Abstract
The folding energetics of the mono-N-glycosylated adhesion domain of the human immune cell receptor cluster of differentiation 2 (hCD2ad) were studied systematically to understand the influence of the N-glycan on the folding energy landscape. Fully elaborated N-glycan structures accelerate folding by 4-fold and stabilize the β-sandwich structure by 3.1 kcal/mol, relative to the nonglycosylated protein. The N-glycan's first saccharide unit accounts for the entire acceleration of folding and for 2/3 of the native state stabilization. The remaining third of the stabilization is derived from the next 2 saccharide units. Thus, the conserved N-linked triose core, ManGlcNAc2, improves both the kinetics and the thermodynamics of protein folding. The native state stabilization and decreased activation barrier for folding conferred by N-glycosylation provide a powerful and potentially general mechanism for enhancing folding in the secretory pathway.
Keywords: glycoprotein stability, N-glycan function, N-glycosylation, protein folding, kinetics
One-third of the eukaryotic proteome traverses the secretory pathway, the majority of which is N-glycosylated cotranslationally, a process that vastly expands the functional repertoire of the proteome. Glycosylation has been linked to intrinsic effects on proteins, such as altering structure and activity, and extrinsic effects, such as modulating macromolecular interactions (1). The ability of N-glycans to extrinsically enhance proper folding and reduce aggregation in vivo by recruiting lectin-type chaperones is well-established (2). N-glycans also stabilize protein structure (3), accelerate folding (4), promote secondary structure formation (5), reduce aggregation (4, 6), shield hydrophobic surfaces (4, 7), make disulfide pairing more efficient (8), and increase folding cooperativity (6), all through an apparent intrinsic chemical mechanism. The saccharide substructure of the N-glycan that confers these intrinsic structure stabilizing properties appears to be part of the highly conserved oligomannose N-glycan added cotranslationally to proteins (Fig. 1A) (9). However, a detailed analysis of the structural features that hasten folding and increase protein stability has not yet been conducted because of the challenge of assessing folding in vivo (9) and the difficulty of expressing and/or synthesizing homogeneous glycoproteins for in vitro studies (10, 11).
Fig. 1.
N-glycan structures examined on hCD2ad. (A) The conserved oligomannose N-glycan structure. Saccharide units are labeled a-n. Branches are labeled A, B, and C. Important substructures are shown by brackets: chitobiose, paucimannose, and oligomannose (5–9 Man units). (B) NMR structure of hCD2ad with N-glycan (green) at Asn-65 (red); Lys-61 and Phe-63 (magenta) can be mutated to the rat CD2ad sequence to stabilize nonglycosylated hCD2ad. (C) Variants of hCD2ad used. Species with abundance >5% are shown, except in the case of 3. Because of the complexity of the mixture of glycoforms that compose variant 3, we show only glycoforms with relative abundance >0.5. The small discrepancy in the extent of fucosylation between variants 4 and 5 is likely due to a slight enrichment of the fucosylated glycoform during affinity purification of the product of the enzymatic reaction. Also, the abundances reported for variant 4 are the averages from 3 expressions from Sf9 cells, and those for variant 5 are the averages from 2 preparations.
In eukaryotes, N-linked glycoproteins are biosynthesized by transferring a conserved triantennary N-glycan precursor (Glc3Man9GlcNAc2, Fig. 1A) en bloc to an asparagine residue within the amino acid sequence N-X-T/S during cotranslational translocation of the protein into the endoplasmic reticulum (ER) (12). The outer Glc residues (n, m, and l) and most of the outer mannose residues are rarely found on secreted proteins. Glc n and m are immediately hydrolyzed and Glc l is consumed in the calnexin/calreticulin (CNX/CRT)-assisted folding cycle (2). This cycle is thought to mediate folding and ER-associated degradation (ERAD) in the case of folding failures, a step facilitated by removal of distal Man residues on the B-/C-branches of the N-glycan (13). Although these extrinsic roles are important, the intrinsic effects of the N-glycan may be even more important for mediating proper folding. For example, the transfer of an N-glycan alone can rescue protein folding when CNX/CRT assistance is absent or precluded in vivo (9, 14). In vitro studies also demonstrate that glycoprotein folding is enhanced by the presence of N-glycans (3–6, 8).
Herein, we systematically evaluate substructures of the highly conserved oligomannose glycan (Fig. 1A) to discern which intrinsically accelerate protein folding and/or stabilize the native state of the adhesion domain of human cluster of differentiation 2 (hCD2ad; Fig. 1B), a member of the Ig superfamily (IgSF) that mediates formation of the immunological synapse (15). Kinetic and thermodynamic analysis of these glycoforms revealed that only the core triose, β-N-(Man-β1,4-GlcNAc-β1,4-N-GlcNAc)(ManGlcNAc2, saccharides a-c in Fig. 1A), is required to speed folding by 4-fold and stabilize the native state by 3.1 kcal/mol, equivalent to the stabilization afforded by the entire glycan. This stabilization energy corresponds to a factor of nearly 200 in the folding equilibrium constant, relative to the nonglycosylated protein.
