Abstract
The Torso (Tor) signaling pathway activates tailless (tll) expression by relieving tll repression. None of the repressors identified so far, such as Capicuo, Groucho and Tramtrack69 (Ttk69), bind to the tor response element (tor-RE) or fully elucidate tll repression. In this study, an expanded tll expression pattern was shown in embryos with reduced heat shock factor (hsf) and Trithorax-like (Trl) activities. The GAGA factor, GAF encoded by Trl, bound weakly to the tor-RE, and this binding was enhanced by both Hsf and Ttk69. A similar extent of expansion of tll expression was observed in embryos with simultaneous knockdown of hsf, Trl and ttk69 activities, and in embryos with constitutively active Tor. Hsf is a substrate of mitogen-activated protein kinase and S378 is the major phosphorylation site. Phosphorylation converts Hsf from a repressor to an activator that works with GAF to activate tll expression. In conclusion, the GAF/Hsf/Ttk69 complex binding to the tor-RE remodels local chromatin structure to repress tll expression and the Tor signaling pathway activate tll expression by modulating a dual transcriptional switch.
INTRODUCTION
Receptor tyrosine kinase (RTK) signaling pathways control various developmental processes and cellular outputs (1,2). Receptor kinases are activated by autophosphorylation once bound by a ligand, and this activates a series of proteins, in the order of Ras, Raf-1, mitogen-activated protein kinase kinase (Mapkk), and Mapk (the Ras/Raf/Mapk cassette). The activated Mapk turns on the expression of target genes (2,3). Studies from several developmental signaling pathways reveal that some signals regulate expression of target genes by utilizing transcriptional switches. In the absence of signaling, target genes are repressed by one or multiple transcription repressors that bind to specific DNA sequences. In the presence of signaling, either the repressor is inactivated, or the co-activator is activated or translocated into the nucleus, allowing the target genes to be switched from the repressive state into the active state (4).
Development of both the anterior and posterior poles, which are the terminal domains of Drosophila embryos, is specified by the maternal terminal system. The torso (tor) gene that encodes a RTK is a key component in this system (5,6). Tor is distributed uniformly throughout the embryonic membrane and its kinase activity is locally activated by a ligand, the C-terminus of Trunk that is localized at both poles. The Ras/Raf/Mapk cassette is then activated, leading to the restricted expression of two zygotic genes, tailless (tll) and huckebein (1,7,8). The tll gene is essential for development of the terminal domains. Larvae homozygous for tll loss-of-functions exhibit no brain and have an abnormal cephalopharyngeal skeleton at the anterior, and no external structures behind abdominal segment 8 (9). The expression patterns of tll at stage 4 are two caps at both poles of embryos. The anterior cap becomes a horse-shoe pattern at stage 5 (10). Cytological and genetic analyses reveal that the posterior expression of tll in embryos lacking tor activity is largely diminished, whereas that posterior expression in embryos with constitutively active Tor is greatly expanded. These observations lead to the conclusion that tll expression is regulated by the Tor pathway (1,7,11).
Genetic and molecular data suggest that the activation of tll expression by the Tor pathway is through relief of transcriptional repression. First, detailed analyses of the tll cis-regulatory region revealed the presence of an 11-bp DNA sequence that acts as a key repression element, which was designated as the tor response element (tor-RE). Mutation of this element results in ectopic tll expression in the middle of embryos where tor is inactive (12–14). Second, a zinc-finger transcription repressor, Tramtrack69 (Ttk69) (15), binds to a TCCT element (TC5) at the 3′ flanking region of the tor-RE and assists in the initiation of tll repression (16). Ttk69 is degraded after being phosphorylated by Mapk (17). Third, tll expression in embryos lacking the maternally contributed HMG-like protein Capicuo or co-repressor Groucho is expanded toward the central domain (18,19). Nonetheless, the degree of this expansion of tll expression is far less than that observed in embryos from mothers with a tor gain-of-function, suggesting that these proteins are components of a large repression complex. However, the nature of this complex remains unknown (7).
Identification of repressors that directly bind to the tor-RE is vital to obtain insights into how the repression complex forms. Using column chromatography, we previously identified that the GAGA factor, GAF encoded by the Trithorax-like (Trl) gene (20), binds to the sequence TGAG in the reverse strand of the tor-RE (13). In agreement with the enhancement of position effect variegation by Trl loss-of-function (20), numerous studies indicate that GAF opens up the local chromatin structure to increase transcription factor binding (21,22), and prevents heterochromatin propagation by interacting with the NURF chromatin remodeling complex to activate the expression of a number of genes (22–24).
Among the cis-regulatory elements of genes bound by GAF, the heat shock element (HSE) that activates hsp70 expression is intriguing. Similar to the tor-RE, HSE contains a triple GAA-inverted repeat and the AGAG sequence that are bound by Heat shock factor (Hsf) and GAF, respectively. The synergistic interaction between Hsf and GAF is required for maximum hsp70 expression (25). In the tor-RE, the sequence bound by GAF is TGAG, and two GAAs in an inverted-repeat fashion (Supplementary Figure S1) locate at the 3′-end and downstream, constituting a potential binding site for Hsf. The differences between the tor-RE and HSE are the relative positions of the GAF and Hsf binding sites, number of the GAA repeats and the spacing between the GAA inverted repeats. Information from the UCSC Genome Bioinformatics (http://genome.ucsc.edu/) shows that the tor-RE plus 12 downstream nucleotides are highly conserved among the divergent Drosophila species, except for the spacing between the GAA inverted repeats (Supplementary Figure S1). The regulatory elements with either 2- or 3-bp spacing are presumably functional in regulating tll expression in these Drosophila species.
Numerous studies have shown that Hsf activates the expression of many genes when cells are subjected to stresses, such as heat, hypoxia, heavy metals and infection (26–28). However, a few reports show that Hsf also acts as a transcriptional repressor (29–31). In this study, we tested whether Hsf forms a complex with GAF (GAF/Hsf) and binds to the tor-RE for tll repression using dosage-dependent genetic interaction and DNA binding experiments. Since the Tor signaling pathway regulates tll expression, the effect of Hsf phosphorylation on tll expression was also investigated.
MATERIALS AND METHODS
Fly stocks and genetics
The fly lines net1 cn1 hsf1/CyO, cn1 hsf4 bw1, w; P{w+mw, FRT}2A and w; P{w+mw, ovoD1}2X P{w+mw, FRT}2A/st1 β-Tub85DD ss1 es/TM3 Sb1 (abbreviated as P{w+mw, FRT}2A ovoD1) were obtained from the Bloomington Stock Center. Lines w; TrlDHΔ34/TM6B Tb1 e, w; TrlR85/TM6B Tb1 e (20) and w*; ttkle11/TM3 Sb1 (32) were generously provided by Drs D.-H. Huang, F. Karch and Y.-N. Jan. Both hsf1 and TrlDHΔ34 are amorph (33; Dr D.-H. Huang, unpublished data). These lines were used to obtain females who were transheterozygous for hsf1 and TrlDHΔ34, hsf1 and ttkle11 or TrlDHΔ34 and ttkle11, and were mated with males carrying the P{w+MC=tll-MRR-lacZ}G11 transgene (G11; Supplementary Figure S2). tll-MRR stands for tll minimal regulatory region (13). Embryos collected from these crosses were stained using in situ hybridization, and X-gal staining to reveal tll expression patterns.
Using meiotic recombination (34), w; P{w+mw, FRT}2A TrlR85/TM6B Tb1 e was generated. Females of w; P{w+mw, FRT}2A TrlR85/P{w+mw, FRT}2A ovoD1 from germ-line clone experiments (GLC) (35) were crossed with G11 males to obtain embryos with reduced maternal Trl activity.
Generation of transgenic flies
For constructing P{w+ MC = UASP-hsf} transgenes, the BamHI/XbaI DNA fragment containing hsf coding region was subcloned into BamHI and XbaI sites of pUASP (36). A cDNA encoding TorD4021 was a generous gift from Dr C. Nusslein-Volhard (8). The coding region was amplified using PCR and subcloned into pUASP.
