Abstract
The leader RNA of the 5′ untranslated region (UTR) of coronaviral genomes contains two stem–loop structures denoted SL1 and SL2. Herein, we show that SL1 is functionally and structurally bipartite. While the upper region of SL1 is required to be paired, we observe strong genetic selection against viruses that contain a deletion of A35, an extrahelical nucleotide that destabilizes SL1, in favor of genomes that contain a diverse panel of destabilizing second-site mutations, due to introduction of a noncanonical base pair near A35. Viruses containing destabilizing SL1-ΔA35 mutations also contain one of two specific mutations in the 3′ UTR. Thermal denaturation and imino proton solvent exchange experiments reveal that the lower half of SL1 is unstable and that second-site SL1-ΔA35 substitutions are characterized by one or more features of the wild-type SL1. We propose a “dynamic SL1” model, in which the base of SL1 has an optimized lability required to mediate a physical interaction between the 5′ UTR and the 3′ UTR that stimulates subgenomic RNA synthesis. Although not conserved at the nucleotide sequence level, these general structural characteristics of SL1 appear to be conserved in other coronaviral genomes.
Abbreviations: UTR, untranslated region; CoV, coronavirus; SARS, severe acute respiratory syndrome; MHV, mouse hepatitis virus; gRNA, genomic RNA; TRS-B, body transcription regulatory sequence; sgRNA, subgenomic RNA; WT, wild type; RT, reverse transcription; 1D, one-dimensional; hpe, hours postelectroporation; DSS, 2,2-dimethyl-2-silapentane-5-sulfonate
Keywords: coronavirus, RNA virus replication, RNA dynamics, genetic interaction, mouse hepatitis virus
Introduction
Coronaviruses (CoVs) comprise a large group of enveloped, single-stranded, positive-sense mRNA viruses classified in the family Coronaviridae of the order Nidovirales.1 CoVs infect many different vertebrate hosts and induce a broad spectrum of diseases, including severe acute respiratory syndrome (SARS).2, 3 CoVs have been classified into three subgroups based on serologic relatedness, genetic organization, and sequence similarity. Mouse hepatitis virus (MHV) is the most extensively studied CoV, is genetically tractable, and provides small animal models for several diseases including SARS,4 a disease caused by the related group 2 SARS CoV.
Like other CoVs, MHV has a large ≈ 32-kb positive-sense viral RNA genome flanked by 5′ and 3′ untranslated regions (UTRs). The 5′ two-thirds of the genome encodes the replicase complex (open reading frame 1), whose expression requires a − 1 1a/1b translational frameshift,5 with downstream open reading frames encoding a variety of structural and accessory proteins required for replication and assembly of infectious viruses.1, 6 MHV transcription generates a set of nested subgenomic mRNAs, which contain 5′ and 3′ regions that are identical with those of the genomic RNA (gRNA).7, 8, 9 The most widely accepted model used to explain this unique discontinuous transcription mechanism posits that subgenomic negative-strand RNAs are synthesized from genome-sized template, and that the subgenomic mRNAs are synthesized from subgenomic minus strands.10, 11, 12 Zuniga et al. have refined this model by providing strong support for the hypothesis that heptameric core intergenic sequences or “body” transcription regulatory sequences (TRS-Bs) signal template switching to the TRS 5′ leader (TRS-L), as well as suggest the existence of RNA–RNA interactions between the 5′ UTR and 3′ UTRs and the proteins bound to these UTRs.13
Many cis-acting sequences in the 5′ UTR have been shown to play an important role in RNA transcription and replication;14, 15, 16, 17, 18 however, insights into the secondary structure of the 5′ UTR were, until recently, limited. We have developed a consensus secondary structure model based on the sequences of nine CoVs, in which the 5′ leader RNA, consisting of the 5′ most 72 nucleotides of the 5′ UTR in MHV, is characterized by two conserved stem–loop structures, denoted SL1 and SL2.19, 20 We have shown that SARS-CoVs SL1 and SL2 can replace their MHV counterparts in an otherwise MHV gRNA; these chimeric viruses are transcriptionally active and replication competent.19 This experiment strongly supports the predicted secondary structural architecture of the 5′ UTR in MHV. SL2 is a highly conserved pentaloop containing structural features consistent with a U-turn architecture that plays a critical role in subgenomic RNA (sgRNA) synthesis, as well as in translation.20
In this report, we extensively characterize the functional and structural properties of SL1. We show that SL1 adopts a bipartite structure with the functional boundary of two consecutive pyrimidine–pyrimidine (pyr–pyr) base pairs in the middle of the SL1 helix.20 The upper region of the stem must be base paired to support viral replication to near-wild-type (WT) levels. In striking contrast, characterization of mutant viruses in which an unpaired nucleotide, A35, is deleted leads to the recovery of a diverse collection of mutant viruses that contain single-nucleotide second-site mutations in the lower half of SL1, all of which are kinetically and thermodynamically destabilizing. In addition, these viruses contain one of two single-nucleotide substitutions in the extreme 3′ UTR of the genome. Nonviable mutants in this region of SL1, like SL2 mutants characterized previously,20 fail to support sgRNA synthesis. These data, taken collectively, suggest that the base of SL1 must adopt an optimized stability or kinetic lability to mediate a long-range interaction between the 5′ UTR and the 3′ UTR that is an obligate step in the transcription of sgRNAs. The nature of this interaction is discussed.
