an increased production of reactive oxygen species (ROS) as a result of a specific mutation (1, 9, 10, 12).
Several of the cancer-associated mutations found in mtDNA result in structural modifications of cytochrome c oxidase (CytcO) (9, 13–15) (Fig. 1A). This enzyme is the final electron acceptor in the electron-transport chain and catalyses the reduction of oxygen to water. In this reaction, 4 electrons per O2 molecule are transferred from the more positively charged (P-) side of the membrane, and 4 protons are transferred from the more negatively charged (N-) side of the membrane. In addition, CytcO pumps on average approximately 1 proton per electron over the membrane, thereby increasing the energy-conservation efficiency, such that a net of 8 charges are transferred across the membrane per reduced O2 [for recent reviews on the structure and function of CytcO, see (16–18)]. CytcO receives electrons from cytochrome c, which binds on the P-side and initially reduces the di-nuclear copper site, CuA. The electrons are transferred consecutively to heme a and then to the catalytic site, which consists of heme a3 and copper B (CuB). When heme a3 and CuB become reduced, O2 binds to heme a3, after which the molecule is reduced in a stepwise process thereby cycling between a number of partly reduced intermediate states. Even though CytcO has not itself been implicated in ROS formation, changes in its structure and function may indirectly affect the remaining part of the respiratory chain [for a recent review, see (19)].
Fig. 1.
The structure of CytcO from R. sphaeroides. (A) Subunits I–IV are shown in different colors as indicated. The heme groups are shown in green and the cooper ions in orange (B). Gly 171 is situated above heme a and Ala 501 below the heme groups (see encircled residues). (B) Hemes a and a3 and CuB and CuA, situated in subunit I. The position of Asp-171 (in the Gly171Asp structural variant) is indicated together with the 2 functionally important arginine residues; 481 and 482. The distance from Asp-171 to the Arg-482 residue and one of the heme a propionates is approximately 2 and 3 Å, respectively. The figures were prepared using the Visual Molecular Dynamics software (57). The energy minimization calculation on the Gly171Asp structural variant CytcO was performed using the HyperChem 7.1 software (Hypercube, Inc.).
In a recent study, Greaves et al. (15) describe 2 mutations, which result in amino acid substitutions in subunit I of CytcO, found in CytcO-deficient colonic crypts from colon cancer patients. One of the mutations, 6277A>G, results in substitution of a well-conserved residue, Gly-125 by an Asp. The other mutation, 7275T>C, was found in another colon cancer patient, and it is equivalent to the Ser458Pro amino acid substitution where Ser-458 is also a conserved amino acid residue. Studies on mtDNA mutations often involve the use of isolated mitochondria or transmitochondrial cybrids. One disadvantage when using these systems is that it is difficult to discriminate between situations where for example, CytcO is less active (inactive), displays a lower pumping stoichiometry, or is expressed at lower amounts. These problems are mainly due to technical difficulties associated with biochemical and functional characterization of the dysfunctions. In the present study, we introduced the mutations discussed above, using site-directed mutagenesis, altering the above-mentioned residues in subunit I of the CytcO (cytochrome aa3) from the bacterium Rhodobacter sphaeroides. This bacterial oxidase is a good model of the mitochondrial counterpart (20) where Gly-125 corresponds to Gly-171 and Ser-458 to Ala-501 (Fig. 1B). The structure of the R. sphaeroides enzyme (subunits I–III) (21, 22) (Fig. 1A) is nearly identical to the corresponding subunits of the mammalian CytcO (23), which form the catalytic core. CytcOs from R. sphaeroides, Pseudomonas denitrificans, and yeast have previously been used to investigate the effect of other disease-related mutations on the function of the enzyme (24–28).
Here, we show that the Gly171Asp CytcO displayed approximately 34% steady-state catalytic activity linked to proton pumping; however, an intrinsic proton leak was found in the enzyme, which implies that the corresponding mtDNA mutation is likely to diminish the energy conservation efficiency of the mitochondrion.
Results
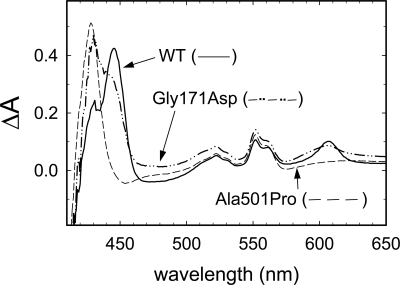
Introduction of the mutation corresponding to the Ala501Pro substitution in R. sphaeroides (cytochrome aa3) resulted in a significantly decreased growth rate, and the cells grew to a low density compared with that of the wild-type. Analysis of the reduced-oxidized difference absorption spectra of the solubilized membrane fractions (Fig. 2) showed that the cells carrying the Ala501Pro substitution did not display any peaks characteristic to CytcO (e.g., at 445 and 605 nm), which indicates that the enzyme was not expressed (or its structure was perturbed such that it could not be reduced). The Gly171Asp mutation, on the other hand, did not have any effect on the growth rate of the R. sphaeroides cells, and the spectral characteristics were similar to those of the wild-type membrane fraction (Fig. 2); there were peaks at 445 and 605 nm. The other components of the spectra originate from contributions of hemes b and c (because the difference spectra are not normalized, they only show the proportions of the different components; e.g., the heme b/heme a ratio is larger in the Gly171Asp than in the wild-type CytcO).