Since the hsp70 basal promoter in pWIZ (37) functions poorly in Drosophila germ lines, the EcoRI–XbaI DNA fragment containing the white intron was excised from pWIZ and inserted into XbaI and KpnI sites of pUASP, after EcoRI and KpnI ends being filled with Klenow enzyme (Roche Applied Science). The resulting vector, pP{w+=UASP-w-intron}HC, was abbreviated as pHC.
Approximately 700-bp DNA fragments were amplified from Trl, hsf or ttk69 cDNA by PCR for generating UASP-RNAi constructs. Primer sets for Trl, hsf and ttk69 were AAACTCTAGACGTATTGGGTAAG/GAGTCTAGAGTTGTATCGTTACC, GACAATCTAGATTCGCGCTTC/CTATCTTCTAGAATATCTCCAGC and GAGCCTCTAGAGTTCAAGTAC/ATGTGCGTCTAGACGCGGG, respectively. XbaI site in each primer is underlined. Amplified DNA fragments were separately inserted into NheI and AvrII sites of pHC in an inverted-repeat fashion.
All resulting plasmids were transformed into fly using the P-element mediated germ-line transformation (38,39). GAL4-GCN4 (40) was used to drive expression of hsf, torD4021 or the three double-stranded RNAs (dsRNAs).
The site-directed mutagenesis
For amino-acid substitution to S378 and T390 in Hsf, the regulatory domain (RD) was subcloned into pGEX-4T (GE/Amersham Biosciences). The resulting plasmid was designated as pGEX4T-Hsf-RD. The site-directed mutagenesis was followed the procedure described in the instruction manual of QuikChange site-directed mutagenesis kit (Stratagene, Inc.). Sequences of primer pairs for replacing either S378 or T390 by Ala or Asp were GGTGCCAAACGCTCCGCCTTATTACGAG/CTCGTAATA AGGCGGAGCGTTTGGCACCTCCTGG (S378A), CCAGGAGGTGCCAAACGATCCGCCTTATTACGAG/CTC GTAATAAGGCGGATCGTTTGGCACCTCCTGG (S378D) and GAATGTGCTTACCGCGCCCATGGTGCGG/ CCGCACCATGGGCGCGGTAACACATTC (T390A). One hundred and twenty-five nanograms of primers for each pair were used to amplify DNA from plasmid pGEX4T-RD using PCR with 2.5 U of Pfu DNA polymerase (Fermentas, Inc.). The amplified DNAs were digested by DpnI. The DpnI-treated DNA was transformed into Escherichia coli XL-1 blue. The resulting plasmids were designated as pGEX4T-hsf-RS378A, pGEX4T-hsf-RS378D and pGEX4T-hsf-T390A. DNA fragments from these clones were later used to replace the same DNA fragment in pP{w+MC = UASP-hsf} (41).
Bacterial synthesis of GAF, Ttk69, Hsf and various forms of Hsf-RDs
The hsf coding sequence was amplified from an EST clone, LD33470, by PCR and inserted into KpnI and XhoI sites of an expression vector, pET29a. Sequence of the inserted DNA fragment was verified by sequencing. The resulting plasmids and pRSET-ttk69 (16) were individually transformed into E. coli BL21(DE3) pG-Tf2 (TaKaRa Inc.) for the protein synthesis (42,43). For synthesizing GAF, pAR-GAF, a generous gift from Dr W. Soeller, was transformed into E. coli BL21(DE3) pLysS. Except the GST fusion proteins, the bacterially expressed proteins were partially purified using the SP-Sepharose column chromatography. The GST fusion proteins were purified according to the procedure provided by the manufacture (GE/Amersham Bioscience).
Electromobility shift assay and DNaseI footprinting
Sequence of the probe is CGTTCCTTGCTCAATGAATTTTTCG (the tor-RE is underlined). Two complementary oligonucleotides were synthesized and labeled with γ[32P]-ATP and polynucleotide kinase. The 32P-labeled probe was mixed with various amounts of bacterially expressed proteins in a reaction buffer (10 mM Tris–Cl pH8.0, 10 mM KCl, 6 mM β-mercaptoethanol, 3 mM MgCl2, 1 mM EDTA, 10% glycerol, and 4.5 ng salmon sperm DNA) (44). Binding reactions were carried out on ice for 10 min before loading onto a 4% polyacrylamide gel in a TGE buffer (25 mM Tris base, 190 mM glycine and 1 mM EDTA). DNA–protein complexes were visualized by autoradiography using Phosphoimager (Typhoon, GE/Amersham Bioscience) (41). To measure binding affinity, radioactivities of bands were measured using the software provided by manufacturer (GE/Amersham Bioscience).
DNaseI footprinting experiments were performed as described by Kadonaga et al. (45), with modifications. The tll-MRR labeled with γ[32P]-ATP and polynucleotide kinase was served as probe (41). The reaction buffer used in electromobility shift assay (EMSA) was used, instead of the HEPES buffer, pH 7.5 (13). The binding mixture was incubated at room temperature for 10 min. In both EMSA and DNaseI experiments, nonspecific DNA competitors, poly dI-dC, were used.
Shift-western blotting
Volumes of various proteins were the same as those used in EMSA, except quantity of the [32P]-labeled probe and incubation temperature, as indicated in the legend of Figure 3 and Supplementary Figure S4. Shift-western blotting was performed as described previously (46). In brief, DNA–protein complexes were separated in 6% polyacrylamide gels and transferred onto stacked nitrocellulose (Schleicher & Schuell) and nylon membranes (Hybond-XL, GE/Amersham Biosciences). The radiolabeled probe on nylon membrane was detected by autoradiography, whereas the proteins in the complexes on the nitrocellulose membrane were detected by immunoblotting with the chemiluminescence assay kit (47; Western lightingTM, Blossom Biotechnologies, Inc, Taiwan).
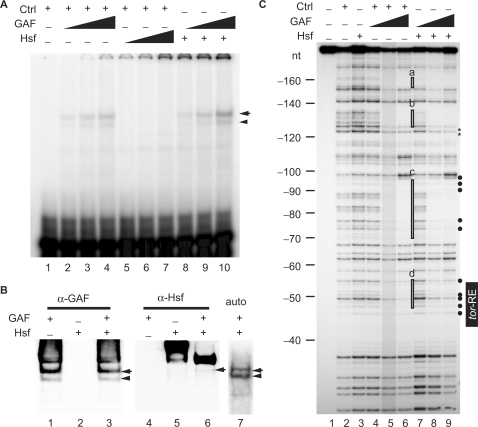
Figure 3.
Hsf alters the DNA-binding property of GAF. (A) EMSA was performed using a 25-bp 32P-labeled probe with various amounts of proteins. The volume of the control was 2 μl (lanes 1–7). Various volumes of Hsf (1.25, 2.5 and 5 μl in lanes 5–7; 5 μl in lanes 8–10) and GAF (1, 2 and 4 μl in lanes 2–4 and 8–10) were used. The DNA–protein complexes were separated in a 4% polyacrylamide gel and detected using a PhosphoImager (GE/Amersham Biosciences). An arrow head and arrow indicate the tor-RE bound by a monomer and dimer, respectively. (B) Shift-western blotting of the complexes formed with GAF (4 μl in lanes 1, 3, 4, 6 and 7), Hsf (5 μl in lanes 2, 3, 5, 6 and 7) and the labeled probe. Quantity of the probe was 5-fold higher than that used in EMSA. GAF and Hsf on the nitrocellulose membrane were detected by immunoblotting with anti-GAF and Hsf antibodies, respectively. The ‘auto’ represents an autoradiogram that shows position of the radiolabeled probe on the nylon membrane. The DNA–protein complexes indicated by an arrow and arrow head are the same as those shown in (A). (C) DNaseI footprinting was used to show where Hsf, GAF or GAF/Hsf binds in the tll-MRR. The control is proteins extracted from bacteria carrying the pET28a vector. The protein concentrations of the control, GAF and Hsf proteins determined by the Bio-Rad protein assay (Bio-Rad Laboratories, Inc.) are 2.24, 1.75 and 0.34 mg/ml, respectively. Various amounts of proteins were incubated with the probe in a final volume of 50 μl. The reaction mixture was incubated at room temperature for 10 min. A constant volume of control (2 μl in lanes 2 and 4–6) and Hsf (10 μl in lanes 3 and 7–9) were used, whereas various volumes of GAF (1, 2 and 4 μl; represented by triangles) were used (lanes 4–6 and 7–9). Brightness of lane 5 is reduced by 15%. Regions protected by GAF alone are highlighted by open rectangles (a–d) at the right of lane 6. Comparing intensities of bands in lanes 9 with those in lane 6, bands with decreased and increased intensity are marked by asterisks and close circles at the right of lane 9, respectively. Numbers at the left indicate distance in nucleotide from the BstNI site (Supplementary Figure S2).
