Abstract
Vitamin C (L-ascorbic acid, AsA) is an essential metabolite for plants and animals. Kiwifruit (Actinidia spp.) are a rich dietary source of AsA for humans. To understand AsA biosynthesis in kiwifruit, AsA levels and the relative expression of genes putatively involved in AsA biosynthesis, regeneration, and transport were correlated by quantitative polymerase chain reaction in leaves and during fruit development in four kiwifruit genotypes (three species; A. eriantha, A. chinensis, and A. deliciosa). During fruit development, fruit AsA concentration peaked between 4 and 6 weeks after anthesis with A. eriantha having 3–16-fold higher AsA than other genotypes. The rise in AsA concentration typically occurred close to the peak in expression of the L-galactose pathway biosynthetic genes, particularly the GDP-L-galactose guanyltransferase gene. The high concentration of AsA found in the fruit of A. eriantha is probably due to higher expression of the GDP-mannose-3′,5′-epimerase and GDP-L-galactose guanyltransferase genes. Over-expression of the kiwifruit GDP-L-galactose guanyltransferase gene in Arabidopsis resulted in up to a 4-fold increase in AsA, while up to a 7-fold increase in AsA was observed in transient expression studies where both GDP-L-galactose guanyltransferase and GDP-mannose-3′,5′-epimerase genes were co-expressed. These studies show the importance of GDP-L-galactose guanyltransferase as a rate-limiting step to AsA, and demonstrate how AsA can be significantly increased in plants.
Keywords: Ascorbate biosynthesis, GDP-L-galactose guanyltransferase, GDP mannose epimerase, gene expression, over-expression, vitamin C
Introduction
L-ascorbic acid (AsA), commonly known as vitamin C, is an essential metabolite for plants and animals although humans, and some other animals, have to obtain their AsA from the foods they eat. Cooked meat and seeds contain low amounts of AsA and so the main dietary sources of AsA for humans are fruit and vegetables. In plants, AsA is a part of the antioxidant system important for photosynthesis and is vital for detoxifying the free radicals generated as side products from this process. AsA is also a cofactor for many enzymes (Arrigoni and De Tullio, 2000), controls cell division, and also affects cell expansion (Arrigoni and De Tullio, 2000; Noctor and Foyer, 1998; Pastori et al., 2003; Smirnoff and Wheeler, 2000). The role of AsA as a cofactor for ACC oxidase suggests it is required for fruit ripening in climacteric fruit (De Tullio et al., 2004; Green and Fry, 2005). It is also a substrate for the production of other fruit acids such as L-tartaric, L-threonic, L-glyceric, and L-oxalic acids (Debolt et al., 2007; Loewus, 1999). In addition, being an important part of the cellular redox system, the AsA redox state may also be important in plant senescence, defence, and stress responses (Barth et al., 2004; Lopez-Carbonell et al., 2006; Noctor, 2006).
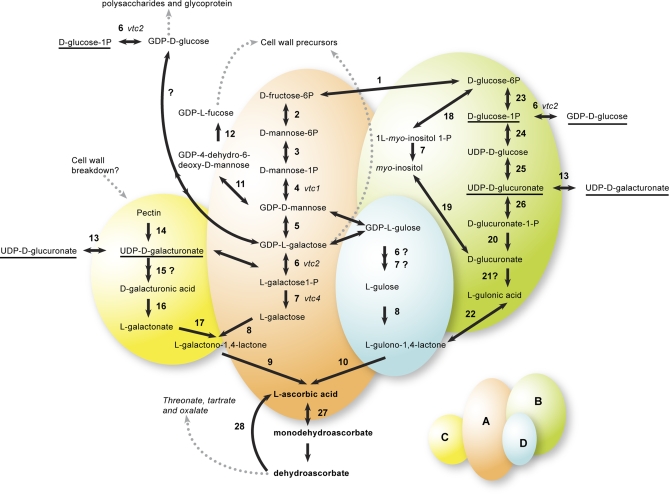
Several biosynthetic routes to AsA have been proposed (Fig. 1) with the pathway through L-galactose the best established (L-galactose pathway; Wheeler et al., 1998). The remaining unknown enzyme in the L-galactose pathway has recently been identified (Dowdle et al., 2007; Laing et al., 2007; Linster et al., 2007). There are other suggested routes to AsA through galacturonic acid (Agius et al., 2003; Loewus, 1999), L-gulose (Wolucka and Van Montagu, 2003, 2007) and myo-inositol (Lorence et al., 2004), but the available evidence from Arabidopsis mutants of the L-galactose pathway genes suggest that the AsA derived from these alternate pathways form a relatively small proportion of the total AsA pool, because they do not compensate for the low levels of AsA seen in L-galactose pathway mutants (e.g. vtc1, vtc2; Conklin et al., 1999; Linster et al., 2007). More recently, from an analysis of double mutants of the two GDP-galactose guanyltransferase genes which are seedling lethal, the authors suggested that the L-galactose pathway is the only significant pathway to AsA in Arabidopsis (Dowdle et al., 2007).
Fig. 1.
Reactions, enzymes, and context of ascorbic acid biosynthesis and regeneration in plants. (A) L-Galactose pathway, reactions 2–9. (B) myo-Inositol/glucuronate pathway, reactions 7, 18–26. (C) Galacturonate pathway, reactions 14–17. (D) L-Gulose pathway, possible reactions 5, 6, 7, 8, and 10. Reactions with question marks after the number are hypothetical and the exact enzyme is yet to be identified. Underlined chemical names are those that appear in more than one position in the diagram. Gene expression of transcripts of numbered enzymes in bold type were analysed. 1, glucose-6-phosphate isomerase; 2, mannose-6-phosphate isomerase; 3, phosphomannomutase; 4, GDP-mannose pyrophosphorylase; 5, GDP-mannose-3′,5′-epimerase; 6, GDP-L-galactose transferase; 7, L-galactose-1-phosphate phosphatase; 8, L-galactose dehydrogenase; 9, L-galactono-1,4-lactone dehydrogenase; 10, L-gulono-1,4-lactone oxidase; 11, GDP-D-mannose-4,6-dehydratase; 12, GDP-L-fucose synthase; 13, UDP-galacturonate epimerase; 14, polygalacturonate 4-α-galacturonosyltransferase; 15, galacturonate-1-phosphate uridylyltransferase and galacturonate-1-phosphate phosphatase (hypothetical); 16, D-galacturonic acid reductase; 17, aldonolactonase; 18, L-myo-inositol 1-phosphate synthase; 19, myo-inositol oxygenase; 20, D-glucurono-1-phosphate phosphatase; 21, glucuronate reductase; 22, gulonolactonase; 23, phosphoglucomutase; 24, UDP-glucose-pyrophosphorylase; 25, UDP-glucose dehydrogenase; 26, glucuronate-1-phosphate uridylyltransferase; 27, monodehydroascorbate reductase; 28, dehydroascorbate reductase; vtc, vitamin C content.
The fruits of the kiwifruit vine (Actinidia spp.) are especially rich sources of AsA and a tremendous variation of AsA content exists within the fruits of this genus, ranging from 40 (2.3 μmol g−1 FW) to over 1500 mg (>85 μmol g−1) AsA per 100 g fruit fresh weight (Ferguson and MacRae, 1992), making kiwifruit an excellent species to investigate the genetic basis of AsA production. For this reason, the basis of the variation in AsA levels seen between the fruits of different kiwifruit genotypes has been investigated (Fig. 2). In one genotype it is shown that exceptionally high concentrations of AsA in the fruit can be correlated with high expression levels of GDP-mannose-3′,5′-epimerase and GDP-L-galactose guanyltransferase. The importance of GDP-L-galactose guanyltransferase as the rate-limiting step to AsA production was confirmed in transgenic plants where over-expression of the kiwifruit GDP-L-galactose guanyltransferase gene resulted in significant increases in AsA levels. The addition of the GDP-mannose-3′,5′-epimerase gene in a transient system resulted in even higher AsA levels. These results have exciting potential for both conventional and molecular breeding purposes.
Fig. 2.
Fruit (nearing maturity) of the four Actinidia genotypes assayed in this study. Mp097 and Mp212 are two mapping population genotypes of A. chinensis, ‘Hayward’ is the green A. deliciosa, and 11-4-18a is A. eriantha.
