Abstract
MicroRNAs (miRNAs) mark a new paradigm of RNA-directed gene expression regulation in a wide spectrum of biological systems. These small non-coding RNAs can contribute to the repertoire of host-pathogen interactions during viral infection. This interplay has important consequences, both for the virus and the host. There have been reported evidences of host-cellular miRNAs modulating the expression of various viral genes, thereby playing a pivotal role in the host–pathogen interaction network. In the hide-and-seek game between the pathogens and the infected host, viruses have evolved highly sophisticated gene-silencing mechanisms to evade host-immune response. Recent reports indicate that virus too encode miRNAs that protect them against cellular antiviral response. Furthermore, they may exploit the cellular miRNA pathway to their own advantage. Nevertheless, our increasing knowledge of the host–virus interaction at the molecular level should lead us toward possible explanations to viral tropism, latency and oncogenesis along with the development of an effective, durable and nontoxic antiviral therapy. Here, we summarize the recent updates on miRNA-induced gene-silencing mechanism, modulating host–virus interactions with a glimpse of the miRNA-based antiviral therapy for near future.
INTRODUCTION
MicroRNAs (miRNAs) are small ∼22 nucleotide (nt) non-coding RNAs (ncRNAs) that play an important role in the post-transcriptional regulation of gene expression in a wide range of organisms from unicellular eukaryotes to multicellular eukaryotes by a variety of mechanisms. Initially, these were discovered in Caenorhabditis elegans (1,2), but now they are known to be widespread in nature (3–8). It therefore comes as no surprise that viruses, which typically employ many components of the host gene expression machinery, also encode miRNAs (9–16). Thus far, 8619 miRNA genes have been annotated from 87 organisms of which Homo sapiens top the list with 695 miRNA genes identified till date. These data are available at microRNA Registry database managed by researchers at University of Manchester and the Wellcome Trust Sanger Institute (http://microrna.sanger.ac.uk/sequences) (17). Over the course of evolution, viruses have developed highly sophisticated mechanisms to exploit the biosynthetic machinery of host cells and elude the cellular defense mechanisms (18,19). Present research advances reveal that the complex interaction between viruses and host cells also involves miRNA-mediated RNA-silencing pathways (20).
Viruses have a more intricate interaction with the host cell, which creates problem in inactivating a virus without doing any harm to the host cell. Combating viral infection by targeting viral proteins and pathways unique to the viral life cycle has become possible for a few viruses without unacceptable host cell toxicity (21). Hence, only a few effective antiviral drugs exist. Viral resistance, sequence diversity and drug toxicity are significant problems for all antiviral therapies. This has lead toward harnessing the potential of RNA interference (RNAi) as an innate antiviral defense mechanism (22,23).
RNAi represents a vital component of the innate antiviral immune response in plants and invertebrate animals. Furthermore, it serves as a host gene-regulation mechanism that is triggered by the expression of highly structured miRNA molecules. However, role of cellular miRNAs in the defense against viral infection in mammalian organisms has thus far remained elusive. Hence, it is important to understand the intricate details regarding the influence of viral replication on the abundance and distribution of miRNAs within the host cell. It has been proposed that cellular miRNAs may have a substantial effect on viral evolution and have the potential to regulate the tissue tropism of viruses in vivo (20). Viruses too exploit miRNA-induced gene-silencing pathway by encoding their own miRNAs (9,16,24–27). Thus, studying the changes in miRNA landscape during viral replication may help us understand the molecular regulation of host defenses and the attempt by viruses to overcome host defense during infection. A wide range of complex interactions is possible through miRNA–mRNA coupling during host–virus interaction (21). In this game of pathogen-host interaction, viruses strive to succeed by effective usage of host machinery and expressing viral proteins, whereas efficient hosts limit viral invasion by putting up innate and adaptive antiviral defenses.
The present review discusses the existing intricate details about the role of miRNAs in virus–host interaction. Furthermore, it discusses its therapeutic implications along with the existing resources needed to study such interaction.
VIRAL miRNAs
Thomas Tuschl and his group at Rockefeller University reported the existence of viral miRNAs for the first time in Epstein-Barr virus (EBV) (9). Till now, 141 miRNA genes have been identified in 15 viruses from three viral families, herpesvirus, polyomavirus and retrovirus. Herpesvirus family with three subfamilies, viz., α-, β- and γ-herpesvirinae express a large number of distinct miRNAs. Among these, γ-herpesvirus encodes maximum number of miRNAs (9,28,29). EBV of γ-herpesvirus subfamily has the highest number of miRNAs (17). Kaposi's sarcoma-associated viruses (KSHV), a member of the γ-herpesvirus subfamily encodes an array of 13 distinct miRNAs, all of which are expressed at readily detectable levels in latently KSHV-infected cells. The remaining three members encoding miRNAs are murine γ-herpesvirus 68 (MHV68) (10,30), Rhesus monkey rhadino virus (RRV) (31) and Rhesus lymphocryptovirus (rLCV) (28).
Furthermore, reports have been published on identification of miRNAs encoded by polyomaviruses, viz., BK polyomavirus (BKV), JC polyomavirus (JCV) and simian virus 40 (SV40) (11,32), Human cytomegalovirus (HCMV) (10,25,33); Herpes Simplex virus-1(HSV-1) (16,26), HSV-2 (34), Murine cytomegalovirus (MCMV) (13,35) and Marek's disease virus type 1 and 2 (MDV-1 and MDV-2) (36–38). The details about the viruses, their pathogenicity in host and miRNAs encoded by them are provided in Table 1.
Table 1.
