Abstract
The end of the Last Glacial Maximum (LGM) dramatically reshaped temperate ecosystems, with many species moving poleward as temperatures rose and ice receded. Whereas reinvading terrestrial taxa tracked melting glaciers, marine biota recolonized ocean habitats freed by retreating sea ice. The extent of sea ice in the Southern Hemisphere during the LGM has, however, yet to be fully resolved, with most palaeogeographic studies suggesting only minimal or patchy ice cover in subantarctic waters. Here, through population genetic analyses of the widespread Southern Bull Kelp (Durvillaea antarctica), we present evidence for persistent ice scour affecting subantarctic islands during the LGM. Using mitochondrial and chloroplast genetic markers (COI; rbcL) to genetically characterize some 300 kelp samples from 45 Southern Ocean localities, we reveal a remarkable pattern of recent recolonization in the subantarctic. Specifically, in contrast to the marked phylogeographic structure observed across coastal New Zealand and Chile (10- to 100-km scales), subantarctic samples show striking genetic homogeneity over vast distances (10,000-km scales), with a single widespread haplotype observed for each marker. From these results, we suggest that sea ice expanded further and ice scour during the LGM impacted shallow-water subantarctic marine ecosystems more extensively than previously suggested.
Keywords: Durvillaea antarctica, genetic, phylogeography, raft
Climate change is a major force driving population extinctions, particularly near the limits of a species' range (1). During glacial maxima, many taxa retreat from the cooling poles into lower-latitude refugia, and subsequent interglacial recolonization of high-latitude habitats can occur rapidly, especially in highly dispersive taxa, as indicated by the genetic homogeneity of reinvading populations (1–3). Although such evolutionary patterns have been established for a variety of Northern Hemisphere species (1–7), relatively little is known about the biotic effects of recent glaciations in the more oceanic Southern Hemisphere. Nevertheless, it seems likely that ice-sensitive marine biota of the subantarctic would have followed comparable extirpation–recolonization patterns as receding sea ice “unlocked” the Southern Ocean. Contemporary ice scouring, for example, is known to purge much of the shallow water benthos within current Antarctic sea ice range (8, 9), and any coastlines within Last Glacial Maximum (LGM) sea ice limits are likely to have been similarly affected.
Sea ice can strongly affect global systems, influencing ocean circulation patterns, levels of reflected radiation (10, 11), and rates of climate change (12). Our understanding of LGM ice conditions in the south, however, remains incomplete. The best current estimate of Southern Hemisphere LGM sea ice cover, by Gersonde et al. (13), is an amalgamation of data from numerous previous studies and highlights substantial information gaps in the subantartic region. To date, most LGM sea ice estimates have been derived from subfossil diatom data [reviewed by Gersonde et al. (13)]. The advent of modern molecular techniques, however, provides an alternative means of assessing organismal history, and thereby shedding light on historical climate conditions (1, 14). Here, in a genetic approach to assessing past sea ice conditions, we use phylogeographical analyses of an ice-sensitive subantarctic macroalga to infer the extent of winter sea ice (WSI) during the LGM.
Southern bull kelp (Durvillaea antarctica) is a keystone species of cool-temperate intertidal ecosystems across much of the Southern Hemisphere (15, 16). One of the world's largest kelps (17), it is found in the intertidal and upper subtidal of almost all subantarctic islands, as well as along the coasts of New Zealand (NZ), Chile, and the Falkland Islands (18). Importantly, however, this species' southern range limits correspond to the northern extent of sea ice (18); it is entirely absent from severely ice-affected areas, such as the shores of South Shetland, South Orkney, South Sandwich, and Bouvet Islands (18), and from glaciated regions of Heard Island (16) and South Georgia (19). Although some macroalgal species may survive in ice-scoured regions, D. antarctica is completely eliminated (20), and its presence or absence has even been suggested as an indicator of ice scour extent (20), making this taxon an ideal model organism for studies of LGM sea ice. In addition, D. antarctica is robust and buoyant, and hence has extremely high dispersal potential: once detached, it may be capable of rafting vast distances (18, 21–24). Here, by using DNA sequence analysis (mitochondrial COI; chloroplast rbcL) to assess circum-subantarctic phylogeographic patterns of D. antarctica (≈300 specimens, 45 localities), we test the hypotheses that: (i) bull kelp was eliminated from ice-affected subantarctic regions during the LGM, resulting in genetic homogeneity of reinvading populations, and (ii) rafting of the buoyant adult plants facilitated rapid recolonization.
