Abstract
To date, the most prevalent model for transport of pre-proteins to plant mitochondria is based on the activity of an N-terminal extension serving as a targeting peptide. Whether the efficient delivery of proteins to mitochondria is based exclusively on the action of the N-terminal extension or also on that of other protein determinants has yet to be defined. A novel mechanism is reported here for the targeting of a plant protein, named MITS1, to mitochondria. It was found that MITS1 contains an N-terminal extension that is responsible for mitochondrial targeting. Functional dissection of this extension shows the existence of a cryptic signal for protein targeting to the secretory pathway. The first 11 amino acids of the N-terminal extension are necessary to overcome the activity of this signal sequence and target the protein to the mitochondria. These data suggest that co-operation of multiple determinants within the N-terminal extension of mitochondrial proteins may be necessary for efficient mitochondrial targeting. It was also established that the presence of a tryptophan residue toward the C-terminus of the protein is crucial for mitochondrial targeting, as mutation of this residue results in a redistribution of MITS1 to the endoplasmic reticulum and Golgi apparatus. These data suggest a novel targeting model whereby protein traffic to plant mitochondria is influenced by domains in the full-length protein as well as the N-terminal extension.
Keywords: Plant mitochondria, secretory pathway, targeting signals
Introduction
In eukaryotic cells, the presence of several distinct organelles generates the need for efficient protein targeting mechanisms of newly synthesized proteins. Targeting of most proteins destined to the secretory pathway is initiated by the binding of the signal recognition particle (SRP) to a signal sequence in a nascent polypeptide chain emerging from a cytosolic ribosome. The nascent polypeptide is then co-translationally inserted in the endoplasmic reticulum (ER) upon recognition of the SRP by an ER-membrane anchored SRP receptor (Nagai et al., 2003). The synthesis of proteins destined to other organelles, such as plastids and mitochondria, generally occurs on free ribosomes and the targeting is post-translational (reviewed in Alder and Johnson, 2004). Protein targeting to mitochondria relies on an N-terminal extension on the protein precursor, the so-called pre-sequence, which directs the protein to the organelles. Pre-sequences do not have a common primary sequence but are generally composed of an N-terminal leader sequence of 20–35 amino acids, enriched in basic, hydrophobic, and hydroxylated residues (Neupert, 1997; Schatz and Dobberstein, 1996). The pre-sequence appears to fold into a defined secondary structure. This folding is essential for the correct distribution of charged and apolar residues and is necessary for efficient protein import (Matouschek et al., 1997; Gaume et al., 1998). The N-terminal part of the pre-sequence forms a positively charged amphiphilic α-helix or β-sheet, whereas the C-terminal region probably serves as a recognition site for matrix proteases (Gavel and von Heijne, 1990; Neupert, 1997). Pre-sequences are generally cleaved off from precursors as they pass through the mitochondrial double membrane via the outer and inner membrane translocases (TOM and TIM complexes, respectively), or once inside the mitochondria by specific peptidases (for reviews see Neupert, 1997; Glaser et al., 1998).
Although it is generally assumed that import mechanisms are conserved in different organisms, import mechanisms into plant mitochondria appear to rely on several peculiarities that include proteins involved in the TIM and TOM complexes and the primary and secondary structures of the pre-sequences (for a review see Millar et al., 2006). Much information on mitochondrial protein targeting mechanisms has been gathered from the functional dissection of the N-terminal pre-sequences (Logan and Leaver, 2000; Chabregas et al., 2001; Duby et al., 2001). There is, however, little information on whether pre-sequences are the exclusive determinants for mitochondrial targeting or if other protein domains influence the efficiency of the process.
The question of whether the functional role of a mitochondrial pre-sequence may be influenced by distal amino acid residues within the full-length protein is addressed here. A novel nuclear-encoded Arabidopsis thaliana protein has been identified, called MITS1 (MItochondrial-Targeting Signal 1), which appears to be targeted to mitochondria. Live cell imaging analyses of the N-terminal extension of MITS1 and a series of MITS1-deletions fused to the yellow fluorescent protein (YFP) indicated that the N-terminal pre-sequence is responsible for the intracellular targeting of the protein. However, in contrast to the full-length peptide, a leaderless pre-sequence (lacking the first 11 amino acids) directed YFP protein fusions to the ER. Furthermore, mutation of a tryptophan residue at position 361 (W361A) resulted in the redistribution of MITS1 to the ER and Golgi apparatus, suggesting that mitochondrial targeting processes in plant cells may rely not only on the composition of the pre-sequence but also on that of other domains within the protein sequence.