List of viruses encoding microRNAs, their genomic location, hosts and diseases caused by these viruses
| DNA virus families | Species | Diseases | Host | Nos. | miRNAs | Genomic location | Functions | References |
|---|---|---|---|---|---|---|---|---|
| α-Herpesviruses | Herpes Simplex Virus-1 | Cold sores | Human | 6 | hsv1-miR-H1 | 450-bp upstream of TSS of LAT region | Maintenance of viral latency | 16,26 |
| hsv1-miR-H(2-6) | Four are encoded in LAT region and fifth one in unknown transcript | |||||||
| Herpes Simplex Virus-2 | Genital herpes | Human | 1 | hsv2-miR-I | Encoded in LAT exon-2 | Controls viral replication in neurons | 34 | |
| Marek's disease virus | Marek's disease (MD); Neurolymphomatosis; Ocular lymphomatosis | Chicken | 14 | mdv1-miR-M (1–13) | Clustered in MEQ and LAT regions | Neoplastic transformation, antiapoptotic | 36,38 | |
| Marek's disease virus type 2 | Nononcogenic; Neurolymphomatosis; Ocular lymphomatosis | Chicken | 17 | mdv2-miR-M (14–30) | Sixteen are clustered in a long repeat region, encoding R-LORF2 to R-LORF5. The single miRNA is within the C-terminal region of the ICP4 homolog. | 37 | ||
| β-Herpesviruses | Human Cytomegalovirus | Congenital CMV infection; CMV mononucleosis; CMV pneumonitis; CMV retinitis | Human | 11 | hcmv-miR-UL22A, hcmv-miR-UL36, hcmv-miR-UL70, hcmv-miR-UL112, hcmv-miR-UL148D, hcmv-miR-US4, hcmv-miR-US5-1, hcmv-miR-US5-2, hcmv-miR-US25-1, hcmv-miR-US25-2, hcmv-miR-US33 | Intergenic and intronic | Act as immune-response inhibitor, help in viral replication | 10,25,30,33,39 |
| Mouse Cytomegalovirus | Murine | 18 | mcmv-miR-M23-1, mcmv-miR-M23-2, mcmv-miR-M44-1, mcmv-miR-M55-1, mcmv-miR-M87-1, mcmv-miR-M95-1, mcmv-miR-m01-1, mcmv-miR-m01-2, mcmv-miR-m01-3, mcmv-miR-m01-4, mcmv-miR-m21-1, mcmv-miR-m22-1, mcmv-miR-m59-1, mcmv-miR-m59-2, mcmv-miR-m88-1, mcmv-miR-m107-1, mcmv-miR-m108-1, mcmv-miR-m108-2 | Organized into five genomic regions and three clusters | Could influence viral infection and latency | 13,35 | ||
| γ-Herpesviruses | Kaposi's sarcoma-associated Herpesvirus | Kaposi's sarcoma, primary effusion lymphoma (PEL), multicentric Castleman's disease | Human | 13 | kshv-miR-K12-(1-9), kshv-miR-K12-10(a,b), kshv-miR-K12-(11-12) | Located within 3.6-kb intergenic region and Kaposin gene | May regulate kaposin gene; down-regulates thrombospondin 1, which has anti-proliferative, anti-angiogenic activity | 10,27,40–42 |
| Epstein-Barr Virus | Pfeiffer's disease; Burkitt's lymphoma; gastric carcinoma; Nasopharyngeal carcinoma | Human | 23 | ebv-miR-BART(1-20), ebv-miR-BHRF1-(1-3) | Twenty genes are within the introns of BARTs and remaining in BHRF1 | Tumorigenesis of EBVaGCs; regulates LMP1 expression (Nasopharyngeal Carcinoma); down-regulate the viral DNA polymerase BALF5; | 9,28,29,43–45 | |
| Rhesus Lymphocryptovirus | Lymphadenopathy | Simian | 16 | rlcv-miR-rL1-(1-16) | Clustered in BART and BHRF1 | 28 | ||
| Rhesus Monkey Rhadinovirus | Multicentric lymphoproliferative disorder | Simian | 7 | rrv-miR-rR1-(1-7) | Located within a single cluster at the same genomic location as of KSHV | 31 | ||
| Mouse Gamma Herpesvirus 68 | Infectious mononucleosis | 9 | mghv-miR-M1-(1-9) | Intergenic and clustered | 10,30 | |||
| Polyomaviruses | Simian Virus 40 | Tumors | Simian | 1 | sv40-miR-S1 | Down-regulate the expression of viral early genes | 11 | |
| BK Polyomavirus | Polyomavirus-associated nephropathy | Human | 1 | bkv-miR-B1 | -do- | 32 | ||
| JC Polyomavirus | Progressive Multifocal Leuko-encephalopathy | Human | 1 | jcv-miR-J1 | -do- | 32 | ||
| RNA virus families | ||||||||
| HIV | Human immunodeficiency virus 1 | AIDS | Human | 3 | hiv1-miR-H1, hiv1-miR-N367, hiv1-miR-TAR | Found in nef gene and TAR element | Supress Nef function; supress HIV-1 virulence | 15,46,47 |
TSS: transcription start site; LAT: latency-associated transcript; BART: BamA rightward transcript; BHRF1: BamH1 fragment H rightward open reading frame 1; TAR: trans-acting responsive element; nef: negative factor.
miRNA-biogenesis pathway (Figure 1) poses some serious problem for RNA viruses and a group of DNA viruses (poxviruses) to encode miRNAs (4). However, it is possible to overcome these problems by adopting nonconventional ways. Omoto et al. (46) have reported the presence of miRNAs in HIV-1 (which is an RNA virus) infected cells, although extensive studies by Pfeffer et al. (10) as well as Lin and Cullen (47) have failed to confirm the existence of viral miRNAs in HIV. Recently, Ouellet et al. (15) have also identified miRNAs within HIV-1 trans-activation responsive (TAR) element. It is definitely essential and still needs further investigation to find whether the expression of TAR miRNAs influences viral replication or the efficiency of host-antiviral defenses.
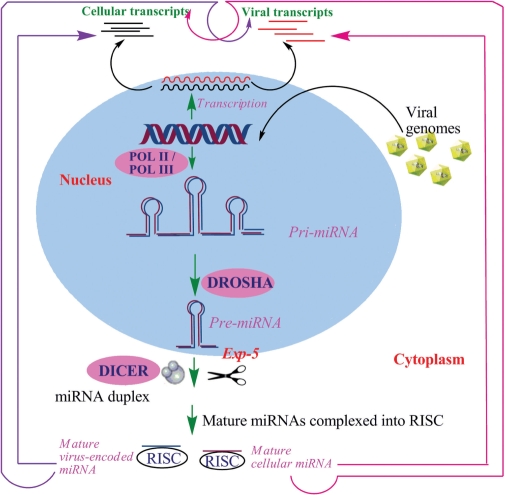
Figure 1.
Biogenesis of microRNAs. Processing of both host miRNAs (violet) and virus-encoded miRNAs (blue) is assumed to occur through the same pathway. The arrows (pink) indicate the effects of cellular miRNAs on their own cellular transcripts as well as on viral transcripts. The violet arrow shows the effects of virus-encoded miRNAs on both cellular and viral transcripts. Both these arrows suggest possible miRNA-mediated interactions between viruses and their hosts.
BIOGENESIS OF miRNAs
Any understanding of the potential role of miRNAs in viral pathogenesis and studies into the entire spectrum of host–virus interactions at the miRNA level requires an appreciation of the genomic location, transcription and processing of miRNAs (48,49).
Organization of miRNA genes
Viral miRNA genes are found as single or clustered transcription units (50–52). The genomic location of the virus-encoded miRNAs is very important and to some extent linked with their function. miRNAs of α-herpesviruses, namely HSV-1, MDV-1 and 2 and those of the γ-herpesviruses, namely KSHV, are located closed to and within the latency-associated transcript. These miRNAs are associated with latent transcription (16,36,38,41). The organization of viral miRNA genes within their genomes is provided in Table 1.
