Abstract
Anthocyanins, flavan-3-ols, and flavonols are the three major classes of flavonoid compounds found in grape berry tissues. Several viticultural practices increase flavonoid content in the fruit, but the underlying genetic mechanisms responsible for these changes have not been completely deciphered. The impact of post-veraison sunlight exposure on anthocyanin and flavonol accumulation in grape berry skin and its relation to the expression of different transcriptional regulators known to be involved in flavonoid synthesis was studied. Treatments consisting of removing or moving aside the basal leaves which shade berry clusters were applied. Shading did not affect sugar accumulation or gene expression of HEXOSE TRANSPORTER 1, although in the leaf removal treatment, these events were retarded during the first weeks of ripening. Flavonols were the most drastically reduced flavonoids following shading and leaf removal treatments, related to the reduced expression of FLAVONOL SYNTHASE 4 and its putative transcriptional regulator MYB12. Anthocyanin accumulation and the expression of CHS2, LDOX, OMT, UFGT, MYBA1, and MYB5a genes were also affected. Other regulatory genes were less affected or not affected at all by these treatments. Non-transcriptional control mechanisms for flavonoid synthesis are also suggested, especially during the initial stages of ripening. Although berries from the leaf removal treatment received more light than shaded fruits, malvidin-3-glucoside and total flavonol content was reduced compared with the treatment without leaf removal. This work reveals that flavonol-related gene expression responds rapidly to field changes in light levels, as shown by the treatment in which shaded fruits were exposed to light in the late stages of ripening. Taken together, this study establishes MYB-specific responsiveness for the effect of sun exposure and sugar transport on flavonoid synthesis.
Keywords: bHLH, flavonoids, grape, leaf removal, MYB12, PAR, sugar, source, sink, WDR
Introduction
Grapes (Vitis vinifera L.), both for fresh and wine consumption, are an important source of flavonoids, including anthocyanins, flavonols, and flavan-3-ols. These molecules are particularly relevant in this fruit species since they define colour (Somers and Evans, 1974) and affect taste (Baxter et al., 1997; Vidal et al., 2003; Hufnagel and Hofmann, 2008). In addition, they possess a high antioxidant capacity and contribute to protection against cardiovascular diseases and cancer (reviewed by Lin and Weng, 2006) when consumed as part of a Mediterranean diet. In order to increase berry flavonoid content in the vineyard, it is fundamental to understand the biosynthesis of these molecules and how this is affected by the environment and different viticultural practices.
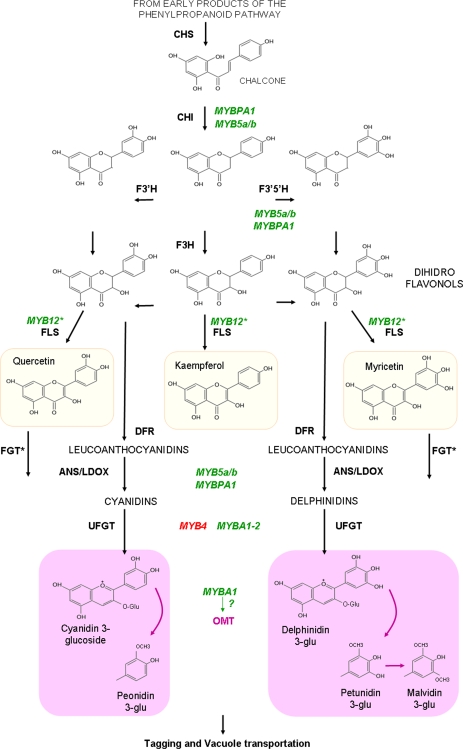
Flavonoid biosynthesis is derived from the phenylpropanoid pathway (Fig. 1), one of the most-characterized secondary metabolic routes in plant systems. Although different groups of proteins are responsible for producing, transporting, and storing flavonoids, the two most-studied classes correspond to the biosynthetic (structural) enzymes and the transcription factors controlling the pathway. Flavonoid transcriptional regulators have been extensively studied in plant species such as maize, petunia, Arabidopsis and, lately, in grapes and apples. From all the possible transcription factors responsible for controlling the pathway, R2R3 MYB, β helix-loop-helix (bHLH), and trypthophan-aspartic acid repeat (WDR) proteins have been the most-extensively analysed (Payne et al., 2000; Baudry et al., 2004).
Fig. 1.
Simplified overview of flavonol and anthocyanin biosynthesis within the phenylpropanoid pathway and its regulation in grape by characterized MYB genes (flavan-3-ols are not shown in this pathway). The repressor MYB4 is shown in red, while all MYB activators are shown in green. Abbreviations: CHS, chalcone synthase; CHI, chalcone isomerase; F3H/F3′H/F3′5′H, flavonoid hydroxylases; DFR, dihydroflavonol-4-reductase; ANS/LDOX, anthocyanidin synthase/leucoanthocyanidin dioxygenase; UFGT, UDP glucose:flavonoid-3-O-glucosyltransferase; FLS, flavonol synthase; and OMT, O-methyltransferase. (This figure is available in colour at JXB online.)
In grapes, some MYB genes have been shown to be involved in flavonoid metabolism (Fig. 1). In particular, many of the white grape cultivars present in the world today arose from multiallelic mutations of the MYBA1 and MYBA2 genes (Kobayashi et al., 2004; Lijavetzky et al., 2006; Walker et al., 2006, 2007; This et al., 2007), which control the last biosynthetic step of anthocyanin synthesis, a glycosylation reaction mediated by the UDP-GLUCOSE FLAVONOID 3-O-GLUCOSYLTRANSFERASE (UFGT) enzyme (Kobayashi et al., 2002). MYB5a (MYBCS-1; Deluc et al., 2006), MYB5b (Deluc et al., 2008), MYBPA1 (Bogs et al., 2007) and MYBPA2 (Terrier et al., 2009) appear to regulate general branches of the pathway (Fig. 1), together with flavan-3-ol synthesis. In grapes, an AtMYB4 homologue (Genbank accession EF113078), found to be a repressor of UFGT (JT Matus et al., unpublished results) was recently isolated and characterized. A putative flavonol-related MYB transcription factor (MYB12; Genbank accession FJ418175) was also found while performing a genome-wide analysis of grape MYB members (Matus et al., 2008). Co-activators belonging to other transcription factor families (bHLH and WDR) have also been isolated recently (JT Matus et al., unpublished results). From all these genes in grape, only MYBA genes have been studied in terms of their modulation by light and hormonal factors (Jeong et al., 2004), as well as by temperature (Mori et al., 2007).