Materials and methods
Plant material and transient expression systems
Four-week-old Nicotiana tabacum (cv. Petit Havana) greenhouse plants grown at 25 °C were used for Agrobacterium tumefaciens (strain GV3101)-mediated transient expression (Batoko et al., 2000). The bacterial optical density (OD600) used for plant leaf transformation was 0.05 for MITS1:YFP and its mutants and for β-ATPase:GFP, and 0.2 for ERD2:GFP.
Molecular cloning
Standard molecular techniques were used for subcloning (Sambrook et al., 1989). The fluorescent proteins used in this study were based on fusions with either mGFP5 (Haseloff et al., 1997) or EYFP (Clontech Inc., Palo Alto, CA, USA). The spectral properties of mGFP5 allow efficient spectral separation from YFP (Brandizzi et al., 2002). The ER/Golgi marker used in this study was the H/KDEL receptor ERD2 fused to GFP (Boevink et al., 1998). The mitochondrial marker β-ATPase:GFP was a generous gift of Dr DC Logan, University of St Andrews, UK (Logan and Leaver, 2000). The cDNA of MITS1 (At1g52080, Ref. NM_104089) was amplified by PCR from an ABRC clone. Point mutations and deletion mutants were created using site-directed mutagenesis. The binary vector pVKH18En6 (Batoko et al., 2000) was used for all the constructions in this study. All the prepared inserts were spliced upstream of YFP, using the unique XbaI and SalI sites of the vector (daSilva et al., 2004). A methionine residue was added to the N-terminus of the 12–39 deletion mutant to allow translation.
Bio-informatic, sampling, imaging, and quantification
The bio-informatic tools for the prediction of MITS1 targeting to mitochondria were Predotar (Small et al., 2004), iPSORT and PSORTII (Bannai et al., 2002), MitoPred (Guda et al., 2004), and SignalP (Nielsen et al., 1997). The simulation of the Helical Wheel Projection of the MITS1 N-terminal was from http://rzlab.ucr.edu/scripts/wheel/wheel.cgi (D Armstrong and R Zidovetzki).
Imaging was performed using an upright Zeiss Laser Scanning Confocal Microscope LSM510 META (Zeiss, Jena, Germany) with a ×63 water immersion objective. Transformed leaves were analysed 48 h after infection of the lower epidermis. For imaging expression of YFP constructs, GFP constructs or both, the imaging settings as described by Brandizzi et al. (2002) were used. Appropriate controls were used to exclude the possibility of energy transfer between fluorochromes and cross-talk. Images were acquired using non-saturating settings and the same imaging parameters were used. Post-acquisition image processing was carried out using CorelDraw12 software.
Results
MITS1 is efficiently targeted to plant mitochondria
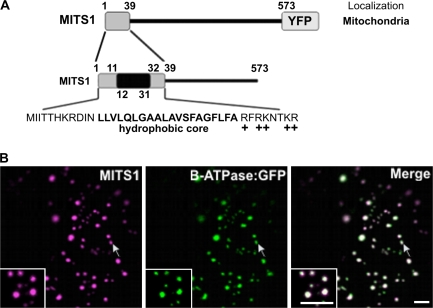
MITS1 (AGI: At1g52080) is a putative actin-binding protein of 573 amino acid residues with a predicted molecular mass of 66 kDa. The N-terminal region of this protein (39 amino acids) contains a hydrophobic stretch of 20 residues (predicted with TMHMM and TMPred (Hofmann and Stoffel, 1993; Krogh et al., 2001) and flanking regions enriched with positively-charged amino acids (Fig. 1A). Because of the predicted secondary structure of this sequence, various publicly available bio-informatics tools suggest targeting of MITS1 to mitochondria (see Materials and methods). To confirm this prediction experimentally, full-length MITS1 was fused to the N-terminus of the yellow fluorescent protein (YFP; Fig. 1A), and the resulting construct was expressed in tobacco leaf epidermal cells for live cell confocal microscopy analyses. For simplicity, this construct was named MITS1. MITS1 appeared in numerous structures of heterogeneous size and shape (Fig. 1B), resembling the previously described appearance of plant mitochondria (Logan and Leaver, 2000). Co-localization analyses of MITS1 with β-ATPase:GFP, a known mitochondrial marker (Logan and Leaver, 2000), confirmed that MITS1 localizes to mitochondria (Fig. 1B), thus providing experimental support for our prediction. These observations prompted us to carry out a functional dissection of the NH2-terminal extension of MITS1 to identify the targeting determinants within this region.
