Abstract
Bacillus subtilis OhrR is a dimeric repressor that senses organic peroxides and regulates the expression of the OhrA peroxiredoxin. Derepression results from oxidation of an active site cysteine which ultimately results in formation of a mixed disulfide with a low molecular weight thiol, a cyclic sulfenamide, or overoxidation to the sulfinic or sulfonic acids. We expressed a single-chain OhrR (scOhrR) in which the two monomers were connected by a short amino-acid linker. scOhrR variants containing only one active site cysteine were fully functional as repressors and still responded, albeit with reduced efficacy, to organic peroxides in vivo. Biochemical analyses indicate that oxidation at a single active site is sufficient for derepression regardless of the fate of the active site cysteine. scOhrR with only one active site cysteine in the amino-terminal domain is inactivated at rates comparable to wild-type whereas when the active site is in the carboxyl-terminal domain the protein is inactivated much more slowly. The incomplete derepression noted for single active site variants of scOhrR in vivo is consistent with the hypothesis that protein reduction regenerates active repressor and that, in the cell, oxidation of the second active site may also contribute to derepression.
INTRODUCTION
Bacillus subtilis OhrR is a homodimeric repressor protein in both the DNA-bound and unbound states (1). When cells are exposed to low levels of organic peroxides, OhrR is functionally inactivated, leading to the derepression of the ohrA-encoded thiol peroxidase (2,3). B. subtilis OhrR is the prototype for a subgroup of organic peroxide-sensing repressors that have a single cysteine residue in each monomer (the 1-Cys family). In contrast, Xanthomonas campestris OhrR is representative of those family members that have an additional cysteine near the carboxy-terminus that can react with the active site cysteine to form an intersubunit disulfide (2-Cys family) (4,5).
OhrR proteins are, at the structural level, the best characterized members of the MarR (multiple-antibiotic resistance) family of winged helix-turn-helix DNA-binding proteins. High resolution structural data are available for B. subtilis OhrR in both the free and DNA-bound states (1) and for X. campestris OhrR in both the oxidized and reduced states (6). Additional members of this family that also use a cysteine-based redox-sensing mechanism include the Staphylococcus aureus MgrA virulence regulator and the Pseudomonas aeruginosa MexR regulator (7–9). More distantly related members of the MarR family for which structural information is available include the Escherichia coli and Methanobacterium thermoautotrophicum MarR repressors (10,11). While these proteins all function as dimeric, DNA-binding proteins that respond to ligands (or oxidants), the stoichiometry and consequences of ligand interaction differ significantly between systems. For example, MarR proteins bind up to two salicylates per monomer, although the physiological relevance of the various binding sites observed in the crystal structures has been controversial (11,12). Conversely, P. aeruginosa MexR binds one molecule of peptide inducer (ArmR) per dimer (9).
Previous biochemical and structural analyses have demonstrated that oxidation of OhrR can lead to a number of chemically distinct outcomes (13). Indeed, oxidation can be either reversible or irreversible depending on the oxidant and the solution conditions (14). Initially, organic hydroperoxides react with the lone cysteine residue in B. subtilis OhrR, C15, to generate the cysteine sulfenic acid (-SOH) derivative (3) which, however, retains DNA-binding activity (13). The functional inactivation of the protein requires further reaction of the sulfenate to form any of a number of possible products. For example, in cells treated with cumene hydroperoxide (CHP), OhrR accumulates as a mixture of S-thiolated protein (mixed disulfides with cysteine and other low molecular weight thiols) and protein that contains a cyclic sulfenamide (a product in which the sulfenate condenses with a neighboring backbone amide). Both of these products can be reduced by reaction with thiol reductants thereby regenerating active repressor (13). In contrast, in cells treated with linoleic acid hydroperoxide (LHP), OhrR accumulates largely as the overoxidized protein in which the sulfenate is irreversibly oxidized to the sulfinic and sulfonic acids (14). We have also demonstrated that introduction of a second cysteine residue into the carboxyl-terminal region of OhrR, at positions analogous to those seen in 2-Cys family members, converts B. subtilis OhrR to a protein that rapidly forms intersubunit disulfides upon exposure to oxidant (15).
We have monitored the products of OhrR oxidation and correlated the oxidation states of the protein with activity using both in vivo and in vitro assays. Our results indicate that the C15 residues in both monomers of the dimer can be simultaneously oxidized. Specifically, mass spectrometry studies indicate (i) quantitative S-cysteinylation of C15 in vitro in protein treated with CHP in the presence of cysteine, (ii) quantitative C15 oxidation in vivo as judged by a lack of residual reactivity with iodoacetamide and (iii) formation of two intersubunit disulfide bonds in protein dimers engineered to contain a second, carboxyl-terminal Cys residue that can trap the C15 sulfenate as a disulfide (13,16). Furthermore, structural analysis of the oxidized X. campestris OhrR revealed the simultaneous formation of two intersubunit disulfide bonds (6). These results indicate that oxidation of OhrR can occur at both active sites and that any conformational changes elicited by the first oxidation event do not preclude a second oxidation event. However, these results do not address the question of whether a singly oxidized protein dimer is functional as a repressor.