Fig. 2.
Difference spectra (reduced-oxidized) of the solubilized membrane fractions. The absorbance peak at 605 nm (typical for a-type hemes) indicates that the Gly171Asp structural variant CytcO is expressed (cf. the difference spectrum of the membrane fraction containing the wild-type CytcO). For the Ala501Pro mutant, however, there is no peak at 605 nm, indicating that the membrane preparation does not contain any detectable CytcO. It should be noted that the absorbance values are not normalized; the spectra were adjusted such that the peaks are of similar size.
The purified Gly171Asp CytcO displayed the same CO-binding kinetics as the wild-type CytcO, which indicates an unperturbed catalytic site. Furthermore, internal electron transfer between the hemes and proton transfer, linked to redox changes at heme a3 (29), were unaffected by the structural substitution (SI Text). The multiple turnover activity of the Gly171Asp CytcO was measured at pH 6.5 and found to be 34 ± 11% of that of the wild-type CytcO.
Proton Pumping Activity.
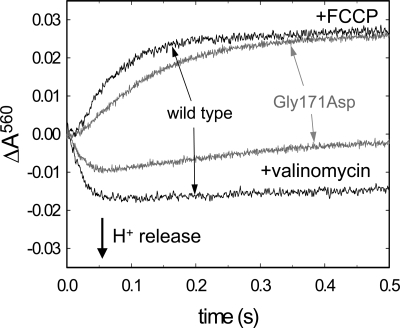
Proton pumping by CytcO was investigated by mixing liposome-reconstituted CytcO with reduced cytochrome c in the presence of O2 and the pH dye phenol red (Fig. 3). The cytochrome c:CytcO ratio used allowed a total of 8 CytcO turnovers. First, the potassium ionophore valinomycin was added to equilibrate the electrical component of the electrochemical proton gradient established by CytcO, and absorbance changes of phenol red were detected at 560 nm to follow changes in pH outside the vesicles. With the wild-type CytcO, a decrease in absorbance is typically observed, with the same rate as that of cytochrome c oxidation, consistent with proton release to the outside of the CytcO vesicles (Fig. 3).
Fig. 3.
Absorbance changes associated with proton uptake and release (pumping). An aerobic solution of CytcO reconstituted in phospholipids vesicles was mixed at a 1:1 ratio in a stopped-flow spectrometer with a solution containing reduced cytochrome c. A decrease in phenol red absorbance in the presence of valinomycin (K+ ionophore used to dissipate the electrical component of the proton gradient) is consistent with acidification, i.e., protons are pumped out of the vesicles. The absorbance increase in the presence of the proton ionophore FCCP is a result of alkalinization of the solution due to proton uptake during CytcO turnover (the protons are taken up from the inside of the vesicles and then equilibrate across the membrane). Experimental conditions: 0.5 μM CytcO reconstituted in lipid vesicles, 16 μM reduced cytochrome c, 100 μM phenol red, 50 μM Hepes-KOH, 45 mM KCl, 44 mM sucrose, 1 mM EDTA at pH 7.6, 250 μM O2.
After addition of the proton ionophore FCCP, the “pumped protons” equilibrate across the membrane, and only the net consumed (substrate) protons contribute to the dye absorbance change, which is seen as an increase in the absorbance (Fig. 3). Because it is known that approximately 4 protons are consumed per CytcO molecule for each turnover, the amplitude of the trace with FCCP can be used to quantify the number of protons pumped per turnover. The data with the wild-type CytcO showed a pumping stoichiometry of 0.75 ± 0.1 H+/e− (SD of 3 measurements) (approximately 0.65 H+/e− in Fig. 3).
The data showed that the Gly171Asp structural variant initially displayed a smaller pumping stoichiometry than the wild-type CytcO; however, the most striking observation is that with the Gly171Asp CytcO, after the initial absorbance decrease associated with acidification of the outside solution (t < 50 ms), the absorbance increased over a time scale of approximately 0.3 s, which indicates that protons rapidly leaked back into the vesicles. This rapid increase in absorbance was not observed with the wild-type CytcO, which indicates that the proton leak is found specifically within the Gly171Asp structural variant CytcO. Furthermore, the apparently smaller initial pumping stoichiometry with the Gly171Asp CytcO is presumably due to the proton leak, which competes with pumping on the time scale of the measurement (see also Discussion).