In situ hybridization
The procedure of in situ hybridization was described by Tautz and Pfeifle (48). Embryos were fixed by 10% paraformaldehyde, 50 mM EGTA in 1× PBS. The fixed embryos were treated with proteinase K to have a better penetration for digoxigenin-labeled RNA. Location of the probe in embryos was detected using an antidigoxigenin antibody conjugated with alkaline phosphatase. The color development was carried out in a staining solution (100 mM Tris, pH 9.5, 100 mM NaCl, 50 mM MgCl2, 0.1% Tween-20 and 1 mM levamisol HCl) containing NBT and X-phosphate. tll and lacZ patterns in embryos were viewed under the DIC light microscope and photographed using CCD camera (CoolSnap, Photomatrics, Inc.).
The X-gal staining
Embryos were collected and dechorionated by 100% bleach. After wash, embryos were soaked in heptane saturated with glutaraldehyde for 15 min. The fixed embryos were incubated with 0.1% X-gal in a ferric phosphate buffer (10 mM NaPO4 buffer, pH 7.2, 3.1 mM K3[FeII(CN)6], 3.1 mM K4[FeIII(CN)6], 150 mM NaCl and 1 mM MgCl2) (49). Stained embryos were mounted in 10% glycerol containing 0.1% Triton X-100 right before observation under the DIC light microscope and photographed using digital camera (Coolpix 995, Nikon, Co.).
RESULTS
hsf is likely to participate in tll repression
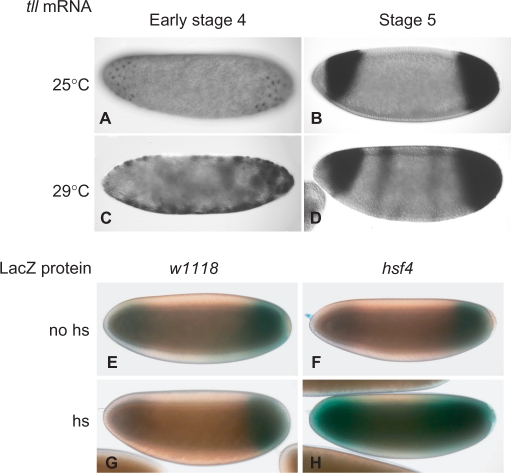
An expanded pattern of tll expression was observed in early stage-4 w1118 embryos at 29°C (Figure 1C) and the expanded expression gradually withdrew back to the normal expression pattern at late stage 5 (Figure 1D). hsf4, encoding a mutant Hsf that cannot bind DNA at 29°C (33), was used to determine whether Hsf played a role in tll repression. A 30 min heat shock at 37°C applied to embryos from hsf4; G11 parents resulted in an expanded expression pattern, as represented by lacZ (Figure 1H). The embryos without heat shock showed the normal expression pattern (Figure 1F). tll expression patterns in w1118 embryos with or without heat shock were normal (Figure 1E and G). Since DNA binding is required for Hsf to activate gene expression (33), these results supported us to initiate investigations into role of Hsf in tll repression.
Figure 1.
hsf activity is likely to participate in tll repression. (A–D) tll mRNA patterns in embryos at early stage 4 (A and C) and stage 5 (B and D) were determined by in situ hybridization with a digoxigenin-labeled antisense tll RNA. w1118 embryos were incubated at either 25 (A and B) or 29°C (C and D). Embryos are arranged in a sagittal view, with the anterior towards the left. (E–H) lacZ protein distribution in embryos from G11 (E and G) or hsf4; G11 (F and H) parents was determined by X-gal staining. The heat-shock treatment (hs) was at 37°C for 30 min (G and H). G11 embryos serve as controls (E and G). Embryos at early stage 5 are shown, with the anterior towards the left.
Both Hsf and GAF are required for the repression and activation of tll
Our previous results showed that base substitutions to the 5′ sequence of the tor-RE result in a uniform lacZ expression pattern (G53 in Supplementary Figure S2). When the GAA sequence at the 3′-end of the tor-RE was substituted by TTA (G54 in Supplementary Figure S2), a uniform expression pattern was also observed, but the level was lower than that driven by G53 (compare Supplementary Figure S2D with S2C). The discrepancy between these two patterns suggested that two different factors bind to the tor-RE and contribute differently to tll expression. While GAF binds to the 5′-end of the tor-RE (13), does Hsf bind to the GAA sequence?
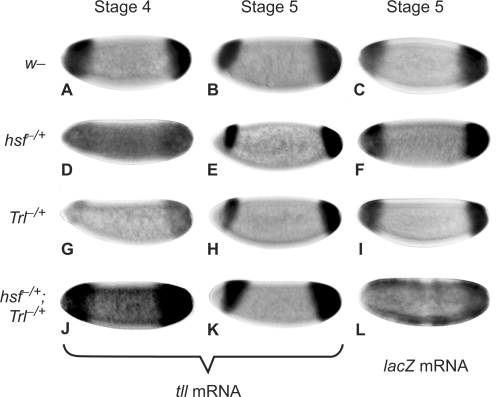
tll expression patterns were examined in embryos lacking either hsf or Trl activity. Since the tll gene is one of the earliest activated zygotic genes, factors that regulate tll expression in stage-4 embryos are provided maternally. GLC is the most common method to obtain females heterozygous for a mutant, i.e. hsf1, that can produce embryos lacking the gene activity. In addition, lacZ expression patterns in embryos from females crossed with G11 males that carry the tll-MRR-lacZ transgene were analyzed to reveal whether Hsf or GAF functions through the tll-MRR. Unfortunately, females from GLC with either hsf1 or TrlDHΔ34 produced no or droplet eggs, consistent with the fact that hsf and Trl activities are required for oogenesis (33,50). Therefore, dosage-dependent genetic experiments with hsf1 and TrlDHΔ34 were used. Embryos from females transheterozygous for hsf1 and TrlDHΔ34 exhibited an expanded tll expression pattern with a high background in the middle of embryos (Figure 2J), as well as an expanded lacZ expression pattern (Figure 2L). The posterior expression domains of tll in embryos with reduced both hsf and Trl activities were measured as percentage of egg length. The averaged domain was 31.5 ± 5.1% that was significantly expanded comparing to that in w1118 embryos (24.8 ± 2.7%) (P < 0.001 determined by Student's t-test; n = 70 embryos). tll expression patterns resumed to normal at stage 5 and after (Figure 2K). By comparing the central domain of stage-5 embryos (Figure 2E) as background with that in stage-4 embryos, 48% of stage-4 embryos with reduced hsf activity (n = 61) appeared to have a higher background of tll expression in the middle of embryos (Figure 2D). A similar ratio (40%) of embryos was observed with a higher background of lacZ expression in the middle of the embryos (Figure 2F). Reduction of Trl activity only affected the expression level of tll, but not the pattern of expression (Figure 2G and H). The de-repression at stage 4 indicated that the genetic interaction of hsf with Trl is important for tll repression, and that Hsf plays a key role in this repression.
Figure 2.
The genetic interaction of hsf with Trl is critical for tll repression and activation. Embryos from females of w1118 (A–C), hsf1/+ (D–F), TrlDHΔ34/+ (G–I) and hsf1/+; TrlDHΔ34/+ (J–L) crossed with G11 males were used to determine the tll expression patterns (A, B, D, E, G, H, J and K) and lacZ (C, F, I and L) using in situ hybridization with digoxigenin-labeled antisense tll and lacZ RNA, respectively. Expression patterns at either late stage 4 or early stage 5 are indicated by stage 4 and 5. Genotypes of females are shown to the left. Embryos are arranged in a sagittal view, with the anterior towards the left.