Here, we report analyses of single-chain OhrR variants (scOhrR) in which the two monomers are fused into a single protein chain by addition of a short amino-acid linker. We have monitored both protein oxidation (using mass spectrometry) and functional inactivation (using a fluorescence-based DNA-binding assay) of wild-type scOhrR (WT-WT), variants that contain only a single active site C15 residue, and a variant that additionally contains an introduced cysteine to enable formation of a protein disulfide. In aggregate, our results indicate that oxidation of a single active site in the scOhrR proteins is sufficient to inactivate the repressor in vitro. However, scOhrR variants with only a single active site do not respond as well as wild-type scOhrR to oxidant in vivo, implying that the second oxidation event is important for generating and/or maintaining the induced state in the cellular environment.
MATERIALS AND METHODS
Expression of scOhrR dimers in B. subtilis
Both monomeric wild-type and the scOhrR variants were expressed in B. subtilis using the integational plasmid pDG1731 (17). Plasmid constructs were integrated by double crossover recombination into the thrC locus and each protein was expressed under the control of its native promoter elements. Monomeric OhrR was expressed from the integrational plasmid pDG1731-ohrR which recombines by double-crossover recombination into the thrC locus.
For expression of scOhrR variants we followed the construction strategy presented by Lee et al. (18). The first ohrR copy with a 5-amino-acid (GGGGS) linker was PCR-amplified using primer #2437 (5′-CTTTCTAAGCTTTTTAAACATGCTATG-3′, HindIII site underlined) and #3147 (5′-TTCCATCGATCCTCCTCCTCCATTTTTTTGATGAAGTGTTTCCAGTAA-3′; complement of ohrR stop codon italicized) with pDG1731-ohrR as template. The resulting product includes 91 bp of DNA upstream of the ohrR coding sequence including the promoter site (3). The second copy of ohrR was amplified using primer #3148 (5′-AAAAATGGAGGAGGAGGATCGATGGAAAATAAATTTGATCATATGAAA-3′, ohrR start codon italicized) and #2621 (5′-GACACTTGAATTCGCTGAATAAATAAA-3′, EcoRI site underlined). A similar strategy was used to introduce a 10-amino-acid linker using primers #3400 (5′-CATCGATCCTCCTCCTCCCGATCCTCCTCCTCCATTTTTTTGATGAAGTGTTTCCAGTA-3′) and #3401 (5′-AATGGAGGAGGAGGATCGGGAGGAGGAGGATCGATGGAAAATAAATTTGATCATATGAA-3′). The fused-ohrR gene was generated in a joining PCR reaction containing both ohrR copies, and primers #2437 and #2621. The reaction was denatured at 95°C for 3 min, followed by 10 cycles of 95°C for 30 s, 55°C for 5 min, and extension at 72°C for 1 min 15 s. This was followed by 20 cycles of 95°C for 30 s, 47°C for 30 s, and extension at 72°C for 2 min 30 s. The desired PCR product was gel purified, digested with HindIII and EcoRI, and cloned into pDG1731 (17). The resulting plasmid was integrated into thrC in strain HB2013 [CU1065 SPβc2Δ2::Tn917::(ohrA-cat-lacZ) ohrR::kan; (3)]. The sequences of the fused-ohrR genes were verified by sequencing. For construction of the C15S scOhrR variants, a previously described site-directed mutant (19) was used as the template for PCR amplification as described above. To construct the C15S,G120C double mutant, the pDG1731-based plasmid expressing the C15S-WT scOhrR protein was used as template for site-directed mutagenesis using previously described primers to introduce the G120C mutation (15).
Transcription fusion analysis
Cells were grown in Luria broth at 37°C with aeration until OD600 ∼ 0.4. The cells were either untreated or treated with 100 μM CHP or 5 µM LHP for 15 min which had previously been determined to be the optimal concentrations for in vivo responsiveness (14). Samples were harvested and assayed for β-galactosidase activity (20).
scOhrR overexpression and purification
The fused-ohrR genes were PCR-amplified by using primers #527 (5′-GGTGAACACCATGGAAAATAAATT-3′; NcoI site containg ohrR start codon underlined) and #528 (5′-CCGGATCCGTTGCTGAATAAATAAA-3′; BamHI site underlined), and were cloned into pET16b (Novagen) and verified by DNA sequencing. The scOhrR proteins were overexpressed in E. coli BL21/DE3(pLysS) (Novagen) and purified as previously described for the dimeric repressor (19). Purified proteins were >90% pure as judged by coomassie-stained SDS–PAGE (Supplementary Figure S1).