Oxidation of the Reduced CytcO.
As indicated above, the steady-state turnover activity of the Gly171Asp CytcO structural variant was approximately one-third of that of the wild-type CytcO. To identify the reaction step(s) responsible for the decreased overall activity, we first investigated the reaction of the reduced CytcO with O2, i.e., the oxidative part of the reaction cycle. The reduced CytcO with the CO-ligand bound at the catalytic site was mixed with an O2-saturated solution after which the ligand was dissociated by a short laser flash, which enabled O2 to bind to the catalytic site. The reaction of the reduced CytcO with O2 was monitored at a number of wavelengths specific to transitions between oxygen intermediates and redox changes of the metal cofactors (Fig. 4). We first describe the reaction sequence with the wild-type CytcO (30) and then point to the differences with the Gly171Asp structural variant CytcO.
Fig. 4.
Absorbance changes associated with the reaction of the fully reduced CytcO with O2. The absorbance changes at 445 nm (A) and 580 nm (B) are mainly due to ligand binding to the catalytic site and to redox reactions of the heme groups; at 445 nm both hemes a and a3 contribute, while at 580 nm we mainly observe redox changes of heme a, and formation and decay of the F state. At 830 nm (C), an increase in absorbance is mainly associated with oxidation of CuA. All traces are scaled to 1 μM reacting CytcO. Results with the wild-type and Gly171Asp CytcO are shown in black and red, respectively. Experimental conditions: approximately 1 μM reacting enzyme, 0.1 M Hepes-KOH, 0.1% dodecyl-β-D-maltoside, and 1 mM O2 at pH 7.4 and 22 °C.
The initial, unresolved increase in absorbance at 445 nm (Fig. 4A) is associated with dissociation of the CO ligand. It is followed in time by an absorbance decrease (τ ≅ 30–50 μs), which is associated with electron transfer from heme a to the catalytic site resulting in formation of a state that is called “peroxy” and denoted PR. The reaction is also seen at 580 nm as an absorbance decrease on the same time scale (Fig. 4B). Next, a proton is transferred from solution to the catalytic site with a time constant of approximately 100 μs forming a state called “ferryl” (F). Formation of the F state is seen most clearly as an absorbance increase at 580 nm (see time range approximately 0–200 μs in Fig. 4B). The reaction is also linked to fractional electron transfer from CuA to heme a, which results in approximately 50% oxidation of CuA, seen as an increase in absorbance at 830 nm (see time range approximately 0–300 μs in Fig. 4C). Formation of the fully oxidized (O) state (τ ≅ 1 ms) involves electron and proton transfer to the catalytic site, and it is seen as a decrease in absorbance at 445 and 580 nm (Fig. 4 A and B) due to oxidation of hemes a and a3 and an absorbance increase at 830 nm (Fig. 4C) due to further oxidation of the fraction CuA that remained reduced after the preceding reaction (PR → F transition, τ ≅ 100 μs).
As seen in Fig. 4, the Gly171Asp structural variant was oxidized over the same time scale as the wild-type CytcO. However, there is one noteworthy difference on the time scale of formation of the F state. In the Gly171Asp structural variant, the absorbance increase at 580 nm was replaced by a plateau, and the absorbance increase at 830 nm was absent. Taken together, these data indicate that the fractional electron transfer from CuA to heme a did not take place on the time scale of F formation, and CuA remained reduced until the final reaction step (F → O) of the reaction where the site became fully oxidized.
Reduction Kinetics.
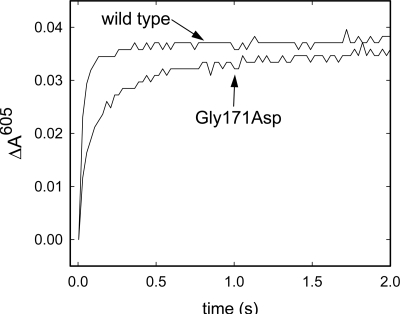
As described above, the Gly171Asp CytcO was oxidized over the same time scale as the wild-type CytcO, which indicates that the lower turnover activity of the structural variant CytcO is due to slowed reduction kinetics. To test this assumption, we investigated also the reductive part of the catalytic cycle. The overall reduction rate was slower with the Gly171Asp than with the wild-type CytcO. Furthermore, inspection of the reduction kinetics at 605 nm (Fig. 5), where heme a has an absorption peak in the reduced minus oxidized difference spectrum, indicates that this slowed reduction kinetics is primarily due to slowed reduction of heme a. In both the wild-type and Gly171Asp CytcO, the reduction kinetics was biphasic; the time constants were 20 ms (90% of the total amplitude) and 0.2 s (10% of the total amplitude) with the wild-type CytcO, and 50 ms (60% of the total amplitude) and 0.3 s (40% of the total amplitude) with the Gly171Asp CytcO. Thus, the amplitude of the slower component was 4 times larger with the Gly171Asp than with the wild-type CytcO.