Both Trl and hsf activities were also required for tll activation, since tll mRNA levels in embryos from mothers heterozygous for hsf1 or TrlDHΔ34 were lower than those in the wild-type embryos (compare Figure 2D and G with 2A). Furthermore, tll expression patterns in embryos from GLC with TrlR85 were normal at stage 4, but the expression levels were much lower (Supplementary Figure S3E). In conclusion, both Hsf and GAF were required for the repression and activation of tll.
Hsf alters the footprinting pattern over the tor-RE protected by GAF
EMSA was performed to reveal whether Hsf binds to the tor-RE. Results showed that Hsf alone was unable to bind to the tor-RE (lanes 5–7 in Figure 3A). Based on the fact that Hsf physically interacts with GAF (25), various quantities of GAF were mixed with Hsf to test whether GAF assists Hsf binding to the tor-RE. The binding of GAF/Hsf to the tor-RE was marginally increased (compare lanes 10 with 4 in Figure 3A). Shift-western blotting was then performed to reveal whether Hsf associates with the complex formed by GAF and the tor-RE. As expected, GAF was detected in the complexes with or without Hsf (lanes 3 and 1 in Figure 3B), whereas Hsf was only detected in the presence of GAF (lane 6 in Figure 3B). These results indicated that Hsf associated with the tor-RE/GAF complex, but not the tor-RE alone.
Espinas and colleagues (51) demonstrate that GAF forms oligomers through the Bric-a-Brac/Tramtrack/Broad complex (BTB) domain at its N-terminus, and one single GAF oligomer interacts with two adjacent GAF binding sites that are separated over 20 bp. Our previous DNaseI footprinting results showing specific binding of GAF to the tor-RE (13) was likely due to an assistance of GAF oligomer binding to high affinity sites upstream the tor-RE. The stable binding of GAF to the tor-RE might then recruit Hsf through the direct interaction with GAF (25). Using DNaseI footprinting to test this idea, four footprints (a–d) in the tll-MRR were bound by GAF (lanes 4–6 in Figure 3C). Intensities of bands in footprint d corresponding to the 5′ half of the tor-RE were diminished, indicating that the tor-RE was a weak site for GAF. Consistent with the EMSA result, no site in the tll-MRR was protected by Hsf (lane 3 in Figure 3C). When GAF and Hsf were added, the patterns of footprints b to d plus their flanking regions were altered (lane 9). In footprint b, intensities of bands in the 3′ flanking region was decreased, as indicated by asterisks. In both sites c and d, the footprinting regions were narrower than those protected by GAF alone. Intensities of bands, indicated by close circles at the right of lane 9, were increased (compare intensities of bands in lane 9 with those in lane 6 in Figure 3C). These results indicated that Hsf altered the DNA-binding property of GAF.
GAF, Hsf and Ttk69 form a protein complex that binds to the tor-RE tightly
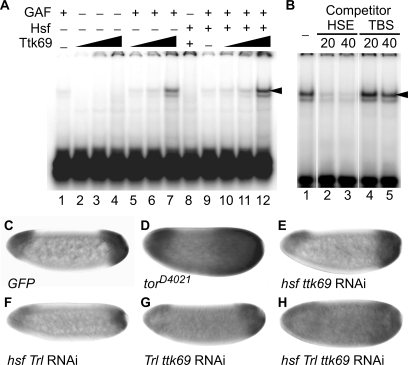
Our previous results showed that Ttk69 acts as an accessory factor to initiate tll repression (16). In addition, Pagans and colleagues (52) show that the interaction with GAF is required for Ttk69 to repress gene expression in the absence of Ttk69 binding sequences. Therefore, Ttk69 might form a complex with GAF/Hsf by interacting with GAF. To test this hypothesis, we examined whether Ttk69 enhances GAF/Hsf binding to the tor-RE using EMSA. The results showed that Ttk69 enhanced GAF and GAF/Hsf binding to the tor-RE by 5- and 18-fold, respectively (compare lanes 7 and 12 with 1 and 9 in Figure 4A). However, Ttk69 alone did not bind to the tor-RE (lanes 2-4), and did not enhance Hsf binding (lane 8) to the tor-RE. A tight association between Ttk69 and GAF/Hsf was revealed by competition experiments using TBS competitor DNA that contains the Ttk69 binding site in the even-skipped promoter (52). A 20-fold excess of HSE competitor DNA abolished the binding of GAF/Hsf/Ttk69 to the tor-RE (lane 2 in Figure 4B), whereas TBS in 40-fold excess did not interfere with GAF/Hsf/Ttk69 binding (lane 5 in Figure 4B). Results from shift-western blotting showed that both Ttk69 and Hsf were present in the DNA–protein complex (Supplementary Figure S4). Taking into consideration the fact that there was no TC5 in the probe used for EMSA and that Ttk69 physically interacts with GAF (53), these results indicated that GAF, Hsf and Ttk69 form a protein complex that binds tightly to the tor-RE, independently of TC5.
Figure 4.
GAF, Hsf and Ttk69 form a stable complex on the tor-RE, repressing tll expression. (A) The bacterially expressed Ttk69 (0.15 mg/ml) was used to test whether Ttk69 enhanced GAF/Hsf binding to the tor-RE using EMSA. The probe, GAF and Hsf were the same as those used in Figure 3A. GAF (2 μl in lanes 1, 5–7 and 9–12) and Hsf (3 μl in lanes 8–12) were mixed with various volumes of Ttk69 (1, 2 and 4 μl in lanes 2–4, 5–7 and 10–12; and 2 μl in lane 8). (B) A competition experiment shows association of Ttk69 with GAF/Hsf on the tor-RE. HSE, 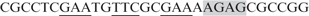 (binding sites for Hsf and GAF are underlined and shadowed, respectively), and a Ttk69 binding site in the even-skipped cis-regulatory region, TCCTCATGGTCCTGCCGAGCAG (TBS), were used as competitor DNAs. The folds in molar excess of the competitor DNAs, HSE and TBS, are shown on top of lanes 2–5. Using a PhosphoImager, the radioactivities of the upper bands indicated by an arrow head in (A and B) were measured for quantitative analysis of DNA binding. (C–H) tll expression patterns in embryos with various combinations of RNAi to knockdown hsf, Trl or ttk69 activities, as shown at bottom of each panel. tll expression patterns in embryos with GFP (C) or TorD4021 (D) expression serve as controls. Embryos are arranged in a sagittal view, with the anterior towards the left.
(binding sites for Hsf and GAF are underlined and shadowed, respectively), and a Ttk69 binding site in the even-skipped cis-regulatory region, TCCTCATGGTCCTGCCGAGCAG (TBS), were used as competitor DNAs. The folds in molar excess of the competitor DNAs, HSE and TBS, are shown on top of lanes 2–5. Using a PhosphoImager, the radioactivities of the upper bands indicated by an arrow head in (A and B) were measured for quantitative analysis of DNA binding. (C–H) tll expression patterns in embryos with various combinations of RNAi to knockdown hsf, Trl or ttk69 activities, as shown at bottom of each panel. tll expression patterns in embryos with GFP (C) or TorD4021 (D) expression serve as controls. Embryos are arranged in a sagittal view, with the anterior towards the left.
The expanded patterns of tll in embryos with simultaneous knockdown of hsf, Trl and ttk69 activities are similar to those in embryos with constitutively active Tor
The attempt to establish lines that were transheterozygous for hsf1 and ttkle11 or hsf1 and TrlDHΔ34 for dosage-dependent genetic experiments was unsuccessful. Thus, the RNAi approach (37) was used with modifications. To express large quantities of dsRNAs in the germ line, RNAi lines carrying UASP-hsf RNAi, UASP-Trl RNAi or UASP-ttk69 RNAi were generated. As mentioned above and in our previous report (16), the activities of these three genes are all required for oogenesis. Consistent with results from GLC, some of the RNAi lines had eggless phenotypes when dsRNAs were produced. The lines that can produce eggs showed low efficiencies of gene knockdown, as represented by a fly line with simultaneous knockdown of all three gene activities (Supplementary Figure S5). Fly lines with knockdown of at least two gene activities were generated for examining tll expression patterns. Embryos with simultaneous knockdown of all three genes exhibited low levels of tll expression, but had greatly expanded patterns (Figure 4H). The degree of the expansion was comparable to that in embryos with a constitutively activated Tor, TorD4021 (Figure 4D). Embryos with knockdown of both Trl and ttk69 gene activities showed a higher level of tll expression in the middle of embryos (compare Figure 4G with C). Embryos with knockdown of both hsf and Trl gene activities had reduced levels of tll expression at both poles, but the expression patterns remained unaffected (Figure 4F), inconsistent with the results shown in Figure 2J. In contrast, knockdown of both hsf and ttk69 activities had no effect on both the pattern and the level of tll expression (Figure 4E).