Results
hCD2ad Variants.
hCD2ad is a representative member of the IgSF, a common protein fold with a β-sandwich topology, which is thought to have evolved to form stable protein modules at the cell surface (16). hCD2ad is small (105 aa); has no prolines or cysteines to complicate its folding kinetics; and has a single N-glycosylation site at Asn-65, which is in a type I β-turn flanked by strands D and E (Fig. 1B). The innermost G1cNAc2 disaccharide of the oligomannose N-glycan at Asn-65 makes contacts with strands B and E of hCD2ad (17), which are likely important, because strands B and E–G form a structural core common to IgSF folds that is thought to nucleate folding (18). The N-glycan in hCD2ad is necessary for both acquiring and maintaining proper structure (17, 19, 20); in contrast, rat CD2ad does not require an N-glycan to fold (21). Introducing elements of the rat sequence (Lys61Glu and Phe63Leu, shown in magenta in Fig. 1B) into human CD2ad near its glycosylation site enables hCD2ad to fold and maintain structure without its N-glycan (17), allowing for a comparison of the influence of mutation(s) vs. glycosylation.
The hCD2ad variants produced for this study include nonglycosylated, glycosylated, and remodeled glycoforms (Fig. 1C). Nonglycosylated wild type and stabilized mutant hCD2ad, incorporating the single Lys61Glu or double Lys61Glu/Phe63Leu rat mutations (see Fig. 1B), were produced in E. coli (1, 2a, and 2b, respectively). Glycosylated variants were prepared by expressing hCD2ad in HEK293 (human kidney) and Sf9 (insect) cells. Expression in HEK293 cells yielded hCD2ad variant 3 as a complicated mixture of hybrid, complex, and oligomannose glycoforms (Fig. 1C and Table S1). Expression in Sf9 cells yielded hCD2ad variant 4 as a mixture of fucosylated and unfucosylated paucimannose (Man3GlcNAc2) and oligomannose glycoforms (see Fig. 1C and Table S2). Key glycoform variants 5, 6, and 7 were produced by enzymatic remodeling of 4 (Figs. S1 and S2). The N-glycan of variant 5 consisted of the first GlcNAc, in both fucosylated and unfucosylated forms (3:1). That of variant 6 comprised fucosylated and unfucosylated Man1–2GlcNAc2 glycoforms. Finally, the N-glycan of variant 7 was a 9:1 mixture of 2 oligomannose glycoforms, namely Man6GlcNAc2 and Man7GlcNAc2 respectively (Fig. 1C and Table S3). Fig. 2A shows the purity of glycosylated and remodeled hCD2ad variants depicted by SDS/PAGE. Far-UV circular dichroism (CD) and/or fluorescence spectra of the stable variants 2-7 were consistent with previously reported spectra of properly folded hCD2ad (Fig. 2 B and C) (17). As expected, spectra for nonglycosylated wild type 1 suggested that it was largely unfolded; however, it could be induced to fold by adding the osmolyte proline (22) (Fig. 2C; see also SI Appendix).
Fig. 2.
Characterization of hCD2ad variants. (A) SDS/PAGE of glycosylated and enzymatically remodeled hCD2ad variants (3–7)—note that treating 4 with PNGase results in deglycosylation. (B) CD spectra of glycosylated (4) and nonglycosylated (2a) variants indicate that they are natively structured, whereas wild type nonglycosylated hCD2ad (1) shows little structure. (C) Fluorescence spectra of stable variants (2a, 2b, 3–7) are similar (λmax near 325 nm), indicating that they have similar native structures. Under native conditions, 1 resembles a denatured glycosylated variant (4 in 5 M urea, λmax 355 nm), as shown in Inset; 1 requires the addition of the osmolyte proline to adopt the native structure.
Folding Kinetics of hCD2ad Variants.