Fig. 5.
Reduction of wild-type and Gly171Asp CytcO. CytcO and the reducing agents were rapidly mixed in a stopped-flow apparatus equipped with a diode array detector. At 605 nm, reduced heme a has an absorption peak where an increase in absorbance is due to reduction of this site. Experimental conditions: 3 μM CytcO, 0.1 M Hepes-KOH, 0.1% dodecyl-β-D-maltoside, 5 mM sodium dithionite, 12 μM Hexa-ammine-ruthenium chloride at pH 7.4 and 22 °C.
Discussion
When Ala 501 was replaced by a proline, essentially no detectable aa3-type CytcO could be found in the R. sphaeroides cell membranes (see Fig. 2), i.e., the structurally modified protein was not inserted into the membrane. This is perhaps not surprising, because Ala 501 is situated in the middle of an α-helix and proline is known to destabilize the helical structure. Assuming the same scenario in the mitochondrion, the mutation could result in accumulation of reducing equivalents in the respiratory chain leading to increased ROS production (see below).
The data show that the Gly171Asp substitution did not have any significant effect on the overall oxidation rate. However, as a result of the structural alteration, the absorbance increase at 580 nm on the time scale of the PR → F transition (τ ≅ 100 μs, Fig. 4B) was replaced by a plateau, and there was no increase in absorbance at 830 nm (cf. oxidation of CuA) on that time scale (Fig. 4C), which indicates that the F state was formed over the same time scale as in the wild-type CytcO, but the reaction was not linked to simultaneous electron transfer from CuA to heme a (cf. no absorbance increase at 580 or 830 nm). These observations indicate that the midpoint potential of heme a is lowered in the Gly171Asp CytcO, which is qualitatively consistent with the introduction of an acidic residue near the site.
We also investigated reduction of the oxidized CytcO (Fig. 5). The results show that the structural alteration resulted in a significantly slowed overall reduction rate, which could be either due to slowed electron or proton transfer, linked to the electron transfer. To discriminate between these 2 possibilities, we investigated proton transfer in a separate experiment and found that the rate of this reaction was unaffected. Thus, the slower reduction rate must be due to slowed electron transfer to the catalytic site. Analysis of the spectral changes during oxidation of CytcO showed that this decrease in reduction rate was mainly due to slowed reduction of heme a, which is consistent with a lower midpoint potential of the site (see above). Residue 171 is located at a distance of approximately 10 Å from the heme a iron and a few Å from the heme ring (Fig. 1B). Introduction of an acidic amino acid residue close to heme a is likely to destabilize the reduced state of this redox site, making electron transfer to heme a less favorable, which would explain the above-discussed results. This explanation is consistent with recent results from studies of the Ser44Asp (Ser 44 is also located near heme a) structural variant of the R. sphaeroides CytcO, where the Asp was found to be deprotonated around neutral pH and the heme a potential was significantly decreased (31).
The most striking result from this study is that the Gly171Asp mutant CytcO appears to leak protons, as evidenced from the rapid alkalinization (absorbance increase) after the initial acidification (absorbance decrease) of the solution outside of the CytcO-vesicles (Fig. 3). The experiments were done such that the 8 turnovers were completed within approximately 50 ms, while the proton leak occurred over a time scale of approximately 0.3 s. We note that the phenotype described here is mechanistically different from an uncoupled oxidase (“uncoupled” meaning that the catalytic oxygen reduction is not linked to proton pumping). An uncoupled CytcO would simply display lower energy-conservation efficiency (50%), because only the electron transfer to O2 and uptake of substrate protons would contribute to the electrochemical proton gradient. Instead, a transmembrane proton leak in an intact system would act to dissipate the electrochemical proton gradient maintained also by complexes I and III.
Proton leaks in structural variants of CytcO are not unexpected. To pump protons across the membrane a proton pump must accommodate transmembrane proton pathways, which span across the entire thickness of the membrane. Proton transfer through these pathways must be regulated to prevent proton leaks from the positive to the negative side of the membrane, often referred to as “gating” [reviewed in ref. 17]. The “gate” may be, for example, an amino acid side chain, but the term may also refer to changes in the overall structure or changes in barrier heights during the coupled electron and proton transfer (32–35). A structural modification of CytcO may result in changes in timing of electron or proton transfer, changes in the equilibrium constant between the different positions of a gate, the dynamics of the gate, or its pKa values in the different conformations. Disease-related mutations have previously been proposed to lead to intrinsic uncoupling (but not specific proton leaks) of CytcO, even though in this case the mutations were found in SU III (28).