Fig. 1.
MITS1 harbours an N-terminal targeting signal and is localized to mitochondria. (A) Schematic representation of MITS1 and of its N-terminal region. Positively-charged residues follow a 20 residue hydrophobic region, characteristic of a mitochondrial targeting sequence. (B) In epidermal cells of tobacco leaves, MITS1:YFP labels punctate structures of various sizes that colocalize with the mitochondrial marker β-ATPase:GFP (arrows). Insets: magnified section of main panels. Scale bars=5 μm.
The N-terminal extension of MITS1 contains three regions that co-ordinate the mitochondrial targeting signal
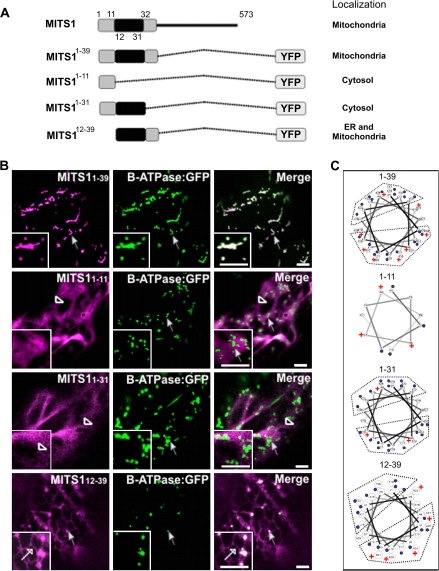
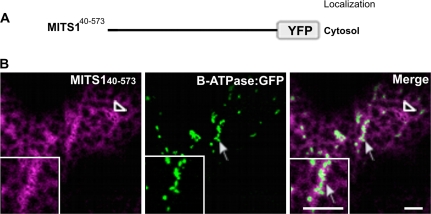
To explore the role of the N-terminal extension of MITS1, several defined segments of the pre-sequence were fused to YFP for confocal microscopy analyses (see Fig. 2A for a schematic representation). It was found that the YFP fusion to the predicted pre-sequence of MITS1 (MITS11–39) was localized to mitochondria, as confirmed by co-expression analyses with β-ATPase:GFP (Fig. 2B). A Helical Wheel Projection analysis of residues 1–39 (see Materials and methods) confirmed the presence of clusters of positive charges on one side of the helix (Fig. 2C), consistent with the known properties of pre-sequences in forming cationic amphipathic helices (Duby et al., 2001). The necessity of this pre-sequence for MITS1 mitochondrial targeting was further reinforced by the evidence that a YFP fusion to MITS1 amino acids 40-573 lacking the entire pre-sequence was cytosolic (Fig. 3).
Fig. 2.
Exploration of the N-terminal 39 residues of MITS1 reveals that a co-ordination of three regions is required for efficient mitochondria targeting. (A) Schematic representation of the N-terminal consecutive domains fused to YFP and their subsequent intracellular localizations. (B) Region 1–39 efficiently targets a YFP to mitochondria (arrow) and the YFP punctate structures fully co-localize with β-ATPase:GFP (arrows). 1–11:YFP (missing the central hydrophobic core and the positively-charged region) and 1–31:YFP (missing the positively-charged region) were localized to the cytosol (empty arrowheads). (C) The Helical Wheel Projection of MITS1 N-terminal pre-sequence shows a cationic cluster in 1–39 and 12–39 sequences (but not in the other pre-sequence truncations) consistent with their localization to mitochondria (blue dots are hydrophobic residues, + indicates positive charge). Insets: magnified section of main panels. Scale bars=5 μm.
Fig. 3.
Residues 1–39 of the N-terminal extension are required for MITS1 to reach mitochondria. (A) A schematic representation of MITS1 lacking the first 39 amino acids. (B) In the absence of the N-terminal pre-sequence, MITS1 was found in the cytosol (empty arrowhead), which in plant cells assumes a diffuse yet reticulated appearance. No co-localization was noticed with the mitochondrial marker, β-ATPase:GFP (arrow). Insets: magnified section of main panels. Scale bars=5 μm.