Northern analysis
Total RNA was isolated from mid-logarithmic cells (OD600 ∼ 0.4), which were either untreated or treated with 100 μM CHP for 15 min, using an RNeasy mini kit (Qiagen) following the manufacturer's protocol. Ten micrograms of total RNA was run on a 1% formaldehyde denaturing gel using NorthernMax denaturing gel buffer and running buffer (Ambion). The RNA was transferred to Zeta-Probe blotting membrane (Bio-Rad) using 10 × SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and prehybridized at 42°C for 1 h. The ohrA probe was amplified with forward primer (#4268; 5′-CTTGAGCTTGATGTCGCAAT-3′) and reverse primer (#4269; 5′-CAATTCTGATGCACTGACTC-3′) and end-labeled with T4 polynucleotide kinase (New England Biolabs) using [γ-32P]ATP. Hybridization of the probe was performed with ULTRAhyd (Ambion) overnight at 42°C, and the membrane was washed twice with a 2 × SSPE (1 × SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7; Ambion) low-stringency wash for 5 min at room temperature and once with a 0.1 × SSPE high-stringency wash for 15 min at 42°C, before detection of the signal using a PhosphorImager (GE Healthcare).
Electrospray ionization–mass spectrometry (ESI–MS) analysis
One milliliter of reaction mixture was mixed with 110 μl of 100% TCA and proteins were recovered by centrifugation at 16 100 × g for 10 min. After additional washing with 10% TCA, samples were dissolved in 30 μl of 2% acetic acid (v/v) and 50% methanol (v/v) and analyzed with an Esquire-LC ion trap mass spectrometer (Bruker, Billerica, MA). The spectra were deconvoluted by using the Daltoniks Data Analysis program (Bruker).
Fluorescence anisotropy (FA)
A 6-carboxyfluorescein-(6F-)-labeled DNA fragment containing the ohrA operator site was generated by annealing 5′-6F-TACAATTAAATTGTATACAATTAAATTGTA-3′ (Integrated DNA technologies) and its unlabeled complementary strand. FA measurements (λex = 495 nM; slit width = 15 nM, λem = 520 nM; slit width = 20 nM, integration time = 1 s) were performed with 50 nM DNA and 300 nM scOhrR in 3 ml of 20 mM Tris (pH 8.0) containing 150 mM NaCl, and 5% (v/v) glycerol. FA values were recorded automatically every 10 s. using a Perkin–Elmer LS55 luminescence spectrometer. The g-factor for the experiments was 1.07 ± 0.01.
RESULTS
Peroxide-sensing by single-chain OhrR repressor proteins (scOhrR)
To determine whether oxidation of one or two active sites (per dimer) is required for the functional inactivation of OhrR we generated a single chain OhrR (scOhrR) in which the two protein monomers were linked by a 5-amino-acid linker (Figure 1) or a 10-amino-acid linker (which yielded comparable results; data not shown). Four different scOhrR variants were characterized. One contained two wild-type OhrR domains (WT-WT), one contained a C15S substitution mutation in the amino-terminal domain (C15S-WT) and another in the carboxyl-terminal domain (WT-C15′S; where the prime indicates residues from the C-terminal, momomer domain), and one contained two mutations in the amino-terminal domain (C15S,G120C-WT) to allow formation of a disulfide bond upon oxidation as previously reported (15). The single-chain repressors were expressed under the control of their native promoter and regulatory elements from a plasmid integrated in single copy into the thrC locus.
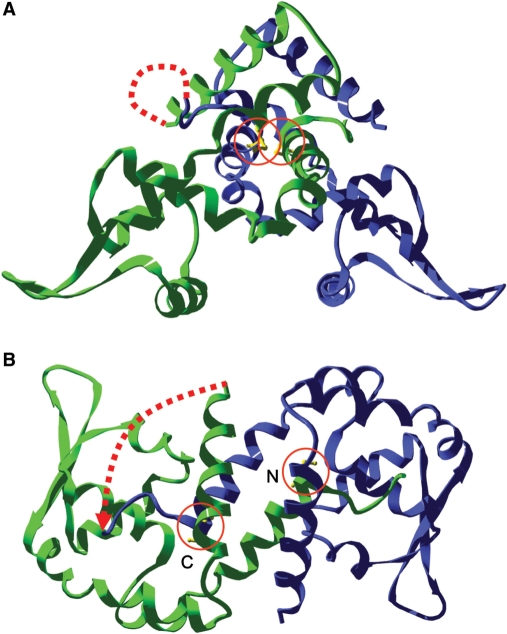
Figure 1.
Ribbon structure of scOhrR. The protein structure, based on the solved crystal structure (1) is shown in a side view (A) and top view (B) with the DNA-binding helices located at the bottom in (A). The first monomer is shown in green and the second in blue. The active site C15 residues are in yellow and circled. The dotted line represents a linker between the C-terminus of the first monomer and the N-terminus of the second monomer. Note that only residues Met8 through His144 were visible in the crystal structure (out of 147 residues), so the N- and C-termini are not accurately defined in this model. In (B), the active sites from the N- and C-terminal monomers are indicated. The linker contains five amino acids (GGGGS) (5L) or two repeats of this sequence in the 10L variants.
