Abstract
Circadian rhythms are generated by endogenous central oscillators that respond to input from the environment and regulate rhythmic outputs. In Arabidopsis, more than a dozen components that affect rhythms have been identified and used to propose models of the central oscillator. However, none has been shown to fulfill one of the expected characteristics of an oscillator component: that a pulse of its expression shifts the phase of circadian rhythms. Here we show that a pulse of the proposed oscillator components CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) causes dramatic phase shifts in rhythms of expression of the circadian reporter CAB2∷LUC, as well as of the clock-associated genes TIMING OF CAB EXPRESSION 1 (TOC1) and GIGANTEA (GI). These results demonstrate that pulses of either CCA1 or LHY are capable of resetting the circadian clock. In contrast, a pulse of TOC1 expression did not elicit phase shifts. Control of TOC1 protein level is in part posttranscriptional; thus a pulse of TOC1 protein could be induced only at times when it is already high. Our work also shows that the ethanol-inducible system can be useful for achieving relatively short (<8 h) pulses of gene expression in seedlings.
Keywords: Arabidopsis thaliana, circadian rhythm, central oscillator, phase shift, clock gene, ethanol-inducible system
The CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene was initially characterized as a transcription factor involved in the phytochrome regulation of a gene encoding a light-harvesting chlorophyll a/b-protein (Lhcb/CAB gene; Wang et al., 1997) and was then found to be intimately involved with circadian rhythms (Wang and Tobin, 1998). Not only do both its RNA and protein show circadian rhythms of expression, but constitutive expression of CCA1 abolishes all examined circadian rhythms, such as those seen in leaf movement and hypocotyl growth. Constitutive CCA1 expression also results in the repression of the endogenous CCA1 gene, showing it functions in an autoregulatory feedback loop (Wang and Tobin, 1998). Experiments with the closely related homologue, LATE ELONGATED HYPOCOTYL (LHY), gave similar results (Schaffer et al., 1998). Therefore, CCA1 and LHY have been proposed as central components in models of the plant circadian clock (Wang and Tobin, 1998; Schaffer et al., 1998; McClung, 2006).
The consensus model for central oscillator function in eukaryotes involves multiple feedback loops based on gene transcription and translation (Young and Kay, 2001; Dunlap and Loros, 2004). An expected characteristic of an oscillator component is that a pulse of its expression will reset the phase of rhythms (Aronson et al., 1994). This feature has been demonstrated for oscillator components in cyanobacteria (KaiC; Ishiura et al., 1998), Neurospora (FREQUENCY; Aronson et al., 1994), and Drosophila (PERIOD; Edery et al., 1994). In Arabidopsis, the proposed core feedback loop of the oscillator consists of CCA1 and LHY, along with TIMING OF CAB EXPRESSION 1 (TOC1) (also known as PSEUDO-RESPONSE REGULATOR 1 (PRR1; Strayer et al., 2000; Makino et al., 2002)). In this proposed loop, TOC1 acts as a positive element to promote the expression of CCA1 and LHY through an unknown mechanism, while CCA1 and LHY proteins function as negative elements to repress TOC1 transcription through direct binding to its promoter (Alabadí et al., 2001). Recently, other genes such as GIGANTEA (GI), LUX ARRHYTHMO (LUX; also known as PHYTOCLOCK 1 (PCL1)), EARLY FLOWERING 4 (ELF4), TIME FOR COFFEE (TIC), and PRR3/5/7/9 have also been proposed to be part of the plant central oscillator (reviewed in McClung, 2008). However, the biochemical functions of the encoded proteins have remained elusive and none has been tested to see the effect of a pulse of its expression on rhythms. Here we sought to determine whether pulses of CCA1, LHY, or TOC1 expression reset the circadian clock.
MATERIALS AND METHODS
Plant lines and growth conditions
The Alc lines generated in this study, as well as toc1-4 (Hazen et al., 2005) and PRR1-OX (Makino et al., 2002), are in the Col ecotype.
Seeds were stratified for 3 days in the dark at 4 °C and then sown onto MS medium (Research Products International Corp., Mt. Prospect, IL) containing 1.5% (w/v) agarose (MS1.5). Seedlings were grown on 90-mm tissue culture plates. For ethanol (EtOH)-pulse experiments the seeds were sown on either 80-mm- or 15-mm-diameter nylon net filter circles (20 mm pore size; Millipore Corp., Bedford, MA) upon MS1.5. The filter served to facilitate the transfer of grown seedlings to fresh medium after EtOH treatment (see “EtOH pulse” section). Seedlings were grown under a 12-h fluorescent light (40 µmol · m−2 · sec−1):12-h dark (12:12) photoperiod at a constant temperature of 22 °C.
DNA constructs for the EtOH-inducible gene expression system
EtOH-inducible DNA constructs were derived from the binary vector pbinSRNACatN (Syngenta, Berkshire, UK). The effector construct pAlcA∷Catalase:TNOS was removed from pbinSRNACatN by HindIII digestion and discarded. The remaining vector sequence harboring the regulator construct 35S∷ AlcR:TNOS was subsequently religated to yield pbinSRN.