Results from a number of experimental and theoretical studies indicate that Arg-481 and Arg-482, together with the heme D-propionates are involved in controlling proton access to either side of the membrane (36–39). Because the Gly-171 residue is located very close to these Arg residues, it is likely that the structural modification would act to perturb the proton-gating machinery of CytcO. Furthermore, the results from a recent study indicate that the Gly-171 residue is part of a loop, consisting of residues 169–175, which switches between different conformations during turnover thereby controlling proton/water access to CytcO (40). Specifically, this loop was found to undergo a major conformational change during the P to F reaction, which is linked to proton pumping and would involve opening of the exit channel to the outside.
As noted above, one possible link between a mutation in the mtDNA and development of disease is an increased production of ROS. In mitochondria, ROS are primarily formed at complexes I and III of the electron transport chain (41, 42) and CytcO is normally not directly involved in release of ROS (43). Nevertheless, it is likely that inhibition of CytcO activity, such as that observed with the Gly171Asp CytcO, would result in an increase in ROS production at complexes I and III due to accumulation of reducing equivalents at these complexes (19, 44). Furthermore, a slowed intramolecular electron transfer to the catalytic site in CytcO would result in more long-lived partly reduced oxygen intermediates and protein-derived radicals, which could result in release of ROS also at complex IV.
Another link between the structure and function of the Gly171Asp CytcO and the disease state may arise from the proton leak in the structural variant, which would act to diminish the energy efficiency of the respiratory chain and perturb transport processes such as protein, ion, and metabolite trafficking. In addition, other consequences of such a specific leak may also be significant. The proton motive force (electrochemical proton gradient), Δp, in respiring mitochondria has a value of 150–200 mV, where the electrical component, Δψ, contributes with approximately 70% of the total Δp (45). This distribution between Δψ and ΔpH is determined by all ion fluxes through transporters and channels across the membrane as well as by ion leaks. Introduction of a specific proton leak in a non-equilibrium system, where different ions flow across the membrane, is likely to act to alter the ratio of Δψ and ΔpH, such that the relative fraction of Δψ would presumably increase. A change in the Δψ/ΔpH ratio would, for example, influence transport into the mitochondrion of Ca2+, which regulates the respiratory chain (19, 46), thereby further perturbing the redox states of the respiratory-chain complexes. Furthermore, it has been suggested that an increase in Δψ beyond approximately 150 mV would not accelerate ATP production, but would act to increase ROS production due to inhibition of complexes I and III (19). In addition, a tight regulation of the Δψ value is critical for tissue homeostasis (47, 48), and increased Δψ values have been found to be a characteristic feature of colonic tumor cells (49) and are linked to an increased probability for tumor growth and development (50, 51).
In summary, we have found that the cancer-associated mutation 6277A>G (Gly125Asp or Gly171Asp in R. sphaeroides CytcO subunit I) leads to a number of functional alterations: (i) a decrease in the CytcO activity due to (ii) slowed intramolecular electron transfer to the catalytic site and (iii) a specific proton leak through the enzyme. The leak would not only act to diminish the overall energy-conversion efficiency, but may also alter the Δψ/Δ pH ratio. Collectively, these functional alterations may provide the link between the mutation and generation of disease. Of course, the data are not sufficient to support a definitive statement as the 2 mutations Gly125Asp and Ser458Pro lead to different effects. Nevertheless, we believe that the current study may provide some clues to the link between functional changes at the molecular level and development of disease.
Materials and Methods
Site-Directed Mutagenesis and Purification of CytcO.
To construct the Gly171Asp and the Ala501Pro mutations in R. sphaeroides, the pUC-based plasmid pJS3-SH, containing the gene encoding subunit I of CytcO, was used. The mutations were introduced using the Quick-Change site-directed mutagenesis kit (Stratagene) and verified by sequencing. The mutated fragment was restricted and ligated into a pRK-based vector suitable for expression in R. sphaeroides cells and containing subunits I–III of CytcO. Because Escherichia coli cells were used during the mutagenesis procedure, the final step was to conjugate the pRK vector containing the mutation into the R. sphaeroides cells using established methods (20). The CytcO was purified from the cell membranes using a Ni2+-NTA affinity column essentially as described in ref. 52.
Steady-State Activity.
The steady-state activity was measured using a Clark oxygen electrode. Purified CytcO was diluted to 10 nM in 50 mM K+-phosphate at pH 6.5 and 0.05% DDM. After the addition of 983 μL of phosphate buffer and 7 μL of 3.6 mM reduced cytochrome c to the oxygraph chamber, 10 μL of the diluted enzyme was added. The oxygen consumption was measured over time.
Reconstitution of CytcO into Liposomes.