Materials and methods
Genotypes chosen for analysis
Samples were collected from two A. chinensis mapping population siblings with similar mature fruit weights but varying ascorbate levels. These plants are closely related to the commercial yellow kiwifruit in that the father of the mapping population and ‘Hort16A’ (also sold as ZESPRI™ GOLD) are siblings. In addition, tissues from the most widely consumed green kiwifruit variety (A. deliciosa ‘Hayward’), as well as from a very high AsA, but relatively unpalatable, species (A. eriantha) were also collected (Fig. 2). Fruit and leaf samples were collected from the mapping population block at the HortResearch Te Puke research orchard, Te Puke, Bay of Plenty, New Zealand, during the 2003–2004 growing season. Mapping population individual vine locations were 32-11-17f for Mp097 and 32-13-04a for Mp212. Actinidia eriantha fruit were collected from the vine at position 11-4-18a. At least nine fruit were collected from each vine for every time point, and from all parts the vine. From 6 weeks after anthesis, the fruit were weighed before being processed. Fruit were cut into longitudinal quarters, and one quarter, including skin and seeds, was chopped into smaller pieces with a sharp knife and frozen in liquid nitrogen. Young (1–3 cm in diameter) and mature fully expanded leaves were also collected for some of the time points. Leaves were sampled from different parts of the vine when collected. All samples were ground to a fine powder in liquid nitrogen and stored at –80 °C.
RNA extraction and cDNA synthesis
Total RNA was extracted using a modified silica dioxide method. A 500 mg aliquot of frozen powder was transferred a 50 ml Falcon tube (Sarstedt) containing 6.75 ml extraction buffer (4.5 M guanidine HCl, 0.2 M Na acetate, 25 mM EDTA, 1 M K acetate, 2.5% (w/v) PVP-40, pH 5.2), 0.75 ml 10% (w/v) SDS and five glass beads (5–7 mm diameter). This was vortexed at maximum speed for 30 s and then incubated for 10 min at 70 °C. The liquid minus glass beads was transferred to a 50 ml Oakridge centrifuge tube (Nalgene®) and incubated on ice for 5 min. The tubes were then centrifuged for 10 min at 20 000 g at 4 °C, after which 6 ml supernatant was transferred to a new tube and then 6 ml NaI solution (5 M NaI, 0.1 M Na2SO3), 6 ml ethanol, and 550 μl of a silica milk suspension (1:1 w/v SiO2 to water, pH 2.0) were added. The samples were gently mixed by rolling at room temperature for 10 min. Following this, they were centrifuged for 1 min at 400 g at room temperature, and the supernatant was discarded. The pellet was resuspended in 10 ml wash buffer (10 mM TRIS-HCl, pH 7.5; 0.05 mM EDTA, 50 mM NaCl, 50% v/v ethanol) and then centrifuged for 1 min at 400 g, at room temperature. This wash cycle was repeated once more and then the pellet was air-dried for 10 min before being resuspended in 5 ml TE buffer (10 mM TRIS-Cl, 1 mM EDTA, pH 8). The RNA was unbound from the silica suspension by incubation at 70 oC for 4 min followed by centrifugation at 20 000 g for 5 min at room temperature. The supernatant was transferred to a fresh tube and precipitated by the addition of one-third volume 8 M LiCl and incubating at –20 °C for 1 h, followed by centrifugation for 20 min at 20 000 g at 4 °C. The RNA pellet was washed three times with 2.5 ml cold (–20 °C) 75% (v/v) ethanol (with centrifugation at 20 000 g for 10 min at 4 °C), then air-dried for 10 min, resuspended in DNAse buffer [77 μl H2O+10 μl 10× DNAse buffer (Sigma)+3 μl RNAseOUT (Invitrogen)], then transferred to a 1.5 ml Eppendorf tube. To this, 10 μl DNAse I (Sigma) was added and the reaction was incubated at room temperature for 20 min. RNA integrity was checked by agarose gel electrophoresis and DNAse treatment was repeated if DNA was seen (by incubating longer); otherwise 11 μl 50 mM EDTA (pH 8.0) was added and the mix was incubated at 70 °C for 10 min. RNA concentration and quality was determined using a 2100 Bioanalyzer (Agilent Technologies). Complementary DNA was synthesized from 1 μg total RNA using SuperscriptIII reverse transcriptase (Invitrogen) with oligo dT20 (Invitrogen), following the manufacturer's instructions. After synthesis, 10 μl cDNA was diluted 100 times in water. Between 1 μl and 2.5 μl was used per 15 μl PCR reaction.
Primer design
Where possible, DNA of the full-length open reading frame of each EST sequence was aligned to a pre-alignment of the open reading frame and genomic DNA sequence of its best matching Arabidopsis protein hit from the TAIR database (http://www.arabidopsis.org/). Primers were then designed using Primer3 software (Rozen and Skaletsky, 1998) so that at least one of the primer pairs spanned an intron–exon junction. Where this was not possible, primers were designed to lie on either side of an intron splice site. Primer design specifications were: an amplicon size of 100/110/120 (minimum/optimum/maximum); optimum primer length of 20 bp; primer Tm of 59/60/61 °C (minimum/optimum/maximum); primer GC% of 45% min and 50% max; with all other parameters left at default. Primer sequences are listed in Supplementary Table S1 at JXB online.
Quantitative real-time PCR (qPCR)
The relative expression of each transcript was determined in triplicate by qPCR using a 7500 Real-Time PCR System (Applied Biosystems). Total reaction volumes of 15 μl contained 7.5 μl Power SYBR™ Green PCR Master Mix (Applied Biosystems), 200 pM forward and reverse primers, and between 1 μl and 2.5 μl pre-diluted cDNA. Thermocycling parameters were 10 min at 95 °C, then 40 cycles of 95 °C for 15 s, then 60 °C for 1 min, with data collection during annealing and extension. After each run, dissociation curves were run to check amplicon purity. Data from the individual runs were collated using 7500 Fast System SDS Software (Applied Biosystems) and the background subtracted cycle threshold (CT) and well component data were exported. The amplification efficiency (Re) of each reaction was calculated from the component data using LinRegPCR (Ramakers et al., 2003) and this was used to calculate relative expression of each gene using the delta CT method (ReCTa-CTb; where ‘a’ and ‘b’ are the CTs of the sample designated to have an expression of 1 and the sample being compared, respectively). The variation in expression values between samples was normalized to the expression of an internal reference gene, the kiwifruit orthologue of At1g13320, a 65 kDa regulatory subunit of protein phosphatase 2A (PP2A). This was chosen from a suite of stable expressed internal reference gene candidates, which had been pre-tested on the entire cDNA set studied here. The choice of internal reference gene candidates was based on the Arabidopsis gene set described by Czechowski et al. (2005) and included orthologues of At1g59830 (PP2A catalyst), At4g27960 (UBC9), At1g13440 (GAPDH), and At5g09810 (actin).
Ascorbate quantification
Total kiwifruit tissue AsA was measured using HPLC on the same liquid nitrogen powdered samples as used for qPCR, as described earlier by Rassam and Laing (2005). Leaf tissue was measured after grinding under liquid nitrogen in the same manner (Laing et al., 2007; Rassam and Laing, 2005). As a measure of the total rate of synthesis of AsA in fruit, total AsA per fruit was calculated as a product of fruit AsA concentration (Fig. 3A) and fruit weight (data not shown). Weight was measured from 6 WAA and estimated using a linear growth assumption up to 6 weeks.
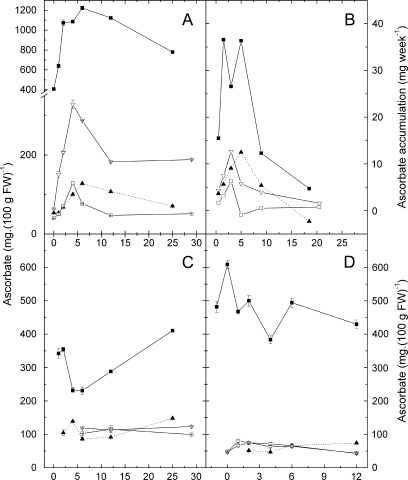
Fig. 3.
Change in L-ascorbic acid (AsA) concentration in Actinidia genotypes during the growing season. (A) AsA concentration in fruit. (B) Rate of change of ascorbate concentration (accumulation) calculated over adjacent time points from (A) and plotted at the mean time interval. Rates were calculated from measured fruit ascorbate and fruit size. Fruit size was measured from 6 weeks and estimated at earlier stages assuming linear growth during this early period. Rates were calculated as the difference in total fruit ascorbate over consecutive time periods divided by the difference in time. (C) AsA concentration in mature leaves (fully expanded leaves ∼20 cm diameter) over the period of a growing season. (D) AsA concentration in young leaves (1–3 cm in diameter) over the period of a growing season. (Open inverted triangles) A. chinensis MP097; (closed squares) A. eriantha; (open squares) A. chinensis MP212; (closed triangles) A. deliciosa. Bars show standard errors.