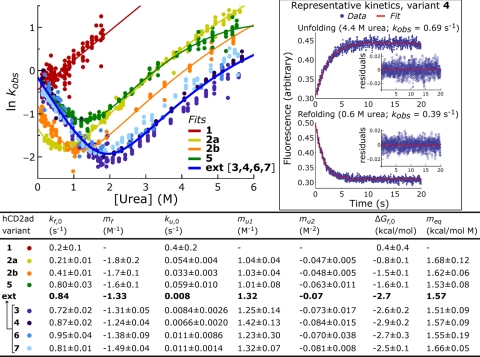
The folding kinetics of the hCD2ad variants were characterized by rapidly mixing protein dissolved in urea solutions with buffer to induce folding, or protein dissolved in buffer with urea solutions to induce unfolding. The conformational changes were followed by tryptophan fluorescence as a function of time. After correcting for photobleaching, the folding kinetics were found to be monoexponential for hCD2ad variants 3–7 (Fig. 3, representative data for 4 in boxed inset; Fig. S3, representative data for other variants) based on the small, generally pattern-free residuals in the fits to the folding kinetics (Fig. 3 and Fig. S3). Even for the few cases in which a slight pattern could be detected in the residuals (see the unfolding kinetics for variant 5 in Fig. S3), adding exponential terms to the fitting function rarely improved the fit substantially. The root mean squared (RMS) residuals improved by ≤1% upon adding a second exponential term for all but 50 of the 1,105 fits performed on the folding time courses of variants 3–7; the RMS residuals improved by ≤5% in all but 3 cases. This behavior demonstrates that the N-glycan heterogeneity present in variants 3–7 does not affect their folding rates; otherwise, the folding kinetics would necessarily be multiexponential. The monoexponential rate constant (kobs, where 1/kobs is the time needed for folding to be 37% complete) is equal to the sum of the folding (kf) and unfolding rate constants (ku). Plots of ln kobs vs. urea concentration for the hCD2ad variants yielded characteristic V-shaped (chevron) plots (Fig. 3). This V-shape arises because kobs is highest at low urea concentrations (because of fast folding) and high urea concentrations (because of fast unfolding), and lowest at moderate urea concentrations (because neither folding nor unfolding is particularly fast) (23). Because glycoform mixture 3 exhibits monoexponential folding and unfolding kinetics, and because the paucimannose substructure (residues a–e in Fig. 1A) is the only substructure common to the constituents of 3, it follows that paucimannose or a substructure thereof dominates the contribution of the N-glycan to hCD2ad folding. Likewise, variants 4-7 also display monoexponential kinetics, indicating that the contribution of the smallest glycan in their respective populations must equal that of the largest. For illustration, in the kinetics of variant 5, the protein bearing GlcNAc folds at the same rate as that with GlcNAcFuc—thus, the appended fucose does not affect the folding rate, at least in the β-turn context in which the hCD2ad glycan is attached.
Fig. 3.
Folding energetics of hCD2ad variants. Chevron plots are depicted for all variants. Data points represent experimentally determined monoexponential rates (kobs) for folding/unfolding at various concentrations of urea, [urea]. Chevron plots were fit to determine: ΔGf,0, kf,0 and ku,0, the free energy of folding, and the folding and unfolding rate constants, respectively, under native conditions; mf and mu1 the linear slopes of the dependences of folding and unfolding on [urea]; and mu2 a small quadratic term accounting for the slight curvature in plots of variants 2–7 at high [urea]. To characterize the folding energetics of 1, which only exhibits an unfolding arm, equilibrium and kinetic methods were combined, as described in SI Appendix. (Inset) Representative kinetic curves for 4.
Because variants 3, 4, 6, and 7 have similar chevron plots (Fig. 3), their kinetic and thermodynamic parameters are also similar. Indeed, the individual kf,0 and ku,0 values of these variants differ by <30% from the average of their values, the equivalent of 0.2 kcal/mol. Similarly, the individual ΔGf,0 values differ by <0.2 kcal/mol from their average. This grouping demonstrates that the effect of their N-glycans on the folding energetics of hCD2ad must arise from their common triose core, ManGlcNAc2 (a–c in Fig. 1A), and that differences in core fucosylation and distal saccharides have little, if any, influence on the folding free energy landscape. This observation is consistent with the conclusions made above based on the monoexponential folding kinetics of these variants. Thus, description of hCD2ad variants 3, 4, 6, and 7 will be based on their average parameters (Fig. 3, average data bolded in table). This grouping is called “ext-hCD2ad” because they all have extended glycans.
The total contribution of the N-glycan to hCD2ad folding can be assessed by comparing the folding of ext-hCD2ad to 1, the nonglycosylated wild type hCD2ad (the folding energetics of 1 were determined differently from those of the other variants, as described in Methods and SI Appendix, because it gives an incomplete chevron plot and requires osmolytes to fold). This comparison reveals that the N-glycan stabilizes the protein by 3.1 kcal/mol, a 200-fold increase in the folding equilibrium constant (Kf increases from 0.5 to 100). Approximately 3/4 of this stabilization (2.3 kcal/mol) is due to the N-glycan slowing the unfolding rate by 50-fold, whereas the remaining 1/4 (0.8 kcal/mol) is due to increasing the folding rate by 4-fold. We speculate on the origins of these changes in rate constants in the Discussion.