Maturation and processing of miRNAs
miRNA biogenesis initiates with the transcription of a pri-miRNA precursor, typically of length ranging from ∼200 nt to several thousand nts by RNA polymerase II (Pol II) (53). On the contrary, work of Borchert et al. (54) showed a miRNA cluster (C19MC) in the human chromosome 19, interspersed among Alu repeats requires RNA polymerase III (Pol III) for transcription (54).Viral miRNAs too undergoes similar processing by Pol II except in a few cases, namely MHV68 miRNAs, which are transcribed by Pol III (10).
Next step in miRNA processing involves the recognition and nuclear cleavage of pri-miRNAs by RNase III enzyme Drosha acting in concert with the double-stranded RNA-binding protein DGCR8 (DiGeorge-syndrome critical-region protein 8) in vertebrates (55,56). This generates ∼60-nt pre-miRNA hairpin, which is transported into the cytoplasm by exportin-5 complexed with Ran-GTP (Figure 1). Here, GTP hydrolysis results in the release of the pre-miRNA.
Drosha-mediated processing of miRNA genes located within the open-reading frames and translation of these protein-coding transcripts must be mutually exclusive. As in the case of KSHV miRNAs, processing of miR-K10 and miR-12 within the nucleus must be modulated in such a way, so that a substantial percentage of KSHV mRNA is able to exit the nucleus before Drosha cleavage (40). Hence, it is possible that the regulation of viral mRNA expression by modulation of Drosha cleavage efficiency has a role in several different virus replication cycles.
Drosha independent miRNA processing has been previously observed in the case of mirtrons (57). The TAR element in HIV-1 having structural similarities with human pre-miRNA let-7a-3 (15) is too short for Drosha processing whose pri-miRNA substrates contain a stem of approx. three helical turns (∼33 bp). Therefore, experimental evidences suggest Drosha independent processing of HIV-1 TAR miRNAs.
The pre-miRNA resulting from Drosha processing is cleaved thereafter by cytoplasmic RNase III enzyme Dicer acting in concert with its cofactor TRBP (transactivating region RNA-binding protein) (58). The terminal loop is removed, generating the miRNA duplex intermediate. Dicer facilitates assembly of the miRNA strand (having weaker 5′ bp) of the duplex into the miRNA effector complexes, called RNA-induced silencing complexes (RISCs) (58). The unincorporated strand termed as ‘passenger strand’ is released and degraded. Although the composition of RISC is not completely defined, the key constituents of it are miRNA and one of the four Argonaute (Ago) proteins (59). The miRNA then directs RISC to complementary mRNAs (60), which is either cleaved or undergone translational repression depending on the degree of complementarity between the RISC-bound miRNA and the target mRNA. A seed sequence within the miRNA (nts 2–8) is known to be critical for binding and target recognition. Perfect complementarity results in mRNA degradation/cleavage, which is rare in animals but not in plants. Such an example is exhibited by the polyoma virus SV40 miRNAs, which are perfectly complementary to early mRNAs transcribed antisense to the pre-miRNA and direct the RISC-mediated cleavage of these early transcripts, responsible for generating strong cytotoxic T-cell (20). In major instances, imperfect/partial complementarity with the target is observed, leading to translational repression of the mRNA transcripts by miRNA-RISC (61,62). In addition to repressing translation, miRNA interactions can lead to deadenylation or target decapping, leading to rapid mRNA decay (63–66).
miRNA editing influences processing pathway
The levels of mature miRNAs expressed within a cell are not simply determined by the transcription of miRNA genes; rather it depends on one or more steps in the processing pathway (67) like RNA editing of pri-/pre-miRNAs. Edited pri-/pre-miRNAs do not undergo Drosha or Dicer cleavage, which eventually reduces the production of mature miRNAs. In certain cases, pri-miRNAs are transported out of the nucleus into the cytoplasm where Drosha fails to process them and they are destroyed (68). Adensosine deaminase editing of specific pri-miRNAs has been reported. This A-I editing event leads to decreased processing of the miRNA by Drosha and increases turnover by the Tudor-SN nuclease, a component of RISC and also a ribonuclease specific to inosine-containing dsRNAs (69). The human and mouse pre-miRNAs of miR-22 are edited at several positions, including sites in the mature miRNA, which are predicted to influence its biogenesis and function (70). Notably, the viral miRNA, kshv-miR-K12-10, with a single adenosine residue substituted by guanosine (miR-K12-10b) is frequently detected among cDNA isolates identified by the small RNA-cloning method. The editing of this particular site does not inhibit pri-miR-K12-10 RNA processing, but leads to expression of mature miRNA with the edited sequence (10). Evidence of RNA editing has also been observed in miR-M7 of MDV, although its effect is unknown (38). However, RNA editing in the seed sequence of a miRNA could re-direct it to a new set of targets (71). All these indicate that the miRNAs originated from the same pre-miRNA may target more corresponding complementary mRNA, making the fine-tuning of the virus-host interaction network more complicated.
EVOLUTIONARY ASPECTS OF CELLULAR VERSUS VIRAL miRNA GENES
Cellular miRNAs and their target sequences are frequently conserved (72), which facilitate computational biologists toward in silico prediction of cellular miRNAs and their targets. Interestingly, viral miRNAs, unlike their vertebrate counterparts do not share a high level of homology, even within members of the same family or with that of the host. However, miRNAs of closely related viruses such as RRV and KSHV are encoded in the same genomic region but do not exhibit sequence homology (31). The miRNAs encoded by chicken α-herpesviruses MDV-1 and MDV-2 are clustered in homologous regions of the viral genomes, which are transcribed during viral latency, but are not homologous in sequence (36–38). In contrary to these, Cai et al. (28) have shown that eight of EBV miRNAs are conserved with rLCV miRNAs, thus arguing for their importance in viral life cycle.
The lack of conservation in viral miRNA sequences attributes to the higher rate of mutations and faster evolution in viruses when compared to eukaryotes. This would mean an evolutionary advantage for rapid adaptation to the host and environmental conditions. However, it offers a challenge to computational biologists as most of the algorithms for miRNA prediction rely heavily on conservation and would prove inadequate in case of viruses. Even a single-point mutation in the seed region can lead to a dramatic shift in miRNA function due to the loss or acquisition of a large number of cellular or viral mRNA targets.
