Abstract
The toxicity of acrolein, an α,β-unsaturated aldehyde produced during lipid peroxidation, is attributable to its high reactivity toward DNA and cellular proteins. The major acrolein−DNA adduct, γ-hydroxypropano-2′-deoxyguanosine (γ-HOPdG), ring opens to form a reactive N2-oxopropyl moiety that cross-links to DNA and proteins. We demonstrate the ability of γ-HOPdG in a duplex oligonucleotide to cross-link to a protein (EcoRI) that specifically interacts with DNA at a unique sequence. The formation of a cross-link to EcoRI was dependent on the intimate binding of the enzyme to its γ-HOPdG-modified recognition site. Interestingly, the cross-link did not restrict the ability of EcoRI to cleave DNA substrates. However, stabilization of the cross-link by reduction of the Schiff base linkage resulted in loss of enzyme activity. This work indicates that the γ-HOPdG−EcoRI cross-link is in equilibrium with free oligonucleotide and enzyme. Reversal of cross-link formation allows EcoRI to effect enzymatic cleavage of competitor oligonucleotides.
Introduction
Noncovalent interactions between DNA and proteins are essential for proper cellular function. The recognition of specific DNA sequences or structures by DNA-binding proteins is a crucial regulatory step during DNA replication, control of gene expression, and the response to and repair of DNA damage. Disruption of these interactions poses a significant threat to genome integrity and cell viability. Protein and DNA damage induced by endogenous electrophiles is associated with a number of human diseases, including cancer, cardiovascular disease, and neurological disorders (1−3). For example, oxidation products of polyunsaturated fatty acids comprise a series of cytotoxic α,β-unsaturated aldehydes, including malondialdehyde, 4-hydroxynonenal, and acrolein, that readily form covalent adducts with cellular macromolecules, contributing to their toxicity (reviewed in ref (4)).
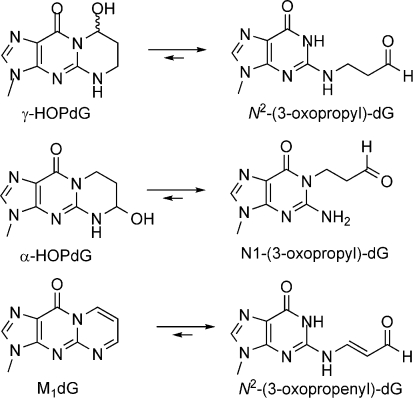
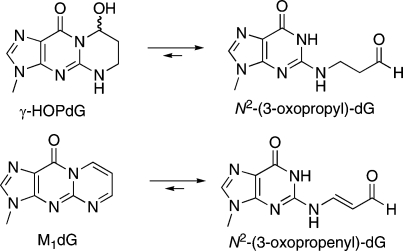
Acrolein is a mutagen and a tumor initiator produced endogenously via lipid peroxidation and myeloperoxidase-catalyzed amino acid oxidation (5−8). It is also an ubiquitous pollutant found in automobile exhaust and cigarette smoke (9). Acrolein exhibits facile reactivity with proteins (8) and represents a significant source of endogenous DNA damage by forming cyclic adducts with DNA bases and DNA−protein cross-links (10,11). The major products of the reaction of acrolein with DNA are exocyclic adducts of deoxyguanosine, (8R/S)-3-(2′-deoxyribos-1′-yl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2a]purin-10(3H)-one (γ-HOPdG)1 and (6R/S)-3-(2′-deoxyribos-1′-yl)-5,6,7,8-tetrahydro-6-hydroxypyrimido[1,2a]purin-10(3H)-one (α-HOPdG) (10) (Figure 1). α-HOPdG is more mutagenic than γ-HOPdG and induces G→T transversions in site-specific mutagenesis experiments (12). Both γ-HOPdG and α-HOPdG are present in human lung DNA at levels in excess of those typically reported for polycyclic aromatic hydrocarbon−DNA adducts (13).
Figure 1.
Structures of DNA adducts and their ring-opening products.
While γ-HOPdG contributes to acrolein toxicity by directly affecting DNA replication (12,14), the adduct itself is a reactive molecule. In duplex DNA, γ-HOPdG undergoes a spontaneous ring-opening reaction to generate N2-oxopropyl-2′-deoxyguanosine that can react with primary amines on cellular macromolecules (15). γ-HOPdG positioned within a Cp*G sequence context in duplex DNA undergoes ring opening and subsequent reaction with a guanine base in the complementary strand to form interstrand DNA−DNA cross-links (16). The ring-opened γ-HOPdG adduct is also reactive toward amino groups of peptides and is capable of forming reversible Schiff base-mediated cross-links to proteins (12).
A role for DNA−protein cross-links during acrolein toxicity has not been fully elucidated. The interaction of adducted oligonucleotides with restriction endonucleases represents a useful model system with which to evaluate the impact of DNA damage on recognition and catalysis of DNA-binding proteins. Previously, our group showed that exocyclic adducts in an EcoRI recognition site blocked binding and cleavage of DNA by that enzyme, although ring-opened adducts were substrates for endonucleolytic cleavage by EcoRI (17). Here, we examine the fate of γ-HOPdG and consequently its ring-opened analogue, when incorporated into duplex substrates of EcoRI. We describe the ability of γ-HOPdG to cross-link to EcoRI and affect enzyme activity. The results suggest that the DNA−protein cross-link is unstable and either undergoes hydrolysis to free oligonucleotide and enzyme or can be reduced to a stable, irreversible cross-link.