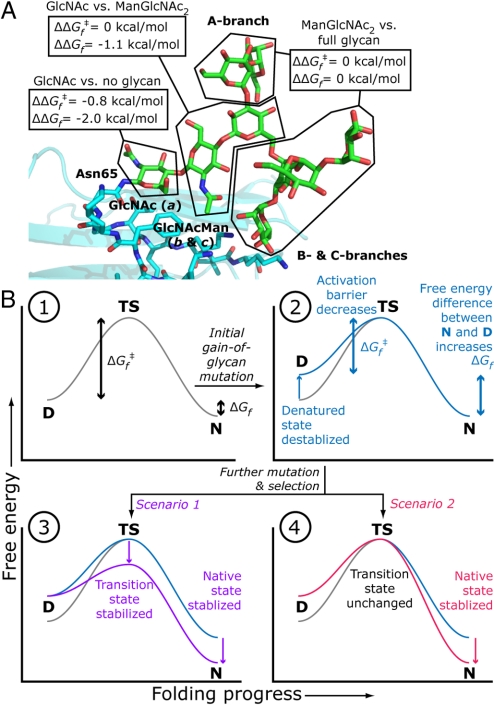
The intrinsic contribution of the N-glycan to protein folding energetics can be further parsed by adding variant 5, with its single GlcNAc, to the comparison. Variant 5 is more stable than 1 by 2.0 kcal/mol (corresponding to a 30-fold increase in Kf), and less stable than ext-hCD2ad by 1.1 kcal/mol (Figs. 3 and 4A). These data show that the majority (65%) of the fully elaborated oligosaccharide's contribution to hCD2ad stability is from the first GlcNAc residue. This observation is consistent with the structural biology of hCD2ad, which shows that this GlcNAc is the minimum glycan component required for stability and that it makes most of the N-glycan-protein contacts (17). Because we found that saccharide units other than a-c do not intrinsically contribute to protein folding (Figs. 3 and 4A), the remaining stabilization (1.1 kcal/mol) from the N-glycan can be attributed to the flanking ManGlcNAc (saccharide units b and c, Fig. 4A). Because the folding rate constant, kf,0, is nearly the same for 5 and ext-hCD2ad, it is clear that virtually all of the N-glycan's effect on the folding rate, which amounts to 0.8 kcal/mol, comes from the first GlcNAc (Fig. 4A). In the parlance of protein folding, this GlcNAc has a Φ value of 0.4. Φ values are formally defined by the equation Φ = ΔΔGf,0‡/ΔΔGf,0, where ΔΔG0 (= 0.8 kcal/mol) and ΔΔGf,0 (= 2.0 kcal/mol) are the differences in transition state and native state energies, respectively, for protein variants of interest (in this case, hCD2ad variants 1 and 5) (24). Φ values quantify the relative effects of a protein substructure on protein folding kinetics and thermodynamics: a Φ value of 1 indicates that the substructure affects kinetics and thermodynamics equally, and that it therefore is natively structured and/or forms its native contacts in the transition state; a Φ value of 0 indicates that it affects thermodynamics but not kinetics, and that it therefore is unstructured and does not form contacts in the transition state. The first GlcNAc's Φ value of 0.4 places it between these two extremes.
Fig. 4.
The effect of the N-glycan on folding energetics. (A) The contributions of the parts of the N-glycan of hCD2ad to the change in transition state (ΔΔGf‡) and native state energies (ΔΔGf). (B) An illustration of how a gain-of-glycosylation mutation and subsequent evolution could affect the folding free energy profile of a protein. D, denatured state; TS, transition state; N, native state; ΔGf‡, activation barrier for folding; ΔGf, free energy of folding. (1) Wild type protein. (2) Initial gain of glycosylation mutant with destabilized denatured state, which is thermodynamically equivalent to stabilized transition and native states. (3 and 4) Mature gain-of-glycosylation mutant in which mutation and selection after the gain-of-glycosylation mutation stabilize both the transition state and the native state (3) or just the native state (4).