Results
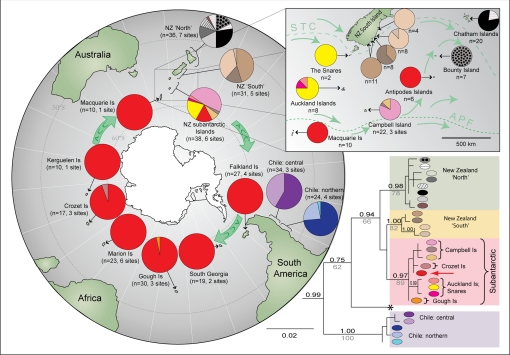
Mitochondrial and chloroplast markers (COI; rbcL) both revealed substantial genetic diversity within D. antarctica (uncorrected distances up to 5.4% for COI), with well-supported clades (or divergent haplotypes) corresponding to northern NZ, southern NZ, Chile, and the subantarctic region [Fig. 1, COI; and supporting information (SI) Fig. S1, rbcL]. Although the clustering of samples into these 4 major geographic clades was essentially identical across both data sets (posterior probabilities 0.97–1.00 for COI; Fig. 1), deeper nodes had weaker phylogenetic support, particularly for the less-informative rbcL marker, and tree topologies differed. The morphologically and genetically distinct “cape” D. antarctica lineage, recently identified from southern NZ, the Snares, and the Auckland Islands (indicated by asterisks in Fig. 1 and Fig. S1) (25), was clearly distinguishable from all 4 of these geographic clades. This undescribed lineage was not detected at any sites outside the NZ region and is not considered subsequently in this paper. Phylogenetic analyses revealed dramatic geographic contrasts in levels of genetic differentiation among populations. At temperate latitudes (NZ and Chile), strong geographic partitioning of genetic variation was observed across small spatial scales (tens to hundreds of kilometers apart), whereas samples from high latitudes (red haplotype; Figs. 1 and 2) showed genetic homogeneity across vast geographic distances.
Fig. 1.
Phylogeographic relationships within D. antarctica based on COI data. The phylogenetic tree (lower right corner) indicates haplotype relationships, with Bayesian PP values above branches and ML bootstraps below. Outgroup taxa have been removed for clarity. “NZ subantarctic” refers to the Snares, Auckland, Campbell, and Antipodes Islands. The morphologically and genetically distinct “cape” form of D. antarctica, recently identified by Fraser et al. (25) from southeastern NZ, is indicated by an asterisk. The global projection shows haplotype distributions and proportions at each locality. Diversity in the southern New Zealand region is illustrated at higher magnification (Inset, upper right). Green arrows show major surface currents.
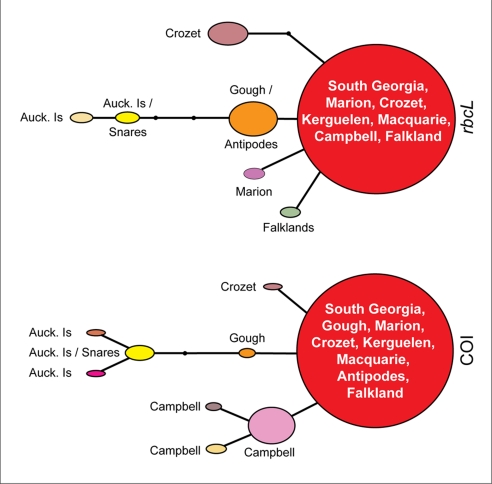
Fig. 2.
Haplotype network diagrams for the “subantarctic” clade of D. antarctica for both mtDNA (COI) and chloroplast (rbcL) datasets. Circle size is scaled according to haplotype frequency. Black dots represent hypothetical haplotypes not detected in the current study.
Levels of population differentiation were particularly high around the coasts of mainland NZ, with geographically proximate samples exhibiting clear genetic divergence (Fig. 1 and Fig. S1). Similarly, Chilean samples were distinguished by considerable genetic diversity among sites and by a north–south phylogeographic break between 36°36′S and 39°49′S for COI (Fig. 1). In contrast, southern subantarctic samples exhibited remarkable genetic homogeneity across both molecular datasets. Of the 174 subantarctic samples sequenced for COI, 140 yielded a single widespread haplotype (red haplotype in Fig. 1). Of the 8 additional subantarctic COI haplotypes, 6 were restricted to the NZ subantarctic region. Similar results were obtained for chloroplast DNA (rbcL), although this less-informative marker exhibited lower overall diversity and weaker deep-node phylogenetic support than COI.