The impact of environmental factors and viticultural practices on the flavonoid content and composition of grape berries has been widely studied in diverse wine-producing regions (reviewed by Downey et al., 2006). Light and all those practices which promote its incidence on berries throughout grape ripening, significantly increase the accumulation of flavonoids (Cortell and Kennedy, 2006) and the expression of their biosynthetic genes (Downey et al., 2004; Jeong et al., 2004). Light-induced flavonoid synthesis requires changes in gene expression mediated by three major classes of photoreceptors: phytochromes, blue/UV-A light receptors, and UV-B light receptors (reviewed by Argüelo-Astorga and Herrera-Estrella, 1998). The best-characterized light receptors in plants are phytochromes (PHY; Quail, 1994). These receptors are able to promote gene expression by three different signal transduction pathways. Of these, the cyclic GMP (cGMP)-mediated pathway regulates genes such as those involved in anthocyanin biosynthesis and CHALCONE SYNTHASE (CHS) was the first gene shown to be dependent on PHY-cGMP signalling (Bowler et al., 1994; Millar et al., 1994; Christie and Jenkins, 1996).
In addition to light, temperature also influences flavonoid production, although in a negative manner. Mori et al. (2007) demonstrated that high temperature increases anthocyanin degradation in grape skin, together with a decrease in expression of flavonoid biosynthetic and MYBA genes. On the other hand, low temperature increases anthocyanin production, as has been observed in grape (Mori et al., 2005; Yamane et al., 2006) and other crop species (maize: Christie et al., 1994; red orange: Lo Piero et al., 2005; apple: Ubi et al., 2006).
Field practices which control vegetative growth of grapevines directly affect the incidence of light on grape clusters. These include shoot, tip, and leaf removal. Leaf removal is generally recommended between the onset of ripening (veraison) and harvest, eliminating approximately one-third of the basal leaves. This practice is applied with the purpose of balancing foliage vigour relative to fruit production, increasing exposure of clusters to sunlight, facilitating ventilation, and diminishing the incidence of fungal diseases. Considering the fact that photosynthetic activity of basal leaves is lower than that of intermediate and apical leaves after berries begin to soften (Hunter and Visser, 1989), post-veraison leaf removal should increase light incidence on the clusters, without significantly affecting the photosynthetic activity of the whole plant. From this period onwards, sugars transported into grapes originate mainly from intermediate and apical leaves (Hunter and Visser, 1989). However, the effect of the time after veraison at which this practice is applied could have a substantial impact on grape physiology, since basal leaves lose their photosynthetic activity gradually during the ripening period.
In addition to this unresolved issue, there are no studies which correlate the changes observed in the content of grape flavonoids under different environmental conditions, with the expression of regulatory genes other than MYBA genes. In this work, the relationship between the expression of members of different transcription factor families and their target genes and flavonoid synthesis was studied under different post-veraison light exposure and leaf removal treatments.
Materials and methods
Experimental design and berry sampling throughout ripening
Different treatments were applied to a commercial Cabernet Sauvignon field at veraison (onset of ripening), located in the Maipo Valley, Chile (33°36’ S, 70°39’ W), during the 2006/2007 growing season. The vines were 10-year-old, drip-irrigated, and grown on their own roots using a bilateral cordon with a vertical shoot positioning trellis system in a north–south row orientation. Plants possessed a medium vigour, as classified by morpho-physiological measurements described in Peña-Neira et al. (2004). Normal commercial irrigation was homogenously applied at 66% potential evapo-transpiration. Nitrogen fertilization was applied during flowering at 66 kg ha−1 and plants were pruned before bud burst leaving two-node spurs per metre.
Veraison was determined as the time at which clusters were 30–50% coloured and sugar concentration reached c. 5° Brix (5% w/w soluble solids). At this stage, treatments T1 (light exposed) and T4 (leaf removal) were imposed in order to increase the sunlight exposure of grape clusters (Fig. 2A). In T1, the basal leaves shading each cluster were moved into a different position by the use of nylon zip-ties (see Supplementary Fig. S1 at JXB online), while in T4, 80% of the basal leaves (those from the first third of each shoot with clusters) were removed. Leaf moving in T2 (delayed sunlight exposure) was applied during the sixth week after veraison (Fig. 2B), while clusters from T3 (shaded cluster) were untreated.
Fig. 2.
(A) Experimental design and data sampling for different light exposure and leaf removal treatments. Coloured clusters represent the grape phenologies observed during the different periods of ripening. Even-numbered weeks (squares) were sampled for HPLC flavonoid analysis, while odd-numbered weeks (asterisks) were sampled for RNA extraction and gene expression quantification. Symbols on the right correspond to each treatment as used in Figs 4–7. (B) Field photograph of grapes before and after T3 treatment (leaves were moved aside but not removed). (This figure is available in colour at JXB online.)
Grape clusters from the east side of each experimental row (exposed to sunlight during the morning until midday) were treated and sampled. In each biological replicate (row), ten grape clusters from six plants were used for treatments T1, T2, and T3. Treatments were imposed altogether in each row. A contiguous plant to each row was exclusively used for the T4 (leaf removal) treatment. A total of 60 berries were sampled weekly from ten grape clusters for 8 weeks after veraison. Weeks 2, 4, 6, and 8 were considered for HPLC analysis while weeks 1, 3, 5, and 7 were for RNA extraction and gene expression quantification by real-time PCR (Fig. 2A). Berries were immediately peeled and deseeded. Berry skins were frozen in liquid nitrogen, and stored at –80 °C until required for RNA extraction.
By the ninth week after veraison (technical maturity or commercial harvest), clusters which hadn't been sampled but were treated, were used for physical and chemical analyses. The weight of 200 berry skins, and the pH and soluble solid content of berry juice were recorded, the latter being determined by means of a temperature compensated digital refractometer (Atago, Japan).
Flavonoid content analysis
Berry phenolics were extracted as in Venencie et al. (1997), with modifications. Berry skin samples (n=60) were weighed and ground with 15 ml distilled water, 20 ml hydroalcoholic solution (EtOH:H2O, 10:90 v/v) and 2.5 g tartaric acid, adjusting the final solution weight to 100 g. Extracts were macerated for 2 h at 30 °C by means of an orbital shaker, centrifuged, and filtered through glass microfibre. Samples were filtered through a 0.45 μm membrane under vacuum at <35 °C. A 2 ml aliquot was used to screen absorbance at 520 nm and 280 nm to quantify anthocyanins and phenolic compounds, respectively, as described by García-Barceló (1990). 150 μl of each sample were then injected into the HPLC-DAD for the analysis of anthocyanin compounds (Peña-Neira et al., 2007).
Non-anthocyanin compounds were extracted from an aliquot (50 ml) of macerated and filtered grape skins, by mixing the sample three times with 20 ml diethyl ether and 20 ml ethyl acetate. The organic fractions were then combined and extracts were evaporated to dryness under vacuum at <35 °C. The residue was dissolved in 1 ml methanol/water (1:1, v/v), and analysed by HPLC-DAD and HPLC-DAD-MS as described by Peña-Neira et al. (2000, 2004). 20 μl of each sample were injected.