The replacement of Lys-61 in the human sequence, which is among a surface cluster of basic amino acids near the N-glycan, with Glu from the rat sequence is especially important for stabilizing the native state of nonglycosylated CD2ad (17). Replacing Phe-63 in the human sequence with Leu from the rat sequence further stabilizes CD2ad (17). However, N-glycosylation (e.g., variants 3-7) more strongly influences protein folding energetics, in both a kinetic and a thermodynamic sense, than introducing these stabilizing amino acid mutations into hCD2ad. Although the 2a mutant (ΔGf = −0.8 kcal/mol for 2a) is nearly 10-fold more stable than 1 (ΔGf = +0.4 kcal/mol for 1), its folding rate is the same, indicating that the Lys61Glu mutation does not affect the folding transition state compared with nonglycosylated wild type (Φ value = 0). In contrast, the presence of any N-glycan affects the thermodynamic stability to a much greater degree in addition to increasing the folding rate, demonstrating a contribution to a higher energy denatured state/lower energy folding transition state for glycosylated versus wild-type hCD2ad, as discussed above. Thus, although hCD2ad may be unusual because it requires glycosylation not only to fold (a general property of N-linked glycoproteins), but to maintain structure [likely because of the glycan's unusual location near several positively charged amino acid side chains (17)], our results suggest that the glycan imparts folding benefits beyond those required simply to stabilize the cluster of positive charges that includes Lys-61.
The stabilized nonglysoylated hCD2ad 2b and the GlcNAcylated variant 5 have nearly identical ΔGf values (−1.5 kcal/mol). However, the folding and unfolding rate constants of the double mutant 2b are each lower by a factor of ≈2. This behavior is consistent with a higher transition state energy and/or a lower denatured state energy for 2b relative to 5. The possibility that the N-glycan raises the energy of the denatured state is of particular interest; this is discussed further below. It is also worth noting that the stabilized nonglycosylated variants 2a and 2b and GlcNAc-bearing variant 5 present similarly higher mf and lower mu values than those of the extended glycans (Fig. 3), where mf and mu are the slopes of the folding and unfolding wings on the chevron plot, respectively. The parameters mf and mu are proportional to the change in exposed surface area during folding and unfolding (25), so this observation suggests that in the ManGlcNAc2 triose core also influences surface area burial during folding, consistent with the observations in ref. 4.
Discussion
Although it is clear that certain N-glycans contribute to the folding and stabilization of glycoproteins (3–7), our results demonstrate and quantify the extent to which the triose core, ManGlcNAc2, stabilizes the native state and lowers the activation barrier for folding of hCD2ad. In particular, our results highlight the strong influence of the first GlcNAc residue on stability and the kinetics of protein folding (65% and 100% of the total N-glycan's contribution to hCD2d folding thermodynamics and kinetics, respectively, Fig. 4A). There are many lines of evidence that support the generality of the stabilizing influence of the N-linked triose core. For example, beyond the paucimannose region of mature N-glycans (Fig. 1), chemical and conformational heterogeneity increase, and few contacts are made with the attached protein, whereas the triose is within the strictly conserved paucimannose core, has a conformation that varies little from glycoprotein to glycoprotein, and makes most of the structurally observed protein-N-glycan contacts (3, 26). The presence of a single GlcNAc residue is known to significantly decrease structural dispersity and increase stability, which has enabled glycoprotein structure determinations (27). This observation substantiates the general importance of this residue to structural integrity, even though it is not always necessary to preserve a folded structure, as it is for hCD2ad (17, 19). Furthermore, the proximal N-linked GlcNAc of model synthetic N-linked glycopeptides is the most influential for rigidifying and biasing conformational space, including promoting β-turn and disulfide bond formation (5, 8, 28–30), consistent with GlcNAc a playing a general kinetic role in glycoprotein folding.
The conserved N-glycan (Fig. 1A) is synthesized and transferred onto proteins at great expense to the cell (2, 12). The conservation of the outer-branch saccharides of oligomannose glycans is explained by their pivotal roles in chaperone interactions (i.e., Glc l in the CNX/CRT-assisted folding cycle) and degradation (i.e., outer branch Man residues in ERAD) (9, 13). To the extent that our results with hCD2ad can be generalized to other N-glycosylated proteins, it is tempting to speculate that the intrinsic benefits of the ManGlcNAc2 triose to folding energetics may contribute to the strict conservation of the inner N-glycan core. Interestingly, some simple organisms lack complicated N-glycans, CNX/CRT, and N-glycan-associated ERAD, and transfer simple N-glycans (some truncated down to GlcNAc2) onto their glycoproteins (31), perhaps enabling them to maintain the beneficial intrinsic effects of N-glycans on folding energetics while saving cellular resources. In higher eukaryotes, the development of inner and outer substructures within the N-glycan to carry out intrinsic and extrinsic functions in protein folding likely emphasizes the biological importance of robust protein folding and stability, which are vital for folding and export of glycoproteins (32).