Arabidopsis transformation and transient expression in tobacco
Arabidopsis was transformed using the Agrobacterium kiwifruit GGT in a pGreen construct as described elsewhere (Laing et al., 2007) using the floral dipping method (Clough and Bent, 1998). Seed were collected and kanamycin-resistant lines selected. Eight lines were chosen for further study and were taken on to the T3 generation. Plants were checked by growing on kanamycin plates for the presence of the selectable marker and were shown to be kanamycin-resistant. Gene expression was measured as described above. Tobacco was transiently transformed using the same Agrobacterium cloned genes (Hellens et al., 2005; Laing et al., 2007).
Results
AsA in fruit and leaves
Vitamin C (AsA) levels were measured by HPLC in fruit, leaf, and flower samples collected from four kiwifruit (Actinidia) genotypes representing three different species with a range of fruit AsA concentrations: A. deliciosa (∼80 mg per 100 g fresh weight AsA), A. chinensis (MP097 and MP212; two individuals from a population segregating for AsA concentration; ∼50 mg and ∼200 mg, respectively), and A. eriantha (11-4-18a; ∼800 mg) (Fig. 2). In fruit, the concentration of AsA quickly increased after fertilization, peaking around 4 weeks after anthesis (WAA) in the two A. chinensis mapping population individuals and at 6 WAA in A. eriantha and A. deliciosa, before decreasing as the fruit progressed toward maturity (Fig. 3A). The rate of accumulation of AsA on a mg week−1 basis was highest between 2 and 6 WAA (Fig. 3B). This rate of change in total fruit ascorbate was calculated as the difference in total fruit ascorbate over consecutive time periods divided by the difference in time (units mg week−1). The periods of highest accumulation were species-specific in four genotypes. The two A. chinensis genotypes accumulated AsA at their highest rate at 4 WAA and A. deliciosa peaked later at 6 WAA. Actinidia eriantha exhibited a bimodal pattern in accumulation, with the greatest rate of AsA accumulation occurring at 2 WAA and at 5 WAA. At maturity, all genotypes except A. deliciosa were still accumulating AsA, albeit at a rate no higher than 5 mg week−1 (<∼12% of their maximum rate) while A. deliciosa was actually losing ascorbate (Fig. 3B).
As well as having the highest fruit AsA concentration, A. eriantha also had a significantly higher AsA concentration in both mature leaves (up to 3.5-fold) (Fig. 3C) and young leaves (up to approximately 10-fold) (Fig. 3D) than the other genotypes. The AsA concentration in young leaves remained reasonably constant throughout the growing season (Fig. 3D), but in mature leaves of A. eriantha and A. deliciosa, there was a drop in leaf AsA concentration during the middle part of fruit development, corresponding with a period of strong AsA accumulation in the fruit (Fig. 3C).
Selection of genes assayed by qPCR
There are three processes that could determine fruit AsA levels: de novo biosynthesis, turnover (including recycling and degradation), and transport. The genes selected to cover these three processes and the evidence to verify their role in AsA biosynthesis is shown in Supplementary Table S2 at JXB online. Two transport genes were selected, three turnover genes, and 15 biosynthesis related genes. Expression of the last L-galactose AsA biosynthesis gene, L-galactono-1,4-lactone dehydrogenase, could not be measured because there were no kiwifruit orthologues available in the HortResearch Actinidia EST database. There is only one EST for galactose dehydrogenase and none for galactono-1,4-lactone dehydrogenase found in the Actinidia EST database while there are 11–69 ESTs for the earlier steps (Crowhurst et al., 2008). Examination of the Arabidopsis eFP Browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Light_Series) shows the L-galactono-1,4-lactone dehydrogenase gene is the lowest expressed gene of all the genes in the L-galactose pathway to ascorbate. Two potential candidates (GenBank accessions FG441979 and FG431619) for the six proposed L-gulono-1,4-lactone dehydrogenase genes (Wolucka and Van Montagu, 2003, 2007) are present in the HortResearch EST database (Crowhurst et al., 2008) representing only two different Arabidopsis genes. However, there is no known direct experimental evidence proving what their function is and because there were only two candidates from six potential genes, the expression of these genes was not studied.
For the recycling enzymes, Arabidopsis has five DHAR and MDAR genes (Chew et al., 2003). However, alignment of the available Actinidia DHAR ESTs with Arabidopsis DHAR genes showed each species clustered into separate gene families and so selection of genes representing different Arabidopsis members could not be done. Two divergent Actinidia ESTs were chosen to represent the DHAR family. In the case of MDAR and AO, only one EST was selected to quantify gene expression. The transport genes were selected based on their homology to the nucleobase-ascorbate family, although again there is no direct functional evidence (Maurino et al., 2006).
The position of 18 of the selected genes in the three biosynthetic/recycling pathways is shown in Fig. 1.
Genes influencing the de novo biosynthesis of AsA
Expression of L-galactose pathway genes and a competing pathway:
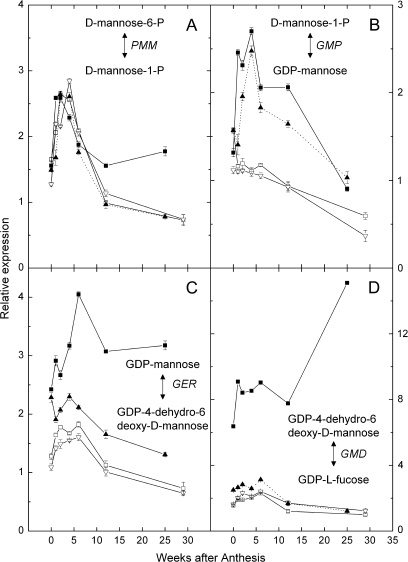
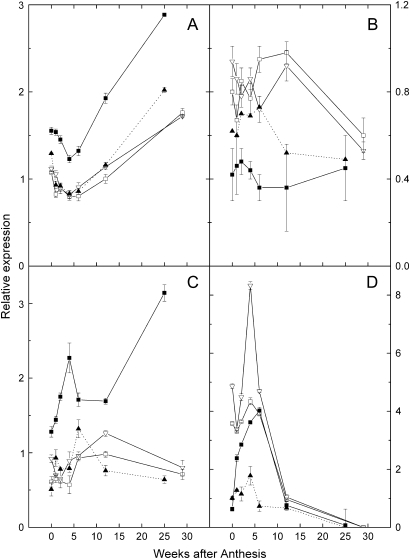
The expression of the six genes in the L-galactose pathway and two genes that divert material from ascorbate in two sets were assayed. The first set of four genes analysed are centred round GDP-mannose, the major branch point in the L-galactose pathway, which can also be utilized for the synthesis of fucose and cell wall precursors. The relative expression levels of GMP (gene encoding GDP-mannose pyrophosphorylase; enzyme 4 in Fig. 1) as well as two genes that encode enzymes that use GDP-D-mannose in fucose biosynthesis [GMD (gene encoding GDP-D-mannose-4,6-dehydratase; enzyme 11 in Fig. 1) and GER (gene encoding GDP-L-fucose synthase; enzyme 12 in Fig. 1)] were all significantly higher in A. eriantha than the other three genotypes (Fig. 4). The expression of PMM (gene for phosphomannomutase, enzyme 3 in Fig. 1) was similar in all genotypes during early fruit development when AsA accumulated most rapidly. Although the enzymes encoded by PMM and GMP feed into the L-galactose pathway, they are not wholly committed to AsA biosynthesis (Seifert, 2004).
Fig. 4.
Relative expression in a developmental series of kiwifruit fruit of genes encoding enzymes leading up to and from GDP-Mannose. (A) PMM (enzyme 3 in Fig. 1); (B) GMP (4); (C) GER (11); and (D) GMD (12). Results are expressed relative to the calibrating internal control gene: kiwifruit orthologue of At1g13320 [65 kDa regulatory subunit of protein phosphatase 2A (PP2A)]. Expression of the target gene in sample 12 weeks after anthesis was assigned the expression value of 1. Symbols as for Fig. 3.