Results
SL1 folds in solution and adopts a functionally bipartite structure
A series of NMR studies of SL1-containing RNA constructs from both HCoV-OC43 and MHV document the key predicted structural features of SL1 in our model,20 the most notable of which are two consecutive pyr–pyr base pairs, U12•C32 and U13•U31, in the middle of the helix, flanked on either side by Watson–Crick A11-U33 and G14-C30 base pairs (Fig. 1 a). In addition, these NMR studies also suggested that either A35 or A36 is an unpaired extrahelical nucleotide and that the alternative pairing of U9 with A35 or A36 introduces a local destabilization in this region of SL1 stem.20 Interestingly, the predicted SL1s in other coronaviral genomes also appear to conserve this general feature of a rather weakly paired region at the base or middle of SL1, containing either or both noncanonical base pairs and extrahelical nucleotides, suggesting that this might be important for the replication of MHV and perhaps other CoVs. This served as the motivation for the construction of a mutant MHV genome harboring a single-nucleotide deletion of A35 (SL1-ΔA35)‡.
Fig. 1.
Predicted secondary structure model of MHV-A59 SL1 constructs discussed here. (a) WT;20 (b) WT*, which corresponds to SL1-Δ(C16/C19/C20) with a 5′-GA overhang required for initiation of in vitro transcription by SP6 polymerase; and (c) second-site revertant SL1s derived from infection with the SL1-ΔA35 virus (ΔA35 RNA sequence shown; recovered single-nucleotide substitutions highlighted in red). The constructs labeled WT*, ΔA35, ΔA35/U33C, ΔA35/C34U, and ΔA35/A36U RNAs (b and c) were used for the NMR and thermal unfolding studies. The 5′ g is a non-native nucleotide, shown in lower case.
Mutational analysis of MHV SL1 base pairing
Since NMR data supported the existence of SL1 in 5′-leader-containing RNA fragments,20 we first investigated the functional importance of SL1 in viral replication. One set of mutations targeted the upper portion of SL1 above the pyr–pyr base pairs, while a second set targeted the lower region of SL1 (Table 1 ). Plaque size phenotypes (Table 1, Supplementary Fig. 1) and one-step growth curves (Fig. 2 , Supplementary Fig. 2) were measured for all viable viruses. Deletion of each of the three cytidines unique to MHV (C16, C19, and C20, with C16 predicted to be extrahelical), either singly or altogether, gave rise to viable viruses with WT-like growth characteristics (Supplementary Fig. 2a). Thus, the C residues are dispensable for viral replication, justifying our use of the WT model (denoted WT*) in physical studies presented below (see Figs. 1b, Fig. 5, Fig. 6, Fig. 7). Deletion of these cytidines results in three consecutive G-C base pairs in the upper segment of the stem–loop (Fig. 1b).
Table 1.
Effect of mutations in SL1 on plaque production
| Virus name | Mutation recovered | Intended effect of mutation on SL1 | Plaque diameter (mm)a |
|---|---|---|---|
| SL1 stem mutants | |||
| MHV-A59 1000 | WT | 2.37 ± 0.05 | |
| SL1-A | G14C/G15U/G17C | Destroys base pairing in the upper region of the stem | Not viable |
| SL1-B | C28G/C29A/C30G | Destroys base pairing in the upper region of the stem | 1.82 ± 0.04 |
| SLI-AB | C28G/C29A/C30G G14C/G15U/G17C | Maintains base pairing in the upper region of the stem | 2.16 ± 0.04 |
| SL1-C | G6C/A7U/G8C | Destroys base pairing in the lower region of the stem | 1.55 ± 0.03 |
| SL1-D | C37G/U38A/C39G | Destroys base pairing in the lower region of the stem | 1.77 ± 0.05 |
| SL1-CD | G6C/A7U/G8C | Maintains base pairing in the lower region of the stem | Not viable |
| C37G/U38A/C39G | |||
| Cytidine deletion mutants | |||
| MHV-A59 1000 | WT | 2.14 ± 0.06 | |
| SL1-ΔC16 | ΔC16 | Removes a bulged C from the upper region of the stem | 2.03 ± 0.05 |
| SL1-ΔC19/C20 | ΔC19/C20 | Removes two bulged Cs from the upper region of the stem | 2.08 ± 0.09 |
| SL1-ΔC16/C19/C20 | ΔC16/C19/C20 | Removes all bulges from the upper region of the stem | 1.87 ± 0.06 |
| SL1-ΔA35 mutant and second-site suppressor mutants | |||
| MHV-A59 1000 | WT | 2.74 ± 0.06 | |
| SL1-ΔA35-PL6-ps1 | ΔA35/3′A29G | Removed bulged A from the lower region of the stem | 2.47 ± 0.09 |
| SL1-ΔA35-PL6-ps2 | ΔA35/G10A/3′A29G | Removed bulged A from the lower region of the stem | 2.53 ± 0.08 |
| SL1-ΔA35-PL12-ps2 | ΔA35/C34U/3′A78G | Removed bulged A from the lower region of the stem | 2.13 ± 0.08 |
| SL1-ΔA35-PL4-ps1 | ΔA35/U33C/3′A29G | Removed bulged A from the lower region of the stem | 2.57 ± 0.09 |
| SL1-ΔA35-PL1-ps1 | ΔA35/A36U/3′A29G | Removed bulged A from the lower region of the stem | 2.46 ± 0.07 |
| SL1-ΔA35-PL3-ps1 | ΔA35/C37U/3′A29G | Removed bulged A from the lower region of the stem | 2.80 ± 0.10 |
| SL1-ΔA35-PL14-ps1 | ΔA35/C37U/3′A78G | Removed bulged A from the lower region of the stem | 2.54 ± 0.07 |
| Mutants that target the lower SL1 stem | |||
| MHV-A59 1000 | WT | 2.2 ± 0.05 | |
| SL1-A5G/U40C | A5G/U40C | Increased stability of lower stem | Not viable |
| SL1-U40C | U40C | Control for A5G/U40C | 2.1 ± 0.06 |
| SL1-A5G | A5G | Control for A5G/U40C | 1.8 ± 0.06 |
| SL1-A5C/U40C | A5C/U40C | Control for A5G/U40C | 2.1 ± 0.06 |
| SL1-A7G/U38C | WT | Increased stability of the lower region of the stem | Unstable |
Data are from four separate experiments. A WT MHV-A59 is included for comparison with each experiment.