A CCA1-containing effector construct was generated as follows: CCA1 (1.8 kb) was amplified from Arabidopsis cDNA by PCR using the primers CCA1-EcoRI3F, 5′-GAATTCATGGAGACAAATTCG-3′ and CCA1-BamHIR, 5′-GACGGATCCTCATGTGGAAGC-3′ and sequenced. The CCA1 ORF was inserted into the EcoRI/BamHI sites of the binary vector pEGAD (Cutler et al., 2000), creating pEGAD-CCA1. The 35S promoter and EGFP sequences were subsequently removed by StuI + EcoRI digestion and replaced with the 287-bp AlcA promoter sequence (HindIII/SalI fragment from pbinSRNACatN) by blunt-ended ligation. Correct orientation of the promoter was confirmed by sequencing. The vector was called pEGAD-pAlcA∷CCA1.
Effector constructs for LHY and TOC1 were made in a similar manner to pEGAD-pAlcA∷CCA1. LHY was cloned using the primers LHY-MfeIF, 5′-CAATTGATGGATACTAATACA-3′ and LHY-XhoIR, 5′-CTCGAGTCATGTAGAAGCTTC-3′. TOC1 was cloned using TOC1-F, 5′-CTGATCATGGATTTGAACG-3′ and TOC1-R, 5′-GCCTTAGAGACAACTCGATAT-3′.
The regulator and effector constructs were independently transformed into WT Arabidopsis via the Agrobacterium-mediated floral dip method (Clough and Bent, 1998). Established effector lines for CCA1, LHY, and TOC1 were crossed with plants carrying the regulator construct to yield Alc∷CCA1, Alc∷LHY and Alc∷TOC1.
CAB2∷LUC circadian reporter
A translation enhancer sequence, Ω (Gallie et al., 1987), was created by annealing the following complementary DNA oligos: Omega(+), 5′-AGCTTATTTTTACAACAATTACCAACAACAACAAACAACAAACAACATTACAATTACTATTTACAATTAC-3′ and Omega(−), 5′-CATGGTAATTGTAAATAGTAATTGTAATGTTGTTTGTTGTTTGTTGTTGTTGGTAATTGTTGTAAAAATA-3′. Double-stranded Ω was inserted into the XhoI/HindIII sites of pGL3-Promoter Vector (Promega Corp., Madison, WI), yielding an Ω:LUC fusion. Ω:LUC was then subcloned into the BamHI/XbaI sites of the plant binary vector pKYLX71 (Schardl et al., 1987) (the 35S promoter within pKYLX71 had been previously removed by EcoRI + XhoI digestion). The resulting promoter-less vector was called pKYLX-Ω:LUC. To create the CAB2∷LUC reporter construct, the −222 to +7 region (relative to transcription start) of the CAB2 promoter was amplified from Arabidopsis gDNA by PCR using the primers CAB2p_F, 5′-GGTACCGTTGAAGTATTCAG-3′, and CAB2p_R, 5′-CTCGAGAGTGATTAAAACTG-3′. The amplified CAB2 promoter fragment was ligated upstream of Ω in pKYLX-Ω:LUC at the SacI/XhoI sites, yielding pKYLXCAB2∷LUC. CAB2∷LUC was transformed into WT, Alc∷CCA1, Alc∷TOC1, and Alc∷LHY plants.
RNA quantitation
Total RNA was extracted from 8-day-old seedlings as described (Chang et al., 1993). The RNA was treated with RQ1 DNase (Promega Corp., Madison, WI) and reverse transcribed using random hexamer primers and M-MLV Reverse Transcriptase (Invitrogen Corp., Carlsbad, CA) following the manufacturer’s instructions. Quantitative RT-PCR reactions were carried out on an MX3000P machine (Stratagene, La Jolla, CA) using general PCR reagents (Denville Scientific, Metuchen, NJ) supplemented with 0.5X SYBR Green I (diluted from a “10,000 X” stock; Invitrogen Corp.). RNA HELICASE 8 (RH8; Schaffer et al., 2001) was used as a noncycling control. Reactions were performed in triplicate.