The second set of four genes analysed encode enzymes that convert GDP-L-galactose to the precursor of AsA. GDP-L-galactose is thought to be mainly used for ascorbate biosynthesis although there are reports of L-galactose in cell walls (Roberts and Harrer, 1973; Zablackis et al., 1996) and mucilages (Anderson, 1933; Naran et al., 2008). GDP-mannose-3′,5′-epimerase (gene GME; enzyme 5 in Fig. 1), reversibly catalyses the conversion of GDP-D-mannose to GDP-L-galactose and also to GDP-L-gulose (Wolucka et al., 2001; Wolucka and Van Montagu, 2003; Major et al., 2005). This enzyme has been suggested to be a regulatory step in ascorbate acid biosynthesis (Wolucka and Van Montagu, 2003). Expression of this gene peaked between 1 and 2 WAA and was most highly expressed in A. eriantha fruit at 2 WAA, about 70% higher than in the A. chinensis genotypes (Fig. 5A). After the peak, expression in all the genotypes waned as the fruit developed to maturity, although expression integrated over time was higher in A. eriantha than in the other genotypes (see Supplementary Table S3 at JXB online).
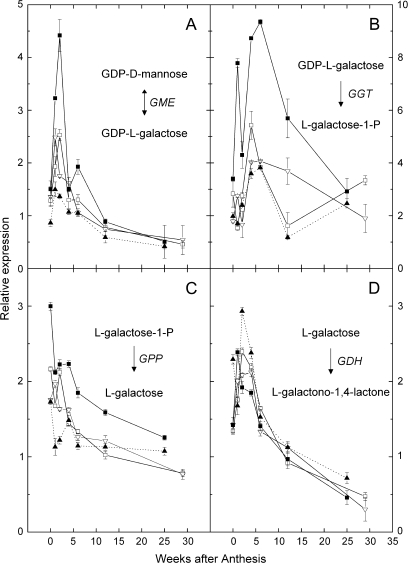
Fig. 5.
Relative expression in a developmental series of kiwifruit fruit of genes encoding enzymes of the L-galactose pathway. (A) GME (5); (B) GGT (6); (C) GPP (7); (D) GDH (8). Symbols as for Fig. 3. In (C), the same enzyme also catalyses the conversion of myo-inositol-1-P to myo-inositol. The gene sequence of the final enzyme leading to ascorbate [L-galactono-1,4-lactone dehydrogenase (9)] was not available and so no PCR results are available. Calibrating internal control gene: kiwifruit orthologue of At1g13320. Expression of the target gene in sample 12 weeks after anthesis was assigned the expression value of 1. The L-myo-inositol-1-P phosphatase that converts L-myo-inositol-1-P to myo-inositol (reaction 7 in Fig. 1) is shown in Fig. 5C. Symbols as for Fig. 3.
The expression of the kiwifruit transcript of GDP-L-galactose guanyltransferase (GGT; enzyme 6 in Fig. 1) (Laing et al., 2007) peaked between 4 and 6 WAA (Fig. 5B) which corresponded to the peak in AsA concentration. In A. eriantha, GGT transcripts were 2-fold to nearly 4-fold higher in the fruit than those in the other genotypes at 1, 4, and 6 WAA. The A. eriantha GGT expression showed an early peak at 1 WAA then fell at 2 WAA, after which a second expression phase occurred (Fig. 5B). A similar pattern was seen in AsA concentration (Fig. 3B). Integrated over fruit development, expression of GGT in A. eriantha was up to 2.5 times higher than in other genotypes (see Supplementary Table S3 at JXB online).
Comparing relative levels of GGT expression between genotypes other than A. eriantha did not explain the AsA variation observed. For instance, the low AsA A. chinensis Mp212 genotype had higher expression than its high AsA sibling, Mp097, up to and including the 4 WAA stage. After 4 WAA the high AsA Mp097 then had a constant level of expression between 4 WAA and 12 WAA, whereas expression in Mp212 fell after its maximum at 4 WAA to a minimum at 12 WAA, but then increased again after this stage. A. deliciosa had a somewhat similar GGT expression pattern to the low AsA Mp212, but peaked later at 6 WAA, corresponding with its peak in AsA concentration. Thus the high concentration of AsA found in the fruit of one genotype (A. eriantha) is probably due to higher expression of GME and GGT.
The kiwifruit enzyme downstream of GGT is L-galactose-1-phosphate phosphatase (GPP; enzyme 7 in Fig. 1) (Laing et al., 2004). The expression of this gene (Fig. 5C) showed a similar trend in all the genotypes and was highest in young fruit and highest in A. eriantha. The gene for L-galactose dehydrogenase (GDH; enzyme 8 in Fig. 1) showed even smaller genotype differences (Fig. 5D).
Expression of the last AsA biosynthesis gene, L-galactono-1,4-lactone dehydrogenase, could not be measured because there were no kiwifruit orthologues available in the HortResearch Actinidia EST database (Crowhurst et al., 2008) and attempts to clone the gene from kiwifruit using degenerate PCR primers failed.
Expression of genes in the galacturonate pathway:
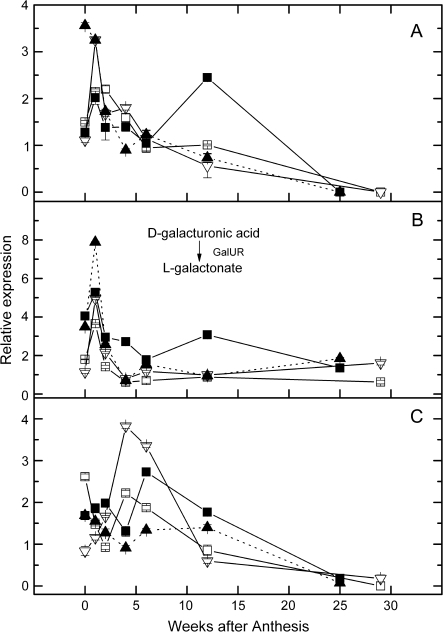
The galacturonate pathway to AsA uses D-galacturonic acid reductase (GalUR; enzyme 16 in Fig. 1), to reduce D-galacturonic acid to L-galactonic acid which must then be converted to L-galactono-1,4-lactone by an as yet unknown enzyme. There were at least three different candidates in the HortResearch EST database with homology to D-galacturonic acid reductase. Three of these shared greatest identity with a proven GalUR (AY663110) from strawberry [Fragaria×ananassa; (Agius et al., 2003)] and orthologues in Arabidopsis (At1g59950 and At1g59960) (see Supplementary Table S2 at JXB online). Over-expression of the strawberry GalUR in Arabidopsis led to a 2–3-fold enhancement in AsA content (Agius et al., 2003).
Gene expression of the three GALUR genes is shown in Fig. 6. There was no obvious relationship between gene expression and AsA levels in the different genotypes, although the gene expression of all three genes peaked early in fruit development. Expression of GALUR2 in mature leaves was significantly raised over young expanding leaves (see Supplementary Table S4 at JXB online). These genes have not been functionally characterized as to substrate specifity and may use substrates other than D-galacturonic acid.
Fig. 6.
Relative expression in a developmental series of kiwifruit fruit of galacturonic acid reductase genes. (A) GalUR1 (16); (B) GalUR2; (C) GalUR3. Calibrating internal control gene: kiwifruit orthologue of At1g13320. Expression of the target gene in sample 12 weeks after anthesis was assigned the expression value of 1. Symbols as for Fig. 3.
Expression of genes in the myo-inositol pathway:
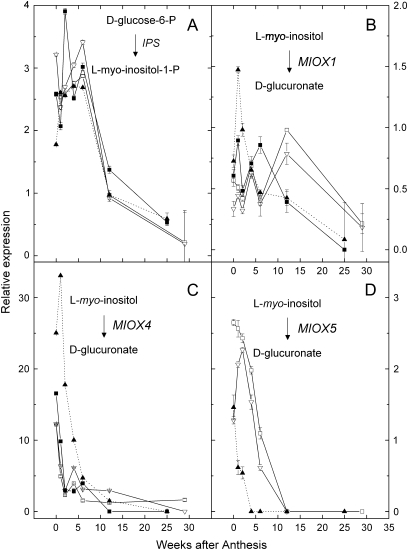
Myo-inositol is an important metabolite in A. deliciosa accumulating early in fruit development and declining after about 7 weeks (Klages et al., 1998).The first step of myo-inositol biosynthesis is the conversion of D-glucose-6-phosphate to L-myo-inositol 1-phosphate by L-myo-inositol 1-phosphate synthase (gene IPS; enzyme 18 in Fig. 1) (Johnson and Sussex, 1995; Loewus and Loewus, 1983). L-myo-inositol 1-phosphate is then dephosphorylated by L-myo-inositol-1-P phosphatase (Gillaspy et al., 1995) [also L-galactose-1-phosphate phosphatase (GPP) (Laing et al., 2004)]. A myo-inositol oxygenase (MIOX; enzyme 19 in Fig. 1) then oxidizes L-myo-inositol to form D-glucuronate, which can be used for the production of AsA (Lorence et al., 2004), or else channelled to cell wall uronosyl and pentose units (Loewus and Loewus, 1983).