Plaque diameters were calculated as an average of at least 50 plaques, as described previously.21
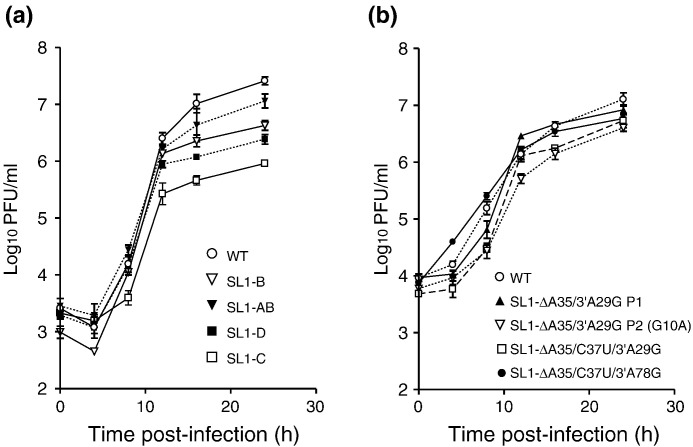
Fig. 2.
One-step growth curves of viable mutant and WT (MHV-A59 1000) viruses. (a) Viruses containing mutations in the upper (SL1-B and SL1-AB) and lower (SL1-C and SL1-D) portions of SL1 (see Table 1 for sequences). (b) Viruses recovered from infection with SL1-ΔA35 viruses.
Fig. 5.
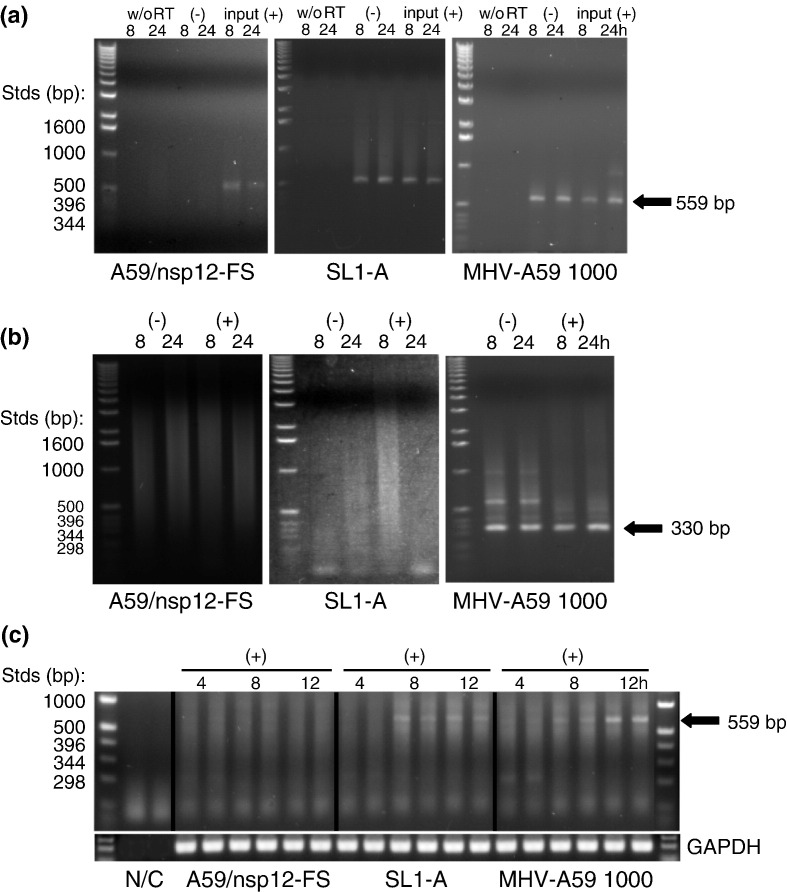
Analysis of MHV-specific RNA synthesis. In all cases, BHK-R cells were electroporated with in vitro transcripts corresponding to the genomes of MHV-A59 1000 (WT), SL1-A, or a A59/nsp12-FS frameshift mutant incapable of directing the synthesis of viral RNAs. Total RNAs were extracted at the times indicated (4 hpe, 8 hpe, and 12 hpe) and analyzed by RT-PCR (see Materials and Methods) for (a) negative-sense anti-gRNA; (b) negative- and positive-sense sgRNA 7; or (c) gRNA. Note that total RNA is used as the template for RT in (a) and (b); poly(A) + RNA was used as the template in (c), and duplicate samples were analyzed. The arrows indicate the position of the amplicons expected for each RNA species. GAPDH, RNA recovery control.