Quantitative RT-PCR primers
| CCA1_F | 5′-GGGGTGTGAATGATGGAAAAGA-3′ |
| CCA1_R | 5′-CGATCTTCATTGGCCATCTCAG-3′ |
| CCR2_F | 5′-GCTCTTGAGACTGCCTTCGCTC-3′ |
| CCR2_R | 5′-CTCGTTAACAGTGATGCTACGG-3′ |
| GI_F | 5′-AAGCGACTTTTAGACGATGTGCTG-3′ |
| GI_R | 5′-GATTGGAAGAATGACTGCATGACC-3′ |
| LHY_F | 5′-GACAACGCGGTTCAAGATGTTC-3′ |
| LHY_R | 5′-CCAAGGGTAGTTTTGCATGCTG-3′ |
| LUX_F | 5′-AGCCGTACGATTCAGGATGTGA-3′ |
| LUX_R | 5′-CTCGATCTTCCTCCTCCACGTT-3′ |
| PRR9_F | 5′-ATTGACAAGCGTCCTGATAGTATTTAT-3′ |
| PRR9_R | 5′-AACTTACAGAGCAAGATCTTTTCAGAG-3′ |
| RH8_F | 5′-CAATGGCTTCGAAGAGGTCAGA-3′ |
| RH8_R | 5′-TGGGTCGATAACTTCGGATTGA-3′ |
| TOC1_F | 5′-GTGGTCTTGGTGCTGATGGAAC-3′ |
| TOC1_R | 5′-GACCCAGCAAGATGATCCAATG-3′ |
Anti-TOC1 antibody production
A fragment of the TOC1 open reading frame encoding amino acid residues 164 to 294 was amplified by PCR using the primers 5′-GGGATCTGAATTCAGTGATCCAAACAC-3′ and 5′-AAGATGACTCGAGCCATTTCCATCGAC-3′. The fragment was introduced into the EcoRI/XhoI sites of the vector pGEX-4T-1 (Pharmacia, Uppsala, Sweden). Recombinant GST-TOC1(164–294) was expressed in E. coli BL21/DE3 cells and purified using glutathione sepharose (GE Healthcare, Buckinghamshire, UK) as described (Wang et al., 1997). Recombinant proteins were used to generate polyclonal anti-TOC1 antiserum (Harlan Bioproducts for Science, Inc., Indianapolis, IN). Antiserum was subjected to immunoaffinity purification against GSTTOC1-fragment immobilized to a nitrocellulose membrane as described (Olmsted, 1981).
Western blotting
Protein from 8-day-old seedlings was extracted in 1× urea/SDS buffer (4 M urea, 2.5% SDS, 20 mM Tris-HCl, pH 6.8, and 0.05 mM EDTA, pH 8.0) containing 1× protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN), 2 mM PMSF, 50 µM MG115, and 50 µM MG132. Protein concentrations were measured with a DC protein assay reagent (Bio-Rad, Hercules, CA) and 50 µg of total proteins was separated on 8% (w/v) SDS-polyacrylamide gels. Proteins were electrophoretically transferred to PVDF membranes (Millipore, Bedford, MA). Immunoblotting procedures were performed as described (Lu and Hrabak, 2002) using antibodies to CCA1 (Wang and Tobin, 1998), LHY (Daniel et al., 2004), TOC1, or actin. Quantitation of immunodetected proteins was performed using ImageJ software (http://rsb.info.nih.gov/ij/).
EtOH pulse
Transient activation of the EtOH-inducible system was achieved by short exposures (≤ 20 min) of 8- to 11-day-old seedlings to EtOH vapor. The tissue culture plates served as convenient vapor chambers. For seedlings on 90-mm plates, inductions were carried out by wetting a paper filter (83-mm diameter; Chromatography Paper (thick); Fisher Scientific Inc., Hampton, NH) with diluted EtOH (in water) and adhering it to the underside of the plate lid. For seedlings on 24-well plates, paper filters of dimensions 10.7 × 13.5 cm were adhered to the inner lid of a P5000 pipette tip box (Denville Scientific, Metuchen, NJ). The concentration of the EtOH was 1% (v/v) unless stated otherwise. The seedlings were moved to fresh MS1.5 after treatment to avoid continuous induction from residual EtOH.
Bioluminescence assays
Groups of approximately 20 CAB2∷LUC seedlings were grown on 15-mm-diameter nylon net filters (20 mm pore size; Millipore Corp., Bedford, MA) on MS1.5 for 6 days in 12:12. The seedlings were then transferred to wells of a 24-well tissue culture plate (#3047 Falcon; Becton Dickinson & Co., Franklin Lakes, NJ) that contained 0.6 mL of MS1.5 and 1 mM D-luciferin (catalog number L-8220; Biosynth AG, Switzerland). The seedlings were incubated in 12:12 for 2 more days before transfer to continuous light (LL) for the experiment.
Six-minute exposures were acquired with a CCD camera (Princeton Instruments VersArray 512B; Roper Scientific, Inc., Trenton, NJ) every 60 min. The LL light source was a mixture of red (λmax = 660 nm) and blue (λmax = 470 nm) light-emitting diodes with a combined fluence rate of approximately 30 µmol · m−2 · sec−1. Luciferase activity was quantified using ImageJ software. Rhythmic period was measured using the freeware program PEANUTS (http://www.vu-wien.ac.at/i128/Peanuts.htm) that calculates Lomb-Scargle periodograms.