CytcO-containing lipid vesicles were prepared essentially as described in ref. 53. Briefly, purified CytcO was diluted to 4 μM in 0.1 M Hepes at pH 7.4 and 4% sodium cholate. Soybean lecithin was dissolved in 0.1 M Hepes at pH 7.4 and 2% cholate to 40 mg/mL. The lipid solution was sonicated and mixed with the CytcO solution at a 1:1 ratio. The cholate was gradually removed using Bio-Beads SM-2 Adsorbent (Bio-Rad Laboratories). The buffer was exchanged for a 0.1 M KCl solution at pH 7.4, using a PD10 column (GE Healthcare Life Sciences). Using the above-mentioned lipid-to-CytcO ratio, each vesicle typically contains at most 1 CytcO molecule. Approximately 75% of the CytcO molecules are oriented with the cytochrome c-binding site toward the outside solution, i.e., in the same direction as in the native membrane (54).
Proton Pumping.
Liposome-reconstituted CytcO at a concentration of 0.5 μM in 50 μM Hepes-KOH, 45 mM KCl, 44 mM sucrose, 1 mM EDTA, and 100 μM phenol red at pH 7.6 was mixed (1:1 mixing ratio) with 16 μM reduced cytochrome c in 50 μM Hepes-KOH, 45 mM KCl, 44 mM sucrose, 1 mM EDTA, and 100 μM phenol red at pH 7.6 in a stopped-flow spectrophotometer. Absorbance changes of the pH dye phenol red were measured at 560 nm. In the presence of the K+ ionophore valinomycin (used to equilibrate the electrical component of the electrochemical gradient), these absorbance changes are due to pH changes and reflect proton pumping from the inside of the vesicles to the outside. After addition of the proton ionophore carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), the net consumption of protons during enzyme turnover was detected.
Preparation of Fully Reduced CO-Bound CytcO and Flow-Flash Measurements.
CytcO in 0.1 M Hepes at pH 7.4 and 0.1% DDM was diluted to a concentration of 7 μM and transferred to an anaerobic cuvette. The redox mediator PMS was added at a concentration of 1 μM, and the atmosphere in the cuvette was exchanged for N2. The enzyme was reduced by adding 2 mM ascorbate. Complete reduction of CytcO was verified from an analysis of the absorption spectrum. The N2 atmosphere was exchanged for CO, which results in formation of the CytcO-CO complex where the ligand is bound to heme a3.
To study the reaction of the CytcO with O2, fully reduced CO-bound CytcO was rapidly mixed, at a 1:5 ratio, with an O2-saturated solution of 0.1 mM Hepes at pH 7.4 and 0.1% DDM in a stopped-flow spectrophotometer (Applied Photophysics) (55). Approximately 300 ms after mixing, the CO molecule was dissociated from the heme a3-CuB site by means of a short laser flash (Quantel, Brilliant B, approximately 200 mJ at 532 nm), allowing oxygen to bind to the reduced catalytic site. The reaction was followed in time by recording the absorbance changes at single wavelengths (see Figure Legends).
Reduction Kinetics.
The reduction rate of the fully oxidized CytcO was monitored using a stopped-flow spectrophotometer (Applied Photophysics) essentially as described in ref. 56. A solution of 7 μM CytcO in 0.1 M Hepes at pH 7.4 and 0.1% DDM was rapidly mixed at a 1:1 ratio with a solution containing 25 μM hexa-ammine-ruthenium chloride, 10 mM sodium dithionite, 0.1 M Hepes at pH 7.4, and 0.1% DDM. Absorbance changes reflecting reduction of heme a and the heme a3-CuB sites were monitored at a number of different wavelengths simultaneously using a diode-array detector.
Supplementary Material
Acknowledgments.
We thank Robert W. Taylor at the University of Newcastle for valuable discussions and Håkan Lepp who performed the energy minimization calculations. This work was supported by grants from the Swedish Cancer Society and the Knut, Alice Wallenberg Foundation, and the Center for Biomembrane Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811450106/DCSupplemental.