Having established that the pre-sequence of MITS1 is sufficient to redistribute a fluorescent protein to mitochondria, the next aim was to establish the functional role of different domains within this pre-sequence. When a YFP fusion to the first 11 amino acids preceding the central hydrophobic region of the pre-sequence, MITS11–11 (Fig. 2A) was expressed, it was found that the protein was distributed in the cytosol rather than to mitochondria (Fig. 2B). This suggests that this region is insufficient to target YFP to organelles efficiently.
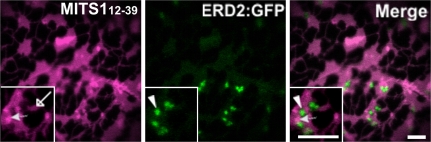
A fusion of the first 31 amino acids of the MITS1 to YFP, which lacked the positively charged region at the C-terminus of the pre-sequence, MITS11–31 (Fig. 2A), was also found in the cytosol (Fig. 2B). This is consistent with the required presence of the positively charged region in functional mitochondrial pre-sequences (Duby et al., 2001). Interestingly, however, the MITS112–39 peptide fusion (Fig. 2A), which was made by adding a methionine residue upfront to allow translation, and contained the hydrophobic core region and the positively-charged flanking region, was localized to the ER as well as the mitochondria (Figs 2B, 4). This suggests that the 12–39 amino acid region of MITS1 can function as a promiscuous targeting signal for microsomal and mitochondrial membranes.
Fig. 4.
MITS112–39 does not localize at Golgi bodies. To exclude the possibility that the punctate structures labelled by MITS112–39 peptide fusion were Golgi bodies, cells were cotransformed with the ER/Golgi marker, ERD2:GFP (Boevink et al., 1998) and MITS112–39. As shown in this figure, MITS112–39 labelled the ER (empty arrow), and dots (full arrow), which did not colocalize with the Golgi (arrowhead). These dots corresponded to mitochondria as shown in Fig. 2. Insets: magnified section of main panels. Scale bars=5 μm.
Taken together, these data indicate that the first 39 amino acids of MITS1 constitute a pre-sequence for efficient mitochondrial targeting. They also suggest that the first 11 amino acids of the pre-sequence alone appear to be necessary for directing the protein to mitochondria rather than to the ER, despite being insufficient for targeting a YFP fusion to mitochondria.
The data presented above mirrored the Helical Wheel Projection analyses of each peptide that showed that peptide 12–39 has the most cationic charges on the side of the helix after peptide 1–39, followed by peptides 1–31 and 1–11 (Fig. 2C). Consistent with these Helical Wheel projections, peptides 1–39 and 12–39 were capable of targeting YFP to the mitochondria, although a subcellular pool of the 12–39 YFP fusion was also directed to the ER. On the other hand, the remaining peptides (1–11 and 1–31) were not sufficient for directing YFP to any organelle, most likely due to the absence of a defined cationic side on the helical structure.
Mutation of tryptophan 361 redistributes MITS1 to the ER and Golgi apparatus
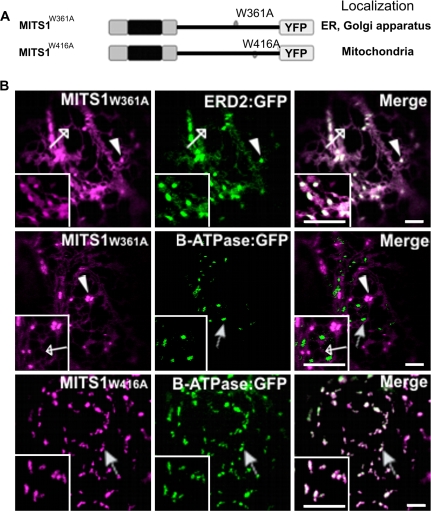
Having demonstrated that the N-terminal extension of MITS1 functions alone as an efficient targeting signal to mitochondria, the next aim was to determine whether other domains of the full-length protein could influence the activity of this pre-sequence. Within the MITS1 sequence two tryptophan residues (W361 and W416) were found. Tryptophan is a rare amino acid that is known to be involved in the targeting and stability of some proteins (Garcia et al., 1992; Kleinberger-Doron and Kanner, 1994; Hoffman et al., 2006). Therefore it was decided to test whether mutation of either of these residues in the full-length MITS1 would affect the mitochondrial targeting of MITS1. Each of the two tryptophan residues was changed to alanine (W361A and W416A, see Fig. 5A for a schematic representation) and the constructs were expressed as YFP-fusions in plants. Confocal microscopy analyses revealed that the mutant MITS1W361A (Fig. 5B) was localized at the ER and the Golgi apparatus, as demonstrated by co-expression of the mutant with the known ER and Golgi marker, ERD2:GFP (Boevink et al., 1998); Fig. 5B). Colocalization analyses with β-ATPase:GFP further excluded the possibility that the punctate structures labelled by MITS1W361A were mitochondria (Fig. 5B). On the other hand, a MITS1 bearing mutation of the tryptophan residue at position 416 (MITS1W416A) was localized at mitochondria labelled with β-ATPase:GFP (Fig. 5B) in the same manner as wild-type MITS1 (Fig. 1B). These data suggest that the integrity of specific amino acids that are distal from the N-terminal domain of a mitochondrial protein affect the function of a pre-sequence in a dominant fashion.