The native state stabilizing and folding accelerating attributes of N-glycans may account for their high rate of mutational introduction into proteins and their low rate of mutational ablation (33). Fig. 4B outlines an intriguing, if speculative, narrative for the molecular evolution of the subset of N-glycosylation sites that intrinsically enhance folding energetics. When a gain-of-N-glycosylation mutation arises, the glycan can increase the energy of the denatured state by restricting backbone conformational space (28–30), as mentioned above. Denatured state destabilization would accelerate folding by decreasing the folding activation barrier (ΔGf‡), while also stabilizing the native state by increasing the free energy difference between the denatured and native states (ΔGf becomes more negative) (Fig. 4B, panels 1 and 2). Although the amino acids near the N-glycosylation site are unlikely to provide optimal glycan-protein contacts initially, further mutation and selection could yield stabilizing contacts (like those frequently observed between proteins and their N-linked triose) (3, 26) especially if they enhance protein function. For proteins that are vulnerable to ERAD because they fold slowly (32), mutations that are selected for to increase function may stabilize the transition state and the native state so that they further accelerate protein folding (Fig. 4B, panel 3, scenario 1). For fast-folding but unstable proteins, the selected sequence that improves function need only enhance thermodynamic stability (32). This stability enhancement could arise by either accelerating folding or by slowing unfolding or both; the latter mechanism, which, as noted above, is responsible for 3/4 of the stabilization imparted by the glycan to hCD2ad, is illustrated in Fig. 4B, panel 4, scenario 2. By extension, protein stabilization by the introduction of an N-glycosylation site may be important for evolving protein function when the residues required for function are not optimal for stability. This may be the case for human CD2ad, which has a different binding partner than its unglycosylated rat ortholog (34).
Far from being a passive pedestal for displaying distal saccharides to recruit the CNX/CRT-chaperone system, the ManGlcNAc2 triose core of the N-glycan can directly participate in stabilizing the native state and decreasing the folding activation barrier in a protein like hCD2ad (compare variants 1 to 3-7). Although N-glycosylation is not always required for protein folding and function, mutational gain of an N-glycosylation site seems to represent an elegantly simple, efficient, and potentially general strategy for enhancing folding energetics relative to mutagenesis of the polypeptide sequence alone (for intrinsic benefits alone, compare variant 1 to 2a and 2b vs. 1 to 3-7). The extrinsic biology of folding and intrinsic enhancement of folding energetics conferred by the N-glycan structure provide an excellent explanation for why the process of N-glycosylation is strictly conserved (2, 31). In conclusion, although the accumulated data are consistent with our hypothesis that the core triose of N-linked glycans has a general structure stabilizing effect, critical further support requires synthesizing and characterizing other N-linked glycoproteins, a substantial future undertaking that is underway.
Methods
General.
Unless otherwise noted, chemicals and products were purchased from Fisher Scientific or Sigma–Aldrich. Protein was concentrated using Amicon centrifugation devices, MWCO 10kDa (Millipore). Final hCD2ad concentrations were determined by BCA assay or by absorbance at 280 nm (calculated extinction coefficients; Vector NTI, Informax). Phosphate-buffered saline (PBS) was prepared from tablets (SIGMA P-4417) and maintained at pH 7.2. Urea denaturant was prepared in PBS and concentrations were determined by index of refraction. All buffer solutions were filtered (Millipore 0.2μm) before use. Data were fitted in Mathematica 5.2 or 6.0 (Wolfram Research). The hCD2ad NMR structure was rendered with PyMOL (pdb ID: 1gya).
hCD2ad Variants.
Cloning, expression, and purification are described in detail in SI Appendix. Glycoform variants 5-7 were produced from 4 by enzymatic remodeling, as described below and outlined in Fig. S1, and purity and glycoform abundance are reported in Fig. 2, Fig. S2, and Tables S1–S3. Briefly, the N-glycan of variant: 5 (cut by endo-β-N-acetylglucosaminidase F and D, or Endos F and D), consisted of GlcNAc a, both fucosylated and unfucosylated forms (3:1); 6 (remodeled by Jack bean α-mannosidase) comprised fucosylated and unfucosylated Man1–2GlcNAc2 glycoforms; and, 7 (the uncleaved N-glycan portion from Endo D treatment purified by Con A) was a mixture of the oligomannose glycoforms, Man6GlcNAc2: Man7GlcNAc2 (9:1).
Enzymatic Remodeling.
Concanavilin A (ConA) lectin chromatography was performed with the ConA Glycoprotein Isolation Kit (Pierce), following the manufacturer's protocols. As a finishing step, all variants were dialyzed into PBS, purified by gel filtration (BioSep Sec-S 3000; Phenomenex), and concentrated. Final products were verified by SDS/PAGE (Fig. 2A and Fig. S2) and LCMS. Detailed procedures for enzymatic remodeling are given in SI Appendix. Briefly, remodeling conditions were as follows:
For variant 6: 4 (0.5 mg/mL) was treated with proteomics grade jack bean mannosidase (Sigma, 0.5U jbm/mg 4) at 37 °C, overnight. Reaction mixtures were subsequently passed through ConA resin collecting eluate.