Phase shift measurements
Groups of T2 CAB2∷LUC seedlings in homozygous WT, Alc∷CCA1, Alc∷LHY, and Alc∷TOC1 backgrounds were treated with EtOH vapor at different times during the 4th cycle in LL (72 to 96 h). Rhythmic luciferase activity was subsequently monitored for 4 days. Relative phase shifts in CAB2∷LUC rhythms between control (untreated) and EtOH-treated groups were measured by cross-correlation analysis (Levine et al., 2002). Data from the first 24 h after treatment were not included in the analysis due to a temporary increase in CAB2∷LUC activity caused by EtOH directly and/or movement of seedlings to fresh medium. Phase shifts and times of EtOH treatment were converted to circadian time (CT) by multiplying by (24/free running period (h)). Average free running period ± SEM of CAB2∷LUC rhythms in various backgrounds: WT, 25.84 ± 0.14 h (n = 8); Alc∷CCA1, 25.44 ± 0.13 h (n = 15); Alc∷LHY, 25.58 ± 0.29 h (n = 6); and Alc∷TOC1, 25.72 ± 0.21 h (n = 24).
RESULTS AND DISCUSSION
To give a pulse of putative oscillator components, we utilized the EtOH-inducible system from Aspergillus nidulans (Caddick et al., 1998; Roslan et al., 2001). The reasons for choosing this system were three-fold. First, the system showed promise for being capable of generating short pulses (<24 h) of gene expression (Deveaux et al., 2003). Second, it is a relatively simple system as it comprises just two expression constructs (Fig. 1a). Third, it was unlikely that the inducer would affect the circadian clock mechanism because very low concentrations of EtOH are sufficient for induction (Roslan et al., 2001). The requirement that the inducer must not affect circadian rhythms ruled out the option of using a heat-shock inducible system because the Arabidopsis circadian clock can use temperature changes as input for entrainment (Somers et al., 1998).
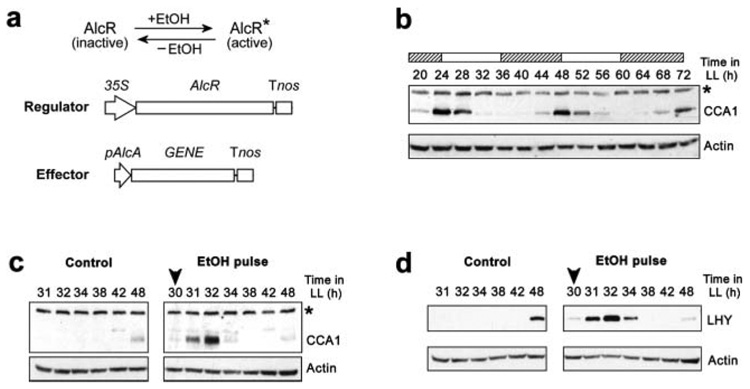
Figure 1. Ethanol (EtOH)-mediated pulses of CCA1 and LHY expression in plants.
(a) DNA constructs for the EtOH-inducible system. The ability of AlcR to function as a transcriptional activator is dependent on the presence of EtOH. Active AlcR binds the inducible promoter, pAlcA, and promotes transcription of the gene-of-interest. Tnos = terminator of the NOS gene; 35S = constitutive promoter. (b) Western blot showing the rhythm of CCA1 abundance in WT plants under continuous light (LL). (c, d) Western blots showing pulses of protein following an EtOH vapor treatment at 30 h in LL. (c) Pulse of CCA1 protein in Alc∷CCA1 seedlings. (d) Pulse of LHY protein in Alc∷LHY seedlings. Treatment time is marked by the arrowhead. Control plants were not treated. Asterisk = nonspecific band. Subjective day and subjective night are denoted by white and hatched bars, respectively.
CCA1, LHY, and TOC1 were used as effectors (inducible genes; Fig. 1a) and introduced into plants transformed with the regulator construct. The resulting plant lines were called Alc∷CCA1, Alc∷LHY, and Alc∷TOC1. The circadian oscillations of endogenous CCA1 and LHY protein show peaks of expression at subjective dawn (Fig. 1b, and Daniel et al., 2004). EtOH treatment of Alc∷CCA1 or Alc∷LHY seedlings in the middle of the subjective day, when endogenous CCA1 and LHY levels are low, resulted in a substantial increase in their RNA (Fig. S1a, b; Figures S1–S6 available online at http://jbr.sagepub.com/supplemental/) and protein (Fig. 1c, d). EtOH-induced CCA1 RNA peaked 1 h after treatment and then rapidly declined. The peak level of induced CCA1 exceeded the peak level of endogenous CCA1 RNA by approximately four-fold (Fig. S1a). Induced CCA1 protein peaked 2 h after treatment and also exceeded its endogenous peak level at 48 h in LL (Fig. 1c). Similar results were obtained for EtOH treatment on Alc∷LHY plants, except that the peak level of induced LHY was less than two-fold higher than the peak level of endogenous LHY RNA (Fig. S1b). By treating with EtOH at various times in LL, we found that induction of either protein can be carried out at any point in the circadian cycle (Fig. S2).