Fig. 5.
Tryptophan 361 influences the activity of the MITS1 N-terminal pre-sequence. (A) Schematics of the mutations within MITS1 fusions to YFP. (B) Confocal images of tobacco leaf epidermal cells coexpressing a MITS1:YFP mutant and either ERD2:GFP or β-ATPase:GFP. MITS1W361A:YFP was found in the ER (empty arrow) and Golgi apparatus (arrowhead) as confirmed by the ER/Golgi apparatus marker ERD2:GFP (ER, empty arrow; Golgi apparatus, arrowhead). The mutation of tryptophan 416 to alanine did not affect the distribution of MITS1 to mitochondria (arrow). Insets: magnified section of main panels. Scale bars=5 μm.
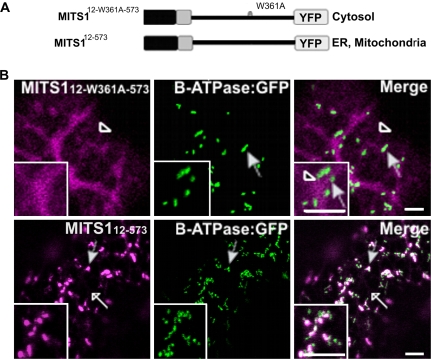
Finally, since it was shown that the pre-sequence lacking the first 11 amino acids (peptide 12–39) is capable of directing a YFP fusion to the ER and mitochondria (Fig. 2B), we wanted to explore the possibility of whether the W361A mutation, which affects the activity of the pre-sequence (Fig. 5), would also interfere with the targeting properties of peptide 12–39. To investigate this, a YFP fusion was constructed of the MITS1W361A mutant lacking the first 11 residues of full-length MITS1 (MITS112-W361A-573; Fig. 6A). Differently from the ER/mitochondria targeting of MITS112–573 (Fig. 6B; see Supplementary Fig. S1 at JXB online), the resulting chimera was found in the cytosol (Fig. 6B). As incorporation of the alanine residue in position 361 must occur after the synthesis of the N-terminal 12–39 sequence which is responsible for ER and mitochondria targeting, these data further strengthen our hypothesis that distal protein residues may influence targeting properties of an N-terminal sequence.
Fig. 6.
Tryptophan 361 mutation influences the behaviour of a truncated MITS1. (A) Schematic representation of the MITS112–573 constructs. (B) Confocal images of tobacco leaf epidermal cells show distribution of MITS112-W361A-573:YFP in the cytosol (empty arrowhead) but no colocalization with β-ATPase:GFP. MITS112–573 was found in the ER (empty arrows) and dots. Most of these colocalized with mitochondria (full arrows) but not with the Golgi (see Supplementary Fig. S1 at JXB online). Insets: magnified section of main panels. Scale bars=5 μm.