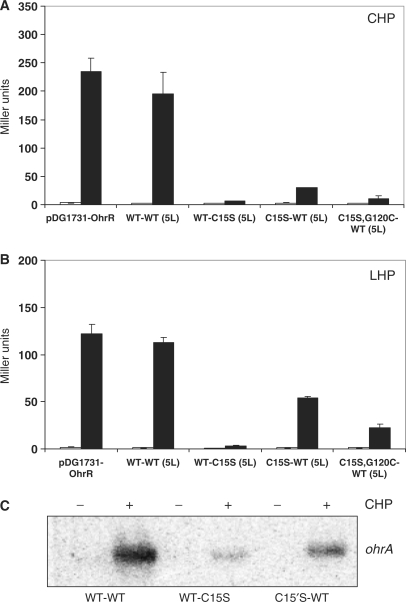
To monitor the activity of the scOhrR proteins, we used an integrated ohrA-lacZ reporter fusion as described (3,16). All four scOhrR repressors were fully functional as repressors as judged by complementation of the ohrR mutant strain (Figure 2). The WT-WT scOhrR responded normally to organic peroxides in vivo as compared to the monomeric OhrR (expressed from the integrational plasmid pDG1731-OhrR). In contrast, those scOhrR variants that retained only a single active site cysteine were significantly reduced in their ability to respond to CHP. This relatively poor responsiveness was observed regardless of which monomer domain contained the active site C15 residue (Figure 2A) and was observed with both the 5 and 10 amino-acid linkers (data not shown). The scOhrR with the active site within the C-terminal domain responded better to oxidant under all conditions tested. When cells were treated with LHP, a similar pattern of responsiveness was observed, and in this case the C15S-WT scOhrR variant allowed approximately 50% derepression relative to the WT-WT scOhrR and monomeric WT OhrR controls (Figure 2B). This pattern of responsiveness was further confirmed by using northern blot analyses to directly monitor expression of the ohrA mRNA (Figure 2C).
Figure 2.
Peroxide-responsiveness of scOhrR in vivo. β-Galactosidase and northern analyses of strains expressing scOhrR and containing an ohrA-cat-lacZ fusion. (A) Cells were either untreated (empty bars) treated (filled bars) with 100 μM CHP for 15 min. (B) Cells were either untreated (empty bars) treated (filled bars) with 5 μM LHP for 15 min. Error bars represent the SD (n = 3). (C) Northern blot analysis of the ohrA transcript in cells with and without CHP treatment, as in (A).
It is not obvious how to interpret these partially responsive phenotypes. The fact that significant responsiveness is still observed with scOhrR repressors containing a single active site suggests that oxidation at one site is sufficient for sensing and responding to peroxides. On the other hand, the reduced responsiveness (particularly for the WT-C15S scOhrR) suggests that full derepression may normally require oxidation at two active sites (at least for the scOhrR protein).
Functional inactivation of scOhrR in response to a single S-cysteinylation event
To more closely monitor the correlation between protein oxidation and DNA-binding, we purified each of the scOhrR repressors (Supplementary Figure S1) and monitored DNA-binding using a FA-based assay (Figure 3A). As reported previously for the wild-type dimeric protein, the WT-WT scOhrR was completely inactivated by treatment with 6 μM CHP in buffer containing 10 μM free cysteine (13). As expected, protein inactivation was due to S-cysteinylation since activity was rapidly and quantitatively restored by addition of dithiothreitol (DTT).
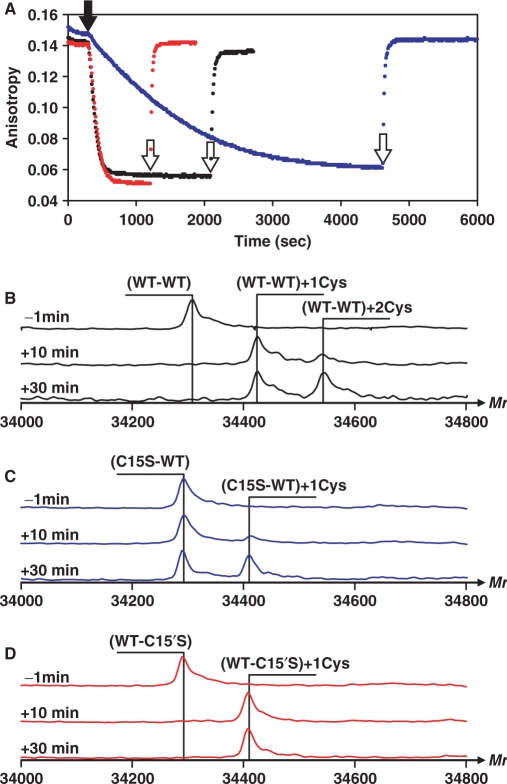
Figure 3.