The chromatographic system for HPLC-DAD analysis of anthocyanins consisted of an HPLC equipped with a 991 photodiode-array detector (Waters Corp. Milford, MA, USA) using a Chromolith Performance RP-18 (4.6×100 mm) column. The detection was carried out by scanning from 210 to 600 nm. The elution gradient consisted of the following solvents. Solvent A: water; solvent B: water/formic acid (5%, v/v), solvent C: acetonitrile, starting from 0 to 10 min, 77–50% B; 3–30% C; 10–12 min, 100% C at a constant flow of 3 ml min−1. The same liquid chromatography system equipment was used for non-anthocyanin compound analysis. Separation was performed on a reverse-phase Waters Nova-Pack C18 (300×3.9 mm ID) with 4 packing. Two mobile phases were employed for elution. (A) Water/acetic acid (98:2 v/v) and (B) water/acetonitrile/acetic acid (78:20:2 by vol.). The gradient profile was 0–55 min, 100–20% A; 55–70 min, 20–10% A; 70–90 min, 10–0% A. Detection was performed by scanning from 210 to 360 nm with an acquisition speed of 1 s. Samples were analysed in duplicate.
The identification of derivate flavonol and anthocyanin compounds (see Supplementary Fig. S2 at JXB online) was carried out by comparison of their spectra and retention time with those obtained by Peña-Neira et al. (2004, 2007). The standards were purchased from Apin Chemicals (Abingdon, Oxford, UK), Sigma Chemicals (Poole, Dorset, UK), and Merck (Darmstadt, Germany): for flavonols, myricetin-3-O-galactoside, myricetin-3-O-glucoside, isorhamnetin-3-O-galactoside, quercetin-3-galactoside, quercetin-3-rutinoside, quercetin-3-glucoside, quercetin-3-rhamnoside, kaempferol-3-galactoside, and kaempferol-3-glucoside were used. For malvidin-3-glucoside the standard was purchased from Extrasynthése (Lyon, France). Quantitative determinations were performed using the external standard method with commercial standards. The flavonol and anthocyanin calibration curves were obtained at 280 nm and 520 nm, respectively, by injection of different volumes of standard solutions under the same conditions as for the samples analysed. Flavonol glycosides were quantified with the curve of quercetin-3-O-glucoside. Anthocyanins were quantified with the curve of malvidin-3-O-glucoside.
Nucleic acid extraction and cDNA synthesis
Total RNA was isolated from berry skins according to the procedure of Reid et al. (2006), using a CTAB-Spermidine extraction buffer. For cDNA synthesis, one μg of total RNA was reverse transcribed with random hexamer primers in an 18 μl reaction mixture using the StrataScript® reverse transcriptase (Statagene, USA) according to the manufacturer's instructions.
Quantitative comparison of gene expression throughout berry skin development
Relative transcript quantification of isolated genes was performed by real-time RT-PCR, using the Brilliant® SYBR® Green QPCR Master Reagent Kit (Stratagene) and the Mx3000P detection system (Stratagene) as described in the manufacturer's manual. Amplification of a fragment of the UBIQUITIN1 gene (99 bp; TC53702, TIGR database, VvGi5) was used for normalization (Downey et al., 2003). PCR conditions, standard quantification curves for each gene, primer efficiency values (see Supplementary Table SI at JXB online) and relative gene expression calculations were conducted according to Poupin et al. (2007). Briefly, standard quantification curves with serial dilutions of PCR products were constructed for each gene to calculate amplification efficiency according to:
| (1) |
This value was then used to obtain an accurate ratio between the expression of the gene of interest (GOI) and the housekeeping gene, using Equation (2):
| (2) |
Gene expression levels were normalized to the expression of the first sample for the full shaded treatment (T4), in order to obtain a calibrated ΔCt for each gene.
In all cases, R2 values of standard curves were above 90%. Ct values for UBIQUITIN varied no more than one unit between all samples analysed for each real time experiment. All experiments were performed with three biological replicates and three technical replicates. Reaction specificities were tested with melt gradient dissociation curves, electrophoresis gels, and cloning and sequencing of each PCR product.
Statistical analysis
Flavonoid composition and expression profile data were statistically analysed by two-way ANOVA to test the significance of the effects of treatments at the different stages of berry ripening. Tukey media comparison analysis was performed to compare the treatments at the same berry ripening stage. Statistical differences between means were based on the least significant method when F values were significant with P <0.05.
Results and discussion
Cluster light exposure levels have no effect on general chemical parameters at harvest
Vineyard row orientation has a pronounced effect on the photosynthetically active radiation (PAR) received by the two sides of the rows (Grifoni et al., 2008). In addition, radiation and temperature are different on each side of a north–south oriented row (Pereira et al., 2006). The vineyard used in this study had a north–south orientation and clusters on the eastern side were considered for analysis in this study.
A PAR measurement device was set up within the grape bunch, simulating the position of a single berry in the cluster (Fig. 3A). For 10 h, incident PAR was recorded every 5 min (Fig. 3B). Photographs of the experimental rows were taken until 14.00 h at 4 WAV (see Supplementary Video S1 at JXB online). PAR measurements revealed that exposed and shaded clusters received different intensities of radiation (Fig. 3). While exposed fruits were subjected to almost incident PAR levels between 11.00 h and 13.00 h (above 1500 μmol m−1 s−1), shaded clusters received between 100–400 μmol m−1 s−1 during the same period of the day (Fig. 3B).
Fig. 3.
PAR measurements taken at 4 weeks after veraison for shaded and exposed clusters from the east side of one of the experimental rows. Incident PAR is included. (A) Arrow indicates the position of the PAR meter in each cluster. (B) Daily measurements from 08.00 h to 18.00 h.
By harvest time (9 WAV), the remaining clusters did not show any differences in skin weight, pH or sugar concentrations in berry juice (Table 1), suggesting that, by the end of ripening, neither shade nor leaf removal affected final sugar or acid content. In addition, cluster morphology and size was not affected by any treatment, except for a possible premature lignification of peduncles in clusters from treatments T1 and T4 (see Supplementary Fig. S3 at JXB online).
Table 1.
General physical and chemical analyses of Cabernet Sauvignon grape berry samples from each light exposure treatment, taken at 9 weeks after veraison
| T1 | T2 | T3 | T4 | |
| Skin weight of 200 berries (g) | 58.7±7.3 | 52.8±0.7 | 55.4±0.4 | 54.0±1.1 |
| pH | 3.68±0.02 | 3.59±0.01 | 3.63±0.01 | 3.66±0.03 |
| Soluble solids (Brix degrees) | 24.7±0.3 | 23.8±0.7 | 24.4±0.5 | 25.1±0.3 |
Standard deviations are shown (±). Using a Tukey test, no significant differences were found in any of these measurements.