Haplotype networks constructed for subantarctic COI and rbcL datasets (Fig. 2) show strong congruence, each characterized by a widespread (red) haplotype common to South Georgia, Marion, Kerguelen, Crozet, Macquarie, and the Falkland Islands. In each case, a “Gough Island” haplotype was one mutation step away and was intermediate to an Auckland Islands/Snares cluster. Interestingly, although the Antipodes Island samples yielded the widespread COI haplotype, the same samples gave a distinct haplotype for rbcL (also shared with Gough Island samples; Fig. 2 and Fig. S1).
The unimodal mismatch distribution for subantarctic COI haplotypes was consistent with the expectations of a sudden population expansion model (26), with no significant deviation from the expected distribution (P > 0.05). The Fu neutrality test (27) also supported the subantarctic population expansion, with stasis rejected (FS = −5.63, P sim FS ≤ obs FS = 0.01).
Discussion
Our circum-subantarctic analyses of DNA variation in D. antarctica support the hypothesis that this kelp species only recently recolonized the subantarctic region; the species exhibits a striking degree of genetic homogeneity in these regions compared with lower-latitude putative glacial refugia, a pattern typical of rapidly expanding, reinvading populations (1–3). Recolonization is likely to have involved a series of long-distance rafting events of the buoyant kelp from remote source populations (21, 24). Our findings strongly suggest that subantarctic kelp populations were eliminated during the LGM, most plausibly via ice scour. Although this interpretation conflicts with recent oceanographic reconstructions, which suggest LGM sea ice did not reach many subantarctic islands (Fig. 3) (13), it should be noted that fine-scale LGM data are still lacking for many Southern Ocean regions (Fig. 3) (13). On the basis of our analysis, therefore, we suggest that LGM subantarctic sea ice extended substantially further north than is currently recognized. More broadly, our study emphasizes the utility of phylogeographic analysis for studying the evolutionary effects of climate change and for potentially elucidating paleoenvironmental history (28).
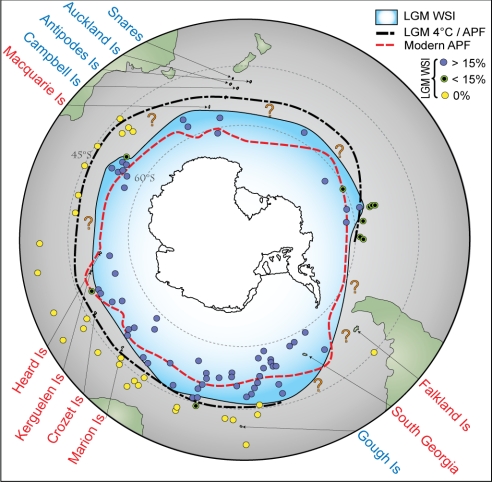
Fig. 3.
Positions of the subantarctic islands in relation to present-day conditions and reconstructed LGM oceanographic features, including LGM WSI extent, modified from Gersonde et al. (13). Dot points indicate sediment core sites used to reconstruct LGM sea ice cover (after ref. 13): yellow dots contained no evidence of sea ice-associated diatoms, green dots indicate (minimal) evidence, and blue dots indicate high proportions of ice-indicator diatoms. Regions demarcated by question marks lack core data (13). Based on D. antarctica genetic data, red labels indicate “recolonized” islands putatively affected by LGM ice scour, whereas blue labels indicate putative “refugial” islands. The LGM 4 °C isotherm has been identified as a feature roughly equivalent, in terms of water properties and gradients, to the modern APF (13).
Recent Recolonization of Bull Kelp in the Subantarctic.
Although the widespread D. antarctica is a genetically diverse species, the vast majority of this variation is restricted to temperate latitudes. Within NZ and Chile, for example, substantial genetic differentiation was detected between populations separated by relatively small geographic distances (e.g., tens of kilometers). In high subantarctic latitudes, by contrast, genetic homogeneity was detected over vast geographic distances. For instance, South Georgia, Marion, Crozet, Kerguelen, Macquarie, and the Falkland Island samples were essentially indistinguishable at both COI and rbcL. This striking latitudinal contrast in diversity strongly suggests recent colonization of the southern islands from more northern refugial population(s), mirroring postglacial recolonization patterns observed in numerous Northern Hemisphere taxa (1–7). Although we are unable to accurately estimate the timing of recolonization because of a lack of robust mutation rate estimates (7), circumpolar genetic homogeneity certainly seems consistent with postglacial origins. Latitudinal variation in phylogenetic diversity was also detected within the subantarctic region itself, with northern “refugial” islands (e.g., Auckland Islands and the Snares) represented by distinct genetic assemblages (Figs. 1 and 2) relative to the homogeneous “recolonized” southern subantarctic populations.