Fig. 6.
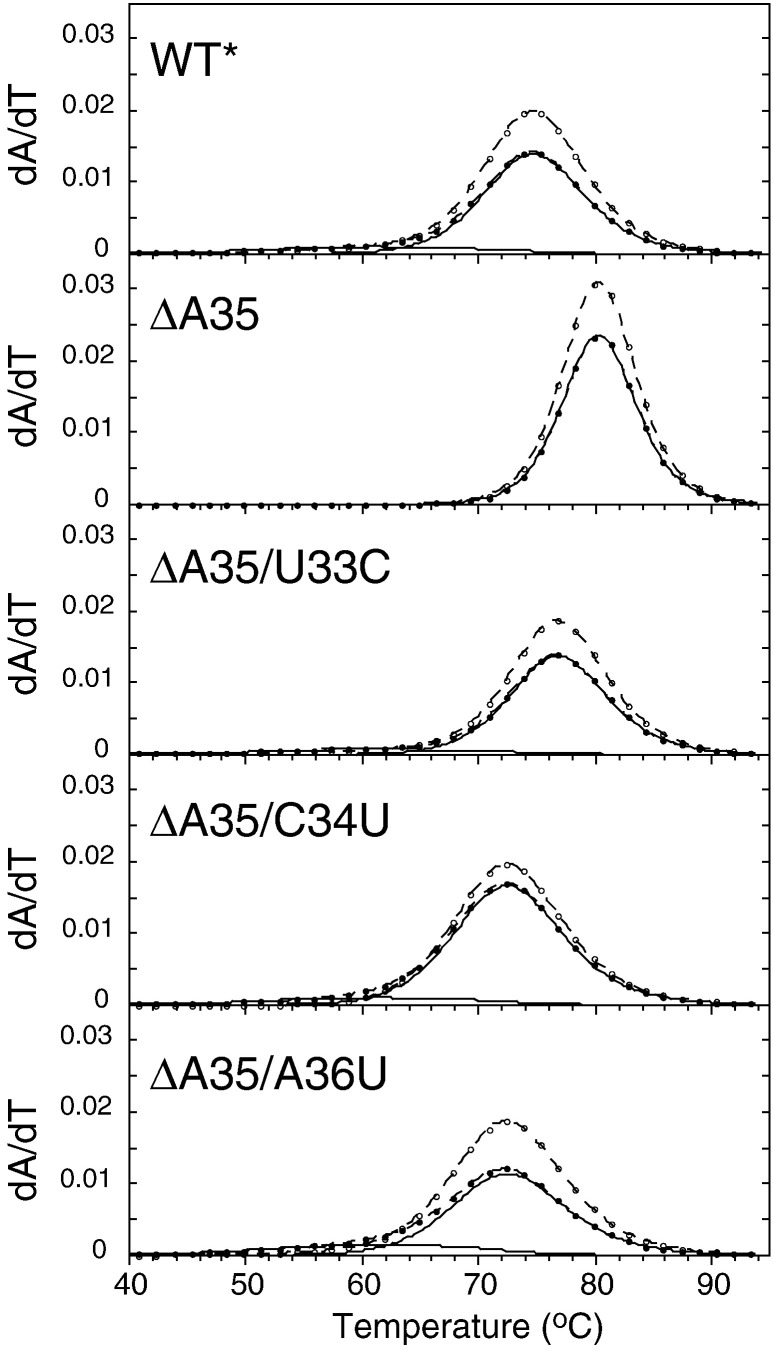
Comparison of the thermal unfolding of the WT*, ΔA35, ΔA35/U33C, ΔA35/C34U, and ΔA35/A36U SL1 RNAs. The experimental optical melting profiles show every fifth data point collected at 260 nm (•) and 280 nm (○), with the calculated fits (dashed lines) shown. For the ΔA35 RNA, the nonlinear least-squares simultaneous composite fit to a single transition unfolding model, and the transition is shown in solid line. For the WT* model and three recovered SL1 mutant ΔA35/U33C, ΔA35/C34U, and ΔA35/A36U RNAs, a nonlinear least-squares simultaneous composite fit to a two-transition unfolding model, and component transitions 1 and 2 (solid lines) are shown. The thermodynamic parameters derived from these fits are compiled in Table 2.
Fig. 7.