Anthocyanin content and genes regulating their synthesis are differentially affected by sunlight exposure and leaf removal
Several reports have shown that anthocyanins and flavonols are directly affected by exposure to sunlight or UV radiation (reviewed by Downey et al., 2006). Light is a fundamental requirement for colour formation in grapes and other fruit crops such as apple. Despite this, differences in the experimental design, the analytical measurements used, the cultivar chosen, the geographical location of the experimental site and many other factors, have produced contradictory results regarding the relationship between anthocyanin content and light in grapes (Hunter et al., 1995; Bergqvist et al., 2001; Spayd et al., 2002; Downey et al., 2004). In addition, no studies have been carried out in which changes in flavonoid content have been correlated with expression of regulatory genes, other than MYBA1 (Jeong et al., 2004).
The sampling approach and timing conducted in this work was designed to detect sequential and/or temporal cause–effect relationships between gene transcript abundances and metabolite levels. Total anthocyanin accumulation was significantly higher in both light-exposed treatments (T1 and T4), including the glycosylated, acylated, and p-coumaroylated derivatives of all anthocyanins (Fig. 4). Anthocyanin glycosides in grapes are based on the di-hydroxylated derivates cyanidin and peonidin, and the tri-hydroxylated derivates delphinidin, petunidin, and malvidin, with the latter being the most abundant in wine grape cultivars (Roggero et al., 1984; Hebrero et al., 1988). When each one of these molecules was analysed, different responses under sunlight exposed, shaded, and leaf removal treatments were observed (Fig. 4; see Supplementary Table SII, at JXB online).
Fig. 4.
Concentration of total and 3-O-glycosylated anthocyanin compounds from the different light exposure treated berry skins, taken from 2–8 weeks after veraison. (filled inverted triangles) T1 exposed; (open circles) T2 delayed; (filled circles) T3 shaded; (open triangles) T4 leaf removal. Anthocyanin concentrations are calculated in malvidin equivalents. Vertical bars indicate the standard deviation (three biological replicates). Different letters indicate significant differences between treatments as calculated by Tukey statistical analysis (P <0.05).
Sunlight exposure (T1) increased the levels of delphinidin-3-O-glucoside (Dp3G), cyanidin-3-O-glucoside (Cy3G), petunidin-3-O-glucoside (Pt3G), peonidin-3-O-glucoside (Po3G), and malvidin3-O-glucoside (Mv3G) throughout all stages of berry ripening (Fig. 4). In the leaf removal treatment (T4), the levels of the most methylated di- and tri-hydroxylated derivates (Po3G and Mv3G, respectively) were significantly lower in the second week after veraison, even when they were compared with the delayed (T2) and shaded (T3) treatments. Dp3G, Pt3G, and Cy3G were not detected in the samples from the leaf removal treatment at this ripening stage. This observation suggests an effect of removing source organs (basal leaves) on flavonoid synthesis during the initial stages of berry ripening. Despite this, and with the exception of Mv3G, all anthocyanin levels in T4 increased to those observed in the fully exposed T1 treatment after the fourth week, suggesting the activation of a compensation process, at least regarding anthocyanin accumulation. Pt3G and Po3G abundances were even higher in the leaf-removal treatment compared to T1 at 8 WAV, prior to harvest. Mv3G levels were significantly higher in the exposed treatment, and although the leaf removal treatment increased Mv3G concentration by the fourth week, it never reached the same levels as in T1.
Although the least abundant anthocyanin observed was Cy3G, its concentration increased in the fully shaded treatment (T3) during ripening when compared to T1, T2, and T4. All other non-acylated glycosides began to decrease during the latter stages of ripening in all treatments. Similar results regarding possible shifts in anthocyanin composition have been suggested, in which low light and cool climates could increase the concentration of non-acylated cyanidin glycosides (Downey et al., 2006).
Since significant differences were observed in the metabolic profiling of anthocyanins, it was reasonable to expect a differential expression in some of the flavonoid biosynthetic genes, under the treatments applied in this study (Fig. 5). Metabolites were analysed in even-numbered weeks, whereas gene expression was determined in odd-numbered weeks. This sampling approach enabled us to observe different possible regulatory mechanisms for flavonoid synthesis in a fruit development stage-specific manner. In general, biosynthetic genes were not affected by treatments during the first week of sampling, although metabolites already showed differences between treatments. This led us to propose a possible non-transcriptional regulation of flavonoid synthesis during the initial weeks of berry ripening. In fact, it has been shown that the activity of several enzymes of the pathway, such as PHENYLALANINE AMMONIA-LYASE (PAL), can be affected by light through a non-transcriptional mechanism (Sreelakshmi and Sharma, 2008). From the third week, biosynthetic genes already differ in expression, depending on each treatment. Regulation at the transcriptional level is thus suggested for the mid and latter stages of berry skin ripening.
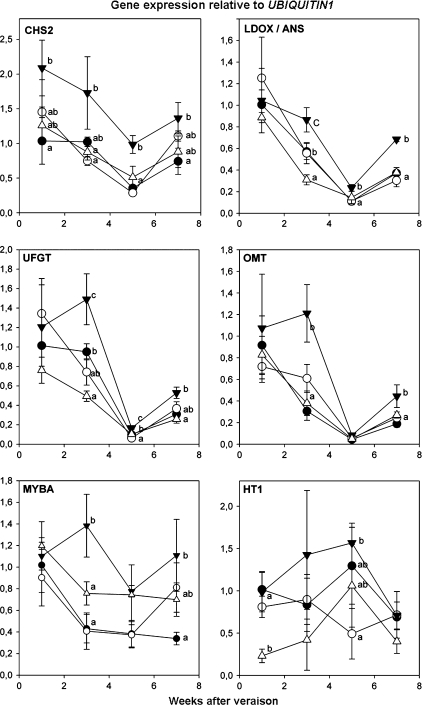
Fig. 5.
Changes in the expression of anthocyanin biosynthetic genes, the MYBA regulator and HEXOSE TRANSPORTER1, under different light exposure or leaf removal treatments. (filled inverted triangles) T1 exposed; (open circles) T2 delayed; (closed circles) T3 shaded; (open triangles) T4 leaf removal. Transcript levels are expressed in relation to the VvUBIQUITIN1 gene. Vertical bars indicate the standard deviation (three biological replicates). Different letters indicate significant differences between treatments as calculated by Tukey statistical analysis (P <0.05).
CHS2 has been reported to be isoform of CHALCONE SYNTHASE which is most affected by light (Jeong et al., 2004), and was used to test the effectiveness of the sunlight exposure treatments in changes of gene expression. In this study, CHS2 expression was increased by light and it appears that this response is concomitant with the expression of LEUCOANTHOCYANIDIN OXIDASE (LDOX), O-METHYLTRANSFERASE (OMT), and UFGT, because of their remarkably similar expression profiles. The expression of these four biosynthetic genes declined throughout ripening until 7 WAV, at which point a small increase was observed. The decrease in gene expression correlates with the decay in the rate of accumulation of many of the anthocyanins studied between weeks 4 and 8, as seen in Fig. 4 with a decrease in the curve slope. At 3 WAV and 5 WAV, a significantly higher expression value for the T1 treatment was observed, indicating that these genes are being regulated by light. In addition, expression was affected negatively by the leaf removal treatment as shown in the T4 treatment (Fig. 5, upper and middle panels).