Discussion
The pre-sequence amphipathicity influences the targeting of MITS1
At present, the biological function of MITS1 remains unknown, but publicly available databases (NCBI and TAIR) indicate MITS1 as a putative actin-binding protein, with an ‘actinin-type actin-binding domain signature 1’ that is similar to a region involved in the actin-binding activity of the chloroplastic actin-binding protein, CHUP1 (Oikawa et al., 2003). In addition, several different types of protein domain prediction software revealed that MITS1 contains an AAA-ATPase motif (55–360; SMART, Schultz et al., 1998; Letunic et al., 2006), and two leucine zipper patterns (279–300 and 324–345; ScanProsite, (Hulo et al., 2008). In contrast with CHUP1, MITS1 is localized to mitochondria. Helical Wheel analysis strongly suggests that the pre-sequence of MITS1 attains a secondary structure with a clustered cationic side that is a known property of mitochondria targeting sequences (Duby et al., 2001). The N-terminal part of the mitochondrial pre-sequence is believed to fold into an amphiphilic α-helix both in a phospholipid environment and in vivo (Roise et al., 1988; Lemire et al., 1989). Consistent with these observations, the MITS1 mutants with a less defined polar cluster on the side of the helical wheel, compared with the 1–39 peptide, showed a reduced ability to target YFP to mitochondria. Although stability of the α-helix is important for maintenance of mitochondrial import (Hammen et al., 1996; Heard and Weiner, 1998), in vivo mutagenesis analysis of a plant pre-sequence from the β-subunit of the F1-ATPsynthase from Nicotiana plumbaginifolia showed that the N-terminal helical structure of the pre-sequence is necessary but not sufficient for efficient mitochondrial import, and that its hydrophobic residues play an essential role in in vivo mitochondrial targeting (Duby et al., 2001). These findings may explain how peptide 12–39, which maintained a similar cluster of cationic amino acids on the side of the helix in comparison to the full-length pre-sequence, showed targeting to the mitochondria and also to other organelles. It is possible that the distribution of the hydrophobic residues of peptide 12–39 may be altered, resulting in a less efficient targeting signal in comparison to the full-length pre-sequence.
The MITS1 pre-sequence contains promiscuous targeting signals
The evidence that peptide 12–39 is distributed to the ER in addition to mitochondria, suggests that the first 11 amino acids of the N-terminal extension are necessary to overcome protein mistargeting to the ER. Whether the putative signal sequence masked by the first 11 amino acids is functional for MITS1 in planta is unknown, but the presence of multiple targeting signals in the same protein sequence has been reported for certain post-translationally targeted proteins containing either a nuclear localization signal (AtLIG1; Sunderland et al., 2006) or peroxisomal signal (FPS protein; Martin et al., 2007). In these cases, the function of the mitochondrial pre-sequence resulted in a dominant effect over the other sequences. It has also been demonstrated that chimeric signals may be functional in directing proteins to different organelles in the same cell. This is the case for chloroplast and plant mitochondrial proteins (Brink et al., 1994; Pujol et al., 2007), which, to some extent, have similar targeting sequences. Interestingly, it has been demonstrated that some chimeric signal sequences may retain the ability for ER and mitochondria targeting. For example, for the biogenesis of the hepatic P4501A1 isoenzyme a microsomal signal sequence may be cleaved to activate a mitochondrial targeting signal (Addya et al., 1997). In this experimental system, we did not observe ER localization of either a pre-sequence-YFP fusion or full-length MITS1-YFP fusion but it will be interesting to determine whether the activation of the microsomal signal sequence in MITS1 may occur in planta (i.e. the activation may be development or stress-regulated), as this may represent a novel protein targeting mechanism.
Targeting activity of the MITS1 pre-sequence is influenced by a distal amino acid residue
Our results indicate that the integrity of an amino acid residue placed distally from the N-terminal extension is a factor that influences the activity of the mitochondrial pre-sequence. This phenomenon was specific to tryptophan in position 361 as the tryptophan in position 416 did not appear to affect MITS1 targeting to mitochondria. These data support the suggestion that, in plant cells, the nature of the mature protein can also affect the targeting properties of the pre-sequence (Lee and Whelan, 2004). As mitochondrial targeting is a process that occurs post-translationally and involves interaction of the newly synthesized proteins with cytosolic chaperones (Young et al., 2003; Yano et al., 2004), it is possible that the integrity of distal domains of pre-proteins is important for a productive surface interaction with these chaperones for efficient targeting.