Experimental Procedures
Reagents
T4 polynucleotide kinase and PvuII were obtained from New England Biolabs (Beverly, MA). EcoRI was purified as described (18). EcoRI for some cross-linking experiments was purchased from MBI Fermenetas (Hanover, MD). NaCNBH3 and NaBH4 were purchased from Sigma (St. Louis, MO). [γ-32P]-ATP was from ICN Biomedicals, Inc. (Costa Mesa, CA).
Oligonucleotides
The 21-mer oligonucleotides contained a unique EcoRI restriction site (5′-GAATTC-3′) in the sequence 5′-TATCATGTCTNAATTCCCGGT-3′, where N = dG, γ-HOPdG, or pyrimido[1,2-α]purin-10(3H)-one (M1dG). The sequence of the complementary 27-mer oligonucleotide was 5′-AGACCGGGAATTCAGACATGATACGGA-3′. The scrambled EcoRI sequence was 5′-TATCATGTCTNTATACCCGGT-3′, where N = γ-HOPdG; it was annealed to its complementary sequence. γ-HOPdG-modified oligonucleotides were synthesized and purified as previously described (19). The M1dG-modified oligonucleotide was synthesized using the method previously described (20) and purified by polyacrylamide gel electrophoresis (21). Unmodified 21-mer and 27-mer oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Trapping of Covalent DNA−Protein Complexes Using NaCNBH3
Modified and unmodified 21-mer oligonucleotides were 5′-end-labeled in a phosphorylation buffer of 50 mM 4-morpholinepropanesulfonic acid (MOPS) (pH 7.3), 10 mM MgCl2, 100 μM EDTA, and 5 mM dithiothreitol containing 5 μM oligonucleotide, 0.5 mM [γ32P]-ATP, and 10 units of T4 polynucleotide kinase. Oligonucleotides were purified from free [γ32P]-ATP by passage through Bio-Spin P-6 columns (Bio-Rad, Hercules, CA). For restriction endonuclease studies, substrates were generated by annealing radiolabeled γ-HOPdG- or M1dG-modified 21-mer (2 pmol) to a complementary 27-mer (10 pmol) in 50 mM MOPS (pH 7.4) and 100 mM NaCl (50 μL total volume) by heating to 90 °C for 3 min, followed by slow cooling to room temperature. DNA−protein cross-links were detected essentially as described with slight modifications (12). 32P-labeled substrate (160 pM) was incubated with EcoRI or PvuII restriction enzyme (1.6 nM) or bovine serum albumin (BSA) (5000 nM) in 50 mM MOPS (pH 7.4), 10 mM NaCl, and 0.1 mM ethylenediaminetetraacetic acid (EDTA), at room temperature for 2 h. NaCNBH3 (50 mM) was added immediately following the addition of enzyme. Because Mg2+ is required for EcoRI cleavage activity but not required for the specific binding of enzyme to DNA, Mg2+ was omitted from the incubation. The reactions were terminated by addition of Laemmli sample buffer, then boiled for 5 min, and separated by 10% sodium dodecyl sulfate−polyacrylamide gel electrophoresis (SDS-PAGE) (22). Cross-linked bands were visualized using a FujiFilm FLA-5000 phosphorimager analyzer.
Restriction Digestion of Adducted Oligonucleotide Substrates
Radiolabeled substrates were generated by annealing adducted or unmodified 5′-32P-labeled 21-mer oligonucleotide with an equimolar amount of 5′-32P-labeled 27-mer complement. Annealing of 2.5 pmol of each oligonucleotide was performed in buffer (50 mM MOPS, pH 7.4, and 100 mM NaCl) by heating to 90 °C for 3 min, followed by slow cooling. Nonradiolabeled substrates were generated by annealing adducted or unmodified 21-mer with complementary oligonucleotide under the conditions described above. Restriction digests (10 μL) contained 5 nM 32P-labeled substrate, 495 nM unlabeled substrate, 50 mM MOPS (pH 7.9), 10 mM MgCl2, 100 mM NaCl, and 5 nM EcoRI restriction endonuclease. The reactions were carried out for specified times at 37 °C, then quenched by the addition of 10 mM EDTA in 90% formamide. The products of the reaction were resolved on a 20% denaturing gel using the Ultrapure Sequagel system (National Diagnostics, Atlanta, GA). The positions of the bands were visualized using a FujiFilm FLA-5000 phosphorimager analyzer. Cleavage of the EcoRI substrate generated two 5′-32P-labeled products of different sizes. The 21-mer oligonucleotide 5′-cleavage product was an 11-mer, whereas the 27-mer substrate’s 5′-cleavage product was an 8-mer. Thus, by monitoring the appearance of the 11-mer and the 8-mer, cleavage of both strands of the substrate duplex could be monitored simultaneously. The extent of cleavage was determined by integrating the radioactivity of each band, then dividing the radioactivity of the band corresponding to cleavage product by the sum of the radioactivity of cleaved and uncleaved oligonucleotide to give the percent cleavage.