Expression of MYBA1, as a direct regulator of UFGT expression (Kobayashi et al., 2002), is regulated by light (Jeong et al., 2004; Fig. 5, bottom). In apple, a MYB factor responsible for anthocyanin synthesis has also been described as affected by light (Takos et al., 2006), but in addition, it is shown here that MYBA1 was also affected in the leaf removal treatment at the third week after veraison (Fig. 5, bottom). As recently reported, this work supports a role for MYBA co-regulating, either directly or indirectly, the expression of genes other than UFGT, such as OMT (MC Cutanda-Perez et al., 2009).
Changes in anthocyanin synthesis after leaf removal are related to the expression of a hexose transporter
Since both anthocyanin profiling and expression of anthocyanin-related genes were affected in the leaf removal treatments, it was possible that leaf organ removal itself repressed flavonoid synthesis. Since source organs were being removed in this treatment, a possible gene being affected could be a sugar transporter, which incorporates sugars exported from the leaves into the berries, contributing to the sink–source balance. The grape HEXOSE TRANSPORTER1 (HT1) is expressed in all berry tissues and leaves and is involved in sugar import into the berry (Fillion et al., 1999). As seen in Fig. 5 (bottom), HT1 expression was affected only in the early stages of berry skin ripening (1 WAV), exclusively in the leaf removal treatment. This down-regulation could be due to a decrease in source organ potential in the period in which leaves are highly photosynthetically active (first weeks after veraison).
In addition to a possible role of HT1 in the first stages of mesocarp berry growth, before veraison (Vignault et al., 2005; Conde et al., 2006), removing source organs diminished HT1 expression in berry skin, suggesting an important role of this transporter in this particular berry tissue. HT1 is also regulated post-transcriptionally by a hexokinase-independent mechanism (Conde et al., 2006). Therefore, it is necessary to study the effect of this practice on the expression and regulation of invertases, sucrose transporters, and recently-isolated hexose transporters (Hayes et al., 2007), to understand the global impact on sugar accumulation.
Even though mesocarp cells are specialized in incorporating and hydrolysing sucrose into glucose and fructose for storage during grape ripening, exocarp cell sugar transport could directly affect skin colour pigmentation in the same cells, as there is a direct relationship between sugar content and anthocyanin synthesis in grape (Vitrac et al., 2000) and other species (Solfanelli et al., 2006). This correlation has been proven to be mediated by MYB factors in Arabidopsis (Teng et al., 2005). In grapes, biosynthetic genes such as LDOX and DFR, possess ‘sucrose boxes’ in their promoters (Gollop et al., 2001, 2002). These are regulatory elements which determine sugar-specific gene expression responsiveness and are also found in the promoter of grape HT1 (Atanassova et al., 2003). By this means, at 1 WAV, a decrease in HT1 expression under the leaf removal conditions could diminish sugar import into exocarp cells, with the consequent repression of flavonoid-related expression at 3 WAV. Hormone signalling (e.g. ethylene or abscisic acid) may be playing an additional role in this response since organ removal constitutes a stress event to the plant. This issue should also be addressed in the future.
Genes affecting other branches of the flavonoid biosynthetic pathway are less affected by light exposure
In model species such as Arabidopsis, genes belonging to both the MYB and bHLH families, which modulate flavonoid content in different plant organs, have been reported to be differentially modulated by environmental conditions (e.g. by light in Arabidopsis; Cominelli et al., 2008). In addition, a WDR factor regulating anthocyanin synthesis in Perilla frutescens was found to be up-regulated by light (Sompornpailin et al., 2002). In this work, the expression of different members of the grape MYB, bHLH, and WDR families, which are able to control flavonoid synthesis when expressed in homologous or heterologous systems was also studied.
MYB5a (Deluc et al., 2006), MYB5b (Deluc et al., 2008), and MYBPA1 (Bogs et al., 2007) are capable of activating the grapevine promoters of several biosynthetic genes of the flavonoid pathway, including LDOX. They also regulate flavan-3-ol synthesis by controlling LEUCOANTHOCYANIDIN REDUCTASE (LAR) and ANTHOCYANIDIN REDUCTASE (ANR) expression. The differences observed in LDOX expression under the light exposure treatments were only related to the differences observed in MYB5a, as seen in Fig. 6. In addition, MYB5a expression was affected at 1 WAV for the leaf removal treatment, suggesting a much faster response than MYBA1 following changes in sugar transport. The differences observed in anthocyanin content could not be explained by alterations in MYBPA1 expression, as the accumulation of transcripts of this gene were not affected in the different light exposure treatments. MYB5b expression was very similar to the expression previously reported by Deluc et al. (2008), and levels did not vary under the treatments applied in this study. It has been suggested that MYB5a and MYB5b, which regulate similar structural genes, exert their regulatory effect in different periods of berry ripening, with MYB5a predominating in the early stages and MYB5b towards the later stages (Deluc et al., 2008). It is possible that MYB5a is being co-regulated with MYBA1 and that it could be affected by light and sugar transport into the berry.
Fig. 6.
Changes in transcript levels of MYB, MYC, and WDR regulators of different branches of flavonoid synthesis, under different light exposure or leaf removal treatments. (filled inverted triangles) T1 exposed; (open circles) T2 delayed; (filled circles) T3 shaded; (open triangles) T4 leaf removal. Transcript levels are expressed in relation to the VvUBIQUITIN1 gene. Vertical bars indicate the standard deviation (three biological replicates). Different letters indicate significant differences between treatments as calculated by Tukey statistical analysis (P <0.05).
Some MYB factors also possess repressor activities and inhibit phenolic compound synthesis (Jin et al., 2000; Aharoni et al., 2001). FaMYB1 is an anthocyanin repressor found in strawberry. It was suggested that its function was to regulate the excess levels of flavonoids which could have a cytotoxic effect if they over-accumulate in the cytoplasm and are not efficiently transported into vacuoles (Aharoni et al., 2001). In Arabidopsis, AtMYB4 is also a repressor, regulating one of the first steps of the phenylpropanoid pathway, controlled by the CYNAMATE-4-HYDROXYLASE (C4H) enzyme, necessary to synthesize sinapate esters in response to UV light (Jin et al., 2000). This gene is down-regulated by UV-B in order to increase the content of these protective UV-screening molecules only under stress conditions. Recently, a grape MYB4 homologue repressor was isolated and characterized (JT Matus et al., unpublished results). Despite its similarity to AtMYB4, this gene negatively regulates UFGT expression, thus it is a direct repressor of anthocyanin synthesis. Despite its importance and the fact that is up-regulated during berry ripening (Matus et al., 2008), it seems that light or changes in sugar import do not affect MYB4 expression throughout ripening (Fig. 6).