A similar explanation, however, is not sufficient to explain the cytosolic distribution of the MITS1 mutant that lacks the first 11 amino acids of the N-terminal extension and bears a mutation of the tryptophan in position 361 (MITS112-W361A-573). This is because peptide 12–39 and MITS112–573 were found to be sufficient to target YFP to the ER. As the synthesis of the distal portion of MITS1 containing tryptophan 361 should occur after import of the N-terminal region into the ER during co-translational translocation, the tryptophan 361 mutation should not interfere with the distribution of the mutant to the ER. One possibility is that the 12–39 peptide associates with the cytosolic face of the ER, rather than facilitating translocation through the ER membrane.
In conclusion, a novel aspect of mitochondrial protein targeting in plants has been demonstrated that encompasses functional co-ordination of the pre-sequence and the integrity of a distal amino acid residue. Further studies in an endogenous context are required to gain an understanding of whether the signal sequence that appears in the N-terminal extension of MITS1 depends on mechanisms that are known to affect targeting of a protein to different organelles [i.e. alternative splicing and/or differential initiation of translation (Ma and Taylor, 2008; Christensen et al., 2005)].
Supplementary data
Supplemental material can be found at JXB online.
Supplementary Fig. S1. MITS112–573 does not localize to Golgi bodies.
Supplementary Material
Acknowledgments
This work was supported by grants awarded to FB from the Department of Energy at Michigan State University, the Canada Foundation for Innovation (CFI), the Canada Research Chair fund, and the Natural Science and Engineering Research Council of Canada (NSERC). LAM was supported by an NSERC post-doctoral fellowship. ASJ was supported by an NSERC undergraduate summer research award. We thank Dr DC Logan (University of St Andrews, UK) for providing β-ATPase:GFP DNA.
References
- Addya S, Anandatheerthavarada HK, Biswas G, Bhagwat SV, Mullick J, Avadhani NG. Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2. Journal of Cell Biology. 1997;139:589–599. doi: 10.1083/jcb.139.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder NN, Johnson AE. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. Journal of Biological Chemistry. 2004;279:22787–22790. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I. A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. The Plant Cell. 2000;12:2201–2218. doi: 10.1105/tpc.12.11.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. The Plant Journal. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C. Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. The Plant Cell. 2002;14:1293–1309. doi: 10.1105/tpc.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink S, Flugge UI, Chaumont F, Boutry M, Emmermann M, Schmitz U, Becker K, Pfanner N. Preproteins of chloroplast envelope inner membrane contain targeting information for receptor-dependent import into fungal mitochondria. Journal of Biological Chemistry. 1994;269:16478–16485. [PubMed] [Google Scholar]
- Chabregas SM, Luche DD, Farias LP, Ribeiro AF, van Sluys MA, Menck CF, Silva-Filho MC. Dual targeting properties of the N-terminal signal sequence of Arabidopsis thaliana THI1 protein to mitochondria and chloroplasts. Plant Molecular Biology. 2001;46:639–650. doi: 10.1023/a:1011628510711. [DOI] [PubMed] [Google Scholar]
- Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. The Plant Cell. 2005;17:2805–2816. doi: 10.1105/tpc.105.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva LL, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. The Plant Cell. 2004;16:1753–1771. doi: 10.1105/tpc.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G, Oufattole M, Boutry M. Hydrophobic residues within the predicted N-terminal amphiphilic alpha-helix of a plant mitochondrial targeting presequence play a major role in in vivo import. The Plant Journal. 2001;27:539–549. doi: 10.1046/j.1365-313x.2001.01098.x. [DOI] [PubMed] [Google Scholar]
- Garcia JC, Strube M, Leingang K, Keller K, Mueckler MM. Amino acid substitutions at tryptophan 388 and tryptophan 412 of the HepG2 (Glut1) glucose transporter inhibit transport activity and targeting to the plasma membrane in Xenopus oocytes. Journal of Biological Chemistry. 1992;267:7770–7776. [PubMed] [Google Scholar]
- Gaume B, Klaus C, Ungermann C, Guiard B, Neupert W, Brunner M. Unfolding of preproteins upon import into mitochondria. EMBO Journal. 1998;17:6497–6507. doi: 10.1093/emboj/17.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Engineeringineering. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- Glaser E, Sjoling S, Tanudji M, Whelan J. Mitochondrial protein import in plants. Signals, sorting, targeting, processing, and regulation. Plant Molecular Biology. 1998;38:311–338. doi: 10.1023/a:1006020208140. [DOI] [PubMed] [Google Scholar]