In some experiments, incubation mixtures were prepared in which 500 nM unlabeled adducted substrate or unmodified substrate was incubated for 2 h with 5 nM EcoRI. Then, 500 nM unmodified substrate, spiked with 32P-labeled unmodified substrate, was added to the reaction mixture, and cleavage was monitored for 2 h. For other experiments, reactions were prepared under noncleavage conditions, that is, in the absence of Mg2+. γ-HOPdG-modified substrate or unmodified substrate (comprising 5 nM 32P-labeled DNA and 495 nM nonradiolabeled DNA) was incubated in Mg2+-free buffer (50 mM MOPS, pH 7.9, and 100 mM NaCl) for 2 h with EcoRI (5 nM) in the presence or absence of NaCNBH3. Following preincubation, 10 mM MgCl2 was added to the reaction mixture, and cleavage was monitored for 1 h.
Results
Detection of a γ-HOPdG-Mediated Cross-Link to the Restriction Endonuclease, EcoRI
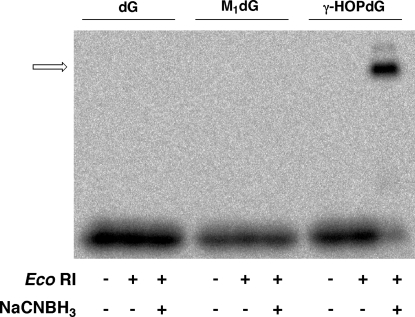
EcoRI binds the palindromic sequence, 5′-GAATTC-3′, and sequentially cleaves both strands of the DNA duplex at the phosphodiester bond positioned 3′ to deoxyguanosine. 32P-labeled oligonucleotide duplexes were prepared containing a unique EcoRI recognition site that was modified with γ-HOPdG. Substrate DNA was incubated for 2 h with EcoRI (molar ratio of DNA duplex: EcoRI = 1:10) in the presence of NaCNBH3, as described in the . NaCNBH3 does not react with EcoRI or γ-HOPdG but is capable of reducing an imine cross-link between the free aldehyde form of γ-HOPdG and a lysine residue. EcoRI reacted with γ-HOPdG-modified substrates to form a DNA−protein cross-link observable by SDS-PAGE analysis (Figure 2). Formation of the gel-shifted band corresponding to the protein−DNA complex was dependent on stabilization of the cross-link by reduction with NaCNBH3. A weaker, second band was observed that migrated more slowly than the major γ-HOPdG−EcoRI band. The identity of this second band is unknown.
Figure 2.
γ-HOPdG forms a cross-link to EcoRI. 32P-labeled γ-HOPdG- or M1dG-modified or unmodified DNA substrates (160 pM) were incubated with EcoRI (1.6 nM) at room temperature for 2 h in the presence or absence of 50 mM NaCNBH3. The products were resolved by SDS-PAGE and visualized by phosphorimager analysis. This is a representative autoradiogram from at least five independent experiments. The cross-linked product is indicated by the arrow.
γ-HOPdG ring opens to form an aldehydic moiety that is capable of reacting with proteins (15). The malondialdehyde−deoxyguanosine adduct, M1dG, similarly ring opens in duplex DNA to form a potentially reactive oxopropenyl moiety, N2-oxopropenyl-dG (23). The presence of M1dG in the EcoRI recognition site does not prevent recognition of the site by EcoRI (17). Therefore, we tested whether M1dG is also able to form cross-links to EcoRI. In contrast to γ-HOPdG, no cross-link was observed when M1dG-modified substrates were incubated with EcoRI with or without NaCNBH3 reduction (Figure 2).
Specificity of γ-HOPdG−EcoRI Cross-Link Formation
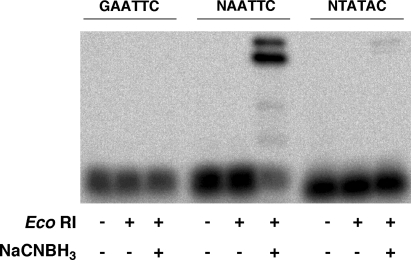
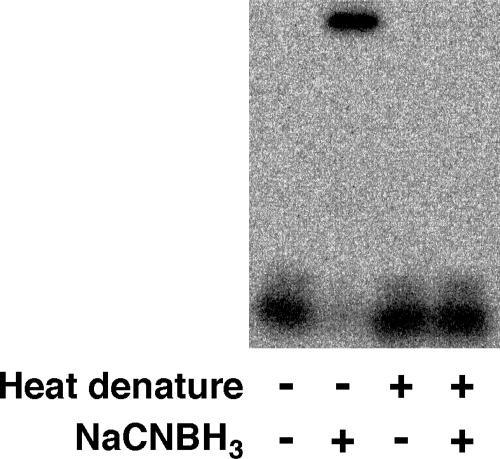
DNA substrates were prepared in which γ-HOPdG was positioned in a scrambled EcoRI recognition site (5′-GTATAC-3′). No EcoRI−DNA cross-link was observed when γ-HOPdG was in the scrambled sequence (Figure 3). When EcoRI was inactivated by heat (65 °C for 20 min) prior to the reaction with γ-HOPdG-modified substrate, only an extremely faint band at the position of the DNA−protein cross-link was detected after 2 h (Figure 4). Furthermore, γ-HOPdG did not cross-link to a restriction enzyme, PvuII, that does not specifically recognize the sequence of DNA where the adduct was positioned. Also, γ-HOPdG did not form cross-links with BSA, a common stabilizing agent added to commercial preparations of EcoRI, even at a DNA:BSA molar ratio = 1:5000 (data not shown). These results indicate that the cross-link formed between γ-HOPdG and EcoRI is most likely formed as a result of specific binding of the EcoRI recognition sequence and probably involves amino acids in close proximity to the adduct in the DNA−protein complex.