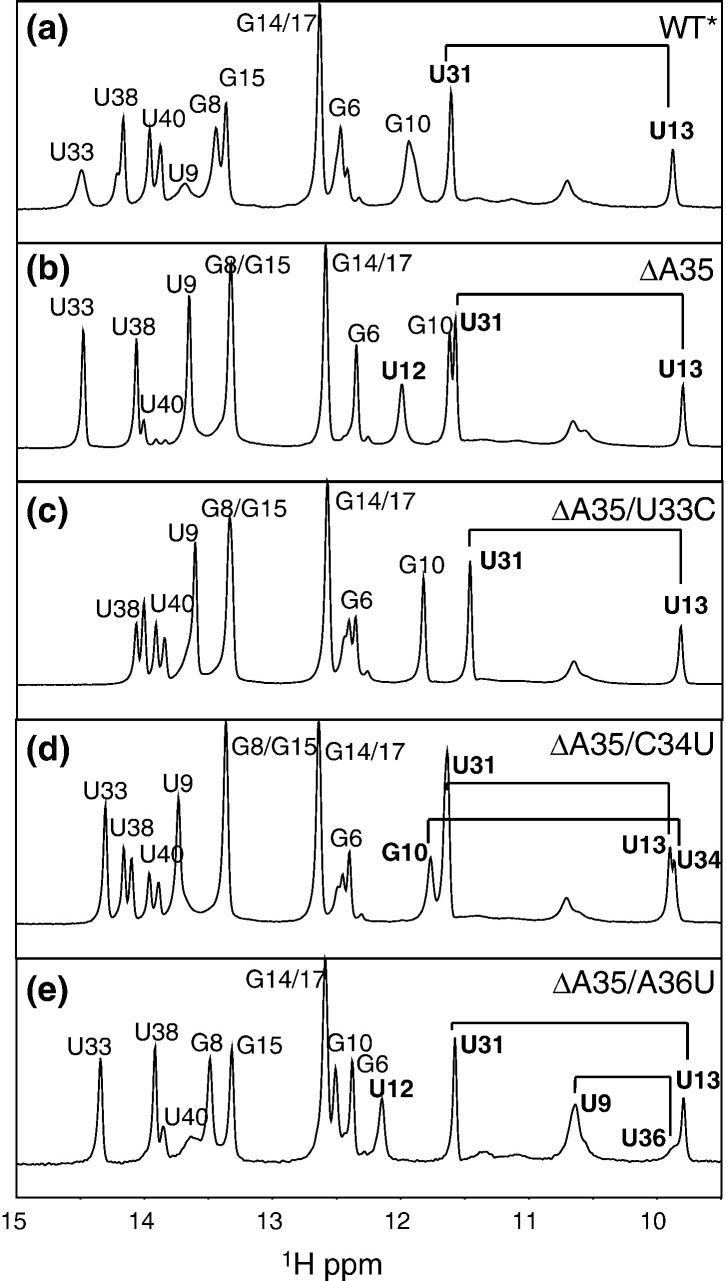
Imino proton regions of 1D jump–return echo spectra acquired at 10 °C and 10 mM KPi (pH 6.0) for WT* (a), ΔA35 (b), ΔA35/U33C (c), ΔA35/C34U (d), and ΔA35/A36U (e). Imino protons corresponding to noncanonical base pairs are shown in bold. Note that some spectra (WT*, ΔA35/U33C, and ΔA35/C34U) are characterized by slow conformational heterogeneity at the base portion of SL1 (A5-U40, G6-C39, and A7-U38 base pairs).
Mutations that disrupt the three consecutive G-C base pairs in the WT genome either were nonviable (SL1-A) or resulted in a virus with a moderately smaller plaque size (SL1-B). Computer-assisted modeling with Mfold and ViennaRNA (see Kang et al. 19) indicates that the SL1-B mutant may adapt an alternative stable folding that largely maintains the SL1 structure, whereas the SL1-A mutant does not (data not shown). In contrast, the compensatory mutant SL1-AB—containing both the SL1-A and the SL1-B mutations and predicted to restore base pairing—possessed a similar plaque size and had almost identical growth kinetics as the WT virus (Table 1, Fig. 2a, Supplementary Fig. 1). These data reveal that the upper portion of SL1 must be base paired in order to support efficient virus replication.
In contrast, the mutations introduced below the two consecutive pyr–pyr base pairs gave rise to completely opposite results. Two viral genomes carrying mutations that destroy base pairing in this region (SL1-C and SL1-D) were found to be viable, albeit with significantly smaller plaque sizes and slower one-step growth curves than the WT virus (Table 1, Fig. 2a, Supplementary Fig. 1). Surprisingly, the corresponding double transversion mutant predicted to restore base pairing here (SL-CD) was not viable (Table 1).