For variants 5 and 7: 4 (0.5 mg/mL) was treated with 50 milliunits/mL Endo D (EMD Biosciences) and/or 50 units/mL Endo Hf (New England Biolabs) under appropriate native conditions (Endo D: 100 mM Mes, pH 6.5; Endo Hf: 50 mM sodium citrate, pH 5.5; or double digests, using Endo D buffer). Digestions were performed overnight at 37 °C. Con A was used to separate hCD2ad variant 5 (collected in the flow through), and protein bearing uncleaved N-glycan variant 7 (collected after washing and elution).
Spectroscopy.
Measurements were made in quartz cuvettes, at 25 °C, in PBS (pH 7.2). CD spectra (10–50 μM variant) were recorded on an Aviv Model 202SF CD spectrometer and are reported as mean per residue ellipticity, [θ]. Fluorescence emission spectra (318–380 nm, normalized to 354 nm; excitation at 280 nm) of variants (5–20 μg/mL) were collected on a CARY Eclipse fluorometer (Varian).
Kinetics.
Kinetics data were obtained as described in SI Appendix and fit to a monoexponential (Eq. 1) containing the rate constant for folding/unfolding (kobs) and corrective photobleaching (kpb) component.
Monoexponential rate constants (kobs) were obtained from stopped-flow measurements fit to Eq. 1 (representative kinetic data, fits, and residuals, Fig. 3 Inset and Fig. S3). Chevron plots (log kobs vs. [urea]) (Fig. 3) were fit to Eq. 2 to determine: kf,0 and ku,0, the folding and unfolding rate constants under native conditions; mf and mu1, the linear slopes of the dependences of the folding and unfolding rate constants on urea concentration, and mu2, the small quadratic coefficient accounting for slight curvature exhibited in the chevron plots at high [urea], possibly because of structural changes in the folding transition state (35).
The complete set of observed rate constants for all hCD2ad variants is available in Dataset S1. The free energy of folding (ΔGf,0) under native conditions was determined using Eq. 3. Notably, ΔGf,0 values determined by kinetic and thermodynamic experiments are in good agreement (Fig. S4).
Note that the dependence of ΔGf on urea concentration, or meq, is equal to RT (mu1 − mf) (we exclude the effect of mu2). For unstable variant 1, the value of kobs at 0 M urea (kobs,0) was determined by linear extrapolation, and Kf,0 was determined by equilibrium osmolyte renaturation/urea denaturation (see SI Appendix and Fig. S5). The values of kf,0 and ku,0 were calculated by solving the simultaneous equations Kf,0 = kf,0/ku,0 and kobs,0 = kf,0+ku,0.
Supplementary Material
Acknowledgments.
We thank W. E. Balch, I. A. Wilson, and J. C. Paulson for critically reading this manuscript; T. Foss, R. L. Wiseman, and S. Deechongkit for helpful early discussions on the project; and D. L. Powers for help with fitting the protein folding kinetics data. This work was supported by National Institutes of Health Grant GM51105 (to J.W.K. and E.T.P.) and GM044154 (to C.-H. W.) and predoctoral support from Achievement Rewards for College Scientists (to S.R.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810318105/DCSupplemental.