In conclusion, these results demonstrated that Hsf, GAF and Ttk69 specifically bind to the tor-RE together, and are essential for tll repression in vivo. These results also indicated that Ttk69 acts as a co-repressor for tll repression.
Rpd3 is involved in tll repression
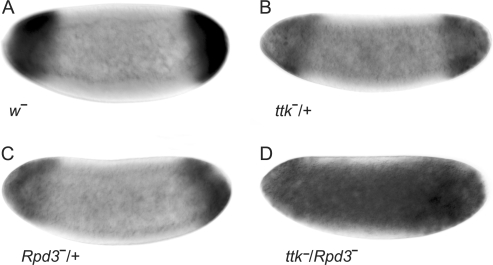
Approximately 75% of late stage-5 embryos (n = 16) from GLC with TrlR85 exhibited the anterior-stripe expression of tll that was expanded posteriorly (Supplementary Figure S3G), indicating that embryos with reduced Trl activity was unable to properly refine the anterior-stripe expression of tll. Several reports pointed out that GAF physically associates with the Polycomb complex proteins to repress the expression of a number of genes (23,54–56). In addition, interaction with Rpd3 is required for Ttk69 to suppress neuronal cell fate (17). Therefore, does GAF/Hsf/Ttk69 recruit chromatin remodeling factors such as Rpd3 (57,58)? As shown in Figure 5D, the tll expression patterns in embryos with reduced ttk and Rpd3 activities were expanded, whereas the expression patterns in embryos with reduction of either ttk or Rpd3 activity were normal (Figure 5B and C). The participation of Rpd3 in tll repression suggested that GAF/Hsf/Ttk69 recruits chromatin remodeling factors that lead to a higher order organization of local chromatin structure, allowing repression of tll expression.
Figure 5.
Rpd3 activity participates in tll repression. Embryos from females of w1118 (A), ttkle11/+ (B), Rpd3303/+ (C) and ttkle11/Rpd3303 (D) crossed with w1118 males were used to determine tll expression patterns using in situ hybridization. The genotypes of the females are indicated at the bottom of each panel. tll expression patterns in embryos at late stage 4 are shown. Embryos are arranged in a sagittal view, with the anterior towards the left.
S378 in Hsf is a major site for Mapk phosphorylation, and phosphorylation converts Hsf from a repressor to an activator of tll expression
tll expression is activated by the Tor pathway by relieving transcriptional repression (13). In vertebrates, the S307 MAPK phosphorylation site in the RD of HSF1s (59) is highly conserved (Figure 6A). In order to investigate whether Hsf is a substrate of Mapk in Drosophila, putative Mapk phosphorylation sites in the RD that are conserved among Drosophila species were identified using the ClustalW program. These sites were tested to determine whether they function in tll repression in vitro and in vivo.
Figure 6.
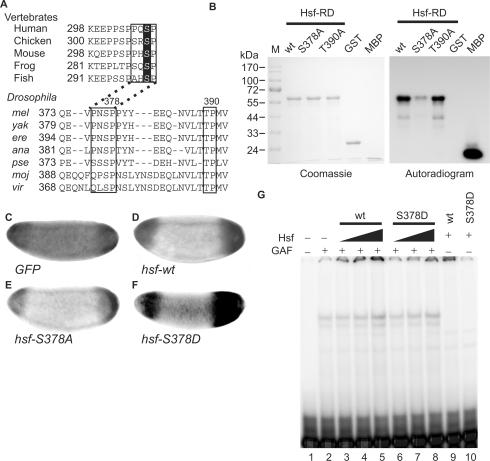
Phosphorylation by Mapk converts Hsf from a repressor to an activator for tll expression. (A) Amino acid sequences of HSF1s in vertebrates were retrieved from Genbank and the regulatory domains of these HSF1s (HSF-RD) were aligned using the ClustalW program to reveal conserved amino acids. Putative Mapk recognition sequences are boxed and S307, a target site of Mapk in HSF1s, is highlighted. The same procedure was applied to Hsfs in Drosophila species including melanogaster (mel), yakuba (yak), erecta (ere), ananassae (ana), pseudoobscura (pse), mojavensis (moj) and virilis (vir). Amino acid sequences in boxes that contain two putative Mapk phosphorylation sites, S378 and T390, are highly conserved among Drosophila species. (B) In an in vitro phosphorylation assay, 100 pmol of GST-Hsf-RD and two mutant forms (GST-RD-S378A and T390A), as well as negative (GST) and positive (myelin basic protein, MBP) controls were used. After incubation with Mapk and γ[32P]-ATP, the proteins were separated in a 12% SDS gel and visualized using Coomassie blue staining, shown at the left. After the gel was dried, autoradiography was used to reveal the phosphorylated proteins, shown at the right. (C–F) tll expression patterns in late stage-4 embryos from mothers transheterozygous for hsf1 and TrlDHΔ34, determined by in situ hybridization, are shown and the anterior is arranged towards the left. Supplementation with UAS-hsf-wt (D), -S378A (E) and -S378D (F) driven by GAL4-GCN4 suppresses the expanded tll expression patterns. GFP expression was used as a negative control (C). (G) Hsf-S378D binding to the tor-RE is enhanced by GAF. The protein concentration of Hsf-S378D is 0.34 mg/ml. The Hsf, GAF and the probe were the same as those used in Figure 3A. GAF (4 μl) was mixed with various volumes of Hsf (wt) or Hsf-S378D (S378D) (2.5 μl in lanes 3 and 6; 5 μl in lanes 4 and 7; and 10 μl in lanes 5 and 8–10) and 32P-labeled probe. The DNA–protein complexes were separated in a 4% polyacrylamide gel and visualized using a PhosphoImager.
Using the minimal consensus sequence phosphorylated by Mapk [Ser/Thr-Pro; (60)], two putative sites S378 and T390 were found, that are conserved among Drosophila species (Figure 6A). The RD of wild-type and two different amino-acid substitutions, S378A and T390A, were fused to GST and expressed in bacteria (abbreviated as Hsf-RD-wt, Hsf-RD-S378A and Hsf-RD-T390A). The bacterially expressed proteins were incubated with Mapk in the presence of γ[32P]ATP. As shown in Figure 6B, myelin basic protein (MBP, a positive control), Hsf-RD-wt and Hsf-RD-T390A were phosphorylated by Mapk, whereas the degree of Hsf-RD-S378A phosphorylation was greatly diminished. These results indicated that S378 in Hsf-RD is the major site of Mapk phosphorylation.
In order to demonstrate that S378 is an important residue regulated by the Tor pathway in vivo, transgenic fly lines carrying UASP-hsf, UASP-hsf-S378A or UASP-hsf-S378D were generated. To facilitate demonstration of the functions of the various hsf transgenes, these transgenes were expressed in embryos that had reduced activities of both hsf and Trl (Figure 6C). The results of these experiments showed that supplementation with wild-type Hsf restored the normal tll expression pattern (Figure 6D). When Hsf-S378A was supplemented, the levels of tll mRNA at both poles was lower, although the tll expression patterns were restored (Figure 6E). These results further supported that Hsf is involved in tll repression, and indicated that S378 was the site in Hsf responsible for the conversion from repressor to activator for tll activation. Role of Hsf phosphorylation involved in tll activation was further supported by the higher level of tll expression and slightly expanded expression patterns when Hsf-S378D was supplemented (Figure 6F), and by the similar binding affinities of Hsf-wt and Hsf-S378D to the tor-RE, which were enhanced by GAF (Figure 6G). These results supported a role for Hsf as a transcriptional switch, which is modulated by the Tor pathway that regulates tll expression.