References
- 1.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 2.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 3.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McFarland R, Taylor RW, Turnbull DM. Mitochondrial disease—its impact, etiology, and pathology. Curr Top Dev Biol. 2007;77:113–155. doi: 10.1016/S0070-2153(06)77005-3. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 6.Carew JS, Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9:657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 8.Abu-Amero K, Alzahrani AS, Zou M, Shi Y. High frequency of somatic mitochondrial DNA mutations in human thyroid carcinomas and complex I respiratory defect in thyroid cancer cell lines. Oncogene. 2005;24:1455–1460. doi: 10.1038/sj.onc.1208292. [DOI] [PubMed] [Google Scholar]
- 9.Petros JA, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polyak K, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 11.Jin X, et al. Relationship between mitochondrial DNA mutations and clinical characteristics in human lung cancer. Mitochondrion. 2007;7:347–353. doi: 10.1016/j.mito.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Lenaz G, et al. Bioenergetics of mitochondrial diseases associated with mtDNA mutations. Biochim Biophys Acta. 2004;1658:89–94. doi: 10.1016/j.bbabio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Gallardo ME, et al. m.6267G>A: A recurrent mutation in the human mitochondrial DNA that reduces cytochrome c oxidase activity and is associated with tumors. Hum Mutat. 2006;27:575–582. doi: 10.1002/humu.20338. [DOI] [PubMed] [Google Scholar]
- 14.Jones JB, et al. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:1299–1304. [PubMed] [Google Scholar]
- 15.Greaves LC, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci USA. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brändén G, Gennis RB, Brzezinski P. Transmembrane proton translocation by cytochrome c oxidase. Biochim Biophys Acta. 2006;1757:1052–1063. doi: 10.1016/j.bbabio.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Brzezinski P, Ädelroth P. Design principles of proton-pumping haem-copper oxidases. Curr Opin Struct Biol. 2006;16:465–472. doi: 10.1016/j.sbi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Wikström M, Verkhovsky MI. Mechanism and energetics of proton translocation by the respiratory heme-copper oxidases. Biochim Biophys Acta. 2007;1767:1200–1214. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Hüttemann M, et al. Regulation of oxidative phosphorylation, the mitochondrial membrane potential, and their role in human disease. J Bionenerg Biomembr. 2008;40:445–456. doi: 10.1007/s10863-008-9169-3. [DOI] [PubMed] [Google Scholar]
- 20.Cao J, Hosler J, Shapleigh J, Revzin A, Ferguson-Miller S. Cytochrome aa3 of Rhodobacter sphaeroides as a model for mitochondrial cytochrome c oxidase. J Biol Chem. 1992;267:24273–24278. [PubMed] [Google Scholar]
- 21.Svensson-Ek M, et al. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 22.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 24.Bratton M, Mills D, Castleden CK, Hosler J, Meunier B. Disease-related mutations in cytochrome c oxidase studied in yeast and bacterial models. Eur J Biochem. 2003;270:1222–1230. doi: 10.1046/j.1432-1033.2003.03482.x. [DOI] [PubMed] [Google Scholar]
- 25.Meunier B. Site-directed mutations in the mitochondrially encoded subunits I and III of yeast cytochrome oxidase. Biochem J. 2001;354:407–412. doi: 10.1042/0264-6021:3540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucioli S, et al. Introducing a novel human mtDNA mutation into the Paracoccus denitrificans COX I gene explains functional deficits in a patient. Neurogenetics. 2006;7:51–57. doi: 10.1007/s10048-005-0015-z. [DOI] [PubMed] [Google Scholar]
- 27.Meunier B, Taanman JW. Mutations of cytochrome c oxidase subunits 1 and 3 in Saccharomyces cerevisiae: Assembly defect and compensation. Biochim Biophys Acta. 2002;1554:101–107. doi: 10.1016/s0005-2728(02)00217-7. [DOI] [PubMed] [Google Scholar]
- 28.Mather MW, Rottenberg H. Intrinsic uncoupling of cytochrome c oxidase may cause the maternally inherited mitochondrial diseases MELAS and LHON. FEBS Lett. 1998;433:93–97. doi: 10.1016/s0014-5793(98)00891-6. [DOI] [PubMed] [Google Scholar]
- 29.Karpefors M, et al. Electron-proton interactions in terminal oxidases. Biochim Biophys Acta. 1998;1365:159–169. doi: 10.1016/s0005-2728(98)00058-9. [DOI] [PubMed] [Google Scholar]
- 30.Ädelroth P, Ek M, Brzezinski P. Factors determining electron-transfer rates in cytochrome c oxidase: Investigation of the oxygen reaction in the R. sphaeroides and bovine enzymes. Biochim Biophys Acta. 1998;1367:107–117. doi: 10.1016/s0005-2728(98)00142-x. [DOI] [PubMed] [Google Scholar]
- 31.Mills DA, et al. Proton-dependent electron transfer from CuA to heme a and altered EPR spectra in mutants close to heme a of cytochrome oxidase. Biochemistry. 2008;47:11499–11509. doi: 10.1021/bi801156s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brzezinski P, Larsson G. Redox-driven proton pumping by heme-copper oxidases. Biochim Biophys Acta. 2003;1605:1–13. doi: 10.1016/s0005-2728(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 33.Wikström M, Verkhovsky MI, Hummer G. Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochim Biophys Acta. 2003;1604:61–65. doi: 10.1016/s0005-2728(03)00041-0. [DOI] [PubMed] [Google Scholar]