Figure 3.
EcoRI cross-links to γ-HOPdG positioned within the EcoRI restriction site. 32P-labeled γ-HOPdG-modified or unmodified DNA substrates (160 pM) were incubated with EcoRI (1.6 nM) at room temperature for 2 h in the presence or absence of NaCNBH3. γ-HOPdG was positioned in either the EcoRI restriction site (GAATTC) or a scrambled sequence (GTATAC). The products were resolved by SDS-PAGE. A representative autoradiogram of the gel is shown.
Figure 4.
Requirement of properly folded enzyme for cross-link formation. EcoRI (1.6 nM), native or denatured at 65 °C for 20 min, was incubated with γ-HOPdG-modified 32P-end-labeled DNA substrate (160 pM) at room temperature for 2 h. NaCNBH3 (50 mM) was added to the reaction to stabilize the Schiff base linkage between protein and DNA. The products were separated by 10% SDS-PAGE.
Time Course of γ-HOPdG−EcoRI Cross-Link Formation
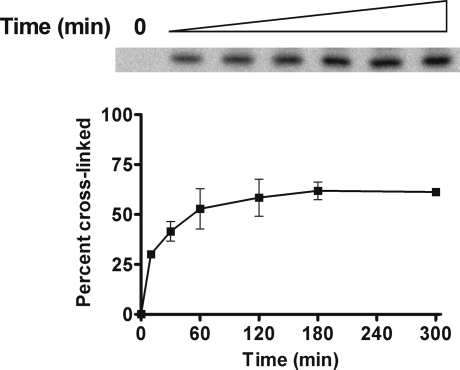
Cross-link formation was monitored over 5 h (Figure 5). Reactions were prepared in the presence of NaCNBH3, and then, a second reducing agent, NaBH4, was added to rapidly quench any remaining aldehydic substrate at specific time points. The γ-HOPdG−EcoRI cross-link was observed within 10 min. The conversion of free DNA to cross-linked product reached a plateau within 3 h.
Figure 5.
Time course of γ-HOPdG cross-linking to EcoRI. γ-HOPdG-modified 32P-end-labeled DNA substrate (160 pM) was incubated with EcoRI (1.6 nM) for up to 5 h at room temperature in the presence of 50 mM NaCNBH3, and then, a second reducing agent, NaBH4 (100 mM), was added to rapidly quench any remaining aldehydic substrate at specific time points. A representative autoradiogram from three independent experiments is presented, which displays the accumulation of the DNA−protein cross-linked band. Percent cross-linking is shown in the graph just below the gel expansion. The values are the means ± standard deviations from three independent experiments.
In Vitro Cleavage of γ-HOPdG-Modified DNA Substrate by EcoRI Restriction Endonuclease
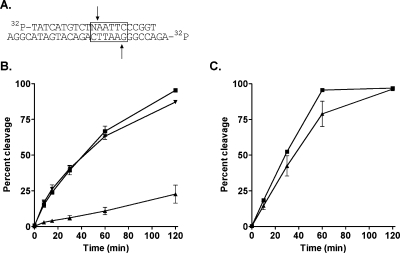
Oligonucleotide duplexes containing a single adduct located within a unique EcoRI recognition site were 5′-32P-end-labeled on both strands. The adducted strand of the duplex was a different length than its complement, and the oligonucleotides were designed to release 32P-end-labeled cleavage products of different lengths. Thus, simultaneous monitoring of cleavage of both strands of the duplex was achieved. The complement strand of the γ-HOPdG-modified substrate was cleaved nearly as efficiently as unmodified substrate (∼90%) (Figure 6B). In contrast, only ∼20% of the γ-HOPdG-modified strand was cleaved by EcoRI within 2 h.
Figure 6.
Effect of γ-HOPdG on EcoRI activity. (A) 5′-Radiolabeled duplex containing a single EcoRI recognition site. The arrows depict cleavage sites. γ-HOPdG was contained on the 21-mer strand, as indicated by “N”. (B) Cleavage reactions containing 5 nM 32P-labeled oligonucleotide duplex, 495 nM nonlabeled oligonucleotide substrate, and 5 nM EcoRI were incubated at 37 °C for various times, then quenched, and subjected to gel electrophoresis and phosphorimager analysis as described in the . Unmodified substrate, 27-mer strand (◼); γ-HOPdG-modified substrate, 27-mer strand (▼); γ-HOPdG-modified substrate, 21-mer strand (▲). (C) Nonlabeled oligonucleotide duplexes modified with γ-HOPdG or unmodified duplexes (500 nM) were incubated with EcoRI endonuclease (5 nM) for 2 h at 37 °C, prior to the addition of fresh unmodified EcoRI substrate, spiked with 32P-labeled unmodified substrate, to a final concentration of 500 nM. Cleavage of the radiolabeled substrate was monitored for 2 h. The data shown represent cleavage of the 27-mer strand of the radiolabeled duplex in the presence of the indicated competitor substrate. The 21-mer strand of each duplex was cleaved similarly to its complement (data not shown). Unmodified competitor substrate (◼); γ-HOPdG-modified competitor substrate (▲). The values are the means ± standard deviations from three independent experiments.