Although a number of factors could potentially explain the apparent LGM extinction of bull kelp in the subantarctic, sea ice scouring remains the most plausible. Low temperatures alone cannot account for its LGM extinction in the subantarctic. Although D. antarctica thrives in the comparatively warm waters of Gough Island (≈12 °C), northern NZ (up to 22 °C), and Chile (up to about 16 °C), the species also currently flourishes in remarkably cold waters, persisting in ice-free areas of partially glaciated Heard Island (16) and South Georgia (20), where water temperatures are near freezing (<2 °C). Additionally, although it might be suggested that the lowered LGM coastlines in some sandy regions (e.g., the Falkland Islands) lacked suitable rocky shore habitat for D. antarctica, this explanation cannot apply to steep volcanic (e.g., Marion, Crozet) and tectonic (Macquarie) islands, which must have offered abundant rocky habitat during glacial periods. Finally, although a few subantarctic islands (South Georgia, Kerguelen) were extensively glaciated during the LGM, most (Macquarie, Falkland, Gough, Crozet, Campbell) had relatively scant terrestrial ice cover (29, 30). In contrast to the above explanations, ice scour has been observed to eliminate intertidal and shallow water benthos in the Southern Ocean (8, 9, 20), and the presence/absence of D. antarctica has even been suggested as an indicator of contemporary ice scour (20).
Ice Scour in the Subantarctic During the LGM.
Credible, data-rich estimates of the extent of sea ice in the Southern Hemisphere during the LGM have existed for only a few decades [reviewed by Armand and Leventer (10) and Gersonde et al. (13)] and are largely based on ice-indicator diatoms from dated sediment cores (13, 32–34). These analyses typically differ in their estimates of LGM winter sea ice, often by several degrees of latitude. Gersonde et al. (13) synthesized these datasets to generate new estimates of LGM sea ice (Fig. 3) but acknowledged that the analyses lacked data for several large areas of the Southern Ocean. In particular, the edge of the LGM sea ice zone (region of 15% ice cover) is poorly constrained for many regions. In the Indian Ocean sector, for example, a single data point (indicating 2.6% ice concentration) constrains the estimated LGM ice edge to the south of Crozet and Marion Islands. Similarly, the Macquarie Island and Falkland Island regions lack adequate data coverage (Fig. 3). On the basis of our genetic data for D. antarctica, we suggest it is premature to discount significant LGM sea ice at such subantarctic localities.
Southern Ocean marine ecosystems are heavily influenced by the Antarctic Polar Front (APF), a circumpolar oceanographic and biogeographic boundary that separates biologically diverse cool-temperate communities from species-poor, cold-water communities (35). Today, the winter sea ice edge roughly coincides with the APF (36). Although there is some debate as to whether the APF itself, which is strongly linked to bathymetric features, shifted northward during the LGM, there is little doubt that the surface isotherm separating cold polar water from warmer, saltier, “subantarctic” water lay considerably further north than at present (36), probably on or about the estimated LGM 4 °C isotherm (13); indeed, Gersonde et al. (13) mark this line as the “LGM APF” (Fig. 3). The 4° isotherm was situated north of most of the “recolonized” (genetically homogeneous) subantarctic kelp populations, whereas the genetically divergent “refugial” subantarctic populations (e.g., Campbell, Auckland, Snares) were all located to the north of this isotherm (Fig. 3). Based on this observation, we suggest that the 4° isotherm (putatively the LGM equivalent of the modern APF) may represent a proxy for the extent of LGM sea ice scour.
Drifting on Shifting Seas: Kelp Dispersal via Rafting.
Rafting of buoyant kelp is likely to be an important ecological phenomenon, facilitating dispersal both of the kelp itself and of associated invertebrate fauna (22, 23, 37–40). Some robust species of brown (phaeophycean) kelp can potentially remain alive and reproductively viable for long periods adrift at sea (e.g., Macrocystis: refs. 41 and 42; Hormosira: ref. 43). The Antarctic Circumpolar Current (ACC; Fig. 1), a surface current driven by strong westerly winds circling Antarctica, is thought to facilitate dispersal of many Southern Ocean marine taxa (24). The sheer numbers of detached D. antarctica rafts in the Southern Ocean, estimated to be in the region of 70 million at any time (21), make effective eastward dispersal via rafting in the ACC a very real possibility for this species (24). In addition, bull kelp in the subantarctic appears to have an extended period of fertility: although D. antarctica in NZ is known to release gametes for 3 to 5 months per year (44, 45), plants on Macquarie Island remain fertile for more than 7 months of the year (16), and similarly long reproductive phases appear to occur on Marion Island, Gough Island, the Antipodes Islands, and South Georgia (45). D. antarctica reproduces in winter months, when water temperatures are coldest, and it is possible that the species' extended period of fertility in the subantarctic is simply due to lower overall water temperatures, permitting longer optimal reproductive conditions; such a characteristic might well increase the chances of successful gamete production in rafting plants, and thereby enhance colonization ability, at high latitudes.