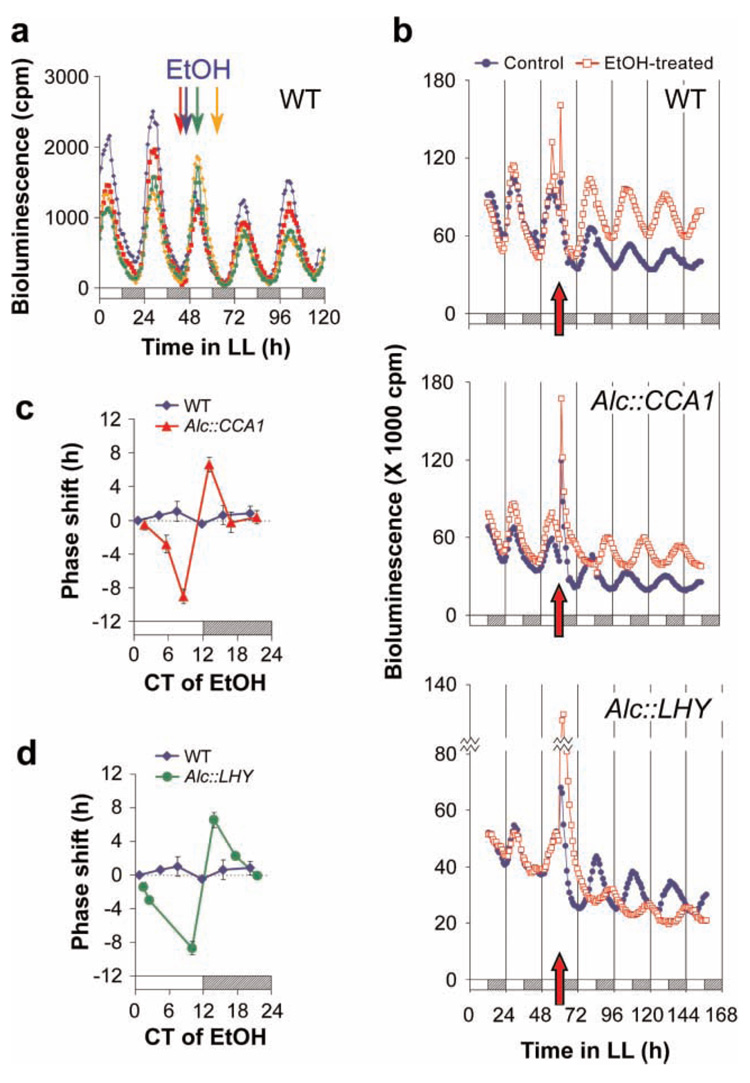
To monitor circadian rhythms over time we utilized the CAB2∷LUC circadian reporter (Millar et al., 1995), which was transformed into the plants containing the inducible Alc constructs. EtOH treatment given at four different times in the circadian cycle did not affect the luciferase rhythms observed in WT plants (Fig. 2a). However, when CCA1 or LHY expression was induced by EtOH at subjective dusk, the luciferase rhythm was shifted by 9 h (Fig. 2b). The phase shifts were evident within 24 h of the treatment and were remarkably stable as they did not revert back to the original entrained phase even after 13 days (Fig. S3). By treating with EtOH at various times we constructed phase-response curves (PRCs) for the CCA1 and LHY pulses (Fig. 2c, d). The PRCs showed that EtOH treatment near subjective dawn (circadian time 0 (CT 0)) did not cause phase shifts. This result was expected because CCA1 and LHY are already abundant at this point in the circadian cycle. Relatively large phase shifts were elicited when the EtOH treatment was given near CT 12, when endogenous CCA1 and LHY levels are low. In contrast, the PRC for WT plants showed no phase shifts for the EtOH treatments. These results demonstrate that a pulse of expression of either CCA1 or LHY is sufficient to reset the phase of the circadian clock. The PRCs for a CCA1 or LHY pulse resemble the published PRC for a red light pulse (Covington et al., 2001), with phase delays and advances occurring in the subjective day and subjective night, respectively. Because CCA1 and LHY expression is induced by red light (Wang et al., 1997; Kim et al., 2003), this result is consistent with the hypothesis that light-induced changes in their expression contribute to the entrainment of the circadian clock by light. Interestingly, an analogous mechanism is present in the Neurospora clock, where resetting of the clock to light is mediated by FREQUENCY, a central oscillator gene whose expression is also induced by light (Crosthwaite et al., 1995).
Figure 2. A pulse of CCA1 or LHY expression is sufficient to reset the circadian clock.