The expression pattern of IPS was similar in all genotypes and was highest in young fruit, after which it fell to low levels (Fig. 7A). Relative expression was similar between leaves and fruit (see Supplementary Table S4 ar JXB online). The expression pattern of the GPP is shown in Fig. 5C. The expression patterns of the three MIOXs tested here were all different from one other (Fig. 7B, C, D) with MIOX4 transcripts being considerably higher in early fruit development than the other MIOX genes (Fig. 7C).
Fig. 7.
Relative expression in a developmental series of kiwifruit fruit of genes involved in inositol metabolism. (A) IPS (18); (B) MIOX1 (19); (C) MIOX4; (D) MIOX5. Calibrating internal control gene: kiwifruit orthologue of At1g13320. Expression of the target gene in sample 12 weeks after anthesis was assigned the expression value of 1 for all except MIOX5, for which no target was detected. In this case the Mp212 6 WAA sample was assigned the expression value of 1. Symbols as for Fig. 3.
There appears to be no relationship between either of these myo-inositol pathway genes and ascorbate levels, especially with respect to the high ascorbate A. eriantha.
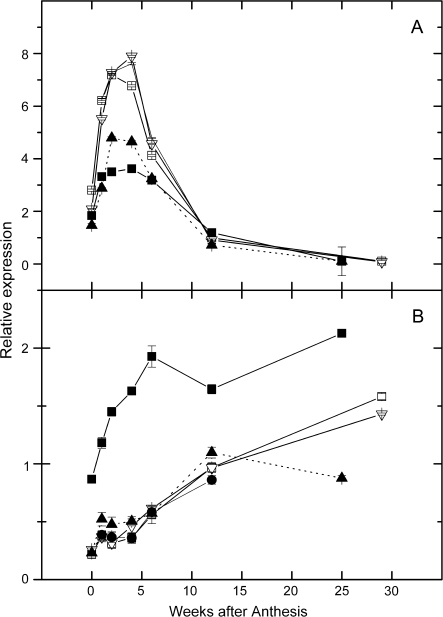
Expression of genes involved in ascorbate regeneration and oxidation:
Oxidized AsA can be regenerated through the action of monodehydro-AsA reductase (gene MDHAR; enzyme 27 in Fig. 1), and dehydro-AsA reductase (gene DHAR; enzyme 28 in Fig. 1). Transcripts of the MDHAR had a ‘hockey stick’-shaped expression pattern in fruit tissues of all the genotypes (Fig. 8A) with expression in A. eriantha higher than in the other three genotypes. Expression of MDHAR was also higher in mature leaves than in young leaves (see Supplementary Table S4 at JXB online).
Fig. 8.
Relative expression in a developmental series of kiwifruit fruit of genes involved in L-ascorbic acid (AsA) recycling and oxidation. (A) MDHAR (27), (B) DHAR1 (28); (C) DHAR2; (D) AO. Calibrating internal control gene: kiwifruit orthologue of At1g13320. Expression of the target gene in sample 12 weeks after anthesis was assigned the expression value of 1. Symbols as for Fig. 3.
The expression pattern of DHAR genes is shown in Fig. 8B and C, with the A. deliciosa and A. chinensis genotypes showing higher expression than A. eriantha over fruit development for DHAR1, but the reverse occurring with DHAR2.
Ascorbate oxidase (AO) showed a strong peak at around 4–6 weeks (Fig. 8D) with the peak in AsA oxidase expression occurring together with or 1 week later than the peak in AsA concentration.
Expression of genes in ascorbate transport:
A number of permease-like/nucleobase-ascorbate transporters have been annotated in plant sequence databases, and a detailed study of 12 putative Arabidopsis genes that were matched by BLAST to a rat nucleobase-ascorbate transporter (NAT) gene family (RnNAT1; SVCT1; AF080453) has been published (Maurino et al., 2006). However, no Arabidopsis gene has definitively been proven to be an ascorbic acid transporter, so two kiwifruit members of this family were chosen that most closely matched animal ascorbate transporters, named T1 and T2. Recently, dehydroascorbate has been demonstrated to be the transported molecule in Arabidopsis cell culture (Horemans et al., 2008), but no genes were identified. For all genotypes, the expression of T1 increased after fertilization to a peak around either 2 WAA or 4 WAA and then fell as the fruit matured (Fig. 9A). Both A. chinensis mapping population individuals had very similar expression patterns, and at their peak they were up to 2-fold higher in terms of relative expression than A. eriantha. The expression of T2 increased as the fruit developed (Fig. 9B). In A. eriantha fruit, T2 transcripts were 3–4-fold higher than in the other genotypes during the period of fast increases in AsA concentration (between 2–6 WAA).
Fig. 9.
Relative expression in a developmental series of kiwifruit fruit of putative L-ascorbic acid (AsA) transporter genes. (A) T1; (B) T2. Calibrating internal control gene: kiwifruit orthologue of At1g13320. Expression of the target gene in sample 12 weeks after anthesis was assigned the expression value of 1. Symbols as for Fig. 3.
Stable over-expression in Arabidopsis and transient expression in tobacco:
The gene expression studies suggested that GGT and GME were key genes involved in the production of high AsA in A. eriantha. To test this, Arabidopsis plants were transformed with the kiwifruit GGT gene. Plants with leaf AsA over four times the control level (Table 1) were observed, but within a line with high AsA progeny, there were always plants with high and low AsA (Table 1) in spite of all plants coming from a high AsA kanamycin-resistant parent, suggesting the occurrence of gene silencing. In some cases the level of AsA fell below the leaf AsA in control plants, suggesting that the endogenous gene was also silenced. A comparison of the DNA sequences between 319998 and the Arabidopsis gene At4g26850 showed that there were stretches of greater than 20 bp that were identical.
Table 1.
Enhancement of Arabidopsis leaf ascorbate (AsA) by the transformation of Arabidopsis with the GGT gene
| Line | Mean AsA if AsA <50 | Mean AsA if AsA >80 | Maximum AsA | Total no. plants | No. AsA <50 | No. with AsA >80 |
| pHEX control | 50±2a | 60 | 20 | 8 | 0 | |
| WT control | 53±2a | 70 | 11 | 5 | 0 | |
| 2.1 | 41±2 | 122±21 | 154 | 11 | 7 | 3 |
| 2.7 | 42±1 | 48 | 12 | 12 | 0 | |
| 6.19 | 43±2 | 140±12 | 184 | 12 | 3 | 6 |
| 6.21 | 44±2 | 101±7 | 124 | 10 | 4 | 5 |
| 8.12 | 42±2 | 65 | 12 | 8 | 0 | |
| 8.7 | 34±4 | 95 | 132 | 11 | 10 | 2 |
| 16.2 | 47±1 | 146±24 | 216 | 12 | 6 | 4 |
| 16.20 | 48 | 128 | 128 | 9 | 1 | 1 |
| 21.2 | 45±2 | 107±7 | 119 | 10 | 5 | 3 |
| 34.5 | 132±15 | 210 | 9 | 0 | 7 | |
| 40.10 | 37±2 | 49 | 11 | 11 | 0 | |
| 44.1 | 42±1 | 64 | 11 | 4 | 0 | |
| 44.12 | 46 | 100±3 | 105 | 11 | 5 | 3 |
| 44.2 | 49 | 164±23 | 204 | 11 | 4 | 2 |
| 44.3 | 41 | 53 | 12 | 11 | 0 | |
| 44.4 | 31±10 | 105 | 105 | 11 | 4 | 1 |
Third generation plants of Arabidopsis (except wild type) were selected as kanamycin-resistant. AsA is in mg 100 g−1 ±standard error. An empty cell means no data apply to that cell.
Mean AsA for all plants.
The third generation plants were also checked for gene expression of the kiwifruit GGT gene (see Supplementary Table S5 at JXB online). In every case, plants with high AsA relative to controls also showed enhanced expression of the kiwifruit gene. In one case, a plant with a low AsA (no plants in this line had high AsA) also showed high levels of the kiwifruit transcript. This may be interpreted as being due to the incomplete processing of the silenced gene transcripts in this line, and hence leaving template for detection by qPCR.
Transient tobacco leaf transformation (Hellens et al., 2005) were also used to measure the effect on leaf AsA of the simultaneous expression of GGT and GME (Table 2). Compared with the P19 control, transformation using the GME gene alone raised AsA by 20%. The GGT by itself resulted in a 4.2-fold increase in AsA. This is a similar result to that reported earlier for GGT (Laing et al., 2007). Simultaneous transformation of tobacco with both genes raised average AsA by 8.6-fold and a maximum of over 12-fold for the oldest leaf, over twice that observed with either gene alone.