- 34.Olsson MH, Warshel A. Monte Carlo simulations of proton pumps: On the working principles of the biological valve that controls proton pumping in cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:6500–6505. doi: 10.1073/pnas.0510860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegbahn PE, Blomberg MR. Energy diagrams and mechanism for proton pumping in cytochrome c oxidase. Biochim Biophys Acta. 2007;1767:1143–1156. doi: 10.1016/j.bbabio.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Qian J, et al. Role of the conserved arginine pair in proton and electron transfer in cytochrome c oxidase. Biochemistry. 2004;43:5748–5756. doi: 10.1021/bi036279o. [DOI] [PubMed] [Google Scholar]
- 37.Puustinen A, Wikström M. Proton exit from the heme-copper oxidase of Escherichioa coli. Proc Natl Acad Sci USA. 1999;96:35–37. doi: 10.1073/pnas.96.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brändén G, et al. The protonation state of a heme propionate controls electron transfer in cytochrome c oxidase. Biochemistry. 2005;44:10466–10474. doi: 10.1021/bi0502745. [DOI] [PubMed] [Google Scholar]
- 39.Seibold SA, Mills DA, Ferguson-Miller S, Cukier RI. Water chain formation and possible proton pumping routes in Rhodobacter sphaeriodes cytochrome c oxidase: A molecular dynamics comparison of the wild type and R481K mutant. Biochemistry. 2005;44:10475–10485. doi: 10.1021/bi0502902. [DOI] [PubMed] [Google Scholar]
- 40.Busenlehner LS, Salomonsson L, Brzezinski P, Armstrong RN. Mapping protein dynamics in catalytic intermediates of the redox-driven proton pump cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:15398–15403. doi: 10.1073/pnas.0601451103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria. Central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Fiskum G, Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J Neurochem. 2002;80:780–787. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- 43.Babcock GT, Wikström M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 44.Dawson TL, Gores GJ, Nieminen A-L, Herman B, Lemasters JJ. Mitochondria as a source of reactive oxygen species during reductive stress in rat hepatocytes. Am J Physiol. 1993;264:961–967. doi: 10.1152/ajpcell.1993.264.4.C961. [DOI] [PubMed] [Google Scholar]
- 45.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York: W.H. Freeman & Co Ltd; 2008. [Google Scholar]
- 46.Newmeyer DD, Ferguson-Miller S. Mitochondria: Releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 47.Augenlicht LH, Heerdt BG. Mitochondria: Integrators in tumorigenesis? Nat Genet. 2001;28:104–105. doi: 10.1038/88800. [DOI] [PubMed] [Google Scholar]
- 48.Heerdt BG, Houston MA, Anthony GM, Augenlicht LH. Mitochondrial membrane potential ([delta psi](mt)) in the coordination of p53-independent proliferation and apoptosis pathways in human colonic carcinoma cells. Cancer Res. 1998;58:2869–2875. [PubMed] [Google Scholar]
- 49.Summerhayes IC, Lampidis TJ, Bernal SD. Unusual retention of rhodamine 123 by mitochondria in muscle and carcinoma cells. Proc Natl Acad Sci USA. 1982;79:5292–5296. doi: 10.1073/pnas.79.17.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heerdt BG, Houston MA, Augenlicht LH. Growth properties of colonic tumor cells are a function of the intrinsic mitochondrial membrane potential. Cancer Res. 2006;66:1591–1596. doi: 10.1158/0008-5472.CAN-05-2717. [DOI] [PubMed] [Google Scholar]
- 51.Heerdt BG, Houston MA, Augenlicht LH. The intrinsic mitochondrial membrane potential of colonic carcinoma cells is linked to the probability of tumor progression. Cancer Res. 2005;65:9861–9867. doi: 10.1158/0008-5472.CAN-05-2444. [DOI] [PubMed] [Google Scholar]
- 52.Zhen Y, et al. Overexpression and purification of cytochrome c oxidase from Rhodobacter sphaeroides. Protein Expr Purif. 1998;13:326–336. doi: 10.1006/prep.1998.0903. [DOI] [PubMed] [Google Scholar]
- 53.Jasaitis A, Verkhovsky MI, Morgan JE, Verkhovskaya ML, Wikström M. Assignment and charge translocation stoichiometries of the major electrogenic phases in the reaction of cytochrome c oxidase with dioxygen. Biochemistry. 1999;38:2697–2706. doi: 10.1021/bi982275l. [DOI] [PubMed] [Google Scholar]
- 54.Faxén K, Brzezinski P. The inside pH determines rates of electron and proton transfer in vesicle-reconstituted cytochrome c oxidase. Biochim Biophys Acta. 2007;1767:381–386. doi: 10.1016/j.bbabio.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 55.Brändén M, et al. On the role of the K-proton transfer pathway in cytochrome c oxidase. Proc Natl Acad Sci USA. 2001;98:5013–5018. doi: 10.1073/pnas.081088398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wikström M, Jasaitis A, Backgren C, Puustinen A, Verkhovsky MI. The role of the D- and K-pathways of proton transfer in the function of the haem-copper oxidases. Biochim Biophys Acta. 2000;1459:514–520. doi: 10.1016/s0005-2728(00)00191-2. [DOI] [PubMed] [Google Scholar]
- 57.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.