VIRUS-ENCODED miRNAs—ORTHOLOGS OF CELLULAR miRNAs
In general, viral miRNAs and cellular miRNAs do not bear seed homology. But presumably, due to the presence of highly evolved gene-regulatory networks, some viral miRNAs have seed homology with cellular miRNAs. Recent report suggests that miR-K12-11 encoded by KSHV shares the first eight nts with hsa-miR-155 (27,73) (refer Figure 2). MiR-155 is processed from a primary transcript, termed as BIC gene (B-cell Integration Cluster), whose upstream region was identified as a common site of integration of the avian leucosis virus (ALV) (74) in lymphomas. miRNA-profiling studies have shown increased expression of miR-155 in a wide range of cancers including lymphomas (75). Gottwein et al. (73) reported that miR-155 and miR-K12-11 regulate similar set of targets including genes with known roles in cell-growth regulation. It has been shown that BACH-1 is one of the predicted mRNAs, targeted by both miR-155 and miR-K12-11 (27). Transient expression of miR-155 occurs in macrophages, T and B lymphocytes and miR-155 knockout mice revealed defects in adaptive immune responses. Furthermore, overexpression of miR-155 in B-cells is associated with the development of B-cell lymphomas in humans, mice and chickens (74) although the mechanism is unknown. Given the apparent role of miR-155 in tumorigenesis and miR-K12-11 being an ortholog of miR-155, it is tempting to speculate that miR-K12-11 may contribute to the development of B-cell tumors seen in KSHV-infected individuals. Inspite of being a distantly related γ-herpesvirus, EBV miRNAs do not bear homology to miR-155 (76). However, previous reports have shown the expression of BIC during EBV infection expressing the full repertoire of EBV latency genes, which implies the role of EBV latency genes in inducing BIC gene (77).
Figure 2.
Virus-encoded microRNAs as orthologs of cellular microRNAs. Encircled boxes show sequence homology of viral miRNAs to human/mouse miRNAs. The homologous bases are shown in blue. (a) Orthologs of cellular miR-155; (b) orthologs of cellular miR-151; (c) orthologs of cellular miR-18a and miR-18b.
Analyzing the entire set of viral miRNAs known till today, such seed homology is observed in a few more cases. One of the interesting cases is the MDV-1 miRNA miR-M4, which bears the same 5/terminal 8 nts as miR-K12-11 and hence might function as an ortholog of miR-155. Since MDV-1 encodes meq oncogene apart from other proteins, miR-M4 might contribute to tumorigenesis in chickens. Furthermore, miR-M1-4 of MHV68 shares 5/terminal 9 nts with murine miR-151. The function of this cellular miRNA is still unknown. Potential cellular orthologs of other viral miRNAs having limited seed homology (nts 2–7) (refer Figure 2), corresponding to the minimal miRNA seed region, include ebv-miR-BART5, rlcv-miR-rL1-8 and mghv-miR-M1-7-5p, which have miR-18a and miR-18b as their cellular counterpart. These two cellular miRNAs are encoded in the miR-17-92 cluster, which has oncogenic function (78).
VIRAL miRNAs-REGULATING GENE EXPRESSION
Regulatory impact on viral transcripts
Viral miRNAs have a regulatory effect on their protein-coding genes. The level of regulation depends on the degree of complementarity of the viral miRNAs with the 3/UTR (untranslated region) of the regulated mRNAs (79). These regulations are beneficiary to the virus toward maintaining its replication, latency and evading the host-immune system (Figure 3).
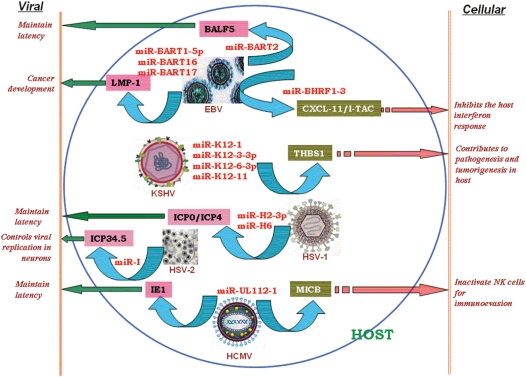
Figure 3.
Effects of virus-encoded microRNAs on viral and cellular transcripts.
MiR-BART2 of EBV exhibits perfect complementarity to the 3/UTR of BALF5, which encodes the viral DNA polymerase (9). Recently, Barth et al. (14) have shown that miR-BART2 down-regulates BALF5. Induction of the lytic viral replication cycle results in a reduction of the level of miR-BART2. Hence, there is a decrease in cleavage of the BALF5 3/UTR. Forced expression of miR-BART2 during lytic replication resulted in a 40–50% reduction of the level of BALF5 protein and a 20% reduction of the amount of virus released from EBV-infected cells. It might be the situation that latently expressed miR-BART2 specifically has evolved to target BALF5, and such an interaction may be essential for maintaining EBV latency. The other EBV miRNAs miR-BART-1p, miR-BART16 and miR-BART17-5p (having imperfect match with the targets) target 3/UTR of the mRNA coding for the latency-associated membrane protein LMP-1 and repress its expression. This regulation decreases LMP-1-mediated activation of nuclear factor-kappa B (NF-κB) as well as apoptosis resistance (44). Hence, these miR-BART miRNA-mediated regulations on LMP1 may explain the discrepancy between LMP1 transcript and protein detection in nasopharyngeal carcinoma. This further highlights the role of the EBV miRNAs in regulating LMP1 downstream signaling to promote cancer development (44).
Grey et al. (79) predicted that miR-UL112-1 of HCMV targets the viral immediate-early protein 1(IE1) mRNA, a transcription factor required for the expression of many viral genes. To test this prediction, mutant viruses were generated that were unable to express miRNA or encoded an IE1 mRNA lacking its target site. Analysis of RNA and protein within infected cells demonstrated that miR-UL112-1 inhibits expression of the major IE1 protein. Such miRNA-mediated suppression of IE genes might be a part of the strategy of these viruses to enter the host and maintain latency.
SV40 miRNAs (miR-S1-5p and miR-S1-3p) are perfectly complementary to early viral mRNAs and target the mRNAs for a protein known as T antigen, leading to its cleavage. On entering into the cell, viral replication is triggered by the production of this T antigen. Furthermore, T antigen serves as a target for host immune (T) cells, which destroys infected cells and prevents the virus from spreading. Thus, the corresponding miRNAs (targeting T antigenic mRNA) accumulate at late times in infection when enough viral replication has been done. Furthermore, it has been shown that cells infected with a mutant virus (that does not produce SVmiRNAs) are more likely to get killed by cytotoxic immune cells rather than the wild-type ones (11). Thus, it is shown that viral evolution has taken advantage of the miRNA pathway to generate effectors that enhance the probability of successful infection.
MDV latency-associated transcripts include miR-M6 to miR-M8 and miR-M10 and miR-M13, which maps to a large intron at the 5′ end (38,80). This is presumably derived from a large 10-kb transcript that maps antisense to the ICP4 gene, which implies a probable role of these miRNAs toward modulating ICP4 transcript to inhibit entry of the virus into the lytic cycle.
In HIV-1-infected and nef-transduced cells, nef-derived miRNA miR-N367 inhibits HIV-1 transcription in human T cells (46), thus facilitating both viral replication and disease progression. Recently, annotated HSV-1 miRNAs, miR-H2-3p and miR-H6 are reported to downregulate the expression of ICP0 and ICP4 proteins, respectively. Such miRNA-induced down-regulation helps HSV-1 to maintain latency (16). Furthermore, miR-I of HSV-2 regulates expression of a key viral neurovirulence factor, thereby affecting the establishment of latency (34).