Table 2.
L-ascorbic acid (AsA) levels in tobacco leaves transiently transformed with either GGT or GME or both together
| Genes used | Leaf | Average AsA (mg 100 g−1) | AsA relative to P19 control | Average over all leaves | Relative AsA to average P19 control |
| Control P19 only | 1 | 34 | 1.0 | 35.4 | 1.0 |
| 2 | 36 | 1.0 | |||
| 3 | 37 | 1.0 | |||
| GME only | 1 | 41 | 1.2 | 42.1 | 1.2 |
| 2 | 42 | 1.2 | |||
| 3 | 44 | 1.2 | |||
| GGT only | 1 | 91 | 2.7 | 150.2 | 4.2 |
| 2 | 145 | 4.0 | |||
| 3 | 215 | 5.9 | |||
| GGT and GME | 1 | 153 | 4.5 | 303.9 | 8.6 |
| 2 | 306 | 8.5 | |||
| 3 | 453 | 12.4 |
Separate Agrobacterium cultures were mixed and transformations injected into the leaf either together or separately. Two replicates were done for each leaf, and the experiment reported is a typical experiment. Leaf 1 is the youngest leaf.
Discussion
Ascorbate in fruit
The rate of total fruit AsA accumulation (in mg per week) showed a maximum early in fruit development in all genotypes (Fig. 3B) and remained positive in the two higher ascorbate genotypes through to maturity. In MP212, the ascorbate accumulation was essentially zero after 4 WAA, and in A. deliciosa accumulation became negative during the last period until harvest. The accumulation of AsA in the fruit is most likely to be through synthesis or transport, except in A. deliciosa, where degradation may have exceeded these processes in the period up to harvest. Our data show that both the concentration and accumulation rates of fruit ascorbate varied markedly between fruit species and time periods, providing an excellent model to investigate gene factors that control AsA (see Supplementary Table S3 at JXB online).
AsA concentration peaked between 4 WAA and 6 WAA. Cell division in ‘Monty’ kiwifruit (A. deliciosa, previously known as A. chinensis) effectively ceases by 33 d after anthesis (4–5 WAA; Hopping, 1976). Therefore, assuming the genotypes that were tested behaved similarly, the rapid fruit increases in AsA concentration occurred mainly during the cell division phase, with the concentration peaking between the end of cell division and up to 2 weeks thereafter. The fresh weight and cell size both increased at their fastest rates between 3 WAA and 6 WAA (Hopping, 1976). This is therefore, a period of very high growth and metabolic activity and the concurrence of peak AsA concentration agrees with purported roles of AsA in cell division, cell elongation, and as a cofactor for many enzymes (Noctor and Foyer, 1998; Arrigoni and De Tullio, 2000; Smirnoff and Wheeler, 2000; Pastori et al., 2003). Even though the lowest AsA concentration genotypes had the highest fruit size by weight, fruit weight is not generally correlated with AsA concentration in this mapping population (R Ferguson et al., unpublished results). After peaking, AsA concentration fell as the fruit matured, presumably due to slowing of biosynthesis and dilution due to cell expansion (Fig. 3B).
Ascorbate in leaves
The young leaves were rapidly expanding, and synthesis of AsA would have needed to be high to maintain their concentration, especially as in three out of the four genotypes AsA was higher in mature leaves than in young leaves. The exception again was A. eriantha, which not only had significantly higher levels of AsA than the other accessions, but also the levels of AsA tended to decline between young and mature leaves (Fig. 3C, D). However, because the magnitude of leaf expansion is so great, A. eriantha would still have needed to accumulate ∼33-fold more ascorbate than the other accessions during this period of leaf expansion.
Fruit gene expression
In general, transcripts for most, but not all, genes measured in fruit showed peak expression early in fruit development between 2 weeks and 12 weeks and declined as the fruit developed. The gene expression data for L-galactose pathway members in particular, tended to coincide with the early period of highest AsA accumulation per fruit (Fig. 3B). This supports the hypothesis that the L-galactose pathway is the predominant route of AsA biosynthesis in the fruit of kiwifruit. A recent review suggests that the pathway through L-galactose and L-gulose are the predominant routes to AsA in plants (Wolucka and Van Montagu, 2007), but currently the relative contributions of each pathway are not clear.
Transcripts of early L-galactose pathway enzymes were higher in A. eriantha than other genotypes, particularly for the genes GME and GGT, during the period of highest increase in AsA concentration. Transient over-expression of GGT in tobacco leaves led to an approximate 3-fold increase in leaf AsA (Laing et al., 2007), and further transient expression experiments presented here, with co-expressed GME and GGT resulted in an 8–12-fold increase in leaf AsA, whereas transient expression of GME alone resulted in little change in leaf AsA. Stable over-expression of GGT in Arabidopsis also resulted in increased leaf AsA by up to ∼4-fold. This suggests that GGT is a significant rate-limiting enzyme of AsA biosynthesis in vivo. This is supported by studies in Arabidopsis where it was shown that GGT and GME gene expression and GGT enzyme activity increased at high light intensities where ascorbate levels were also increased (Dowdle et al., 2007). The same study showed that vtc2/vtc5 double mutants were arrested in their growth past the cotyledon emergence stage, and then died. The very low ascorbate content of the vtc2 mutant shows that GGT (and thus the L-galactose pathway) is an important pathway for biosynthesis of AsA (Conklin et al., 2000). Later enzymes in the L-galactose pathway, especially GDH showed smaller differences in expression between A. eriantha and the other genotypes, suggesting they are not rate-limiting in AsA biosynthesis. This is borne out by the observation that over-expression of GDH in tobacco had no effect on leaf AsA levels (Gatzek et al., 2002).
Unfortunately, there are no kiwifruit sequence data available for the final step of the AsA biosynthetic pathway, L-galactono-1,4-lactone dehydrogenase, so the expression pattern of this gene could not be determined. However, in tomato plants down-regulated for L-galactono-1,4-lactone dehydrogenase (total knockouts were not recoverable), total ascorbate amounts in both the young developing leaves and fruit tissues were unchanged (Alhagdow et al., 2007), although there were differing ratios of reduced to oxidized AsA. Recent L-galactono-1,4-lactone feeding experiments in strawberry and blackcurrant fruit, as well as earlier experiments, suggest that substrate limitation occurs at an earlier stage in the pathway because the inherent fruit L-galactono-1,4-lactone dehydrogenase activity was sufficient to convert an increased supply of substrate to AsA (Baig et al., 1970; Nascimento et al., 2005; Hancock et al., 2007). Taking these observations into account it seems that L-galactono-1,4-lactone dehydrogenase may not exert strong control over ascorbate flux (in fruit tissues at least), even though its expression pattern tends to correlate with AsA content in certain examples (Pateraki et al., 2004; Tamaoki et al., 2003).
L-galactono-1,4-lactone dehydrogenase also has the ability to oxidize L-gulono-1,4-lactone, although with a significantly lower substrate affinity (higher Km) and maximum rate (Yabuta et al., 2000; Leferink et al., 2008). Thus if L-gulose is a significant direct intermediate in AsA biosynthesis (Wolucka and Van Montagu, 2003, 2007), then it is likely to have a specific dehydrogenase enzyme to carry out the reaction. As stated previously, two potential candidates (GenBank accessions FG441979 and FG431619) for the six proposed L-gulono-1,4-lactone dehydrogenase genes exist in the HortResearch EST database (Crowhurst et al., 2008). However, there is no direct experimental evidence proving their function and because there were only two candidates from six potential genes, the choice was made not to study the expression of these two genes until there is more evidence as to which of these genes are actually involved in ascorbate biosynthesis. Over-expression of a L-gulono-γ-lactone oxidase from rat liver converted both L-gulono-1,4-lactone and L-galactono-1,4-lactone to AsA, and raised AsA by 7-fold compared with controls (Jain and Nessler, 2000). Thus, the shunt through L-gulose may contribute to AsA biosynthesis.
GDH can oxidize both L-galactose and L-gulose, although it has a much higher Km for L-gulose than L-galactose (Gatzek et al., 2002; Mieda et al., 2004; Laing et al., 2007). It was calculated that the kiwifruit GDH would have about 8% activity with L-gulose as compared to L-galactose at limiting L-gulose concentrations (Laing et al., 2007). Again this suggests that, if metabolism through L-gulose is a significant route to AsA, then a specific L-gulose dehydrogenase is likely to exist.