Other regulatory factors and co-activators have been found in grape. The Arabidopsis TRANSPARENT TESTA 8 (TT8) grape homologue MYCA1 was not affected by these light treatments. In Arabidopsis, TT8 regulates flavonoid synthesis and is highly up-regulated by light (Cominelli et al., 2008) and other environmental factors. In apple, at least two MYC (also known as bHLH) proteins are needed to induce anthocyanin synthesis (Espley et al., 2007). In silico analysis of the grape genome suggests that grapes may also possess more than one bHLH affecting flavonoid synthesis (data not shown). It is possible that another bHLH gene could be regulated by light conditions. The WDR1 co-activator was only affected at 5 WAV between treatments T1 and T3. Therefore, MYB factors may participate in this regulation more directly than bHLH or WDR factors, in response to environmental conditions such as light. New members from these families should be isolated and tested to resolve this issue. In any case, differences observed between the responsiveness of transcriptional regulators and target gene expression levels may imply the effect of additional regulatory mechanisms.
Flavonols are the most drastically affected flavonoids under shadow treatments, an effect possibly mediated by the MYB12 transcription factor
Among flavonoids, the accumulation of flavonols was the most dramatically affected in berry skins under the treatments applied in this study (Fig. 7A). Flavonols have a high anti-oxidant capacity and have been associated with the velvet-type astringency of red wines (Hufnagel and Hofmann, 2008). Total flavonol content, including flavonol galactosides and glycosides, was significantly higher in T1 and very similar between T2 and T3 during the first 6 weeks. The leaf removal treatment had almost twice the flavonol content than in T2 at 8 WAV, although levels were not significantly different at 4 WAV. It has previously been reported that flavonols are more affected than anthocyanins under different light levels (Downey et al., 2003; Pereira et al., 2006), although this effect has not been studied before in relation to MYB expression.
Fig. 7.
Changes in total berry skin flavonol content (A) and transcription levels of the flavonol biosynthetic gene FLS4 and its putative regulator MYB12 (B) under different light exposure or leaf removal treatments. (filled inverted triangles) T1 exposed; (open circles) T2 delayed; (filled circles) T3 shaded; (open triangles) T4 leaf removal. Vertical bars indicate the standard deviation (three biological replicates). Different letters indicate significant differences between treatments for each ripening stage as calculated by Tukey statistical analysis (P <0.05).
FLS4 and FLS5 are the two most expressed flavonol synthase isoforms in berries (Fujita et al., 2006), and the former is the most affected under low light conditions. From all the genes studied in this work, MYB12 and FLS4 were the most affected by light (Fig. 7B), even during the first week of berry skin ripening. MYB12 was previously identified in the grape R2R3 MYB subfamily as a putative flavonol regulator (Matus et al., 2008), given its close homology to AtMYB12 which controls FLS expression in Arabidopsis (Mehrtens et al., 2005; Stracke et al., 2007). Although no functional analysis has yet been conducted for this gene, considering that MYB12 and FLS4 expressions and response patterns to light were very similar, it is suggested that FLS4 could be a target of MYB12 in grape. Leaf removal also affected the expression levels of both genes, suggesting that sugar import again is responsible for activating this other branch of the phenylpropanoid pathway. In contrast to what was observed with the anthocyanin content, flavonol levels in T4 did not reach those found in T1 during the last stages of ripening, suggesting that the compensation mechanisms previously suggested for anthocyanins do not occur as efficiently for flavonol accumulation.
Flavonol synthesis has been reported to respond rapidly once shaded tissues are exposed to light (Downey et al., 2004). This response was also observed in the T2 (delayed) treatment, in which light exposure applied in the sixth week after veraison quickly increased flavonol synthesis and expression of MYB12 and FLS4 to levels even greater than the T1 treatment. This change in expression was correlated with a 2-fold increase in total flavonol content compared to T3 (Fig. 7A), although this level is still very low compared to T1 or T4 treatments.
Conclusion
Viticultural practices affect the plant directly if they constitute a stress event such as organ removal, but can also affect the plant indirectly as a consequence of the modified microenvironment. Gene regulation of a metabolic pathway under these conditions varies in terms of the intensity and timing of the practice imposed. In this study, it was possible to observe that leaf removal at veraison has an early diminishing effect on sugar transport and flavonoid synthesis, especially on flavonol accumulation, during the ripening of the berry skin. For most anthocyanins, nevertheless, these differences are compensated at harvest. Although leaf removal increases sun exposure of the cluster, it is necessary to evaluate the exact changes in sink–source relationships that may occur in this condition.
Light and sugar are capable of inducing significant changes in flavonoid-related gene expression. In this study, it was shown that MYB genes regulating flavonoid synthesis are differentially affected by light. MYBs regulating the final anthocyanin or flavonol biosynthetic steps are more affected than MYBs controlling several points of the pathway. Other regulatory genes isolated so far do not respond in the same manner as MYB factors. It is important to examine the presence and function of regulatory elements in the promoters of these light responsive genes in order to understand these differences. In addition, it is suggested that other regulatory mechanisms (not related to transcriptional control) could also be governing flavonoid synthesis at least during the initial stages of berry ripening.
This work exemplifies how the flavonoid content and the genes controlling their synthesis are affected and could be manipulated by viticultural practices such as canopy management. New research efforts will be needed fully to understand the interaction between the plant, the environment, and the field practices in order to modify the quality of grapes. As an interesting projection for continuing the study of the regulation of flavonoid synthesis, it is necessary to analyse whether other viticultural practices, such as irrigation regimes, modify the expression of any of the regulatory genes considered in this study. Screening the expression of biosynthetic genes and their transcription factors under different environmental conditions and field practices will increase our understanding of the complex regulatory network under which flavonoids are being synthesized and accumulated.
Supplementary data
Supplementary material is available at JXB online.
Fig. S1. Basal leaf moving for treatments T1 and T2 using nylon zip-ties.
Fig. S2. HPLC chromatogram for (A) anthocyanidinic (Abs 520 nm) and (B) low molecular weight phenolic compounds, which include flavonol derivatives (Abs 280 nm).
Fig. S3. Cluster morphology at 9 weeks after veraison.
Table S1. Primers used for quantification of transcripts by means of real-time quantitative PCR.
Table S2. Concentration of all anthocyanin compounds from the different light exposure treated berry skins, taken from 2–8 weeks after veraison.
Video S1. Daily time-course of the experimental field in which sunlight treatments were imposed (north orientation).