References
- 1.Varki A. Biological roles of oligosaccharides—All of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 3.Wormald MR, Dwek RA. Glycoproteins: Glycan presentation and protein-fold stability. Struct Fold Des. 1999;7:R155–R160. doi: 10.1016/s0969-2126(99)80095-1. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi H. Chaperone-like functions of N-glycans in the formation and stabilization of protein conformation. Trends Glycosci Glycotechnol. 2002;14:139–151. [Google Scholar]
- 5.Imperiali B, O'Connor SE. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr Opin Chem Biol. 1999;3:643–649. doi: 10.1016/s1367-5931(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 6.Mitra N, Sinha S, Ramya TNC, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci. 2006;31:156–163. doi: 10.1016/j.tibs.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Petrescu A-J, Milac A-L, Petrescu SM, Dwek RA, Wormald MR. Statistical analysis of the protein environment of N-glycosylation sites: Implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 8.O'Connor SE, Pohlmann J, Imperiali B, Saskiawan I, Yamamoto K. Probing the effect of the outer saccharide residues of N-linked glycans on peptide conformation. J Am Chem Soc. 2001;123:6187–6188. doi: 10.1021/ja010094s. [DOI] [PubMed] [Google Scholar]
- 9.Trombetta ES. The contribution of N-glycans and their processing in the endoplasmic reticulum to glycoprotein biosynthesis. Glycobiology. 2003;13:77R–91R. doi: 10.1093/glycob/cwg075. [DOI] [PubMed] [Google Scholar]
- 10.Hanson S, Best M, Bryan MC, Wong CH. Chemoenzymatic synthesis of oligosaccharides and glycoproteins. Trends Biochem Sci. 2004;29:656–663. doi: 10.1016/j.tibs.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Sears P, Wong CH. Toward automated synthesis of oligosaccharides and glycoproteins. Science. 2001;291:2344–2350. doi: 10.1126/science.1058899. [DOI] [PubMed] [Google Scholar]
- 12.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 13.Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol. 2007;3:313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 14.Stanley P, Ioffe E. Glycosyltransferase mutants: Key to new insights in glycobiology. FASEB J. 1995;9:1436–1444. doi: 10.1096/fasebj.9.14.7589985. [DOI] [PubMed] [Google Scholar]
- 15.Tangye SG, Phillips JH, Lanier LL. The CD2-subset of the Ig superfamily of cell surface molecules: Receptor-ligand pairs expressed by NK cells and other immune cells. Semin Immunol. 2000;12:149–157. doi: 10.1006/smim.2000.0217. [DOI] [PubMed] [Google Scholar]
- 16.Williams AF, Barclay AN. The immunoglobulin superfamily–Domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 17.Wyss DF, et al. Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science. 1995;269:1273–1278. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]
- 18.Clarke J, Cota E, Fowler SB, Hamill SJ. Folding studies of immunoglobulin-like β-sandwich proteins suggest that they share a common folding pathway. Struct Fold Des. 1999;7:1145–1153. doi: 10.1016/s0969-2126(99)80181-6. [DOI] [PubMed] [Google Scholar]
- 19.Recny MA, et al. N-Glycosylation is required for human CD2 immunoadhesion functions. J Biol Chem. 1992;267:22428–22434. [PubMed] [Google Scholar]
- 20.Arulanandam ARN, et al. The CD58 (LFA-3) binding site is a localized and highly charged surface area on the AGFCC′C″ face of the human CD2 adhesion domain. Proc Natl Acad Sci USA. 1993;90:11613–11617. doi: 10.1073/pnas.90.24.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker MJ, Dempsey CE, Lorch M, Clarke AR. Acquisition of native beta-strand topology during the rapid collapse phase of protein folding. Biochemistry. 1997;36:13396–13405. doi: 10.1021/bi971294c. [DOI] [PubMed] [Google Scholar]
- 22.Bolen DW, Baskakov IV. The osmophobic effect: Natural selection of a thermodynamic force in protein folding. J Mol Biol. 2001;310:955–963. doi: 10.1006/jmbi.2001.4819. [DOI] [PubMed] [Google Scholar]
- 23.Maxwell KL, et al. Protein folding: Defining a “standard” set of experimental conditions and a preliminary kinetic data set of two-state proteins. Protein Sci. 2005;14:602–616. doi: 10.1110/ps.041205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. 1. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 25.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wormald MR, et al. Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem Rev. 2002;102:371–386. doi: 10.1021/cr990368i. [DOI] [PubMed] [Google Scholar]
- 27.Chang VT, et al. Glycoprotein structural genomics: Solving the glycosylation problem. Structure. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wormald MR, et al. The conformational effects of N-glycosylation on the tailpiece from serum IgM. Eur J Biochem. 1991;198:131–139. doi: 10.1111/j.1432-1033.1991.tb15995.x. [DOI] [PubMed] [Google Scholar]
- 29.Live DH, Kumar RA, Beebe X, Danishefsky SJ. Conformational influences of glycosylation of a peptide: A possible model for the effect of glycosylation on the rate of protein folding. Proc Natl Acad Sci USA. 1996;93:12759–12761. doi: 10.1073/pnas.93.23.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connor SE, Imperiali B. A molecular basis for glycosylation-induced conformational switching. Chem Biol. 1998;5:427–437. doi: 10.1016/s1074-5521(98)90159-4. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee S, et al. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA. 2007;104:11676–11681. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE. An adaptable standard for protein export from the endoplasmic reticulum. Cell. 2007;131:809–821. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 33.Vogt G, et al. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat Genet. 2005;37:692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- 34.Arulanandam AR, et al. A soluble multimeric recombinant CD2 protein identifies CD48 as a low affinity ligand for human CD2: Divergence of CD2 ligands during the evolution of humans and mice. J Exp Med. 1993;177:1439–1450. doi: 10.1084/jem.177.5.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.