To examine whether the reduced cleavage efficiency of modified substrate was due to a sequestration or inactivation of EcoRI by γ-HOPdG, nonradiolabeled γ-HOPdG-adducted substrates or unadducted substrates were incubated with EcoRI for 2 h. Then, 32P-labeled unmodified substrate was added to the reaction mixture, and cleavage was monitored. The radiolabeled unmodified substrate was cleaved to a similar extent, regardless of whether unmodified substrate or γ-HOPdG-modified substrate was initially incubated with the enzyme (Figure 6C). Thus, EcoRI retained complete activity following exposure to the DNA adduct.
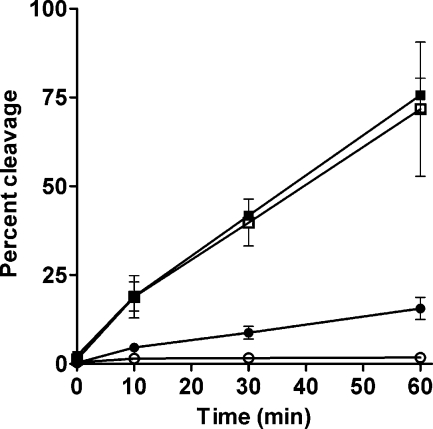
To examine whether a stabilized cross-link could affect EcoRI activity, γ-HOPdG-modified or unmodified substrates were incubated with EcoRI in the absence of Mg2+ and either the presence or the absence of NaCNBH3 for 2 h; then, MgCl2 was added to the reaction mixture to initiate cleavage. Following MgCl2 addition, EcoRI cleavage of the substrate was substantially decreased in samples containing γ-HOPdG-modified DNA and NaCNBH3 as compared to samples containing γ-HOPdG-modified substrate in the absence of reducing agent. There was no effect of NaCNBH3 on cleavage of the unmodified DNA substrate by EcoRI (Figure 7).
Figure 7.
EcoRI cleavage following reduction of the cross-link. 32P-radiolabeled γ-HOPdG-modified substrate or unmodified substrate (500 nM) was incubated with EcoRI (5 nM) in the absence of Mg2+ and either the presence or the absence of NaCNBH3 for 2 h. Following preincubation, MgCl2 (10 mM) was added to initiate cleavage. Reactions were incubated at 37 °C for various times. Cleavage profiles shown are for the 21-mer strand of the DNA duplex. Unmodified substrate, −NaCNBH3 (◼); unmodified substrate, +NaCNBH3 (◻); γ-HOPdG-modified substrate, −NaCNBH3 (●); and γ-HOPdG-modified substrate, +NaCNBH3 incubation (○).
Discussion
The identification of cellular targets of endogenous reactive aldehydes has garnered significant attention in an attempt to understand the genotoxic and cytotoxic mechanisms of these molecules. The products of acrolein’s reaction with cellular macromolecules have been suggested to participate in diseases associated with oxidative stress (24). In addition to forming adducts to protein and adducts to DNA, acrolein induces Schiff base-mediated cross-link formation between DNA and protein (11), and the major acrolein−DNA adduct γ-HOPdG is itself reactive with both DNA and proteins (12,16,25−27). Thus, γ-HOPdG may represent a mode for acrolein’s DNA−protein cross-link formation in vivo. Although a mechanistic link between DNA−protein cross-links and disease has not been elucidated, the formation of cross-links can be detrimental to cell survival. The bulky nature of the cross-links is likely to disrupt DNA metabolism by posing significant blocks to replication and transcription. Consistent with this proposal, aldehyde-derived DNA−protein cross-links block SV40 DNA replication (28). Although little is known about the mutagenic properties of DNA−peptide cross-links, Minko et al. recently described the mutagenic potential of NaCNBH3-reduced−γ-HOPdG-mediated cross-links to peptides (29).
In the present study, we demonstrated the ability of γ-HOPdG to cross-link to a protein that forms intimate interactions with DNA, the restriction endonuclease, EcoRI. Previous work has demonstrated that γ-HOPdG-containing oligonucleotides form cross-links to histone and to T4 pyrimidine dimer DNA glycosylase (12). It appears that γ-HOPdG forms cross-links to EcoRI more rapidly and at a considerably reduced excess of protein to DNA. The differences between the two studies are most likely attributable to the fact that EcoRI has high affinity and specificity for binding duplex oligonucleotides that contain a recognition site and that the γ-HOPdG adduct is present at the first dG residue in the recognition site.
Given that EcoRI was able to recognize and digest γ-HOPdG-modified DNA substrates, γ-HOPdG is presumed to be accommodated in the recognition interface in a manner similar to physiological substrates. Furthermore, the native structure of EcoRI was essential for rapid DNA−protein cross-link formation, and no cross-link was detected when γ-HOPdG was not located within the enzyme’s recognition site. These results suggest that upon binding to the γ-HOPdG-modified cognate site, the reactive N2-oxopropyl moiety forms a cross-link between the 2-amino position of the modified G (NAATTC) and a proximal amino acid at the recognition interface.