Deletion A35 gives rise to second-site suppressor mutations in both SL1 and 3′ UTR
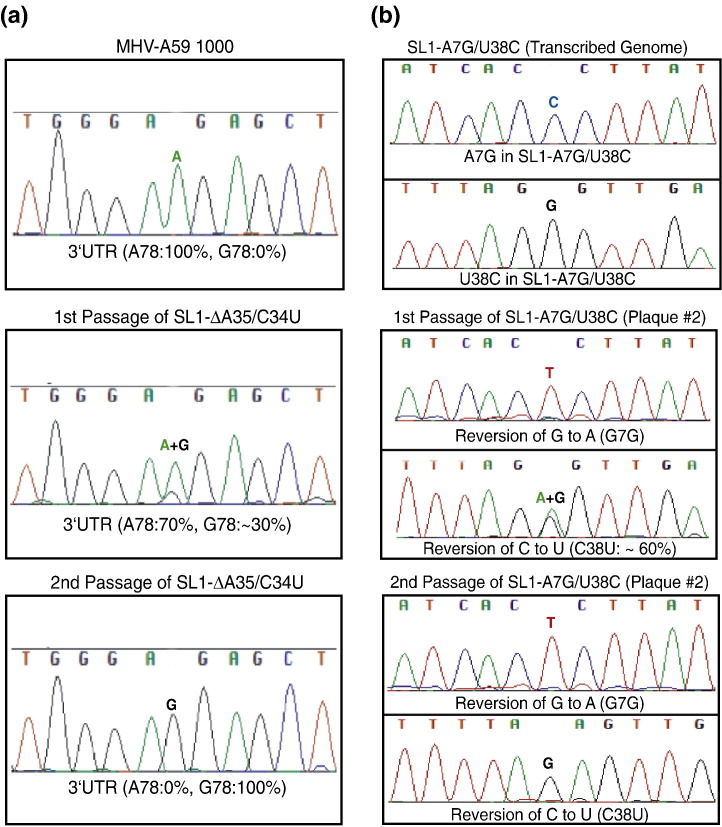
To further investigate the functional importance of the lower SL1 region, we characterized mutant viruses containing a deletion of A35 (SL1-ΔA35). From two completely independent transfections, we recovered 12 plaque-purified viruses. Sequencing of the complete 5′ and 3′ UTRs of the 12 plaque-purified viruses revealed that only 2 of these viruses contained the SL1-ΔA35 sequence; the remaining 10 viruses made up a diverse collection of single-nucleotide second-site substitutions in the immediate vicinity of the deleted nucleotide. Several of these plaque isolates contained identical sequences in their 5′ and 3′ UTRs. Viruses with identical sequences had similar plaque sizes. The plaque phenotypes of one example of each second-site mutation are presented in Table 1. These mutations include C37U, A36U, C34U, and U33C (from the base of SL1 toward the hairpin loop; see Fig. 1c) in the context of the parent SL1-ΔA35 deletion. Additionally, viruses containing the SL1-ΔA35 sequence were completely replaced by viruses containing a second-site mutation (G10A) in SL1 after only one passage in DBT cells, confirming that viruses containing only a deletion of A35 were genetically unstable and were subjected to strong negative selection. Remarkably, although diverse in sequence, all second-site mutants possess a common property: each is predicted to change a Watson–Crick base pair in the WT SL1 to a noncanonical base pair (vide infra).
In addition to these substitutions in the 5′ UTR, all recovered viruses contained one of two second-site single-nucleotide substitutions in the 3′ UTR, 3′-A29G or 3′-A78G [3′, counting from the 3′ nucleotide of the 3′ UTR in the genomic strand, before the poly(A) tail], near the very 3′ end of the genome some 30 kb distant. In one case (SL1-ΔA35/C34U), the passage 1 virus contained a mixture of the 3′-A78G and WT sequences. The WT sequence was lost after one additional passage (Fig. 4a). Both A29G and A78G mutations in the 3′ UTR share the same sequence context, GAGG, relative to the same WT context, GAAG. In addition, in at least one case (C37U), both A29G and A78G substitutions were recovered; this is consistent with the idea that each mutation, which coevolves with the 5′ UTR mutation, is functionally equivalent. These data argue for a physical interaction between 5′ UTR and 3′ UTR in MHV and suggest that it is mediated by a “destabilized” or “dynamic” SL1.
Fig. 4.
Sequence scans of selected mutants. (a) Selection of the mutation at position 3′A29G in the SL1-ΔA35/C34U virus. (b) True reversion of the introduced mutant G7-C38 base pair to the WT A7-U38 base pair. The scans shown correspond to negative-sense sequences.