For the two A. chinensis mapping population genotypes, there were no clear correlations between relative levels of gene expression and AsA concentration. Peaks in expression of GME and GGT coincided with AsA accumulation, but there was no clear difference between the low AsA Mp212 and the high AsA Mp097, and integrated expression was also very similar. Relative expression of GGT was actually higher in the low AsA Mp212 than in the higher AsA genotype at 4 WAA. However, GGT expression in relative terms remained close to its 6 WAA peak through to 12 WAA in the high AsA genotype (Mp097), around 2-fold higher than in the low AsA Mp212 at 12 WAA. Thus, the difference in AsA concentration is likely be a mix of this, different contributions by other biosynthetic routes and transport of AsA into the fruit. Some of these could be the higher expression of GALUR3 in high AsA Mp097 compared with low AsA Mp212, and the 50% higher expression of MIOX4 transcripts in Mp097 at 4 WAA compared with Mp212.
The most striking difference between A. deliciosa and the other genotypes studied here was in the expression of the myo-inositol oxygenases, particularly MIOX4. However, the much higher relative expression of MIOX4 early on during fruit development did not appear to affect AsA concentration, suggesting that myo-inositol is used for something other than AsA biosynthesis in kiwifruit. Myo-inositol is a major carbohydrate of kiwifruit during the early stages of fruit development (Bieleski et al., 1997; Boldingh et al., 2000) and it has been suggested that the oxidized product D-glucuronate is utilized in the production of glucuromannan gum, of which there are high levels in A. deliciosa kiwifruit (Redgwell, 1983). The fall in AsA concentration in mature leaves during the peak period of AsA accumulation in A. deliciosa suggests that translocation from leaves is an important source of AsA in the fruit of this genotype.
Leaf gene expression
The two committed steps in AsA biosynthesis of GGT and GPP had lower relative expression in young leaves, (sink tissues), than mature leaves (source tissues). The converse was true for the pre-L-galactose pathway genes, PMM, GMP, GME, and also the side-pathway GMD and GER. This suggests that in young leaves, flux is directed to producing cell wall polysaccharides, which then shifts toward AsA as the leaf matures. Interestingly, fucose was reported to inhibit GME (Wolucka and Van Montagu, 2003), and the expression of GME transcripts in A. eriantha fruit dropped as GER transcripts peaked. This pattern was not seen in the other genotypes, however. Expression of the putative transporter T1 was also higher in young leaves than in mature leaves, suggesting that sink vegetative tissues might also import AsA.
Conclusions
The expression of the genes GGT and GME correlated with the peak in AsA accumulation in all genotypes and showed the highest expression in A. eriantha, the genotype with the highest AsA. Over-expression through stable transformation in Arabidopsis of GGT or transient expression in tobacco leaves of GME and GGT together confirms the hypothesis that GGT catalyses a major control point of AsA biosynthesis through the L-galactose pathway in plants, and that this in turn creates an earlier rate-limiting step catalysed by GME. It lends credence that the observed GGT and GME expression patterns do indeed have biological relevance to the content of AsA in fruit tissues. This emphasizes the importance of verifying gene expression studies with further physiological experiments, as gene expression does not necessarily reflect enzyme activity. The over-expression results also highlight the significant potential for breeding high AsA content plants by both conventional and transgenic means. Such plants and their fruit would not only be more nutritious, but may also be more resistant to abiotic stresses such as high light, ozone, salt, and drought.
Supplementary data
Supplementary data for this manuscript can be found at JXB online.
Supplementary Table 1. Primers.
Supplementary Table 2. Genes assayed for their expression by qPCR and comparisons against identified enzymes in Arbidopsis.
Supplementary Table 3. Integrated gene expression for each gene in each pathway over the four genotypes from anthesis until fruit maturity.
Supplementary Table 4. Gene expression for leaves and non-pistil floral tissue in the four kiwifruit genetypes.
Supplementary Material
Acknowledgments
We would like to thank Di Barraclough for technical contributions and the HortResearch Marketing department for supplying images. We would also like to thank Ross Ferguson for providing the plant material. Finally, we would like to thank Ross Atkinson for his suggestions for the manuscript. Funded by The New Zealand Foundation of Science, Research and Technology contract C06X0403.
Glossary
Abbreviations
- GER
GDP-D-mannose-4,6-dehydratase
- GMD
GDP-L-fucose synthase
- PMM
phosphomannose mutase
- GMP
GDP-mannose pyrophosphorylase
- GME
GDP-mannose 3′,5′-epimerase
- GGT
GDP-L-galactose transferase
- GPP
L-galactose 1-P phosphatase
- GDH
L-galactose dehydrogenase
- GalUR
D-galacturonic acid reductase
- IPS
inositol-3-phosphate synthase
- MIOX
myo-inositol oxygenase
- MDAR
monodehydroascorbate reductase
- DHAR
dehydroascorbate reductase
- AO
ascorbate oxidase
- T1
permease/sodium-dependent ascorbate transporter
References
- Agius F, Gonzalez-Lamothe R, Caballero JL, Munoz-Blanco J, Botella MA, Valpuesta V. Engineering increased vitamin C levels in plants by over-expression of a D-galacturonic acid reductase. Nature Biotechnology. 2003;21:177–181. doi: 10.1038/nbt777. [DOI] [PubMed] [Google Scholar]
- Alhagdow M, Mounet F, Gilbert L, et al. Silencing of the mitochondrial ascorbate synthesizing enzyme L-galactono-1,4-lactone dehydrogenase affects plant and fruit development in tomato. Plant Physiology. 2007;145:1408–1422. doi: 10.1104/pp.107.106500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E. The preparation of L-galactose from flax seed mucilage. Journal of Biological Chemistry. 1933;100:249–253. [Google Scholar]
- Arrigoni O, De Tullio MC. The role of ascorbic acid in cell metabolism: between gene-directed functions and unpredictable chemical reactions. Journal of Plant Physiology. 2000;157:481–488. [Google Scholar]
- Baig MM, Kelly S, Loewus F. L-ascorbic acid biosynthesis in higher plants from L-gulono-1,4-lactone and L-galactono-1,4-lactone. Plant Physiology. 1970;46:277–280. doi: 10.1104/pp.46.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, Moeder W, Klessig DF, Conklin PL. The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin C-1. Plant Physiology. 2004;134:1784–1792. doi: 10.1104/pp.103.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL, Clark CJ, Klages KU. Identification of myo-inositol as a major carbohydrate in kiwifruit, Actinidia deliciosa. Phytochemistry. 1997;46:51–55. [Google Scholar]
- Boldingh H, Smith GS, Klages K. Seasonal concentrations of non-structural carbohydrates of five Actinidia species in fruit, leaf and fine root tissue. Annals of Botany. 2000;85:469–476. [Google Scholar]
- Chew O, Whelan J, Millar AH. Molecular definition of the ascorbate–glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. Journal of Biological Chemistry. 2003;278:46869–46877. doi: 10.1074/jbc.M307525200. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proceedings of the National Academy of Sciences, USA. 1999;96:4198–4203. doi: 10.1073/pnas.96.7.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Saracco SA, Norris SR, Last RL. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics. 2000;154:847–856. doi: 10.1093/genetics/154.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, et al. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics. 2008;9:351. doi: 10.1186/1471-2164-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tullio MC, Liso R, Arrigoni O. Ascorbic acid oxidase: an enzyme in search of a role. Biologia Plantarum. 2004;48:161–166. [Google Scholar]
- Debolt S, Melino V, Ford CM. Ascorbate as a biosynthetic precursor in plants. Annals of Botany. 2007;99:3–8. doi: 10.1093/aob/mcl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N. Two genes in Arabidopsis thaliana encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. The Plant Journal. 2007;52:673–689. doi: 10.1111/j.1365-313X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Ferguson AR, MacRae EA. Vitamin C in Actinidia. Acta Horticulturae. 1992;297:481–487. [Google Scholar]
- Gatzek S, Wheeler GL, Smirnoff N. Antisense suppression of L-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated L-galactose synthesis. The Plant Journal. 2002;30:541–553. doi: 10.1046/j.1365-313x.2002.01315.x. [DOI] [PubMed] [Google Scholar]