(a) Ethanol (EtOH) does not affect the circadian phase of CAB2∷LUC rhythms in WT seedlings. Each plot represents the mean bioluminescence of 6 individual seedlings. Colored arrows represent times of a 20-min exposure to 10% (v/v) EtOH for their corresponding colored plots. (b) Effect of a 1% (v/v) EtOH pulse on the phase of CAB2∷LUC rhythms in WT, Alc∷CCA1, and Alc∷LHY seedlings. The times of treatment with EtOH are denoted by a red arrow. Plots represent the mean bioluminescence of 20 seedlings. (c, d) Phase-response curves (PRCs) for a 1% (v/v) EtOH pulse on WT plants versus Alc∷CCA1 (c) and Alc∷LHY (d). Seedlings were treated with EtOH at different times in LL and the resulting phase shifts in CAB2∷LUC rhythms were plotted as a function of treatment time. Delays are negative phase shifts and advances are positive. Error bars denote ± SD (n = 2 to 9). CT = circadian time; cpm = counts per minute.
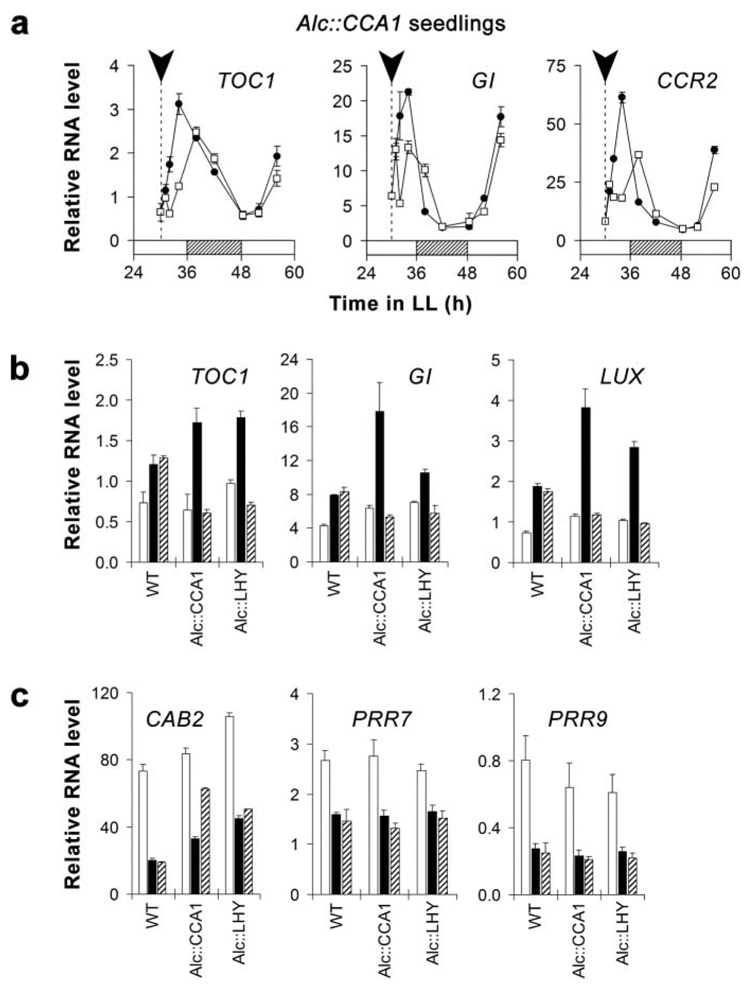
To investigate whether the observed phase shift in CAB2∷LUC rhythms is due to resetting of the central clock, the effect of a CCA1 pulse on the rhythms of other clock-controlled genes was examined. When CCA1 was induced during the subjective day (CT 6), the RNA accumulation of TOC1 and GI was delayed (Fig. 3a). A phase delay was also detected in the RNA accumulation of the evening-phased output gene COLD-CIRCADIAN RHYTHM-RNA BINDING 2 (CCR2; Carpenter et al., 1994; Fig. 3a). Additionally, peaks of TOC1 and CCR2 RNA were shifted by 4 h, which is consistent with the observed shift in CAB2∷LUC rhythms when CCA1 expression was induced at a similar time (compare Fig. 3a with Fig. 2c). A pulse of LHY expression at CT 6 resulted in a similar phase delay in TOC1 expression (Fig. S4). Together, these results demonstrate that a pulse of expression of either CCA1 or LHY can set the phase of the circadian clock.
Figure 3. Effect of induced CCA1 or LHY expression in the middle of the subjective day (CT 6) on the expression of clock-controlled genes.
(a) A pulse of CCA1 expression delays the peak of TOC1, GI, and CCR2 RNA. Alc∷CCA1 seedlings were given an ethanol (EtOH) pulse at the time indicated by the arrowhead. Closed circles = untreated plants; open squares = EtOH-treated. (b) A pulse of CCA1 or LHY represses the accumulation of TOC1, GI, and LUX RNA. (c) Effect of CCA1 and LHY inductions on the accumulation of CAB2, PRR7, and PRR9 RNA. WT, Alc∷CCA1, and Alc∷LHY seedlings were given an EtOH pulse at 30 h in LL (CT 6). White bar = relative RNA level at the time of induction; black bar = level after 2 h (no EtOH); hatched bar = level 2 h after EtOH. RNA abundance was measured by quantitative RT-PCR and normalized to RNA HELICASE 8 (RH8). Error bars denote ± SD (n = 3).