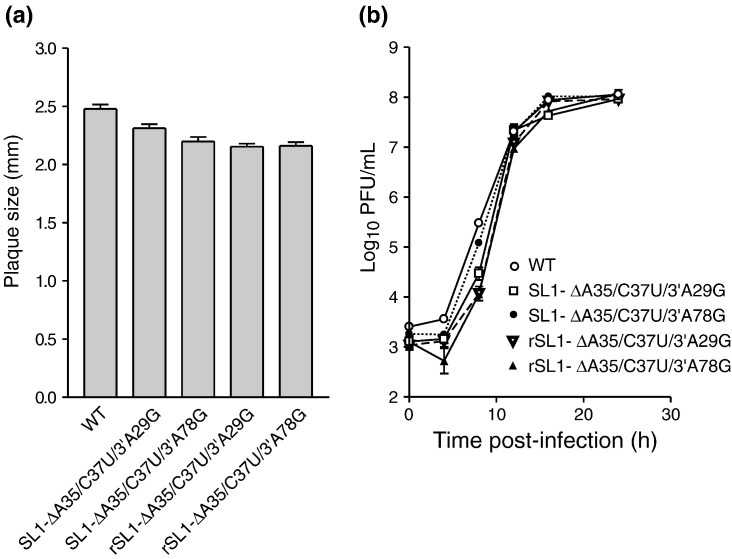
To confirm that the phenotypes of the second-site suppressor viruses that arose after electroporation with the SL1-ΔA35 mutant genome were due to the second-site mutations we detected in the 5′ and 3′ UTRs, and not due to other mutations in regions of the genome that were not sequenced, we examined two mutants, SL1-ΔA35/C37U/3′A29G and SL1-ΔA35/C37U/3′A78G, in more detail. We constructed genomes containing these mutations using our standard reverse genetic system and generated recombinant viruses (designated rSL1-ΔA35/C37U/3′A29G and rSL1-ΔA35/C37U/3′A78G). The sequence of the 5′ and 3′ UTRs of these viruses was verified, and their plaque morphologies and growth phenotypes were compared with the corresponding SL1-ΔA35 second-site suppressor mutants. Each of the six plaque isolates of the recombinant mutant viruses rSL1-ΔA35/C37U/3′A29G and rSL1-ΔA35/C37U/3′A78G contained no additional mutations in their 5′ and 3′ UTRs, and their plaque sizes and morphologies are very similar to those of the originally recovered mutants SL1-ΔA35/C37U/3′A29G and SL1-ΔA35/C37U/3′A78G (Fig. 3 a, Supplementary Fig. 3). These data strongly suggest that the phenotypes of the originally recovered SL1-ΔA35 second-site revertants are indeed due to the mutations in their 5′ and 3′ UTRs, rather than due to other uncharacterized mutations elsewhere in the genome.
Fig. 3.
Phenotypes of recombinant SL1-ΔA35 second-site suppressor viruses. (r) indicates recombinant virus versus those recovered from infection with SL1-ΔA35 virion genomes. (a) Plaque size (mm). (b) One-step growth curves.
Mutations that increase the stability of the lower region of SL1 are not viable
Genomes containing A-U to G-C base-pair substitutions that are predicted to stabilize the lower region of SL1 are either nonviable (SL1-A5G/U40C) or unstable (SL1-A7G/U38C). In the case of SL1-A5G/U40C, single- and double-nucleotide substitutions that destroy the original base pair (SL1-A5G, SL1-U40C, or SL1-A5C/U40C) are all viable and characterized by near-WT-like plaque sizes and growth curve characteristics (Table 1, Supplementary Figs. 1 and 2b). Strikingly, the SL1-A7G/U38C virus is a true revertant, with the A7G substitution repaired in the first passage and with the U38C substitution repaired next to generate the original A7-U38 base pair (Fig. 4 b). These data, taken collectively, suggest that the lower portion of SL1 is weakly base paired or not at all (vide infra), and that this region of the stem plays a critical role in viral replication.
Mutations in SL1 influence RNA synthesis
To determine whether the altered growth phenotypes of MHV SL1 mutant viruses correlated with defects in genome replication or sgRNA synthesis, virus-specific RNAs were metabolically radiolabeled and analyzed by gel electrophoresis. Cells infected with MHV SL1 mutant viruses synthesized both gRNA and the seven sgRNAs in approximately the same ratios as observed for a WT virus infection (Supplementary Fig. 4). SL1-B, SL1-C, SL1-D, SL1-ΔC16/19/20, and SL1-ΔA35/C34U/3′A78G viruses appeared to synthesize the lowest absolute quantities of gRNAs and sgRNAs (< 32% of WT), while the other SL1 mutant viruses directed the synthesis of virus-specific RNAs to levels greater than 55% of those observed with WT virus. One nonviable mutant, SL1-A, was also assayed for the ability to synthesize sgRNA 7 and gRNAs by a series of reverse transcription (RT) PCR assays of RNA extracted from electroporated cells. Previous characterization of nonviable SL2 mutants revealed that the primary defect in these genomes was that they failed to support the transcription of sgRNAs, whereas replication of gRNA was intact,20 with the latter evidenced by a negative-sense gRNA replication intermediate. The same is true of the SL1-A mutant (Fig. 5 ). A comparison of the in vitro translation efficiencies of fusions of WT and several SL1 mutant 5′ UTRs that gave rise to nonviable genomes to luciferase reporters found no effect on translational efficiency (data not shown).