Regulatory impact on cellular transcripts
It is most likely that viral miRNAs should have a modulatory effect on cellular transcripts in order to foster their own agenda. Surprisingly, no viral miRNA exhibits complete complementarity to a cellular transcript so as to induce its degradation (73). Recently, Xia et al. (45) has reported that the inverse effect of BHRF1-3 miRNA expression of EBV is correlated with levels of the IFN-inducible T-cell attracting chemokine CXCL-11/I-TAC, which is a putative cellular target. EBV-miRNAs are known to be expressed in primary lymphomas classically linked to the virus and are associated with the viral latency status. Negative modulation of CXCL-11/I-TAC by a viral-encoded miRNA may serve as an immunomodulatory mechanism in these tumors (45). This may inhibit the host-interferon response by suppressing the above-mentioned target.
KSHV miRNAs downregulate the expression of a multicellular glycoprotein THBS1, having anti-angiogenic and anti-proliferative activity (81). This aids in immune evasion of KSHV-infected cells.
Major histocompatibility complex class I-related chain B (MICB) gene is down-regulated by hcmv-miR-UL112 during viral infection (82). This leads to decreased binding of the natural killer (NK) cell-activating receptor NKG2D with its stress-induced ligand MICB and thus reduces killing of virus-infected cells and tumor cells by NK cells. Thus, HCMV exploits a miRNA-based immunoevasion mechanism for its own survival.
Although, Yao et al. (38) have identified the potential miRNAs in MDV-1, a major challenge lies toward the identification of potential targets for some of the miRNAs overexpressed in these cells. The high level of expression of miR-M2 to miR-M5 and miR-M9 and miR-M12, which are part of same transcriptional unit in the same orientation as Meq gene, suggests that these miRNAs may have major roles in regulating expression of viral and host genes in latent or transformed T cells. Furthermore, this miRNA transcriptional unit being antisense to RLORF8 may also have a regulatory role on expression of RLORF8 transcript. Since miR-M6 to miR-M8 and miR-M10 and miR-M13 are mapped to the LAT region, it is tempting to speculate that they might have antiapoptotic function.
CELLULAR miRNAs-REGULATING GENE EXPRESSION
Cellular miRNAs play a role in building up direct or indirect effect in regulating viral genes. They might be broadly implicated in viral infection of mammalian cells, with either positive or negative effects on replication (83,84).
Viruses exploiting cellular miRNAs to their own advantage
Hepatitis C virus (HCV) is an enveloped RNA virus of the Flavivirus family, which is capable of causing both acute and chronic hepatitis in humans by infecting liver cells. A liver-specific cellular miRNA, miR-122 facilitates the replication of HCV, targeting the viral 5′ non-coding region (85). HCV replication is associated with an increase in expression of cholesterol biosynthesis genes that are regulated by miR-122 (86). This does not seem to occur by a classical miRNA-type function. The virus seems to have co-opted the miRNA in its replication cycle. Such a case suggests the role of miR-122 as a target for antiviral intervention and leaves an interesting issue to check whether other RNA viruses do exhibit similar interactions with cellular miRNAs. On the contrary, Pederson et al. (87) have shown that interferon-modulated downregulation of miR-122 contributes to antiviral effects (87).
Huang et al. (88) have shown that HIV uses miRNA-mediated downregulation of viral protein expression to its own advantage. In ‘latently infected’ resting CD4+ T-cells, HIV-1 is stably integrated into their genome, but does not produce any viral proteins. Hence they cannot be eliminated by the immune system or targeted by any existing anti-HIV drugs, but can rekindle the infection at any time. Huang and colleagues observed that a cluster of cellular miRNAs including miR-28, miR-125b, miR-150, miR-223 and miR-382 are enriched in resting CD4+ T cells when compared with activated CD4+ T cells and that the 3′ ends of HIV-1 messenger RNAs are targeted by this cluster. Inhibition of these miRNAs in resting T-cells, derived from infected individuals on highly active antiretroviral therapy, increased HIV protein expression. This suggests that the HIV-1 uses cellular miRNA to its own advantage by recruiting the resting-cell-enriched miRNAs to control the translation of viral mRNA into protein and thus escapes the host–immune system.
Antiviral effects of cellular miRNAs
Cellular miRNAs effectively restricts the accumulation of the retrovirus, primate foamy virus type 1 (PFV-1) in human cells (83). This is a retro-transcribing virus and has similarity with HIV. Lecellier and his colleagues showed that replication of the wild-type viruses can be inhibited by endogenous cellular miRNA miR-32 expressed in HeLa and BHK21 cells in cell culture through an interaction with a poorly conserved region in the 3′ portion of the PFV genome.
Otsuka et al. (89) observed that impaired miRNA production in mice having variant Dicer1d/d allele makes them hypersensitive to infection by vesicular stomatitis virus (VSV) (89). Lack of cellular miRNAs, miR-24 and miR-93 has been found to be responsible for enhanced VSV replication in such cells. These miRNAs were found to target viral large protein and phosphoprotein genes resulting in Dicer1d/d-mediated suppression of VSV replication in wild-type macrophages. These showed that defect in miRNA processing is responsible for the increased susceptibility to VSV infection in Dicer1d/d mice and hence provides ample genetic evidence supporting the fact that host miRNA can influence viral growth in mammals.
Hariharan et al. (90) have reported candidate targets of various human miRNAs (miR-29a and miR-29b, miR-149, miR-378 and miR-324-5p) to be critical genes of the HIV-1 genome. The putative target genes include nef, vpr, vpf and vpu, which correspond to the entire set of accessory genes of HIV-1. Inspite of being accessory, these genes are important components of the HIV-1 infection and replication cycles. They regulate viral infectivity (vif and nef), viral gene expression (vpr) and progeny virion production (vpu) (91). Gene expression profiles of miRNAs, based on microarray experiments (92), revealed that these miRNAs are expressed at detectable levels in T-cells. Experimental validation of these targets shall strengthen these findings.
Interferon-mediated modulation
Pederson et al. (87) demonstrated interferon (IFN)-mediated modulation of the expression of numerous cellular miRNAs in the treatment of hepatocytes (87) caused by HCV. Expression levels of 30 cellular miRNAs were found to be influenced by IFN α/β or IFNγ. Sequence complementarity analysis of these miRNAs against viral transcripts or viral genomic RNAs revealed matches among several viruses, mostly RNA viruses. Specifically, eight of the miRNAs (miR-1, miR-30, miR-128, miR-196, miR-296, miR-351, miR-431 and miR-448), having nearly perfect complementarity in their seed sequences with HCV RNA genomes, were up-regulated. These miRNAs are capable of inhibiting HCV replication and infection. Similar analysis of the DNA virus, hepatitis B yielded no significant matches.