Although some macroalgal species owe their invasion success to anthropogenic translocation (46), D. antarctica has intrinsically high dispersal potential because of its rafting ability. Given this propensity to disperse, it seems intriguing that D. antarctica exhibits such strong phylogeographic structure along the temperate coasts of NZ and Chile. Disturbances have been shown to facilitate colonization by dispersive weed species (47), and we speculate that D. antarctica is effectively one such opportunistic “weed,” whereby rafting plants permit rapid colonization of disturbed or vacant habitats but have little genetic impact on intact kelp assemblages. Specifically, we predict that in dense stands of D. antarctica, eggs from resident female plants may be almost immediately fertilized by spermatozoa from neighboring male plants, swamping the comparatively small number of immigrant gametes provided by any rafted individuals. The likely role of density blocking by leading-edge migrants of recolonizing populations has been well documented in Northern Hemisphere terrestrial taxa (1), and it could clearly apply to marine systems as well. Future research using microsatellite DNA markers will assess fine-scale genetic connectivity among D. antarctica populations.
Materials and Methods
Sites and Sample Collection.
Tissue samples from fresh frond tips of D. antarctica were collected from 45 localities: 11 from the NZ mainland, 7 from the Chilean mainland, and the remainder from subantarctic islands (Table S1). Wherever possible, several sites were sampled per region (e.g., multiple localities across an island; Fig. 1 and Table S1). Samples were collected by hand from plants growing in the intertidal to subtidal, and an effort was made to sample plants from a range of morphologies and tidal depths at each site.
DNA Sequencing and Analyses.
All DNA extractions and PCRs were carried out per Fraser et al. (25) by using the primers GazF1 and GazR1 (48) to amplify an ≈700-bp region of COI, and the primers KL2 and KL8 (49) for an ≈1,000-bp region of rbcL. Phylogenetic relationships were determined by maximum-likelihood (ML) and Bayesian analyses and included outgroup sequences from Durvillaea willana (Brighton, South Island, NZ) and Durvillaea potatorum (Tathra, Australia), as well as published sequences of Fucus (GenBank accession nos.: AY494079, Fucus vesiculosus for COI; and AF195515, Fucus gardneri for rbcL). ML analyses were performed with an HKY + I + G model for COI (base frequencies: A = 0.2182, C = 0.1586, G = 0.1884, and T = 0.4348; proportion of invariable sites = 0.6785) and a TrN + I + G model for rbcL (base frequencies: A = 0.2974, C = 0.1592, G = 0.2140, and T = 0.3294; rate matrix: A–C = 1.000, A–G = 4.0318, A–T = 1.000, C–G = 1.000, C–T = 10.5062, and G–T = 1.000; proportion of invariable sites = 0.7802) as selected by the AIC (COI) and the (h)LRT (rbcL) of Modeltest 3.06 (50). Relative levels of phylogenetic support for each node were assessed by bootstrapping (51), with heuristic analysis of 1,000 replicate datasets. Bayesian posterior probability (PP) values were calculated by using MRBAYES 3.1.2 (52) with separate substitution models for COI and rbcL determined by Modeltest. Markov Chain Monte Carlo searches were performed, each with 4 chains of 5 million generations, and trees were sampled every 100 generations. The first 10,000 trees sampled were discarded as “burn-in,” as determined by stationarity of lnL and other parameters assessed by using Tracer v1.4 (53), and the remaining trees were used to calculate a consensus topology and posterior probability values. Network analysis was performed by using TCS 1.21 (54) to explore relationships among closely related subantarctic haplotypes. Historic population expansions were assessed in the subantarctic clade by the Fu neutrality test (27) and mismatch distribution analysis (26) using Arlequin version 2.0 (55). The fit of the observed to the expected distribution under the sudden expansion model was tested by using bootstrapping. We are presently unable to accurately estimate the time since population expansion because of the lack of robust mutation rate estimates for marine heterokont algal mtDNA and cpDNA (7). Although some ancient calibration points exist for diatom taxa (see references in ref. 7), time-dependency issues preclude extrapolation of these calibrations to recent (e.g., Holocene or late Pleistocene) biological events (56, 57).