Correlation between scOhrR inactivation and S-cysteinylation. (A) FA assays monitoring inactivation of WT-WT (black), WT-C15′S (red) and C15S-WT (blue). The reactions contain 50 nM DNA, 300 nM scOhrR and 10 μM Cys. At 5 min (300 s), 6 μM CHP was added (filled arrowhead), and 10 mM DTT was added after completion of inactivation (open arrowhead). (B–D) ESI–MS analysis of scOhrR proteins (as indicated) prior to and 10 and 30 min after CHP addition (times indicated by thin arrows in panel A) in reactions parallel to those in (A), but without DTT addition.
Next, we monitored the oxidation state of the scOhrR proteins by ESI–MS at 10 and 30 min after addition of CHP. After 10 min, a time when DNA-binding activity was quantitatively lost, the majority of the WT-WT scOhrR was present as the singly S-cysteinylated protein (Figure 3B). By 30 min an increased amount of the doubly modified protein was detected, although the second S-cysteinylation modification occurred at a much slower rate than the first. A similar rate of S-cysteinylation was observed for the WT-C15′S variant which was also quantitatively inactivated under these conditions (Figure 3D).
When oxidation on the C15S-WT scOhrR was monitored the rate of functional inactivation was greatly reduced with only ∼70% loss of DNA-binding activity after 30 min. of treatment. As expected, ESI–MS analysis of this protein indicated only a single S-cysteinylation event and the amount of S-cysteinylated protein was correlated with the loss of DNA-binding activity (Figure 3C).
Together, these results indicate that oxidation of a single active site to yield the S-cysteinylated species is sufficient for loss of high affinity DNA-binding. Dimeric OhrR normally binds to the ohrA operator site with a Kd ∼5 nM under these conditions (13). Since these FA analyses are conducted with 300 nM scOhrR, this suggests that oxidation leads to at least a 100-fold reduction in DNA-binding affinity. Furthermore, oxidation of the WT-WT scOhrR was biphasic with oxidation of one C15 complete within 10 min. followed by much slower oxidation of the second active site cysteine (protein was not fully oxidized even after 30 min; Figure 3C). These results indicate that the two active sites are no longer functionally equivalent in scOhrR, presumably due to restraints imposed by the linker. Oxidation of C15 (in the amino-terminal domain of the scOhrR) is rapid (comparable to wild-type protein; Figure 3B versus 3D), whereas oxidation of C15′ is significantly slower (Figure 3C). The slow rate of oxidation at C15′ may result from the fact that this Cys residue is closer both in primary sequence and in space to the linker (Figure 1B).
Oxidation of scOhrR by CHP in the presence and absence of free cysteine
We have previously shown that oxidation of OhrR in the presence of free cysteine leads to S-cysteinylation (as in Figure 3), whereas in the absence of cysteine the initially formed protein sulfenate slowly condenses with a backbone amide to generate an inactive protein sulfenamide (13). Like the S-cysteinylated protein, the OhrR sulfenamide was reactivated by incubation with 10 mM DTT, but at a greatly reduced rate as judged using the FA DNA-binding assay.
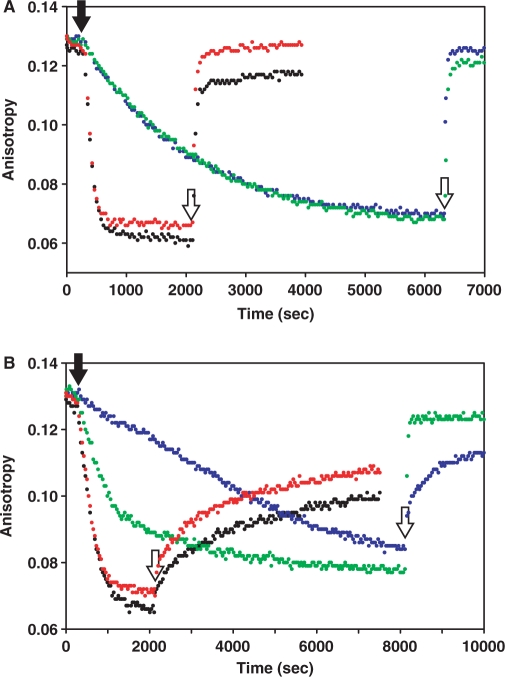
We next compared the extent and rate of inactivation of all four purified scOhrR variants by CHP in the presence and absence of 10 μM free cysteine. In the presence of cysteine, the WT-WT and the WT-C15S proteins were rapidly and reversibly inactivated by CHP (as in Figure 3A) while the C15S-WT scOhrR reacted slowly as noted above (Figure 4A). The C15S,G120C-WT scOhrR also reacted slowly although in this case the additional cysteine residue can capture the initially formed cysteine–sulfenate to generate a protein disulfide.