This has opened the door toward novel host–defense mechanisms that exist in mammalian cells as well as add to the antiviral mechanisms employed by interferons. Furthermore, it might be interesting to check whether similar case exists in other RNA viruses.
Indirect antiviral response of cellular miRNAs
A recent work of Triboulet et al. (93) argues that RNAi limits the replication of HIV-1 in human cells and that cellular miRNAs indirectly contributes to this antiviral response by repressing cellular factors that influence these processes. Type III RNases Dicer and Drosha, responsible for miRNA processing, inhibit virus replication both in peripheral blood mononuclear cells from HIV-1-infected donors and in latently infected cells. They have shown how viral infection overcomes the limitations imposed on virus life cycle by the host miRNA-mediated defenses. HIV-1 actively suppressed the expression of the polycistronic cellular miRNA cluster, miR-17/92, comprising miR-17-5p, miR-17-3p, miR-18, miR-19a, miR-20a, miR-19b-1 and miR-92-1, which limit its replication. Their results suggest that miR-17-5p and miR-20a of this cluster target the 3/UTR of histone acetyltransferase Tat cofactor PCAF to inhibit mRNA translation (93). Furthermore, PCAF has been proposed to promote HIV-1 transcriptional elongation (94). Hence, HIV-1 down-regulates the expression of these cellular miRNAs for efficient replication. On the contrary, they may also up-regulate cellular miRNAs that is beneficial to them. Several miRNAs such as miR-122, miR-370, miR-373 and miR-297 have been reported to be up-regulated by HIV-1 infection, with unknown consequences (93,95).
Human papilloma virus (HPV) causes cervical cancer, which is one of the most common cancers in women worldwide (96). miRNA-expression analysis revealed that three miRNAs were overexpressed and 24 underexpressed in cervical cell lines containing integrated HPV-16 DNA compared to the normal cervix. Furthermore, nine miRNAs were found to be overexpressed and one underexpressed in integrated HPV-16 cell lines in comparison with the HPV-negative CaCx cell line C-33A. Analysis of Marinez et al. (97) showed that miR-218 was specifically underexpressed in HPV positive cell lines, cervical lesions and cancer tissues containing HPV-16 DNA compared to both C-33A and normal cervix. Furthermore, miR-218 targets the epithelial cell-specific marker LAMB3, and it is well observed that LAMB3 is expressed more in the presence of the HPV-16 E6 oncogene, and this effect is mediated through miR-218 (97). These findings may lead toward better understanding of the molecular mechanisms involved in cervical carcinogenesis, which can be helpful in the development of both cancer therapeutics and diagnostics.
Highly pathogenic avian influenza A virus (HPAIV) of subtype H5N1 cause infection in the lower respiratory tract and severe pneumonia in humans (98). Computational procedures predict the role of two human miRNAs miR-507 and miR-136 in modulating the expression of virulent viral (PB2 and HA) genes, respectively (99). Furthermore, microarray data on miRNA gene expression in different tissues have shown that miR-136 is expressed in lungs (100). These miRNAs were found to be absent in the chicken genome, which might be the cause of making a difference in infectivity and lethality of the virus in chicken and human.
Resources facilitating the study of miRNA-mediated virus–host interaction
Inspite of very little information about miRNA gene structures and selectivity constraints regarding choosing their target genes, several computational approaches have been used to identify these important molecules and their targets (101,102).
Computational tools to predict miRNAs and their targets
Computational approaches have been an efficient strategy for predicting miRNA genes and their targets (5). The prediction algorithms range from custom-made programs for searching hairpin loops and energetic stability to advanced algorithms using machine-learning approaches (103–105). Most of these algorithms rely mainly on sequence homology and conservation of seed matches (106), which reduce their prediction accuracy in the case of viral miRNAs and their targets as they do not share close homology. Although there have been some notable exceptions of viral miRNAs having cellular orthologs, their cellular targets may not be conserved. Furthermore, considering the imposition of the constraint regarding accessibility of the predicted sites (targets) due to mRNA folding, application of matching criterion only could result in a large number of false positive predictions.
A better understanding of the sequence and structural components of precursor hairpins involved in miRNA biogenesis (107) has improved the miRNA gene-prediction algorithms. Scaria et al. (99) have developed a miRNA prediction method in viruses based on Support Vector Machines. They rationalized that virus-encoded pre-miRNA hairpins would share the sequence and structure features with that of host as they share the same miRNA-processing machinery. Target prediction algorithms too have improved considerably on incorporating certain novel constraints. In addition to seed pairing, 3/miRNA-mRNA pairing at nts 13–16 and few more determinants have been reported by Bartel and his group (108). Such determinants have been shown to enhance site efficacy in mammals. Some of the widely used target prediction algorithms are Targetscan, PicTar, RNAhybrid and MiRanda (72,108–111). Recently, Bartel and Rajewsky laboratories have established proteomics approaches termed as SILAC (stable isotope labeling with amino acids in cell culture) and pSILAC (pulsed SILAC), respectively, to detect and quantify miRNA-induced changes in protein accumulation. Such approach proves a powerful tool for the identification of miRNA targets (112,113).
Microarray analysis of cellular gene expression upon ectopic expression of individual or multiple miRNAs or after inhibition of miRNA function using antisense reagents has been suggested as an alternative approach toward target identification (114,115). Inspite of certain disadvantages, this approach has the potential to reveal indirect downstream consequences of miRNA-mediated regulation on cellular gene expression.
Sources of miRNA data
Several databases related to miRNAs and their targets have been developed, which act as a constant resource for daily experimental research in the biological laboratories. Table 2 lists some of the available important miRNA databases.
Table 2.
Database resources on virus-encoded microRNAs
| Resources | Websites | Descriptions |
|---|---|---|
| ViTa | http://vita.mbc.nctu.edu.tw/ | Database of viral microRNAs and predicted host targets |
| miRBase | http://microrna.sanger.ac.uk/ | Comprehensive miRNA resource including virus-encoded microRNAs |
| Vir-Mir db | http://alk.ibms.sinica.edu.tw/ | Viral miRNA prediction database |
| MicroRNAdb | http://bioinfo.au.tsinghua. edu.cn/micrornadb/ | Comprehensive database for MicroRNAs |
| Ambion microRNA Resource | http://www.ambion.com/techlib/resources/miRNA/ | Resource for microRNA and their targets |
| siVirus | http://sivirus.rnai.jp/ | Website for efficient antiviral siRNA design |
MicroRNA expression profile—a biomarker for viral diseases
Recent advances in miRNA research have initiated the development of novel technologies that enable the measurement of expression levels for all known miRNAs, thereby providing an opportunity for global miRNA profiling. This helps in classifying cancer subtypes more accurately compared to transcriptome profiling of entire sets of known protein-coding genes. Characterizing abnormal miRNA-expression signatures can lead toward development of tissue and biofluid-specific diagnostic markers and new types of oligonucleotide-based drugs. Golub and his colleagues were the first to publish their results on the comparison of global-expression profiling for human miRNA in multiple cancers. They used microbead technology and obtained expression data on several common human cancers (100). Their results present the groups of miRNAs that were differentially expressed in each type of cancer. They showed that each cancer type could be classified based on the expression signature of a relatively small number of miRNAs. Lee et al. (116) have reported a miRNA-expression signature for pancreatic cancer (116).