Supplementary Material
Acknowledgments.
We are enormously grateful to the numerous individuals and organizations who assisted with and funded this work. We thank the South African National Antarctic Program for supporting field trips to Marion and Gough Islands. For field support/sample collection, we thank: Isabelle Ansorge and Rob Anderson (University of Cape Town, Cape Town, South Africa); Paul Brickle, Judith Brown and Ian Strange for field assistance in the Falkland Islands; Marc Lebouvier, Sylvain Gutjahr, Sébastien Mallol, Jean-Philippe Orts, and David Renault (French Polar Institute, IPEV program 136) for collections from Kerguelen and Crozet Islands; Rachel Hadden and Charles Swift (British Antarctic Survey), and the government of South Georgia for collections from South Georgia; Simon Banks (Department of Conservation, Wellington, New Zealand), Gary Wilson and Amélie Augé (University of Otago, Dunedin, New Zealand) for additional collections from Campbell and Enderby Islands; Annelise Wiebkin, James Doube and Robb Clifton (Australian Antarctic Division) for collections from Macquarie Island; Robyn Dunmore (Cawthron Institute, Nelson, New Zealand) for Raramai collections; David Yáñez Jaramillo, Erasmo Macaya Horta, and Martin Thiel for help with Chilean fieldwork; Cameron Hay for advice and for facilitating many NZ collections; and the numerous other volunteers who helped in the field. For laboratory assistance and advice, we thank Tania King, Martyn Kennedy, and Yuri Springer; and for comments on geological aspects, Dave Craw. We thank Heritage Expeditions for supporting trips to the New Zealand subantarctic islands via an Enderby Scholarship. This work was funded by Marsden Contract 07-UOO-099, Department of Zoology and University of Otago Research grants (to J.M.W.); a Shackleton Scholarship (to C.I.F.); a Department of Zoology funding allocation (to C.I.F.); and Allan Wilson Centre for Molecular Ecology and Evolution funds (to H.G.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.R.C. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ550086-FJ550119 and FJ550121-FJ550129).
This article contains supporting information online at www.pnas.org/cgi/content/full/0810635106/DCSupplemental.
References
- 1.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 1996;58:247–276. [Google Scholar]
- 2.Hewitt GM. The genetic legacy of Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 3.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc London B Biol Sci. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marko PB. ‘What's larvae got to do with it?’ Disparate patterns of post-glacial population structure in two benthic marine gastropods with identical dispersal potential. Mol Ecol. 2004;13:597–611. doi: 10.1046/j.1365-294x.2004.02096.x. [DOI] [PubMed] [Google Scholar]
- 5.Edmands S. Phylogeography of the intertidal copepod Tigriopus californicus reveals substantially reduced population differentiation at northern latitudes. Mol Ecol. 2001;10:1743–1750. doi: 10.1046/j.0962-1083.2001.01306.x. [DOI] [PubMed] [Google Scholar]
- 6.Provan J, Wattier RA, Maggs CA. Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol Ecol. 2006;14:793–803. doi: 10.1111/j.1365-294X.2005.02447.x. [DOI] [PubMed] [Google Scholar]
- 7.Hoarau G, Coyer JA, Veldsink JH, Stam WT, Olsen JL. Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol Ecol. 2007;16:3606–3616. doi: 10.1111/j.1365-294X.2007.03408.x. [DOI] [PubMed] [Google Scholar]
- 8.Barnes DKA, Conlan K. Disturbance, colonization and development of Antarctic benthic communities. Philos Trans R Soc London B Biol Sci B. 2007;362:11–38. doi: 10.1098/rstb.2006.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutt J. On the direct impact of ice on marine benthic communities, a review. Polar Biol. 2001;24:553–564. [Google Scholar]
- 10.Armand LK, Leventer A. In: Sea Ice: An Introduction to Its Physics, Chemistry, Biology and Geology. Thomas DN, Dieckermann GS, editors. Oxford: Blackwell; 2003. pp. 333–372. [Google Scholar]
- 11.Clark PU, Alley RB, Pollard D. Northern hemisphere ice-sheet influences on global climate change. Science. 1999;286:1104–1111. [Google Scholar]