Figure 4.
Sensitivity of scOhrR to CHP with and without 10 μM cysteine. FA assays monitoring inactivation of WT-WT (black), WT-C15′S (red), C15S-WT (blue) and C15S, G120C-WT (green). The reactions contained 50 nM DNA, 300 nM scOhrR with (A) or without (B) 10 μM Cys. At 5 min, 6 μM CHP was added (closed arrowhead), and 10 mM DTT was added to reactivate OhrR as indicated by open arrowheads.
When the scOhrR proteins were inactivated by CHP in buffer lacking free cysteine, the initially formed sulfenate is expected to slowly react to form the inactive protein sulfenamide (13). Both the WT-WT and WT-C15S scOhrR proteins formed sulfenamides as judged by the characteristic (slow) rate of reactivation upon exposure to 10 mM DTT (Figure 4B). The C15S-WT protein also formed an inactive sulfenamide as judged by the rate of reactivation, but protein inactivation was slower than for the WT-WT and WT-C15′S scOhrR proteins. Again, this is consistent with the notion that in this case initial protein oxidation (at the more slowly reacting C15′ residue) is rate-limiting and is slower than the rate of cyclization to the inactive sulfenamide. As expected based on studies of the analogous dimeric protein (15), the C15S, G120C-WT protein was functionally inactivated by formation of a protein-disulfide since it is rapidly and quantitatively reactivated by 10 mM DTT even in the absence of free cysteine (Figure 4B). Interestingly, the rate of inactivation of the C15S,G120C-WT protein was faster than that observed for the C15S-WT scOhrR. This might reflect a change in the rate of oxidation of the slowly reacting C15′ residue or, alternatively, a change in reaction pathway. In this scOhrR, for example, CHP could initially oxidize the C120 residue to the sulfenate which could then form a disulfide by reaction with the C15′ thiolate. Regardless of the reaction pathway, these results indicate that formation of a single disulfide bond is sufficient to mediate protein dissociation.
Oxidation of scOhrR by LHP in the presence and absence of free cysteine
Unlike CHP, LHP reacts rapidly both with both C15 and the initially formed C15-sulfenate to generate overoxidized OhrR sulfinic (and sulfonic) acid derivatives (14). The overoxidation of OhrR by LHP can be prevented, in part, by high concentrations of cysteine (which compete for the initial sulfenate product with LHP) or by addition of a second cysteine at position 120 to allow trapping of the sulfenate as a protein disulfide (14,15).
Exposure of either the WT-WT or the WT-C15′S scOhrR proteins to LHP in the presence of 1 mM cysteine led to rapid and quantitative inactivation of the protein (Figure 5A). Under these conditions (1 mM cysteine) protein inactivation is due to S-cysteinylation at C15 as also noted with CHP treatment (Figure 4A). However, the very high concentrations of cysteine needed to compete for the transiently formed C15-sulfenate also serves as a reductant of the S-cysteinylated protein, as shown previously for WT OhrR (13). This can account for the slow regeneration of active protein prior to the addition of DTT. Upon addition of 10 mM DTT, the protein is fully reactivated suggesting that there is little if any irreversible overoxidation under these conditions. The C15S-WT and C15S, G120C-WT scOhrR proteins are only partially inactivated under these conditions. This likely reflects the fact that oxidation at C15′ is much slower than at C15 and the presence of high concentrations of free cysteine leads to protein re-reduction that now occurs on the same time scale as oxidation.
Figure 5.
Sensitivity of scOhrR to LHP with and without 1 mM cysteine. FA assays monitoring inactivation of WT-WT (black), WT-C15′S (red) and C15S-WT (blue) and C15S, G120C-WT (green). The reactions contained 50 nM DNA, 300 nM scOhrR with (A) or without (B) 1 mM Cys. At 5 min, 1 μM LHP was added (closed arrowhead), and 10 mM DTT was added to reactivate OhrR as indicated by open arrowheads.
When these same reactions were repeated in the absence of added cysteine, both the WT-WT and WT-C15′S proteins were rapidly inactivated. In this case, addition of 10 mM DTT was able to restore only a small fraction of the DNA-binding activity. The poor recovery of DNA-binding activity with the WT-WT protein is consistent with irreversible protein overoxidation (14). The inactivation of the WT-C15′S protein under these conditions suggests that overoxidation of a single active site is sufficient for protein inactivation. The recovery of activity was slightly greater (∼40%) with the WT-C15′S protein which suggests that in this case the rate of sulfenamide formation was comparable to the rate of protein overoxidation. Under these same conditions, the C15S, G120C-WT protein was quantitatively inactivated and, in this case, activity could be fully and rapidly restored by 10 mM DTT (Figure 5B). This indicates that formation of a single intraprotein disulfide bond is sufficient to trigger protein dissociation, consistent with the results with CHP (Figure 4B). Finally, we note that the C15S-WT protein was inactivated very slowly under these conditions and the inactive protein appears to be largely in the form of the protein sulfenamide. This is consistent with a model in which access to the active site C15′ residue is impeded in the fused dimer leading to both a slow initial oxidation, and an ineffective second oxidation event, thereby allowing ample time for the cyclization of the initial sulfenate to the sulfenamide.