Lysine residues have been implicated in DNA−protein cross-link formation by acetaldehyde (30) and may also participate in γ-HOPdG-mediated cross-links. The stabilization of the EcoRI−DNA cross-links by NaCNBH3 is consistent with this in that NaCNBH3 preferentially reduces imine bonds formed by reaction of carbonyl groups with lysine residues (31). There are several amino acids involved in the interaction of EcoRI with its recognition sequence, which contain primary amines that could be involved in cross-link formation. Although Lys-148, Lys-117, and Lys-89 are in the vicinity, a likely candidate is Lys-113, which not only polarizes one of the oxygens of the scissile phosphate but also, along with Arg-145, may stabilize the phosphorane intermediate in the transition state (32). The Lys-113 side chain is disordered in the apoenzyme but becomes ordered upon binding DNA substrate (33). In molecular dynamics simulations, Lys-113 moves to different positions depending on the presence or absence of Mg2+(32).
The impact of cross-link formation on EcoRI catalytic activity was unexpected. The conditions used for the enzymatic cleavage assay provided DNA substrate in 100-fold molar excess to EcoRI. Seemingly all of the endonuclease is involved in cross-link formation. Nevertheless, no loss of activity was observed without NaCNBH3 reduction and stabilization of the cross-link; treatment of EcoRI alone with NaCNBH3 did not reduce endonucleolytic activity. The loss of activity observed following treatment of the EcoRI−γ-HOPdG−DNA complex with NaCNBH3 correlates to the appearance of the stable cross-link band on gel electrophoresis. Thus, it appears that the formation of the putative imine-mediated protein−DNA cross-link is readily reversible so that the protein−DNA cross-link hydrolyzes to EcoRI and γ-HOPdG-containing oligonucleotide during the time course of the incubation. Thus, no loss of enzyme activity is apparent without reductive stabilization.
The proposed mechanism for γ-HOPdG-mediated formation of DNA−protein cross-links involves ring opening of γ-HOPdG to N2-(γ-oxopropyl)-dG, followed by reaction with a primary amine. Kurtz and Lloyd observed that γ-HOPdG cross-links occurred only with the NH2 termini of peptides containing lysine and arginine residues (27). This is consistent with the lower pKa and stronger nucleophilic nature of an α-amine as compared to the ε-amine of lysine. In related chemistry, the aldehydic form of an abasic site cross-linked to the catalytically active N-terminal threonine of T4 PDG and the imine was trapped by NaCNBH3 reduction (34). It is unlikely that the γ-HOPdG−EcoRI cross-link occurs at the NH2 terminus of the protein. A revised model of EcoRI in solution places the N-terminal regions at a subunit interface distal to the DNA, and removal of the first four residues has no effect on catalytic activity, suggesting that the N termini are spatially remote from the EcoRI active sites (35,36). It is more likely that cross-linking occurs to a Lys residue in the interior of the protein. Exclusion of water from the recognition interface could lower the pKa of a Lys side chain, thereby increasing the fraction of deprotonated nucleophilic amine.
γ-HOPdG readily formed cross-links to EcoRI, which were trappable by NaCNBH3 reduction, but the M1dG adduct did not. Although M1dG undergoes rapid hydrolytic ring opening in duplex DNA, the aldehyde of the product, N2-(3-oxopropenyl)-dG, is conjugated to the exocylic amino group, which reduces its reactivity [i.e., N2-(3-oxopropenyl)-dG could be considered a vinylogous amide] (37). Furthermore, N2-(3-oxopropenyl)-dG exhibits a pKa of 6.9 so that at physiological pH it exists predominantly as the negatively charged conjugate base (37,38). This is anticipated to significantly reduce its reactivity to nucleophiles. Thus, M1dG does not appear to be a good candidate to form cross-links with DNA binding proteins. Malondialdehyde-mediated DNA−protein cross-links have been documented in vitro and in cells treated with MDA (39). In that study, cross-linking only occurred if the protein−MDA adduct was formed first and then exposed to DNA. This behavior is consistent with the present observation that a DNA molecule containing the malondialdehyde adduct, M1dG, does not form cross-links to EcoRI.
Although γ-HOPdG−protein cross-links can be generated and stabilized in vitro, the reversibility of the cross-link may limit its effects in vivo. In COS-7 cells, replication of plasmid DNA adducted with a NaCNBH3-reduced-γ-HOPdG-peptide cross-link resulted in approximately 8.4% mutation frequency, consisting of primarily G to T transversions, which is a higher frequency of mutation than that observed following replication of the γ-HOPdG adduct (29). The excision of DNA−protein cross-links by the UvrABC system was shown to proceed with moderate efficiency (40). Thus, secondary modifications to the adduct structure are capable of modulating the mutagenicity of γ-HOPdG and underscore the likelihood that DNA replication, transcription, or repair may be disrupted dependent on the longevity of the γ-HOPdG−protein cross-link. To date, the stability of Schiff base-mediated cross-links in vivo has not been elucidated. It will be interesting to determine what biological role, if any, γ-HOPdG-mediated DNA−protein cross-links play in acrolein-mediated toxicity in vivo.