- 12.Keeling RF, Stephens BB. Antarctic sea ice and the control of Pleistocene climate instability. Paleoceanography. 2001;16:112–131. [Google Scholar]
- 13.Gersonde R, Crosta X, Abelmann A, Armand L. Sea-surface temperature and sea ice distribution of the Southern Ocean at the EPILOG Last Glacial Maximum–a circum-Antarctic view based on siliceous microfossil records. Quat Sci Rev. 2005;24:869–896. [Google Scholar]
- 14.Clarke A, Johnston NM, Murphy EJ, Rodgers AD. Introduction. Antarctic ecology from genes to ecosystems: The impact of climate change and the importance of scale. Philos Trans R Soc London B Biol Sci. 2007;362:5–9. doi: 10.1098/rstb.2006.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haxen PG, Grindley JR. In: Antarctic Nutrient Cycles and Food Webs. Siegfried WR, Condy PR, Laws RM, editors. Berlin: Springer; 1985. pp. 637–640. [Google Scholar]
- 16.Klemm MF, Hallam ND. Standing crop of Durvillaea antarctica (Chamisso) Hariot (Phaeophyta) on the Australian sub-Antarctic Macquarie and Heard islands. Phycologia. 1988;27:505–509. [Google Scholar]
- 17.Stevens CL, Hurd CL, Smith MJ. Field measurement of the dynamics of the bull kelp Durvillaea antarctica (Chamisso) Heriot. J Exp Mar Biol Ecol. 2002;269:147–171. [Google Scholar]
- 18.Hay C. A phytogeographical account of the southern bull kelp seaweeds Durvillaea spp. Bory 1826 (Durvilleales Petrov 1965). Proceedings of the International Symposium of Marine Biogeography and Evolution in the Southern Hemisphere; Auckland, New Zealand: Scient Ind Res; 1979. pp. 443–454. [Google Scholar]
- 19.Hay CH. The occurrence of Durvillaea antarctica (Durvillaeales, Phaeophyta) at South Georgia, South Atlantic Ocean. Phycologia. 1988;27:424–427. [Google Scholar]
- 20.Pugh PJA, Davenport J. Colonisation vs. disturbance: The effects of sustained ice-scouring on intertidal communities. J Exp Mar Biol Ecol. 1997;210:1–21. [Google Scholar]
- 21.Smith SDA. Kelp rafts in the Southern Ocean. Glob Ecol Biogeogr. 2002;11:67–69. [Google Scholar]
- 22.Donald KM, Kennedy M, Spencer HG. Cladogenesis as the result of long-distance rafting events in South Pacific topshells (Gastropoda, Trochidae) Evolution. 2005;59:1701–1711. [PubMed] [Google Scholar]
- 23.Thiel M, Gutow L. The ecology of rafting in the marine environment. I. The floating substrata. Oceanogr Mar Biol. 2005;42:181–264. [Google Scholar]
- 24.Waters JM. Driven by the West Wind Drift? A synthesis of southern temperate marine biogeography, with new directions for dispersalism. J Biogeogr. 2008;35:417–427. [Google Scholar]
- 25.Fraser CI, Hay CH, Spencer HG, Waters JM. Genetic and morphological analyses of the southern bull kelp Durvillaea antarctica (Phaeophyceae: Durvillaeales) in New Zealand reveal cryptic species. J Phycol. 2009 doi: 10.1111/j.1529-8817.2009.00658.x. in press. [DOI] [PubMed] [Google Scholar]
- 26.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 27.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craw D, Burridge C, Norris R, Waters J. Genetic ages for Quaternary topographic evolution: A new dating tool. Geology. 2008;36:19–22. [Google Scholar]
- 29.Chown SL, Gremmen NJM, Gaston KJ. Ecological biogeography of Southern Ocean islands: Species-area relationships, human impacts, and conservation. Am Nat. 1998;152:562–575. doi: 10.1086/286190. [DOI] [PubMed] [Google Scholar]
- 30.Hall K. In: Quaternary Glaciations–Extent and Chronology. Part III. Ehlers J, Gibbard PL, editors. Amsterdam: Elsevier; 2004. pp. 339–345. [Google Scholar]
- 31.CLIMAP Project Members. Map Series, Technical Report MC-36. Boulder, CO: Geological Society of America; 1981. Seasonal Reconstructions of the Earth's Surface at the Last Glacial Maximum; pp. 1–18. [Google Scholar]
- 32.Cooke DW, Hays JD. In: Antarctic Geoscience. No 4. Craddock C, editor. Madison, WI: Univ Wisconsin Press; 1982. pp. 1017–1025. IUGS Series B. [Google Scholar]
- 33.Crosta X, Pichon JJ, Burckle LH. Application of the modern analog technique to marine Antarctic diatoms: Reconstruction of maximum sea ice extent during the Last Glacial Maximum. Paleoceanography. 1998;13:284–297. [Google Scholar]