Furthermore, miRNA-expression profile serves as a useful biomarker for virus-infected cells. Using high-throughput microarray method, it has been shown that the expression of HIV-1 genes changes the miRNA profiles in human cells (95). miRNA expression in EBV is distinct for different stages of infection. Differential regulation of EBV miRNA expression implies distinct roles during infection of different human tissues (28). Furthermore, recent reports suggest the role of miRNA-expression profiles to prognosticate disease outcomes in virus infected (117,118). Current miRNA research has posed a serious challenge to decipher the code of miRNA regulation in normal and cancerous cells in order to find a connection between miRNA profiling results and specific phenotypic changes caused by these aberrations.
Hence, comprehensive miRNA databases serve as a source of latest information available about miRNA genes, their structure, function, potential target genes and expression data. This serves as a valuable platform for quick and efficient access to important information about the miRNAs of virus-infected cells. Table 3 lists the available miRNA-expression profile resources.
Table 3.
MicroRNA expression profile resources
| Resources | Websites | Descriptions |
|---|---|---|
| microRNA.org | http://www.microrna.org | A resource for predicted microRNA targets and expression |
| miRAS (miRNA Analysis System at Tsinghua University) | http://e-science.tsinghua.edu.cn/miras | A data processing system for miRNA-expression profiling study. |
| miRNAMap | http://miRNAMap.mbc.nctu.edu.tw/. | Collection of experimental-verified microRNAs and experimental- verified miRNA target genes in human, mouse, rat and other metazoan genomes and expression profiles of human miRNAs in 18 major normal tissues in human. |
| Argonaute | http://www.ma.uni-heidelberg.de/apps/zmf/argonaute/interface | A database for gene regulation by mammalian microRNAs |
| miRGator | http://genome.ewha.ac.kr/miRGator/ | Differential expression analysis for a compendium of miRNA- expression data |
THERAPEUTIC IMPLICATIONS
The discovery of RNAi has resulted in the development of a broad new class of targeted therapeutics called ‘short interfering RNAs’ (siRNAs). Commercialization of siRNA-based therapeutics has been taken up by different companies, which can be extended to miRNAs, which are naturally occurring counterparts of siRNAs. An intense activity in the biomedical research community is witnessed due to an increasing awareness of the importance of miRNAs. It is presumed that miRNAs are the next breakout class of therapeutic molecules following closely behind siRNA. Furthermore, miRNAs will have significant advantages over siRNAs in many applications (23). Professor Eugenia Wang at the University of Louisville has already developed miRNA microarrays targeting viral-specific miRNAs. Huang and his colleagues demonstrate that HIV uses miRNA-mediated down-regulation of viral protein expression to its own advantage. They have shown that strategies based on antisense miRNA constructs may provide therapeutic benefit against HIV (88,119). The novel concept of ‘miRNA Replacement Therapy’ has recently evolved, which involves synthetic miRNAs or miRNA mimetics toward treatment of diseases. With RNAi therapeutics on the cusp of entering the clinic, lots of companies have launched commercial venture toward development of miRNA-based drugs through the steps of feasibility testing to preclinical studies (120). Although there remain many obstacles to achieve success in targeting all latently infected cells in viral infections, that day is not too far when miRNA-mimics and miRNA antisense constructs could help in designing a new generation of antiviral drugs.
CONCLUSIONS
Past few years have witnessed tremendous progress towards understanding various aspects of miRNAs. The integral role of miRNAs in controlling various complex regulatory networks within a cell is gradually coming into limelight. It is seen that host-encoded miRNAs have both positive and negative modulatory effect on viral replication. On the contrary, virus uses their own miRNA-induced gene-silencing machinery to protect them against the cellular antiviral RNAi response and may even affect cellular gene expression. Furthermore, they often use the cellular miRNA pathway to their own advantage.
Discovery of the first virally encoded miRNA has been a recent event and essential information regarding miRNA-induced virus-host interaction related to viral pathogenesis is gradually coming up. Although miRNA-biogenesis pathway poses some serious problem for RNA viruses along with a group of DNA viruses (poxviruses) to encode miRNAs (4), it is possible to circumvent these problems by adopting arcane ways. The advent of mirtrons revealed the process of miRNA biogenesis that uses the splicing machinery to bypass Drosha cleavage in initial maturation (57). A thorough study of these mirtrons and their initial maturation process may lead us toward those nonconventional processes these RNA and poxviruses may adopt to process their miRNAs. However, presence of miRNAs in HIV-1(RNA virus) encoded by the TAR element makes it an attractive problem to find the impact of the HIV-1 TAR miRNAs on gene-expression programming of infected cells and elucidating their role in viral pathogenesis. Furthermore, identification of these miRNAs leads us to think in a new way to investigate the presence of miRNAs in other RNA viruses.
Viral miRNAs can function as orthologs of cellular miRNAs and thereby regulates the expression of numerous cellular mRNAs by means of target sites that are generally evolutionarily conserved. Like KSHV miR-K12-11, being an ortholog of cellular miR-155, the other suggested orthologs need to be investigated further for their functional analysis, which might contribute to more information regarding virus-host interaction.
Relentless effort is being incorporated to make advancement for elucidating miRNA-mediated virus-host interaction so as to develop certain therapeutic approaches to deliver antagomirs into cells for tackling a variety of diseases. Since certain viral miRNAs help them to evade host immune system, related antagomirs might influence viral tropism and viral replication.
All these reveals that miRNA-induced gene-silencing approach holds great promise for selectively inhibiting virus-specific genes or host genes for the treatment of viral infections. Hence, the most fundamental challenge lies toward understanding the entire landscape of the miRNA-mediated host–virus interaction at the molecular level, which will lead toward the development of effective non-toxic antiviral therapy.
FUNDING
Funding for open access charge: Indian Association for the Cultivation of Science, Kolkata (Department of Science and Technology, Government of India).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the reviewers for their useful comments and suggestions to improve the manuscript. We also thank Dr Angshuman Bagchi (Indiana University School of Medicine, Indianapolis, USA), Avijit Guha Roy (University of Buckingham, UK) and our colleagues for critical reading of the manuscript.
REFERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32(Database Issue):109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen BR. Viruses and microRNAs. Nat. Genet. 2006;38:S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh Z, Chakrabarti J, Mallick B. miRNomics—the bioinformatics of microRNA genes. Biochem. Biophys. Res. Commun. 2007;363:6–11. doi: 10.1016/j.bbrc.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 6.Mallick B, Ghosh Z, Chakrabarti J. MicroRNA switches in Trypanosoma brucei. Biochem. Biophys. Res. Commun. 2008;372:459–463. doi: 10.1016/j.bbrc.2008.05.084. [DOI] [PubMed] [Google Scholar]
- 7.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;8:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 8.Zhang R, Su B. MicroRNA regulation and the variability of human cortical gene expression. Nucleic Acids Res. 2008;36:4621–4628. doi: 10.1093/nar/gkn431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 10.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 12.Xing L, Kieff E. Epstein-Barr virus BHRF1 Micro- and stable RNAs during latency III and after induction of replication. J. Virol. 2007;81:9967–9975. doi: 10.1128/JVI.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in Murine Cytomegalovirus. J. Virol. 2007;81:13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiertz E, Hill A, Tortorella D, Ploegh H. Cytomegaloviruses use multiple mechanisms to elude the host immune response. Immunol. Lett. 1997;57:213–216. doi: 10.1016/s0165-2478(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan CS. New roles for large and small viral RNAs in evading host defences. Nat. Rev. Genet. 2008;9:503–507. doi: 10.1038/nrg2349. [DOI] [PubMed] [Google Scholar]
- 20.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berkhout B, Haasnoot J. The interplay between virus infection and the cellular RNA interference machinery. FEBS Lett. 2006;580:2896–2902. doi: 10.1016/j.febslet.2006.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi P, Han J, Lu Y, Wang C, Bu F. Virus-encoded microRNAs: future therapeutic targets? Cell. Mol. Immunol. 2006;3:411–419. [PubMed] [Google Scholar]
- 23.Sall A, Liu Z, Zhang HM, Yuan J, Lim T, Su Y, Yang D. MicroRNAs-based therapeutic strategy for virally induced diseases. Curr. Drug Discov. Technol. 2008;5:49–58. doi: 10.2174/157016308783769478. [DOI] [PubMed] [Google Scholar]
- 24.Nair V, Zavolan M. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 2006;14:169–175. doi: 10.1016/j.tim.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 2005;7:1684–1695. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 26.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schafer A, Cai X, Bilello JP, Desrosiers RC, Cullen BR. Cloning and analysis of microRNA encoded by the primate gamma herpes virus rhesus monkey rhadino virus. Virology. 2007;364:21–27. doi: 10.1016/j.virol.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J. Virol. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 2005;79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl Acad. Sci. USA. 2008;105:10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dölken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J. Virol. 2007;81:13771–13782. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burnside J, Bernberg E, Anderson A, Lu C, Meyers BC, Green PJ, Jain N, Isaacs G, Morgan RW. Marek's disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J. Virol. 2006;80:8778–8786. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Sewer A, Zavolan M, Nair V. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 2007;81:7164–7170. doi: 10.1128/JVI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Watson M, Nair V. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J. Virol. 2008;82:4007–4015. doi: 10.1128/JVI.02659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grey F, Nelson J. Identification and function of human cytomegalovirus microRNAs. J. Clin. Virol. 2008;41:186–191. doi: 10.1016/j.jcv.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral micro-RNAs in latently infected cells. Proc. Natl Acad. Sci. USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim DN, Chae H.-S, Oh ST, Kang J.-H, Park CH, Park WS, Takada K, Lee JM, Lee W.-K, Lee SK. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J. Virol. 2007;81:1033–1036. doi: 10.1128/JVI.02271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo AKF, To KF, Lo KW, Lung RWM, Hui JWY, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl Acad. Sci. USA. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 49.Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 51.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4467. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee Y, Kim M, Han J, Yeom K.-H, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 55.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 56.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 57.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 61.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 62.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 63.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebra fish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 65.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 66.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 67.Smalheiser NR. Regulation of mammalian microRNA processing and function by cellular signaling and subcellular localization. Biochim. Biophys. Acta. 2008;1779:678–681. doi: 10.1016/j.bbagrm.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barthelson RA, Lambert GM, Vanier C, Lynch RM, Galbraith DW. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics. 2007;8:340. doi: 10.1186/1471-2164-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Habig JW, Dale T, Bass BL. MiRNA editing—we should have Inosine this coming. Mol. Cell. 2007;25:792–793. doi: 10.1016/j.molcel.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 73.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rai D, Karanti S, Jung I, Dahia PL, Aguiar RC. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet. Cytogenet. 2008;181:8–15. doi: 10.1016/j.cancergencyto.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang J, Lee EJ, Schmittgen TD. Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes Cancer. 2006;45:103–106. doi: 10.1002/gcc.20264. [DOI] [PubMed] [Google Scholar]
- 78.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grey F, Meyers H, White EA, Spector DH, Nelson J. A Human Cytomegalovirus-Encoded microRNA regulates expression of multiple viral Genes involved in replication. Plos Pathogens. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cantello JL, Parcells MS, Anderson AS, Morgan RW. Marek's disease virus latency-associated transcripts belong to a family of spliced RNAs that are antisense to the ICP4 homolog gene. J. Virol. 1997;71:1353–1361. doi: 10.1128/jvi.71.2.1353-1361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McClure LV, Sullivan CS. Kaposi's sarcoma herpesvirus taps into a host microRNA regulatory network. Cell Host Microbe. 2008;3:1–3. doi: 10.1016/j.chom.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 82.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 84.Berkhout B, Jeang KT. RISCy business: MicroRNAs, pathogenesis, and viruses. J. Biol. Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 85.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 86.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl Acad. Sci. USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pedersen IM, Cheng G, Wieland S, Volinia V, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–923. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4 + T lymphocytes. Nat. Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 89.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 90.Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem. Biophys. Res. Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 91.Emerman M, Malim MH. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 92.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 94.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang K.-T, et al. HIV-1 Tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- 97.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, et al. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 99.Scaria V, Hariharan M, Maiti S, Pillai B, Brahmachari SK. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3:68. doi: 10.1186/1742-4690-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 101.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2006;34:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein M, Tuschl T, van Nimwegen E, Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nam JW, Shin KR, Han J, Lee Y, Kim VN, Zhang BT. Human micro-RNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res. 2005;33:3570–3581. doi: 10.1093/nar/gki668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xue C, Li F, He T, Liu GP, Li Y, Zhang X. Classification of real and pseudo microRNA precursors using local structure sequence features and support vector machine. BMC Bioinformatics. 2005;6:310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rajewsky N. MicroRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 107.Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J. Biol. Chem. 2004;279:42230–42239. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 108.Grimson AG, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, Piedade I, Gunsalus KC, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 110.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 114.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 115.Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 116.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin. Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 119.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat. Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Blow N. Small RNAs: delivering the future. Nature. 2007;450:1117–1120. doi: 10.1038/4501117a. [DOI] [PubMed] [Google Scholar]