Acknowledgments
We are grateful to Carol A. Rouzer for a critical reading of this manuscript and helpful editorial comments. The National Institutes of Health supported this work through research grants (R37CA87819, L.J.M.; R37GM029207, L.J.-J.), a program project grant (P01ES05355, T.M.H.), training grant (T32ES07028, L.A.V.), and center grant (P30ES00267, L.J.M. and T.M.H.). L.J.M. is the Mary Geddes Stahlman Professor of Cancer Research.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: BSA, bovine serum albumin; EDTA, ethylenediaminetetraacetic acid; γ-HOPdG, γ-hydroxy-2′-propanodeoxyguanosine; M1dG, pyrimido[1,2-α]purin-10(3H)-one; MOPS, 4-morpholinepropanesulfonic acid; MS, mass spectrometry; NER, nucleotide excision repair; SDS-PAGE, sodium dodecyl sulfate−polyacrylamide gel electrophoresis.
References
- Marnett L. J. (2000) Oxyradicals and DNA damage. Carcinogenesis 21, 361–370. [DOI] [PubMed] [Google Scholar]
- Berliner J. A.; Zimman A. (2007) Future of toxicology-lipidomics, an important emerging area for toxicologists: Focus on lipid oxidation products. Chem. Res. Toxicol. 20, 849–853. [DOI] [PubMed] [Google Scholar]
- Sayre L. M.; Perry G.; Smith M. A. (2008) Oxidative stress and neurotoxicity. Chem. Res. Toxicol. 21, 172–188. [DOI] [PubMed] [Google Scholar]
- Marnett L. J.; Riggins J. N.; West J. D. (2003) Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J. Clin. Invest. 111, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett L. J.; Hurd H. K.; Hollstein M. C.; Levin D. E.; Esterbauer H.; Ames B. N. (1985) Naturally occurring carbonyl compounds are mutagens in Salmonella tester strain TA104. Mutat. Res. 148, 25–34. [DOI] [PubMed] [Google Scholar]
- Cohen S. M.; Garland E. M.; St. John M.; Okamura T.; Smith R. A. (1992) Acrolein initiates rat urinary bladder carcinogenesis. Cancer Res. 52, 3577–3581. [PubMed] [Google Scholar]
- Anderson M. M.; Hazen S. L.; Hsu F. F.; Heinecke J. W. (1997) Human neutrophils employ the myeloperoxidase-hydrogen peroxide-chloride system to convert hydroxy-amino acids into glycolaldehyde, 2-hydroxypropanal, and acrolein. A mechanism for the generation of highly reactive alpha-hydroxy and alpha,beta-unsaturated aldehydes by phagocytes at sites of inflammation. J. Clin. Invest. 99, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. (1999) Current status of acrolein as a lipid peroxidation product. Trends Cardiovasc. Med. 9, 109–113. [DOI] [PubMed] [Google Scholar]
- Marnett L. J. (1988) Health effects of aldehydes and alcohols in mobile source emissions. In Air Pollution, the Automobile, and Public Health (Watson A. Y., Bates R. R., and Kennedy D., Eds.) pp 579−603, Health Effects Inst., National Academy Press, Washington, DC. [PubMed] [Google Scholar]
- Chung F.-L.; Young R.; Hecht S. S. (1984) Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 44, 990–995. [PubMed] [Google Scholar]
- Costa M.; Zhitkovich A.; Harris M.; Paustenbach D.; Gargas M. (1997) DNA-protein cross-links produced by various chemicals in cultured human lymphoma cells. J. Toxicol. Environ. Health 50, 433–449. [DOI] [PubMed] [Google Scholar]
- Sanchez A. M.; Minko I. G.; Kurtz A. J.; Kanuri M.; Moriya M.; Lloyd R. S. (2003) Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived α-HOPdG and γ-HOPdG regioisomeric deoxyguanosine adducts. Chem. Res. Toxicol. 16, 1019–1028. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Villalta P. W.; Wang M.; Hecht S. S. (2007) Detection and quantitation of acrolein derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 20, 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko I. G.; Washington M. T.; Kanuri M.; Prakash L.; Prakash S.; Lloyd R. S. (2003) Translesion synthesis past acrolein-derived DNA adduct, γ-hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. J. Biol. Chem. 278, 784–790. [DOI] [PubMed] [Google Scholar]
- de los Santos C.; Zaliznyak T.; Johnson F. (2001) NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct γ-OH-1-N2-propano-2′-deoxyguanosine. J. Biol. Chem. 276, 9077–9082. [DOI] [PubMed] [Google Scholar]
- Kozekov I. D.; Nechev L. V.; Moseley M. S.; Harris C. M.; Rizzo C. J.; Stone M. P.; Harris T. M. (2003) DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 125, 50–61. [DOI] [PubMed] [Google Scholar]
- VanderVeen L. A.; Druckova A.; Riggins J. N.; Sorrells J. L.; Guengerich F. P.; Marnett L. J. (2005) Differential DNA recognition and cleavage by EcoRI dependent on the dynamic equilibrium between the two forms of the malondialdehyde-deoxyguanosine adduct. Biochemistry 44, 5024–5033. [DOI] [PubMed] [Google Scholar]
- Cheng S. C.; Kim R.; King K.; Kim S. H.; Modrich P. (1984) Isolation of gram quantities of EcoRI restriction and modification enzymes from an overproducing strain. J. Biol. Chem. 259, 11571–11575. [PubMed] [Google Scholar]