It has been suggested that CCA1 and LHY function as negative elements of the central oscillator to inhibit transcription of positive elements (Alabadí et al., 2001). Therefore, we examined the effect of a pulse of CCA1 or LHY on the expression of proposed central oscillator genes TOC1, GI, and LUX. As shown in Figure 3b, a pulse of either CCA1 or LHY at a time when TOC1, GI, and LUX RNA levels are increasing (CT 6) caused a virtually complete inhibition of the increase within 2 h of the EtOH treatment. This result is consistent with CCA1 and LHY functioning as transcriptional repressors in the central oscillator. In addition to their roles as repressors, CCA1 and LHY have been proposed to activate expression of CAB2, PRR7, and PRR9 (Wang et al., 1997; Farré et al., 2005). Figure 3c shows that a CCA1 or LHY pulse at CT 6 inhibited the decline of CAB2 RNA. The fact that the inhibition caused by the LHY pulse was less than that for CCA1 is likely to be because of the weaker induction of LHY relative to its circadian peak level (Fig. 1c, d; Fig. S1a, b). However, neither PRR7 nor PRR9 expression was affected by the pulses (Fig. 3c); thus neither CCA1 nor LHY alone are sufficient to activate their expression. Together these results support the idea that CCA1 and LHY are involved in both the activation and the repression of clock-associated genes.
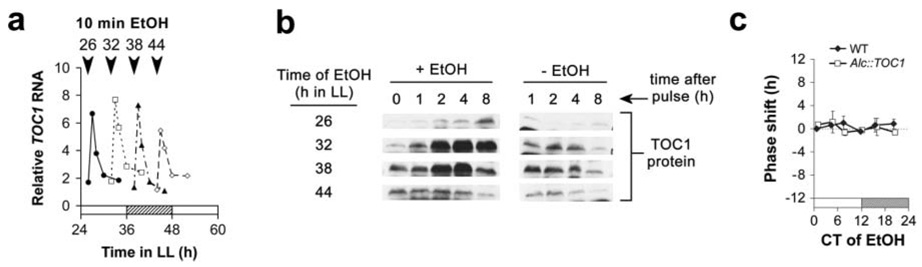
In the core feedback loop of the central oscillator, TOC1 has been proposed to act as an indirect activator of CCA1 and LHY expression (Alabadí et al., 2001). To determine whether a pulse of TOC1 can also reset the phase of the circadian clock, Alc∷TOC1 plants were generated. An EtOH treatment is able to induce TOC1 RNA to approximately 4-fold the level of its endogenous peak, which is comparable to CCA1 induction in Alc∷CCA1 plants (Fig. S1a, c). To confirm that the RNA induction leads to accumulation of TOC1 protein, TOC1-antibodies were generated (Fig. S5). While TOC1 RNA was induced to a similar level at any point in the circadian cycle (Fig. 4a), TOC1 protein was not (Fig. 4b). When EtOH treatment was given near subjective dusk (32 and 38 h in LL), at a time of high endogenous TOC1 expression, TOC1 protein peaked 2 to 4 h after treatment and exceeded the peak level of the endogenous protein (Fig. 4b). However, only a weak accumulation of TOC1 was detected when the inductions were performed near subjective dawn (26 and 44 h in LL), antiphasic to its normal circadian peak (Fig. 4b). These results indicate that the synthesis and/or stability of TOC1 protein is temporally regulated, perhaps by the circadian clock itself. TOC1 protein stability is known to be modulated by ZEITLUPE (ZTL), an F-box protein that also functions as a circadian photoreceptor (Más et al., 2003; Kim et al., 2007). ZTL interacts with TOC1 and targets it for degradation through the 26S proteasome pathway (Más et al., 2003). PRR3, whose expression also peaks in the evening, was recently proposed to function as an inhibitor of this interaction, thereby making TOC1 more stable (Para et al., 2007). It is possible that the induced TOC1 protein in Alc∷TOC1 seedlings at subjective dawn is degraded rapidly through its interaction with ZTL, perhaps due to the lack of protection from PRR3.
Figure 4. Induction of TOC1 protein in Alc∷TOC1 seedlings is restricted to the subjective evening.
(a, b) Alc∷TOC1 seedlings were given a 3% ethanol (EtOH; v/v) pulse at various times (26, 32, 38, and 44 h in LL) and tissue was harvested over time. (a) TOC1 RNA is EtOH inducible at any point in the circadian cycle in the Alc∷TOC1 line. RNA levels were measured by quantitative RT-PCR and were normalized to levels of RH8 RNA. Arrowheads denote times of EtOH treatment for their respective plot. Induction times in LL: circles = 26 h; squares = 32 h; triangles = 38 h; diamonds = 44 h. (b) Western blots showing the accumulation of TOC1 protein following the EtOH pulses. Total protein extracts were isolated from the same tissue as used in (a) and were processed by SDS-PAGE. Blots were probed with purified TOC1 antibodies. (c) Phase-response curve for an EtOH pulse on WT versus Alc∷TOC1 seedlings. Error bars denote ± SD (n = 2 to 9).