Supplementary Material
Acknowledgments
Thanks to Hector Morales for his contribution in HPLC-DAD analysis. This work was supported by the Chilean Wine Consortium 05CTE01-03, the Fruit Consortium, 07Genoma01, Millennium Nucleus for Plant Functional Genomics (P06-009-F), FONDECYT 1080559 and by Fellowships awarded to JTM (MECESUP and CONICYT AT24060171). We also thank Dr Michael Handford (Universidad de Chile) for critically reading the manuscript.
References
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O'Connell AP. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal. 2001;28:319–332. doi: 10.1046/j.1365-313x.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- Argüello-Astorga G, Herrera-Estrella L. Evolution of light-regulated plant promoters. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:525–555. doi: 10.1146/annurev.arplant.49.1.525. [DOI] [PubMed] [Google Scholar]
- Atanassova R, Leterrier M, Gaillard C, Agasse A, Sagot E, Coutos-Thévenot P, Delrot S. Sugar-regulated expression of a putative hexose transport gene in grape. Plant Physiology. 2003;131:326–334. doi: 10.1104/pp.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. The Plant Journal. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Baxter NJ, Lilley TH, Haslam E, Williamson MP. Multiple interactions between polyphenols and a salivary proline-rich protein repeat result in complexation and precipitation. Biochemistry. 1997;36:5566–5577. doi: 10.1021/bi9700328. [DOI] [PubMed] [Google Scholar]
- Bergqvist J, Dokoozlian N, Ebisuda N. Sunlight exposure and temperature effects on berry growth and composition of Cabernet Sauvignon and Grenache in the central San Joaquin valley of California. American Journal of Enology and Viticulture. 2001;52:1–7. [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiology. 2007;143:1347–1361. doi: 10.1104/pp.106.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua NH. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Christie JM, Jenkins GI. Distinct UV-B and UV-A/blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. The Plant Cell. 1996;8:1555–1567. doi: 10.1105/tpc.8.9.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Alfenito MR, Walbot V. Impact of low temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta. 1994;194:541–549. [Google Scholar]
- Conde C, Agasse A, Glissant D, Tavares R, Gerós H, Delrot S. Pathways of glucose regulation of monosaccharide transport in grape cells. Plant Physiology. 2006;141:1563–1577. doi: 10.1104/pp.106.080804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. Journal of Plant Physiology. 2008;165:886–894. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Cortell JM, Kennedy JA. Effect of shading on accumulation of flavonoid compounds in Vitis vinifera (L.) Pinot Noir fruit and extraction in a model system. Journal of Agriculture and Food Chemistry. 2006;54:8510–8520. doi: 10.1021/jf0616560. [DOI] [PubMed] [Google Scholar]
- Cutanda-Perez MC, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L. Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Molecular Biology. 2009 doi: 10.1007/s11103-008-9446-x. In press. [DOI] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiology. 2006;140:499–511. doi: 10.1104/pp.105.067231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiology. 2008 doi: 10.1104/pp.108.118919. DOI:10.1104/pp.108.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey MO, Dokoozlian NK, Krstic MP. Cultural practice and environmental impacts on the flavonoid composition of grapes and wine: a review of recent research. American Journal of Enology and Viticulture. 2006;57:257–268. [Google Scholar]
- Downey MO, Harvey JS, Robinson SP. Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.) Australian Journal of Grape and Wine Research. 2003;9:110–121. [Google Scholar]
- Downey MO, Harvey JS, Robinson SP. The effect of bunch shading on berry development and flavonoid accumulation in Shiraz grapes. Australian Journal of Grape and Wine Research. 2004;10:55–73. [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. The Plant Journal. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillion L, Ageorges A, Picaud S, Coutos-Thévenot P, Lemoine R, Romieu C, Delrot S. Cloning and expression of a hexose transporter gene expressed during the ripening of grape berry. Plant Physiology. 1999;120:1083–1094. doi: 10.1104/pp.120.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A, Goto-Yamamoto N, Aramaki I, Hashizume K. Organ-specific transcription of putative flavonol synthase genes of grapevine and effects of plant hormones and shading on flavonol biosynthesis in grape berry skins. Bioscience, Biotechnology and Biochemistry. 2006;70:632–638. doi: 10.1271/bbb.70.632. [DOI] [PubMed] [Google Scholar]
- García-Barceló J. Técnicas analíticas para vinos. 1990. FAB Ed. Barcelona. Spain. 133. [Google Scholar]
- Gollop R, Even S, Colova-Tsolova V, Perl A. Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. Journal of Experimental Botany. 2002;53:1397–1409. [PubMed] [Google Scholar]
- Gollop R, Farhi S, Perl A. Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Science. 2001;161:579–588. [Google Scholar]
- Grifoni D, Carreras G, Zipoli G, Sabatini F, Dalla Marta A, Orlandini S. Row orientation effect on UV-B, UV-A, and PAR solar irradiation components in vineyards at Tuscany, Italy. International Journal of Biometeorology. 2008 doi: 10.1007/s00484-008-0168-1. doi:10.1007/s00484-008-0168-1. [DOI] [PubMed] [Google Scholar]
- Hayes MA, Davies C, Dry IB. Isolation, functional characterization, and expression analysis of grapevine (Vitis vinifera L.) hexose transporters: differential roles in sink and source tissues. Journal of Experimental Botany. 2007;58:1985–1997. doi: 10.1093/jxb/erm061. [DOI] [PubMed] [Google Scholar]
- Hebrero E, Santos-Buelga C, Rivas-Gonzalo JC. High performance liquid chromatography-diode array spectroscopy identification of anthocyanins of Vitis vinifera variety Tempranillo. American Journal of Enology and Viticulture. 1988;39:227–233. [Google Scholar]
- Hufnagel JC, Hofmann T. Quantitative reconstruction of the non-volatile sensometabolome of a red wine. Journal of Agriculture and Food Chemistry. 2008;56:9190–9199. doi: 10.1021/jf801742w. [DOI] [PubMed] [Google Scholar]
- Hunter JJ, Ruffner HP, Volschenk CG, Le Roux DJ. Partial defoliation of Vitis vinifera L. cv. Cabernet Sauvignon/99 Richter: effect on root growth, canopy efficiency; grape composition, and wine quality. American Journal of Enology and Viticulture. 1995;46:306–314. [Google Scholar]
- Hunter JJ, Visser JH. The effect of partial defoliation, leaf position and developmental stage of the vine on leaf chlorophyll concentration in relation to the photosynthetic activity and light intensity in the canopy of Vitis vinifera L. cv. Cabernet Sauvignon. South African Journal of Enology and Viticulture. 1989;10:67–73. [Google Scholar]