DISCUSSION
A model to illustrate how the Tor pathway regulates tll expression
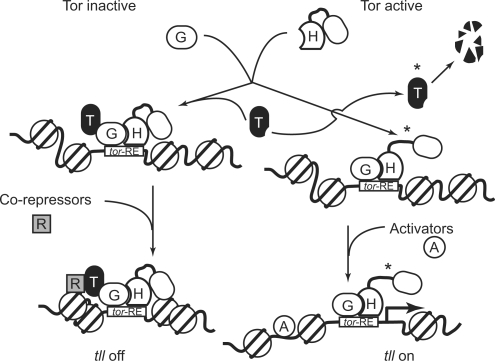
This study showed that the interaction of GAF with Hsf and Ttk69 plays a critical role in regulation of tll expression. At locations where the Tor pathway is inactive, GAF, Hsf, and Ttk69 constitute a protein complex that binds to the tor-RE tightly. The protein complex recruits other co-repressors and chromatin remodeling factors containing Rpd3 to the organization of a high-order local chromatin structure. tll expression is off in this scenario (Figure 7).
Figure 7.
A model illustrates how GAF, Hsf and Ttk69 form a repression complex on the tor-RE to initiate tll repression and how the Tor pathway regulates transcriptional switches. Letters A, H, G, R and T inside the cartoons represent activators, Hsf, GAF, co-repressors and Ttk69. Proteins phosphorylated by Mapk are indicated by asterisks.
We and others have shown that both Hsf and Ttk69 are substrates of Mapk (this study; 17). At locations where the Tor pathway is active, activated Mapk phosphorylates both Hsf and Ttk69. The phosphorylated Ttk69 is degraded (17). The phosphorylation converts Hsf into an activator that leads to an increase in tll transcription. In addition, the expanded lacZ expression patterns in hsf4 embryos at the nonpermissive temperature suggested that other activators, such as Stat (61), are apparently required for full activation of tll transcription (Figure 7).
Multiple factors binding to the tor-RE and its flanking regions constitute a large repression complex
We showed that a protein–protein interaction network, including GAF, Hsf, Rpd3 and Ttk69, was required for tll repression. Studies show that Capicuo interacts with Groucho, which associates with Rpd3 and Sin3A to repress tll expression (18,19). Furthermore, CtBP is essential for Ttk69 to suppress neuronal cell fate (17). Therefore, these factors may also be recruited to repress tll expression.
Multiple GAFs and Ttk69s bound to the flanking regions of the tor-RE. Our results from the DNaseI footprinting experiments showed that GAF binds to four sites in the tll-MRR, including the tor-RE. DNA sequences in footprints a and b match the consensus sequence bound by GAF, 3.5 GA repeats (62). Although DNA sequence in footprint c does not match the consensus sequence, this site contains a GAGA tandem repeat with 1-bp spacing. These three sites are well protected by GAF from DNaseI digestion, without or with little influence by Hsf. Therefore, GAF oligomer binds to these sites with high affinity and likely assists its own binding to the tor-RE. Similarly, binding of Even-skipped protein to low-affinity sites was assisted by its own binding to high-affinity sites at a distance (63).
Ttk69 acted as a co-repressor to increase GAF/Hsf binding to the tor-RE. In another EMSA experiment, a probe containing both the tor-RE and TC5 was used (data not shown). The binding efficiency of GAF/Hsf/Ttk69 to this probe was about 3-fold higher than to the probe used in the experiments shown in Figure 4A, indicating that Ttk69 binds to TC5 and assists GAF/Hsf/Ttk69 binding to the tor-RE. This is consistent with our previous report that base substitution in TC5 clearly affects the initiation of tll repression (16). Additionally, the binding of Ttk69 to TC2 (16) might facilitate tll repression.
Results from the DNaseI footprintings showed different patterns over the tor-RE protected by either GAF/Hsf or GAF alone. Unexpectedly, these different tor-RE complexes showed the same mobility in results from EMSA experiments, which might be explained by the altered binding property of GAF in the presence of Hsf. Ahmad and colleagues report that BTB domains in GAF and human Promyelocytic Leukemia Zinc Finger (PLZF) proteins belong to the Ttk subfamily, and that BTB homodimer is a unit of PLZF oligomer (64). Since GAF oligomer presumably assisted itself in binding to the tor-RE, GAF homodimer could bind the 5′-end as well as the 3′-end of the tor-RE, consistent with the band-shift patterns shown in Figure 3A. When Hsf was added in addition to GAF, the interaction of GAF with Hsf (25) influenced GAF binding to the tor-RE, leading to the alteration of the footprinting pattern. This explanation is supported by the following data. Results from shift-western blotting showed some high molecular weight Hsfs above the DNA–protein complex that is presumably Hsf oligomer based on the finding reported by Farkas and colleagues (65), in which mouse Hsf1 forms oligomer that is broken down to trimers to activate gene expression when it is phosphorylated. Thus, the discrete band above the DNA–protein complex detected by anti-Hsf antibody is likely a result of Hsf oligomer disrupted by interacting with GAF. On the other hand, GAF oligomer is likely affected by Hsf as shown by the results from shift-western blotting showing diminished intensity of bands immediately above the DNA–protein complex in the presence of Hsf (compare intensity of bands above the DNA–protein complex of lane 3 with that of lane 1 in Figure 3B). Therefore, the disruption of GAF oligomer by Hsf provides an explanation to why the pattern of footprint d was altered when Hsf was added to GAF. However, it remains unclear whether Hsf contacts DNA when it exists in the DNA–protein complex. Since association of Hsf with GAF homodimer would increase the mass of DNA–protein complex, we expected to observe a slower mobility of the complex. However, we did not. The DNA sequence flanking footprint d became unprotected in the presence of both GAF and Hsf, suggesting that the negative charges on the DNA probe, less masked by the proteins, might compensate the decreased mobility due to increased mass. Nevertheless, our results from shift-western blotting clearly demonstrated the presence of both GAF and Hsf in the DNA–protein complex.
Displacement of GAF monomer by Ttk69 to form a GAF/Hsf/Ttk69 complex might explain the unchanged band-shift patterns when the three proteins were added to the binding mixture. Molecular weights between GAF and Ttk69 are slightly different and BTB domains in both GAF and Ttk69 proteins belong to the same subfamily (64). Molecular modeling with crystal structure of the BTB domain in human BACH1 (PDB code: 2ihc) as a template (http://swissmodel.expasy.org/SWISS-MODEL.html) was used to test the displacement hypothesis. Results showed that buried areas and free energies (ΔGs) of engaged interfaces among GAF and Ttk69 homodimers and GAF/Ttk69 heterodimer were similar (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html), suggesting that formation of a GAF/Ttk69 heterodimer is possible. This explanation is partially supported by the detection of both Ttk69 and Hsf in the DNA–protein complex. In addition, Pagans and colleagues show that Ttk69 inhibits GAF activation in the absence of Ttk69 binding sites, and addition of Ttk69 significantly increases GAF binding to the 173-bp probe that contains multiple GAF binding sites. However, results from EMSA show that mobility of the DNA–protein complex is slightly affected (52). In conclusion, these data plus the existence of GAF, Hsf and Ttk69 in the DNA–protein complex indicated that the interaction of GAF with Hsf and Ttk69 form a protein complex binding to the tor-RE tightly.
In summary, multiple factors bind to the tor-RE and its flanking regions to form a large repression complex, consistent with the finding that the 240-bp cis-regulatory region of tll (Supplementary Figure S2), but not the tor-RE itself, silences transcription from a heterologous promoter (13).
Low and high-affinity sites bound by GAF/Hsf function in tll repression and activation, respectively
Several studies on gene repression, i.e. bacterial bipA and eno genes (66,67), fly decapentaplegic (68) and human Iε and GP91phox genes (69,70), have revealed that multiple low-affinity sites are bound by a repressor to regulate gene expression. Likewise, multiple weak binding sites that cluster within a short range of a cis-regulatory region are reported to facilitate the cooperative binding of transcription factors, leading to a sharp definition in expression patterns. Reduction of repressor concentration leads to a loss in the definition at the edge of expression domains (71,72). In this study, the binding affinity of GAF to the tor-RE is low, and multiple tor-REs are present in the tll cis-regulatory region (13,16). This explains the poorly-defined boundary of the tll expression patterns in embryos that have reduced hsf and Trl activities.