To examine whether the weak induction of TOC1 protein near subjective dawn and the increased peak level near subjective dusk in Alc∷TOC1 plants affect the phase of the circadian clock, we generated a PRC, again using CAB2∷ LUC as a circadian reporter. We could not detect any phase shifts in rhythms following TOC1 induction at different CTs (Fig. 4c). To rule out the possibility that the induced TOC1 protein is inactive, we induced its expression continuously and measured the period length of the CAB2∷LUC rhythm. Increased TOC1 dosage has been shown to result in long-period rhythms (Más et al., 2003). Figure S6, in the supplementary information, shows that constant induction of TOC1 expression did result in a period lengthened by 6 h, demonstrating that the induced TOC1 is functional. This finding is in agreement with previous reports that found that TOC1 protein level is significantly affected in both prr3 and ztl mutants (Más et al., 2003; Para et al., 2007), suggesting that TOC1 is important for the regulation of circadian period by the clock.
In this work, we have demonstrated that a pulse of CCA1 or LHY expression in plants is sufficient to reset the circadian clock, showing that both proteins function as phase-setting components of the Arabidopsis circadian clock and fulfill one of the proposed criteria for central oscillator components. In addition, the observation that the induced TOC1 protein accumulates only during the subjective evening suggests that TOC1 expression is tightly regulated by the circadian clock and that its protein must exceed a certain level for it to perform its clock-associated function(s). Moreover, we show that the use of the EtOH-inducible system can provide a means for experimentally testing the effect of a pulse of virtually any gene on a biological pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jackie Paine at Zeneca for providing us with the initial plasmid and guidelines for using the EtOH-inducible system, Dr. Steve Kay’s group for providing us with toc1-4 seeds, and Dr. Takeshi Mizuno for providing PRR1-OX seeds. We also thank Drs. Chentao Lin and Candace Webb for thoughtful suggestions and critical reading of this manuscript, and Carole Gorin for her excellent administrative assistance. This work was supported by NIH grant GM23167 to E.M.T. and the George Laties Fellowship to S.M.K.
Footnotes
The online version of this article can be found at: http://jbr.sagepub.com/cgi/content/abstract/23/6/463
REFERENCES
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: Autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- Caddick MX, Greenland AJ, Jepson I, Krause KP, Qu N, Riddell KV, Salter MG, Schuch W, Sonnewald U, Tomsett AB. An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat Biotechnol. 1998;16:177–180. doi: 10.1038/nbt0298-177. [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13:1305–1315. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Loros JJ, Dunlap JC. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci U S A. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:3292–3297. doi: 10.1073/pnas.0400163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveaux Y, Peaucelle A, Roberts GR, Coen E, Simon R, Mizukami Y, Traas J, Murray JA, Doonan JH, Laufs P. The ethanol switch: A tool for tissue-specific gene induction during plant development. Plant J. 2003;36:918–930. doi: 10.1046/j.1365-313x.2003.01922.x. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ. The Neurospora circadian system. J Biol Rhythms. 2004;19:414–424. doi: 10.1177/0748730404269116. [DOI] [PubMed] [Google Scholar]
- Edery I, Rutila JE, Rosbash M. Phase shifting of the circadian clock by induction of the Drosophila period protein. Science. 1994;263:237–240. doi: 10.1126/science.8284676. [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Sleat DE, Watts JW, Turner PC, Wilson TM. A comparison of eukaryotic viral 5′-leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res. 1987;15:8693–8711. doi: 10.1093/nar/15.21.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Borevitz JO, Harmon FG, Pruneda-Paz JL, Schultz TF, Yanovsky MJ, Liljegren SJ, Ecker JR, Kay SA. Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 2005;138:990–997. doi: 10.1104/pp.105.061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- Kim JY, Song HR, Taylor BL, Carré IA. Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 2003;22:935–944. doi: 10.1093/emboj/cdg075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM. An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol. 2002;128:1008–1021. doi: 10.1104/pp.010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002;43:58–69. doi: 10.1093/pcp/pcf005. [DOI] [PubMed] [Google Scholar]
- Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15:223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. Comes a time. Curr Opin Plant Biol. 2008;11:514–520. doi: 10.1016/j.pbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- Olmsted JB. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981;256:11955–11957. [PubMed] [Google Scholar]
- Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA. PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–3473. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, White MR, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, et al. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 2001;28:225–235. doi: 10.1046/j.1365-313x.2001.01146.x. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DF, Hunt AG. Design and construction of a versatile system for the expression of foreign genes in plants. Gene. 1987;61:1–11. doi: 10.1016/0378-1119(87)90359-3. [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AA, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.