- 34.Gersonde R, et al. Last glacial sea surface temperatures and sea-ice extent in the Southern Ocean (Atlantic–Indian sector): A multiproxy approach. Paleoceanography. 2003;18:1061. [Google Scholar]
- 35.Bergstrom DM, Chown SL. Life at the front: History, ecology and change on southern ocean islands. Trends Ecol Evol. 1999;14:472–473. doi: 10.1016/s0169-5347(99)01688-2. [DOI] [PubMed] [Google Scholar]
- 36.Moore JK, Abbott MR, Richmand JG, Nelson DM. The Southern Ocean at the last glacial maximum: A strong sink for atmospheric carbon dioxide. Global Biogeochem Cycles. 2000;14:455–475. [Google Scholar]
- 37.Edgar GJ. Dispersal of faunal and floral propagules associated with drifting Macrocystis pyrifera plants. Mar Biol. 1987;95:599–610. [Google Scholar]
- 38.Waters JM, Roy MS. Out of Africa: The slow train to Australasia. Syst Biol. 2004;53:1063–5157. doi: 10.1080/10635150490264671. [DOI] [PubMed] [Google Scholar]
- 39.Thiel M, Gutow L. The ecology of rafting in the marine environment. II. The rafting organisms and community. Oceanogr Mar Biol. 2005;43:279–418. [Google Scholar]
- 40.Thiel M, Haye P. The ecology of rafting in the marine environment. III. Biogeographical and evolutionary consequences. Oceanogr Mar Biol. 2006;44:323–429. [Google Scholar]
- 41.Macaya EC, et al. Presence of sporophylls in floating kelp rafts of Macrocystis spp. (Phaeophyceae) along the Chilean Pacific coast. J Phycol. 2005;41:913–922. [Google Scholar]
- 42.Hernández-Carmona G, Hughes B, Graham MH. Reproductive longevity of drifting kelp Macrocystis pyrifera (Phaeophycaea) in Monterey Bay, USA. J Phycol. 2006;42:1199–1207. [Google Scholar]
- 43.McKenzie PF, Bellgrove A. Dispersal of Hormosira banksii (Phaeophycaea) via detached fragments: Reproductive viability and longevity. J Phycol. 2008;44:1108–1115. doi: 10.1111/j.1529-8817.2008.00563.x. [DOI] [PubMed] [Google Scholar]
- 44.Hay CH, South GR. Experimental ecology with particular reference to proposed commercial harvesting of Durvillaea (Phaeophyta, Durvillaeales) in New Zealand. Botanica Marina. 1979;22:431–436. [Google Scholar]
- 45.Hay C. In: Biology of Economic Algae. Akatsuka I, editor. The Hague: SPB Academic Publishing; 1994. pp. 353–384. [Google Scholar]
- 46.Voisin M, Engel CR, Viard F. Differential shuffling of native genetic diversity across introduced regions in a brown alga: Aquaculture vs. maritime traffic effects. Proc Natl Acad Sci USA. 2005;102:5432–5437. doi: 10.1073/pnas.0501754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hobbs RJ, Huenneke LF. Disturbance, diversity, and invasion. Implications for conservation. Conserv Biol. 1992;6:324–337. [Google Scholar]
- 48.Saunders GW. Applying DNA barcoding to red macroalgae: A preliminary appraisal holds promise for future applications. Philos Trans R Soc London B Biol Sci. 2005;360:1879–1888. doi: 10.1098/rstb.2005.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lane CE, Mayes C, Druehl LD, Saunders GW. A multi-gene molecular investigation of the kelp (Laminariales, Phaeophyceae) supports substantial taxonomic re-organization. J Phycol. 2006;42:493–512. [Google Scholar]
- 50.Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 51.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 52.Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 53.Rambaut A, Drummond AJ. Edinburgh: Institue of Evolutionary Biology, University of Edinburgh; 2007. Tracer. Version 1.4. Available at http://beast.bio.ed.ac.uk/tracer. [Google Scholar]
- 54.Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 55.Schneider SD, Roessli D, Excoffier L. Geneva: Genetics and Biometry Laboratory, Univ of Geneva; 2000. Arlequin: A software for population genetics data analysis. Version 2.001. Available at http://lgb.unige.ch/arlequin/ [Google Scholar]
- 56.Ho SYW, Phillips MJ, Cooper A, Drummond AJ. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- 57.Burridge CP, Craw D, Fletcher D, Waters JM. Geological dates and molecular rates: Fish DNA sheds light on time dependency. Mol Biol Evol. 2008;25:624–633. doi: 10.1093/molbev/msm271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.