- Nechev L. V.; Harris C. M.; Harris T. M. (2000) Synthesis of nucleosides and oligonucleotides containing adducts of acrolein and vinyl chloride. Chem. Res. Toxicol. 13, 421–429. [DOI] [PubMed] [Google Scholar]
- Schnetz-Boutaud N.; Mao H.; Stone M. P.; Marnett L. J. (2000) Synthesis of oligonucleotides containing the alkali-labile pyrimidopurinone adduct, M1G. Chem. Res. Toxicol. 13, 90–95. [DOI] [PubMed] [Google Scholar]
- Sambrook J. (1989) Molecular Cloning, A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Mao H.; Schnetz-Boutaud N. C.; Weisenseel J. P.; Marnett L. J.; Stone M. P. (1999) Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc. Natl. Acad. Sci. U.S.A. 96, 6615–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calingasan N. Y.; Chun W. J.; Park L. C.; Uchida K.; Gibson G. E. (1999) Oxidative stress is associated with region-specific neuronal death during thiamine deficiency. J. Neuropathol. Exp. Neurol. 58, 946–958. [DOI] [PubMed] [Google Scholar]
- Cho Y. J.; Wang H.; Kozekov I. D.; Kurtz A. J.; Jacob J.; Voehler M.; Smith J.; Harris T. M.; Lloyd R. S.; Rizzo C. J.; Stone M. P. (2006) Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived α-CH3-γ-OH-1,N2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem. Res. Toxicol. 19, 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. J.; Kim H. Y.; Huang H.; Slutsky A.; Minko I. G.; Wang H.; Nechev L. V.; Kozekov I. D.; Kozekova A.; Tamura P.; Jacob J.; Voehler M.; Harris T. M.; Lloyd R. S.; Rizzo C. J.; Stone M. P. (2005) Spectroscopic characterization of interstrand carbinolamine cross-links formed in the 5′-CpG-3′ sequence by the acrolein-derived γ-OH-1,N2-propano-2′-deoxyguanosine DNA adduct. J. Am. Chem. Soc. 127, 17686–17696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz A. J.; Lloyd R. S. (2003) 1,N2-Deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J. Biol. Chem. 278, 5970–5976. [DOI] [PubMed] [Google Scholar]
- Permana P. A.; Snapka R. M. (1994) Aldehyde-induced protein-DNA crosslinks disrupt specific stages of SV40 DNA replication. Carcinogenesis 15, 1031–1036. [DOI] [PubMed] [Google Scholar]
- Minko I. G.; Kozekov I. D.; Kozekova A.; Harris T. M.; Rizzo C. J.; Lloyd R. S. (2008) Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat. Res. 637, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuykendall J. R.; Bogdanffy M. S. (1994) Formation and stability of acetaldehyde-induced crosslinks between poly-lysine and poly-deoxyguanosine. Mutat. Res. 311, 49–56. [DOI] [PubMed] [Google Scholar]
- Nadkarni D. V.; Sayre L. M. (1995) Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem. Res. Toxicol. 8, 284–291. [DOI] [PubMed] [Google Scholar]
- Kurpiewski M. R.; Engler L. E.; Wozniak L. A.; Kobylanska A.; Koziolkiewicz M.; Stec W. J.; Jen-Jacobson L. (2004) Mechanisms of coupling between DNA recognition specificity and catalysis in EcoRI endonuclease. Structure 12, 1775–1788. [DOI] [PubMed] [Google Scholar]
- Grigorescu A. (2003) Structural and energetic determinants of the DNA binding specificity of EcoRI endonuclease. Ph.D. Thesis, University of Pittsburgh.
- Golan G.; Zharkov D. O.; Grollman A. P.; Dodson M. L.; McCullough A. K.; Lloyd R. S.; Shoham G. (2006) Structure of T4 pyrimidine dimer glycosylase in a reduced imine covalent complex with abasic site-containing DNA. J. Mol. Biol. 362, 241–258. [DOI] [PubMed] [Google Scholar]
- Liu W.; Chen Y.; Watrob H.; Bartlett S. G.; Jen.Jacobson L.; Barkley M. D. (1998) N-termini of EcoRI restriction endonuclease dimer are in close proximity on the protein surface. Biochemistry 37, 15457–15465. [DOI] [PubMed] [Google Scholar]
- Jen-Jacobson L.; Lesser D.; Kurpiewski M. (1986) The enfolding arms of EcoRI endonuclease: Role in DNA binding and cleavage. Cell 45, 619–629. [DOI] [PubMed] [Google Scholar]
- Szekely J.; Rizzo C. J.; Marnett L. J. (2008) Chemical properties of oxopropenyl adducts of purine and pyrimidine nucleosides and their reactivity toward amino acid cross-link formation. J. Am. Chem. Soc. 130, 2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggins J. N.; Pratt D. A.; Voehler M.; Daniels J. S.; Marnett L. J. (2004) Kinetics and mechanism of the general acid-catalyzed ring-closure of the malondialdehyde-DNA adduct, N2-(3-oxo-1-propenyl)-deoxyguanosine (N2OPdG-) to 3-(2′-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-a]purin-10(3H)-one (M1dG). J. Am. Chem. Soc. 126, 10571–10581. [DOI] [PubMed] [Google Scholar]
- Voitkun V.; Zhitkovich A. (1999) Analysis of DNA-protein crosslinking activity of malondialdehyde in vitro. Mutat. Res. 424, 97–106. [DOI] [PubMed] [Google Scholar]
- Minko I. G.; Zou Y.; Lloyd R. S. (2002) Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 99, 1905–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]