- Guda C, Fahy E, Subramaniam S. MITOPRED: a genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics. 2004;20:1785–1794. doi: 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]
- Hammen PK, Waltner M, Hahnemann B, Heard TS, Weiner H. The role of positive charges and structural segments in the presequence of rat liver aldehyde dehydrogenase in import into mitochondria. Journal of Biological Chemistry. 1996;271:21041–21048. [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard TS, Weiner H. A regional net charge and structural compensation model to explain how negatively charged amino acids can be accepted within a mitochondrial leader sequence. Journal of Biological Chemistry. 1998;273:29389–29393. doi: 10.1074/jbc.273.45.29389. [DOI] [PubMed] [Google Scholar]
- Hoffman GG, Sona S, Bertin M, Ellington WR. The role of an absolutely conserved tryptophan residue in octamer formation and stability in mitochondrial creatine kinases. Biochimica et Biophysica Acta. 2006;1764:1512–1517. doi: 10.1016/j.bbapap.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase: a database of membrane spanning protein segments. Biological Chemistry Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Research. 2008;36:D245–249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger-Doron N, Kanner BI. Identification of tryptophan residues critical for the function and targeting of the γ-aminobutyric acid transporter (subtype A) Journal of Biological Chemistry. 1994;269:3063–3067. [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lee MN, Whelan J. Identification of signals required for import of the soybean F(A)d subunit of ATP synthase into mitochondria. Plant Molecular Biology. 2004;54:193–203. doi: 10.1023/B:PLAN.0000028787.36766.80. [DOI] [PubMed] [Google Scholar]
- Lemire BD, Fankhauser C, Baker A, Schatz G. The mitochondrial targeting function of randomly generated peptide sequences correlates with predicted helical amphiphilicity. Journal of Biological Chemistry. 1989;264:20206–20215. [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Research. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Leaver CJ. Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. Journal of Experimental Botany. 2000;51:865–871. [PubMed] [Google Scholar]
- Ma Y, Taylor SS. A molecular switch for targeting between endoplasmic reticulum (ER) and mitochondria: conversion of a mitochondria-targeting element into an ER-targeting signal in DAKAP1. Journal of Biological Chemistry. 2008;283:11743–11751. doi: 10.1074/jbc.M710494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Piulachs MD, Cunillera N, Ferrer A, Belles X. Mitochondrial targeting of farnesyl diphosphate synthase is a widespread phenomenon in eukaryotes. Biochimica et Biophysica Acta. 2007;1773:419–426. doi: 10.1016/j.bbamcr.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Matouschek A, Azem A, Ratliff K, Glick BS, Schmid K, Schatz G. Active unfolding of precursor proteins during mitochondrial protein import. EMBO Journal. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Small I. Recent surprises in protein targeting to mitochondria and plastids. Current Opinion in Plant Biology. 2006;9:610–615. doi: 10.1016/j.pbi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Nagai K, Oubridge C, Kuglstatter A, Menichelli E, Isel C, Jovine L. Structure, function and evolution of the signal recognition particle. EMBO Journal. 2003;22:3479–3485. doi: 10.1093/emboj/cdg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annual Review of Biochemistry. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M. Chloroplast unusual positioning1 is essential for proper chloroplast positioning. The Plant Cell. 2003;15:2805–2815. doi: 10.1105/tpc.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C, Marechal-Drouard L, Duchene AM. How can organellar protein N-terminal sequences be dual targeting signals? In silico analysis and mutagenesis approach. Journal of Molecular Biology. 2007;369:356–367. doi: 10.1016/j.jmb.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Roise D, Theiler F, Horvath SJ, Tomich JM, Richards JH, Allison DS, Schatz G. Amphiphilicity is essential for mitochondrial presequence function. EMBO Journal. 1988;7:649–653. doi: 10.1002/j.1460-2075.1988.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. New York: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences, USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Sunderland PA, West CE, Waterworth WM, Bray CM. An evolutionarily conserved translation initiation mechanism regulates nuclear or mitochondrial targeting of DNA ligase 1 in Arabidopsis thaliana. The Plant Journal. 2006;47:356–367. doi: 10.1111/j.1365-313X.2006.02791.x. [DOI] [PubMed] [Google Scholar]
- Yano M, Terada K, Mori M. Mitochondrial import receptors Tom20 and Tom22 have chaperone-like activity. Journal of Biological Chemistry. 2004;279:10808–10813. doi: 10.1074/jbc.M311710200. [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.