- Gillaspy GE, Keddie JS, Oda K, Gruissem W. Plant inositol monophosphatase is a lithium-sensitive enzyme encoded by a multigene family. The Plant Cell. 1995;7:2175–2185. doi: 10.1105/tpc.7.12.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MA, Fry SC. Apoplastic degradation of ascorbate: novel enzymes and metabolites permeating the plant cell wall. Plant Biosystems. 2005;139:2–7. [Google Scholar]
- Hancock RD, Walker PG, Pont SDA, Marquis N, Vivera S, Gordon SL, Brennan RM, Viola R. L-Ascorbic acid accumulation in fruit of Ribes nigrum occurs by in situ biosynthesis via the L-galactose pathway. Functional Plant Biology. 2007;34:1080–1091. doi: 10.1071/FP07221. [DOI] [PubMed] [Google Scholar]
- Hellens R, Allan A, Friel E, Bolitho K, Grafton K, Templeton M, Karunairetnam S, Gleave A, Laing W. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopping ME. Structure and development of fruit and seeds in Chinese gooseberry (Actinidia chinensis Planch) New Zealand Journal of Botany. 1976;14:63–68. [Google Scholar]
- Horemans N, Szarka A, De Bock M, Raeymaekers T, Potters G, Levine M, Banhégyi G, Guisez Y. Dehydroascorbate and glucose are taken up into Arabidopsis thaliana cell cultures by two distinct mechanisms. FEBS Letters. 2008;582:2714–2718. doi: 10.1016/j.febslet.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Nessler CL. Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Molecular Breeding. 2000;6:73–78. [Google Scholar]
- Johnson MD, Sussex IM. 1 L-myo-inositol 1-phosphate synthase from Arabidopsis thaliana. Plant Physiology. 1995;107:613–619. doi: 10.1104/pp.107.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages K, Donnison H, Boldingh H, MacRae E. myo-Inositol is the major sugar in Actinidia arguta during early fruit development. Australian Journal of Plant Physiology. 1998;25:61–68. [Google Scholar]
- Laing WA, Barraclough D, Bulley S, Cooney J, Wright M, Macrae E. A specific l-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proceedings of the National Academy of Sciences, USA. 2004;101:16976–16981. doi: 10.1073/pnas.0407453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Wright MA, Cooney J, Bulley SM. The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an L-galactose guanyltransferase, increases leaf ascorbate content. Proceedings of the National Academy of Sciences, USA. 2007;104:9534–9539. doi: 10.1073/pnas.0701625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leferink NGH, van den Berg WAM, van Berkel WJH. L-Galactono-γ-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis. FEBS Journal. 2008;275:713–726. doi: 10.1111/j.1742-4658.2007.06233.x. [DOI] [PubMed] [Google Scholar]
- Linster CL, Gomez TA, Christensen KC, Adler LN, Young BD, Brenner C, Clarke SG. Arabidopsis VTC2 encodes a GDP-l-galactose phosphorylase, the last unknown enzyme in the Smirnoff–Wheeler pathway to ascorbic acid in plants. Journal of Biological Chemistry. 2007;282:18879–18885. doi: 10.1074/jbc.M702094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA. Biosynthesis and metabolism of ascorbic acid in plants and of analogs of ascorbic acid in fungi. Phytochemistry. 1999;52:193–210. [Google Scholar]
- Loewus FA, Loewus MW. myo-Inositol: its biosynthesis and metabolism. Annual Review of Plant Physiology. 1983;34:137–161. [Google Scholar]
- Lopez-Carbonell M, Munne-Bosch S, Alegre L. The ascorbate-deficient vtc-1 Arabidopsis mutant shows altered ABA accumulation in leaves and chloroplasts. Journal of Plant Growth Regulation. 2006;25:137–144. [Google Scholar]
- Lorence A, Chevone BI, Mendes P, Nessler CL. myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiology. 2004;134:1200–1205. doi: 10.1104/pp.103.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major LL, Wolucka BA, Naismith JH. Structure and function of GDP-mannose-3′,5′-epimerase: an enzyme which performs three chemical reactions at the same active site. Journal of the American Chemical Society. 2005;127:18309–18320. doi: 10.1021/ja056490i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurino VG, Grube E, Zielinski J, Schild A, Fischer K, Flugge U-I. Identification and expression analysis of twelve members of the nucleobase-ascorbate transporter (NAT) gene family in Arabidopsis thaliana. Plant and Cell Physiology. 2006;47:1381–1393. doi: 10.1093/pcp/pcl011. [DOI] [PubMed] [Google Scholar]
- Mieda T, Yabuta Y, Rapolu M, Motoki T, Takeda T, Yoshimura K, Ishikawa T, Shigeoka S. Feedback inhibition of spinach L-galactose dehydrogenase by L-ascorbate. Plant and Cell Physiology. 2004;45:1271–1279. doi: 10.1093/pcp/pch152. [DOI] [PubMed] [Google Scholar]
- Naran R, Chen G, Carpita NC. Novel rhamnogalacturonan I and arabinoxylan polysaccharides of flax seed mucilage. Plant Physiology. 2008;148:132–141. doi: 10.1104/pp.108.123513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento JRO, Higuchi BK, Gómez MLPA, Oshiro RA, Lajolo FM. L-Ascorbate biosynthesis in strawberries: L-galactono-1,4-lactone dehydrogenase expression during fruit development and ripening. Postharvest Biology and Technology. 2005;38:34–42. [Google Scholar]
- Noctor G. Metabolic signalling in defence and stress: the central roles of soluble redox couples. Plant, Cell and Environment. 2006;29:409–425. doi: 10.1111/j.1365-3040.2005.01476.x. [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, Foyer CH. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. The Plant Cell. 2003;15:939–951. doi: 10.1105/tpc.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateraki I, Sanmartin M, Kalamaki MS, Gerasopoulos D, Kanellis AK. Molecular characterization and expression studies during melon fruit development and ripening of L-galactono-1,4-lactone dehydrogenase. Journal of Experimental Botany. 2004;55:1623–1633. doi: 10.1093/jxb/erh186. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rassam M, Laing W. Variation in ascorbic acid and oxalate levels in the fruit of Actinidia chinensis tissues and genotypes. Journal of Agricultural and Food Chemistry. 2005;53:2322–2326. doi: 10.1021/jf048197s. [DOI] [PubMed] [Google Scholar]
- Redgwell RJ. Composition of Actinidia mucilage. Phytochemistry. 1983;22:951–956. [Google Scholar]
- Roberts RM, Harrer E. Determination of L-galactose in polysaccharide material. Phytochemistry. 1973;12:2679–2682. [Google Scholar]
- Rozen S, Skaletsky HJ. Ace primer: automation of PCR primer design based on gene structure. Bioinformatics Application Notes. 1998;18:1538–1539. doi: 10.1093/bioinformatics/18.11.1538. [DOI] [PubMed] [Google Scholar]
- Seifert GJ. Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Current Opinion in Plant Biology. 2004;7:277–284. doi: 10.1016/j.pbi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Wheeler GL. Ascorbic acid in plants: biosynthesis and function. Critical Reviews in Biochemistry and Molecular Biology. 2000;35:291–314. doi: 10.1080/10409230008984166. [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Mukai F, Asai N, Nakajima N, Kubo A, Aono M, Saji H. Light-controlled expression of a gene encoding L-galactono-[γ]-lactone dehydrogenase which affects ascorbate pool size in Arabidopsis thaliana. Plant Science. 2003;164:1111–1117. [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Persiau G, Van Doorsselaere J, Davey MW, Demol H, Vandekerckhove J, Van Montagu M, Zabeau M, Boerjan W. Partial purification and identification of GDP-mannose 3′,5′-epimerase of Arabidopsis thaliana, a key enzyme of the plant vitamin C pathway. Proceedings of the National Academy of Sciences, USA. 2001;98:14843–14848. doi: 10.1073/pnas.011578198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolucka BA, Van Montagu M. GDP-mannose 3′,5′-epimerase forms GDP-l-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. Journal of Biological Chemistry. 2003;278:47483–47490. doi: 10.1074/jbc.M309135200. [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Van Montagu M. The VTC2 cycle and the de novo biosynthesis pathways for vitamin C in plants: an opinion. Phytochemistry. 2007;68:2602–2613. doi: 10.1016/j.phytochem.2007.08.034. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Yoshimura K, Takeda T, Shigeoka S. Molecular characterization of tobacco mitochondrial L-galactono-γ-lactone dehydrogenase and its expression in Escherichia coli. Plant and Cell Physiology. 2000;41:666–675. doi: 10.1093/pcp/41.6.666. [DOI] [PubMed] [Google Scholar]
- Zablackis E, York WS, Pauly M, Hantus S, Reiter WD, Chapple CC, Albersheim P, Darvill A. Substitution of L-fucose by L-galactose in cell walls of Arabidopsis mur1. Science. 1996;272:1808–1810. doi: 10.1126/science.272.5269.1808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.