DISCUSSION
Most structurally characterized DNA-binding regulatory proteins from bacterial systems are rotationally symmetric homodimers and recognize palindromic (inverted repeat) DNA sequences (21). Functionally, the interaction of a homodimer with DNA significantly extends the length of the contacted DNA and thereby increases the specificity of the interaction. A single recognition helix (in a helix-turn-helix unit, for example) typically specifies ∼5–6 bp which is not enough to uniquely define a binding site amidst the complexity of genomic DNA. A dimer, on the other hand, that recognizes two such sites (often called half-sites) with a defined spacing has sufficient selectivity to bind to one or a few sites per genome with high selectivity (22).
One important consequence of the evolution of dimeric DNA-binding regulators is that these proteins generally contain two regulatory domains as well as two DNA-binding domains. One can then pose the question, is ligand binding (or chemical modification) of both monomers required to effect regulation? For some proteins, negative cooperativity between the two ligand binding sites suggests that binding of a single ligand is sufficient to effect regulation. Examples include the cAMP-receptor protein (CRP) (23,24), the mercury-sensing activator MerR (25), and ArsR family metalloregulators M. tuberculosis CmtR and Anabaena AztR (26,27) and S. aureus CzrA (18). In other cases, binding of effectors to both monomers is required to elicit a regulatory response. For example, both active sites in TetR need to be occupied by ligand in order to trigger derepression (28). Similarly, the metal sensors CadC (29) and Synechococcus SmtB (30) must bind metal at both active sites to effect regulation. Within the MarR superfamily of proteins, M. thermoautrotrophicum is thought to bind two salicylate ligands per dimer to effect regulation (11), whereas other members (including S. aureus QacR and P. aeruginosa MexR) respond to the binding of a single effector molecule per dimer (9,31).
In this study, we have used engineered single-chain variants of B. subtilis OhrR (scOhrR) to monitor the DNA-binding activity of proteins oxidized at either one or two active sites per dimer. Fortuitously, the introduction of a flexible linker connecting the C-terminus of the first monomer to the N-terminus of the second significantly reduced the rate of reaction at the C-terminal active site (containing C15′). As a result, even the WT-WT scOhrR variant forms predominantly singly oxidized protein during short times of incubation. ESI–MS analysis of the WT-WT dimers after only 10 min of treatment with CHP (in the presence of cysteine) clearly demonstrated a complete loss of DNA binding at a time when the majority of the protein was singly oxidized (Figure 3). Similarly, the WT-C15S variant is rapidly and reversibly inactivated by CHP under these conditions. In sum, our studies clearly demonstrate that oxidation at a single active site (per dimer) is sufficient to mediate derepression in vitro (Figures 3–5) and this is true regardless of whether the protein is oxidized to the mixed disulfide (with cysteine), an intraprotein disulfide, the cyclic sulfenamide or overoxidized.
Despite the ability of scOhrR variants to be functionally inactivated by oxidation at one active site in vitro, these same variants were not completely inactivated in vivo. One notable difference between the in vitro and in vivo reactions is the presence of reductant in the latter that can regenerate active OhrR. We have suggested previously that the re-reduction of OhrR to an active form is likely to be enzyme catalyzed since the rate of reduction in the presence of physiological levels of cysteine (a major low molecular weight thiol in B. subtilis) is likely too slow to be meaningful (13). Since the physiological reductant for OhrR is not yet known, we were not able to test this hypothesis genetically. However, it is reasonable to propose that oxidation of the second active site [which clearly can and does occur in the native, dimeric protein; (13,15)] would allow the inactivated protein to accumulate in vivo as the doubly oxidized protein and that two cycles of reduction would be required to regenerate functional repressor. In contrast, with those variants that can only become singly oxidized, a single reduction step will immediately regenerate active repressor. This model is consistent with the finding that LHP, which leads to significant overoxidation (thereby blocking enzymatic re-reduction), is a better inducer than CHP in the cell. It is also curious that the scOhrR variant with a single active site that reacts slowest in vitro (C15S-WT) is induced better in vivo than its more reactive counterpart (WT-C15′S) (Figure 2). This might reflect differences not in the rate of inactivation (which is clearly slower), but in the rate of reactivation by cellular reductants (which we postulate is even more dramatically impaired by the steric constraints imposed by the introduced linker region). Further studies will be required to ascertain if these effects account for the relatively poor responsiveness of these scOhrR variants to oxidants in vivo.
FUNDING
This work was supported by a grant from the National Science Foundation (MCB-0640616). Funding for open access charge: NSF (MCB-0640616).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Helmann Laboratory for helpful comments on this manuscript.