Footprint b was marginally affected by supplementing Hsf. The DNA sequence in this site contains one binding site each for GAF and Hsf (−130 GAGAGAG and −115 GAATCCTGCGGAA), located in regions O and P, respectively (referred to Figure 1 in reference 13). The sequence for GAF binding matches the consensus sequence (62). Although this putative Hsf binding sequence does not match the consensus sequence, Yamamoto and colleagues (73) have shown that Hsf is able to bind a sequence with two GAAs spaced by 7 bp. Interestingly, in contrast to the role of tor-RE, deletion of either region O or P from the tll-MRR results in a drastic reduction of lacZ mRNA levels, but no changes in the expression patterns (13). These results not only further support the notion that Hsf and GAF are required for tll activation, but also provide an explanation for (i) the different tll expression patterns in embryos with reduced hsf and Trl activities by either removing one copy of the genes or using RNAi to knockdown activities of the genes, and (ii) the different tll expression levels resulting from either base substitutions in the tor-RE or reduction of gene activities.
Our results showed a moderate effect of Hsf on GAF binding to footprint c. In addition to the GAGA repeat, this site also contains two GAA repeats with the 7-bp spacing, (−86 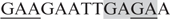 ; the GAGA sequence is shadowed). Results from the DNaseI footprinting experiments showed that GAF bound to this site less efficiently. Furthermore, deletion of this site from the tll-MRR results in a low level and slightly expanded lacZ expression pattern (13). In addition, footprint d at the 5′-end of the tor-RE was a weak site bound by GAF and the footprint pattern protected by GAF was significantly affected by Hsf. Base substitutions to this site severely damaged tll repression (13). These data supported the notion that the low and high affinity sites bound by GAF are the major contributors to tll repression and activation.
; the GAGA sequence is shadowed). Results from the DNaseI footprinting experiments showed that GAF bound to this site less efficiently. Furthermore, deletion of this site from the tll-MRR results in a low level and slightly expanded lacZ expression pattern (13). In addition, footprint d at the 5′-end of the tor-RE was a weak site bound by GAF and the footprint pattern protected by GAF was significantly affected by Hsf. Base substitutions to this site severely damaged tll repression (13). These data supported the notion that the low and high affinity sites bound by GAF are the major contributors to tll repression and activation.
Hsf and Ttk69 act as transcriptional switches regulated by the Tor pathway
All signaling pathways regulate, at least in part, specific transcription factors to activate the expression of target genes (4). In most well-studied signal pathways, the signals directly activate transcription factors (2,3,74,75). In some cases, the signals switch on expression of their target genes from a repressed state. For example, in the absence of Notch, Wnt, Hedgehog or nuclear receptor signaling, the expression of target genes is repressed. There is a common mechanism among these cases. A specific factor or complex, such as Su(H)/CBF1 in the Notch pathway, Lef/Tcf in the Wnt pathway, Ci/Gli in the Hh pathway or the nuclear receptors themselves, bind to a specific DNA sequence to prevent target genes from being transcribed. When the signaling pathway is active, the repression is relieved by the processed receptor, activated activator, co-activators or by the nuclear receptor itself (4,76). Results from this study indicated that both GAF-associated proteins, Hsf and Ttk69, constitute a dual transcriptional switch for tll expression that includes degradation of the Ttk69 co-repressor and conversion of the Hsf repressor into an activator after Mapk phosphorylation, where Mapk is a downstream effector of the Tor pathway (1,7).
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
Aim for the Top University Plan and NSC (94-2752-B-010-005-PAE, 95-2752-B-010-005-PAE and 96-2752-B-010-005-PAE G.J.L.). Funding for open access charge: Aim for the Top University Plan, Department of Education.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr D.-H. Huang for both the TrlDHΔ34 strain and anti-GAF antibody. We would like to acknowledge the service of the fly-food facility in Dr Y. H. Sun's laboratory, the Institute of Molecular Biology, Academia Sinica.
REFERENCES
- 1.Li WX. Functions and mechanisms of receptor tyrosine kinase Torso signaling: lessons from Drosophila embryonic terminal development. Dev. Dyn. 2005;232:656–672. doi: 10.1002/dvdy.20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 3.Hill CS, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 4.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 5.Sprenger F, Stevens LM, Nusslein-Volhard C. The Drosophila gene torso encodes a putative receptor tyrosine kinase. Nature. 1989;338:478–483. doi: 10.1038/338478a0. [DOI] [PubMed] [Google Scholar]
- 6.St Johnston D, Nusslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 7.Furriols M, Casanova J. In and out of Torso RTK signalling. EMBO J. 2003;22:1947–1952. doi: 10.1093/emboj/cdg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprenger F, Nusslein-Volhard C. Torso receptor activity is regulated by a diffusible ligand produced at the extracellular terminal regions of the Drosophila egg. Cell. 1992;71:987–1001. doi: 10.1016/0092-8674(92)90394-r. [DOI] [PubMed] [Google Scholar]
- 9.Strecker TR, Kongsuwan K, Lengyel JA, Merriam JR. The zygotic mutant tailless affects the anterior and posterior ectodermal regions of the Drosophila embryo. Dev. Biol. 1986;113:64–76. doi: 10.1016/0012-1606(86)90108-9. [DOI] [PubMed] [Google Scholar]
- 10.Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR, Lengyel JA. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell. 1990;62:151–163. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- 11.Pignoni F, Steingrimsson E, Lengyel JA. bicoid and the terminal system activate tailless expression in the early Drosophila embryo. Development. 1992;115:239–251. doi: 10.1242/dev.115.1.239. [DOI] [PubMed] [Google Scholar]
- 12.Liaw GJ, Lengyel JA. Control of tailless expression by bicoid, dorsal and synergistically interacting terminal system regulatory elements. Mech. Dev. 1992;40:47–61. doi: 10.1016/0925-4773(93)90087-e. [DOI] [PubMed] [Google Scholar]
- 13.Liaw GJ, Rudolph KM, Huang JD, Dubnicoff T, Courey AJ, Lengyel JA. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 1995;9:3163–3176. doi: 10.1101/gad.9.24.3163. [DOI] [PubMed] [Google Scholar]
- 14.Liaw GJ, Steingrimsson E, Pignoni F, Courey AJ, Lengyel JA. Characterization of downstream elements in a Raf-1 pathway. Proc. Natl Acad. Sci. USA. 1993;90:858–862. doi: 10.1073/pnas.90.3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown JL, Wu C. Repression of Drosophila pair-rule segmentation genes by ectopic expression of tramtrack. Development. 1993;117:45–58. doi: 10.1242/dev.117.1.45. [DOI] [PubMed] [Google Scholar]
- 16.Chen YJ, Chiang CS, Weng LC, Lengyel JA, Liaw GJ. Tramtrack69 is required for the early repression of tailless expression. Mech. Dev. 2002;116:75–83. doi: 10.1016/s0925-4773(02)00143-0. [DOI] [PubMed] [Google Scholar]
- 17.Wen Y, Nguyen D, Li Y, Lai ZC. The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics. 2000;156:195–203. doi: 10.1093/genetics/156.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14:224–231. [PMC free article] [PubMed] [Google Scholar]
- 19.Paroush Z, Wainwright SM, Ish-Horowicz D. Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development. 1997;124:3827–3834. doi: 10.1242/dev.124.19.3827. [DOI] [PubMed] [Google Scholar]
- 20.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 21.Pedone PV, Ghirlando R, Clore GM, Gronenborn AM, Felsenfeld G, Omichinski JG. The single Cys2-His2 zinc finger domain of the GAGA protein flanked by basic residues is sufficient for high-affinity specific DNA binding. Proc. Natl Acad. Sci. USA. 1996;93:2822–2826. doi: 10.1073/pnas.93.7.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adkins NL, Hagerman TA, Georgel P. GAGA protein: a multi-faceted transcription factor. Biochem. Cell Biol. 2006;84:559–567. doi: 10.1139/o06-062. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann M. Anything else but GAGA: a nonhistone protein complex reshapes chromatin structure. Trends Genet. 2004;20:15–22. doi: 10.1016/j.tig.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 25.Mason PB, Jr, Lis JT. Cooperative and competitive protein interactions at the hsp70 promoter. J. Biol. Chem. 1997;272:33227–33233. doi: 10.1074/jbc.272.52.33227. [DOI] [PubMed] [Google Scholar]