- Jeong ST, Goto-Yamamoto N, Kobayashi S, Esaka M. Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Science. 2004;167:247–252. [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO Journal. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Ishimaru M, Hiraoka K, Honda C. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta. 2002;215:924–933. doi: 10.1007/s00425-002-0830-5. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Ruiz-García L, Cabezas JA, De Andrés MT, Bravo G, Ibáñez A, Carreño J, Cabello F, Ibáñez J, Martínez-Zapater JM. Molecular genetics of berry colour variation in table grape. Molecular Genetics and Genomics. 2006;276:427–435. doi: 10.1007/s00438-006-0149-1. [DOI] [PubMed] [Google Scholar]
- Lin JK, Weng MS. Flavonoids as nutraceuticals. In: Grotewold E, editor. The science of flavonoids. New York: Springer Science+Business Media; 2006. pp. 213–238. [Google Scholar]
- Lo Piero AR, Puglisi I, Rapisarda P, Petrone G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. Journal of Agriculture and Food Chemistry. 2005;53:9083–9088. doi: 10.1021/jf051609s. [DOI] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology. 2008;8:83. doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiology. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, McGrath RB, Chua NH. Phytochrome phototransduction pathways. Annual Review of Genetics. 1994;28:325–349. doi: 10.1146/annurev.ge.28.120194.001545. [DOI] [PubMed] [Google Scholar]
- Mori K, Goto-Yamamoto N, Kitayama M, Hashizume K. Loss of anthocyanins in red-wine grape under high temperatura. Journal of Experimental Botany. 2007;58:1935–1945. doi: 10.1093/jxb/erm055. [DOI] [PubMed] [Google Scholar]
- Mori K, Sugaya S, Gemma H. Decreased anthocyanin biosynthesis in grape berries grown under elevated night temperature condition. Scientia Horticulturae. 2005;105:319–330. [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Neira A, Cáceres A, Pastenes C. Low molecular weight phenolic and anthocyanin composition of grape skins from cv. Syrah (Vitis vinifera L.) in the Maipo Valley (Chile): effect of clusters thinning and vineyard yield. Food Science and Technology International. 2007;13:153–158. [Google Scholar]
- Peña-Neira A, Dueñas M, Duarte A, Hernandez T, Estrella I, Loyola E. Effects of ripening stages and of plant vegetative vigor on the phenolic composition of grapes (Vitis vinifera L.) cv. Cabernet Sauvignon in the Maipo Valley (Chile) Vitis. 2004;43:51–57. [Google Scholar]
- Peña-Neira A, Hernández T, García-Vallejo C, Estrella I, Suarez JA. A survey of phenolic compounds in Spanish wines from different geographical origins. European Food Research and Technology. 2000;210:445–448. [Google Scholar]
- Pereira GE, Gaudillere JP, Pieri P, Hilbert G, Maucourt M, Deborde C, Moing A, Rolin D. Microclimate influence on mineral and metabolic profiles of grape berries. Journal of Agriculture and Food Chemistry. 2006;54:6765–6775. doi: 10.1021/jf061013k. [DOI] [PubMed] [Google Scholar]
- Poupin MJ, Federici F, Medina C, Matus JT, Timmermann T, Arce-Johnson P. Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene. 2007;404:10–24. doi: 10.1016/j.gene.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Quail PH. Photosensory perception and signal transduction in plants. Current Opinion in Genetics and Development. 1994;4:652–661. doi: 10.1016/0959-437x(94)90131-l. [DOI] [PubMed] [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biology. 2006;6:27. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggero JP, Ragonnet B, Coen S. Analyse fine des anthocyanes des vins et des pellicules cle raisin par la technique HPLC. Vigne et Vins. 1984;327:38–42. [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiology. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers T, Evans M. Wine quality: correlations with colour density and anthocyanin equilibria in a group of young red wines. Journal of the Science of Food and Agriculture. 1974;25:1369–1379. [Google Scholar]
- Sompornpailin K, Makita Y, Yamazaki M, Saito K. A WD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Molecular Biology. 2002;50:485–495. doi: 10.1023/a:1019850921627. [DOI] [PubMed] [Google Scholar]
- Spayd SE, Tarara JM, Mee DL, Ferguson JC. Separation of sunlight and temperature efects on the composition of Vitis vinifera cv. Merlot berries. American Journal of Enology and Viticulture. 2002;53:171–182. [Google Scholar]
- Sreelakshmi Y, Sharma R. Differential regulation of phenylalanine ammonia lyase activity and protein level by light in tomato seedlings. Plant Physiology and Biochemistry. 2008;46:444–451. doi: 10.1016/j.plaphy.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiology. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiology. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verries C, Cheynier V, Romieu C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in Vitis vinifera L. and suggests additional targets in the pathway. Plant Physiology. 2009 doi: 10.1104/pp.108.131862. 10.1104/pp.108.131862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This P, Lacombe T, Cadle-Davidson M, Owens CL. Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theoretical and Applied Genetics. 2007;114:723–730. doi: 10.1007/s00122-006-0472-2. [DOI] [PubMed] [Google Scholar]
- Ubi BW, Honda C, Bessho H, Kondo S, Wada M, Kobayashi S, Moriguchi T. Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Science. 2006;170:571–578. [Google Scholar]
- Venencie C, Uveira MN, Guiet S. Maturité polyphénolique du raisin mise en place d'une méthode d'analyse de routine. Revue Francaise d'Oenologie. 1997;167:36–41. [Google Scholar]
- Vidal S, Francis L, Guyot S, Marnet N, Kwiatkowski M, Gawel R, Cheynier V, Waters EJ. The mouth-feel properties of grape and apple proanthocyanidins in a wine-like medium. Journal of the Science of Food and Agriculture. 2003;83:564–573. [Google Scholar]
- Vignault C, Vachaud M, Cakir B, Glissant D, Dédaldéchamp F, Büttner M, Atanassova R, Fleurat-Lessard P, Lemoine R, Delrot S. VvHT1 encodes a monosaccharide transporter expressed in the conducting complex of the grape berry phloem. Journal of Experimental Botany. 2005;56:1409–1418. doi: 10.1093/jxb/eri142. [DOI] [PubMed] [Google Scholar]
- Vitrac X, Larronde F, Krisa S, Decendit A, Deffieux G, Mérillon JM. Sugar sensing and Ca2+-calmodulin requirement in Vitis vinifera cells producing anthocyanins. Phytochemistry. 2000;53:659–665. doi: 10.1016/s0031-9422(99)00620-2. [DOI] [PubMed] [Google Scholar]
- Walker AR, Lee E, Robinson SP. Two new grape cultivars, bud sports of Cabernet Sauvignon bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus. Plant Molecular Biology. 2006;62:623–635. doi: 10.1007/s11103-006-9043-9. [DOI] [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DA, Thomas MR, Robinson SP. White grapes arose through the mutation of two similar and adjacent regulatory genes. The Plant Journal. 2007;49:772–785. doi: 10.1111/j.1365-313X.2006.02997.x. [DOI] [PubMed] [Google Scholar]
- Yamane T, Jeong ST, Goto-Yamamoto N, Koshita Y, Kobayashi S. Effects of temperature on anthocyanin biosynthesis in grape berry skins. American Journal of Enology and Viticulture. 2006;57:54–59. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.