REFERENCES
- 1.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Mongkolsuk S, Helmann JD. Regulation of inducible peroxide stress responses. Mol. Microbiol. 2002;45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- 3.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 2001;183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panmanee W, Vattanaviboon P, Eiamphungporn W, Whangsuk W, Sallabhan R, Mongkolsuk S. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 2002;45:1647–1654. doi: 10.1046/j.1365-2958.2002.03116.x. [DOI] [PubMed] [Google Scholar]
- 5.Panmanee W, Vattanaviboon P, Poole LB, Mongkolsuk S. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 2006;188:1389–1395. doi: 10.1128/JB.188.4.1389-1395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Hu J, Chen PR, Lan L, Li Z, Hicks LM, Dinner AR, He C. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc. Natl Acad. Sci. USA. 2008;105:13586–13591. doi: 10.1073/pnas.0803391105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, He C. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 9.Wilke MS, Heller M, Creagh AL, Haynes CA, McIntosh LP, Poole K, Strynadka NC. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc. Natl Acad. Sci. USA. 2008;105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 11.Saridakis V, Shahinas D, Xu X, Christendat D. Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum. J. Mol. Biol. 2008;377:655–667. doi: 10.1016/j.jmb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 2006;8:51–62. [PubMed] [Google Scholar]
- 13.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl Acad. Sci. USA. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soonsanga S, Lee JW, Helmann JD. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol. Microbiol. 2008;68:978–986. doi: 10.1111/j.1365-2958.2008.06200.x. [DOI] [PubMed] [Google Scholar]
- 15.Soonsanga S, Lee JW, Helmann JD. Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J. Bacteriol. 2008;190:5738–5745. doi: 10.1128/JB.00576-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soonsanga S, Fuangthong M, Helmann JD. Mutational analysis of active site residues essential for sensing of organic hydroperoxides by Bacillus subtilis OhrR. J. Bacteriol. 2007;189:7069–7076. doi: 10.1128/JB.00879-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Arunkumar AI, Chen X, Giedroc DP. Structural insights into homo- and heterotropic allosteric coupling in the zinc sensor S. aureus CzrA from covalently fused dimers. J. Am. Chem. Soc. 2006;128:1937–1947. doi: 10.1021/ja0546828. [DOI] [PubMed] [Google Scholar]
- 19.Fuangthong M, Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl Acad. Sci. USA. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 21.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 22.von Hippel PH, Berg OG. On the specificity of DNA-protein interactions. Proc. Natl Acad. Sci. USA. 1986;83:1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanblatt SH, Revzin A. Two catabolite activator protein molecules bind to the galactose promoter region of Escherichia coli in the presence of RNA polymerase. Proc. Natl Acad. Sci. USA. 1983;80:1594–1598. doi: 10.1073/pnas.80.6.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyduk T, Lee JC. Escherichia coli cAMP receptor protein: evidence for three protein conformational states with different promoter binding affinities. Biochemistry. 1989;28:6914–6924. doi: 10.1021/bi00443a021. [DOI] [PubMed] [Google Scholar]
- 25.Shewchuk LM, Verdine GL, Walsh CT. Transcriptional switching by the metalloregulatory MerR protein: initial characterization of DNA and mercury (II) binding activities. Biochemistry. 1989;28:2331–2339. doi: 10.1021/bi00431a052. [DOI] [PubMed] [Google Scholar]
- 26.Liu T, Golden JW, Giedroc DP. A zinc(II)/lead(II)/cadmium(II)-inducible operon from the Cyanobacterium anabaena is regulated by AztR, an alpha3N ArsR/SmtB metalloregulator. Biochemistry. 2005;44:8673–8683. doi: 10.1021/bi050450+. [DOI] [PubMed] [Google Scholar]
- 27.Cavet JS, Graham AI, Meng W, Robinson NJ. A cadmium-lead-sensing ArsR-SmtB repressor with novel sensory sites. Complementary metal discrimination by NmtR AND CmtR in a common cytosol. J. Biol. Chem. 2003;278:44560–44566. doi: 10.1074/jbc.M307877200. [DOI] [PubMed] [Google Scholar]
- 28.Kamionka A, Majewski M, Roth K, Bertram R, Kraft C, Hillen W. Induction of single chain tetracycline repressor requires the binding of two inducers. Nucleic Acids Res. 2006;34:3834–3841. doi: 10.1093/nar/gkl316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Wong MD, Rosen BP. Both metal binding sites in the homodimer are required for metalloregulation by the CadC repressor. Mol. Microbiol. 2002;44:1323–1329. doi: 10.1046/j.1365-2958.2002.02961.x. [DOI] [PubMed] [Google Scholar]
- 30.Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J. Mol. Biol. 2003;333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural mechanisms of QacR induction and multidrug recognition. Science. 2001;294:2158–2163. doi: 10.1126/science.1066020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.