- 26.Clos J, Rabindran S, Wisniewski J, Wu C. Induction temperature of human heat shock factor is reprogrammed in a Drosophila cell environment. Nature. 1993;364:252–255. doi: 10.1038/364252a0. [DOI] [PubMed] [Google Scholar]
- 27.Morimoto RI. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- 28.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 29.Calderwood SK. Regulatory interfaces between the stress protein response and other gene expression programs in the cell. Methods. 2005;35:139–148. doi: 10.1016/j.ymeth.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Inouye S, Fujimoto M, Nakamura T, Takaki E, Hayashida N, Hai T, Nakai A. Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J. Biol. Chem. 2007;282:33210–33217. doi: 10.1074/jbc.M704471200. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong WC, Montell C. tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev. 1993;7:1085–1096. doi: 10.1101/gad.7.6.1085. [DOI] [PubMed] [Google Scholar]
- 33.Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16:2452–2462. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenspan RJ. Fly Pushing: The Theory and Practice of Drosophila Genetics. 2nd edn. New York: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 35.Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- 36.Rorth P. Gal4 in the Drosophila female germline. Mech. Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 37.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 38.Rubin GM, Spradling AC. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 1983;11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spradling AC, Rubin GM. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell. 1983;34:47–57. doi: 10.1016/0092-8674(83)90135-6. [DOI] [PubMed] [Google Scholar]
- 40.Janody F, Reischl J, Dostatni N. Persistence of Hunchback in the terminal region of the Drosophila blastoderm embryo impairs anterior development. Development. 2000;127:1573–1582. doi: 10.1242/dev.127.8.1573. [DOI] [PubMed] [Google Scholar]
- 41.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons, Inc; 1994. [Google Scholar]
- 42.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned gene. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 43.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct the expression of cloned gene. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 44.Zhong M, Wu C. Proteolytic mapping of heat shock transcription factor domains. Protein Sci. 1996;5:2592–2599. doi: 10.1002/pro.5560051222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 46.Demczuk S, Harbers M, Vennstrom B. Identification and analysis of all components of a gel retardation assay by combination with immunoblotting. Proc. Natl Acad. Sci. USA. 1993;90:2574–2578. doi: 10.1073/pnas.90.7.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothwell WF, Sullivan W. In: Drosophila Protocols. Sullivan W, Ashbourner M, Hawley RS, editors. New York: Cold Spring Harbor Laboratory Press; 2000. pp. 141–157. [Google Scholar]
- 48.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 49.Singh A, Kango M, Sinha P. An improved method for chemical devitellinization of X-gal stained Drosophila embryos. Indian J. Exp. Biol. 1995;33:150–152. [PubMed] [Google Scholar]
- 50.Bernues J, Pineyro D, Kosoy A. General, negative feedback mechanism for regulation of Trithorax-like gene expression in vivo: new roles for GAGA factor in flies. Nucleic Acids Res. 2007;35:7150–7159. doi: 10.1093/nar/gkm590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Espinas ML, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, Azorin F. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem. 1999;274:16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 52.Pagans S, Pineyro D, Kosoy A, Bernues J, Azorin F. Repression by TTK69 of GAGA-mediated activation occurs in the absence of TTK69 binding to DNA and solely requires the contribution of the POZ/BTB domain of TTK69. J. Biol. Chem. 2004;279:9725–9732. doi: 10.1074/jbc.M313200200. [DOI] [PubMed] [Google Scholar]
- 53.Pagans S, Ortiz-Lombardia M, Espinas ML, Bernues J, Azorin F. The Drosophila transcription factor tramtrack (TTK) interacts with Trithorax-like (GAGA) and represses GAGA-mediated activation. Nucleic Acids Res. 2002;30:4406–4413. doi: 10.1093/nar/gkf570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aalfs JD, Kingston RE. What does ‘chromatin remodeling’ mean? Trends Biochem. Sci. 2000;25:548–555. doi: 10.1016/s0968-0004(00)01689-3. [DOI] [PubMed] [Google Scholar]
- 55.Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 56.Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 57.Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 59.Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J. Biol. Chem. 1996;271:30847–30857. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- 60.Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 61.Li WX, Agaisse H, Mathey-Prevot B, Perrimon N. Differential requirement for STAT by gain-of-function and wild-type receptor tyrosine kinase Torso in Drosophila. Development. 2002;129:4241–4248. doi: 10.1242/dev.129.18.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granok H, Leibovitch BA, Shaffer CD, Elgin SC. Chromatin. Ga-ga over GAGA factor. Curr. Biol. 1995;5:238–241. doi: 10.1016/s0960-9822(95)00048-0. [DOI] [PubMed] [Google Scholar]
- 63.TenHarmsel A, Austin RJ, Savenelli N, Biggin MD. Cooperative binding at a distance by even-skipped protein correlates with repression and suggests a mechanism of silencing. Mol. Cell Biol. 1993;13:2742–2752. doi: 10.1128/mcb.13.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmad KF, Engel CK, Prive GG. Crystal structure of the BTB domain from PLZF. Proc. Natl Acad. Sci. USA. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farkas T, Kutskova YA, Zimarino V. Intramolecular repression of mouse heat shock factor 1. Mol. Cell Biol. 1998;18:906–918. doi: 10.1128/mcb.18.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mishra M, Deora R. Mode of action of the Bordetella BvgA protein: transcriptional activation and repression of the Bordetella bronchiseptica bipA promoter. J. Bacteriol. 2005;187:6290–6299. doi: 10.1128/JB.187.18.6290-6299.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimada T, Fujita N, Maeda M, Ishihama A. Systematic search for the Cra-binding promoters using genomic SELEX system. Genes Cells. 2005;10:907–918. doi: 10.1111/j.1365-2443.2005.00888.x. [DOI] [PubMed] [Google Scholar]
- 68.Huang JD, Schwyter DH, Shirokawa JM, Courey AJ. The interplay between multiple enhancer and silencer elements defines the pattern of decapentaplegic expression. Genes Dev. 1993;7:694–704. doi: 10.1101/gad.7.4.694. [DOI] [PubMed] [Google Scholar]
- 69.Harris MB, Mostecki J, Rothman PB. Repression of an interleukin-4-responsive promoter requires cooperative BCL-6 function. J. Biol. Chem. 2005;280:13114–13121. doi: 10.1074/jbc.M412649200. [DOI] [PubMed] [Google Scholar]
- 70.Luo W, Skalnik DG. CCAAT displacement protein competes with multiple transcriptional activators for binding to four sites in the proximal gp91phox promoter. J. Biol. Chem. 1996;271:18203–18210. doi: 10.1074/jbc.271.30.18203. [DOI] [PubMed] [Google Scholar]
- 71.Ashe HL, Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–394. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- 72.Lebrecht D, Foehr M, Smith E, Lopes FJ, Vanario-Alonso CE, Reinitz J, Burz DS, Hanes SD. Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proc. Natl Acad. Sci. USA. 2005;102:13176–13181. doi: 10.1073/pnas.0506462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamamoto A, Mizukami Y, Sakurai H. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:11911–11919. doi: 10.1074/jbc.M411256200. [DOI] [PubMed] [Google Scholar]
- 74.Dickson B. Nuclear factors in sevenless signalling. Trends Genet. 1995;11:106–111. doi: 10.1016/S0168-9525(00)89011-3. [DOI] [PubMed] [Google Scholar]
- 75.Rebay I. Keeping the receptor tyrosine kinase signaling pathway in check: lessons from Drosophila. Dev. Biol. 2002;251:1–17. doi: 10.1006/dbio.2002.0806. [DOI] [PubMed] [Google Scholar]
- 76.Gaston K, Jayaraman PS. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell Mol. Life Sci. 2003;60:721–741. doi: 10.1007/s00018-003-2260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.