A low thermostability of SL1 is crucial for virus stability and viability
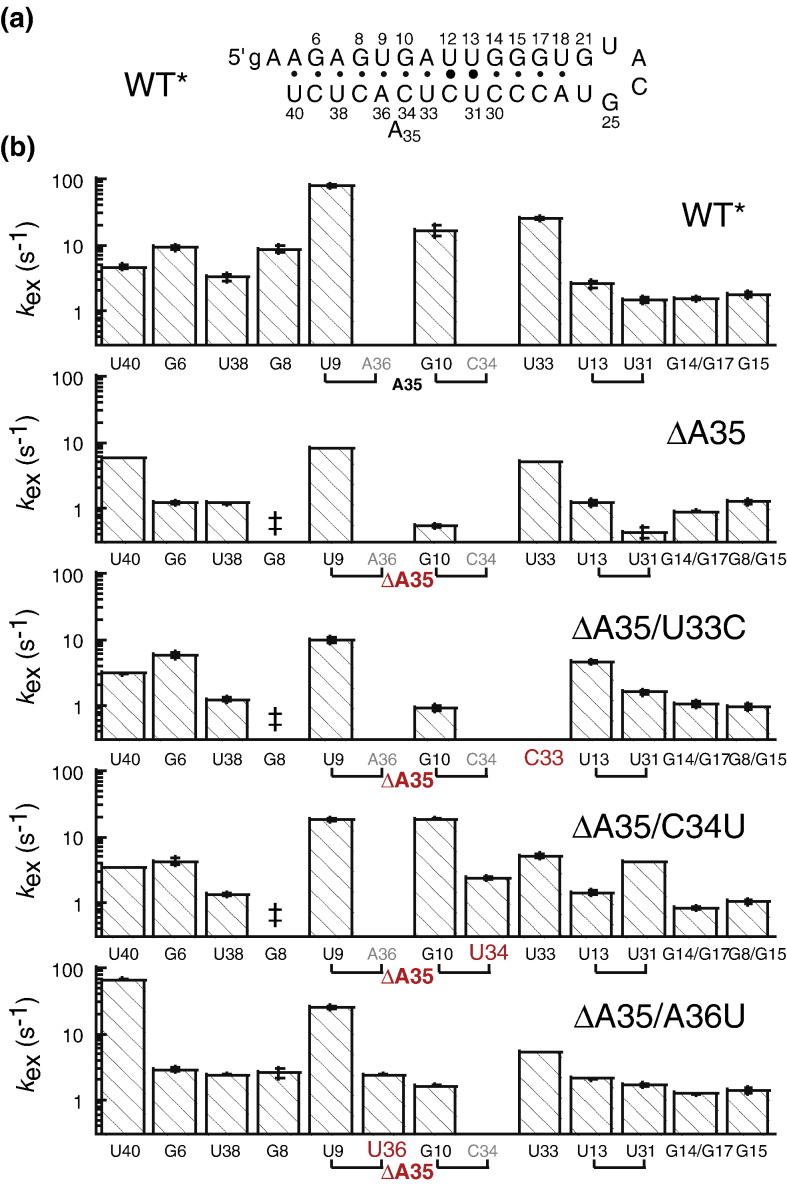
The functional data presented above are consistent with what we term a “dynamic SL1” hypothesis, which posits that the lower region of SL1 must be thermodynamically destabilized and/or dynamically (kinetically) labile in a way that is dependent only on the general physical features of this region of the SL1, rather than on the precise nucleotide sequence, in order to fully support virus replication. In order to test this hypothesis and to gain additional insight into SL1 structure, we synthesized three representative second-site SL1 revertant mutant RNAs (see Fig. 1c), measured their thermodynamic stabilities using quantitative optically monitored thermal denaturation methods (Fig. 6 ), and measured their structural (Fig. 7 ) and dynamic (Fig. 8 ) properties by NMR spectroscopy.
Fig. 8.
Graphical representation of the imino proton solvent exchange rates (kex) for the SL1 WT* and mutants. (a) Secondary structure of the WT* RNA. (b) kex is plotted versus base-pair position (from the bottom to the top of the SL1 helix, from left to right). The mutations are shaded in red. kex could not be unambiguously measured for the G8 for the middle three RNAs due to spectral overlap with G15; the same is true of G14 and G17 imino protons in all spectra. In these cases, the average kex is plotted.
Optical melting profiles (dA/dT) are shown for the WT*, ΔA35, ΔA35/U33C, ΔA35/C34U, and ΔA35/A36U SL1 RNAs in Fig. 6, with the thermodynamic parameters described from a quantitative analysis of these melts compiled in Table 2 . In contrast to all of the other RNAs, the optical melting profile of the ΔA35 SL1 RNA is well-modeled by a single two-state unfolding transition with a t m of 80.3 °C and with a van't Hoff enthalpy of unfolding (ΔH vH) of 118 kcal mol− 1 (Table 2). Thermodynamic parameters derived from melting curves acquired using differential scanning calorimetry are in excellent agreement with the optical melts, as well as the expected ΔH estimated from the nearest-neighbor model.23 In contrast, melting profiles obtained for the WT* RNA, as well as for the three ΔA35 second-site revertant mutant RNAs, are broader than that obtained for the ΔA35 RNA; they also unfold at significantly lower t m. A broadened unfolding transition is consistent with a superposition of two sequential two-state unfolding transitions (e.g., the lower and the upper regions of the SL1 stem) or extreme non-two-state unfolding behavior of the SL1 stem in a single transition. In both cases, fitting of these melting profiles to a single two-state transition would result in insufficient van't Hoff enthalpy of unfolding (ΔH vH) to account for the unfolding of the entire 14-base-pair stem (Fig. 6). This is exactly what we observe, with the apparent ΔH vH of ≈ 70–75 kcal mol− 1 in each case (fits not shown). Invoking a sequential two-state unfolding transition model reveals two unfolding steps with different amplitudes. If we assign the first unfolding step to the denaturation of the lower four base pairs in SL1 (predicted ΔH vH = 38 kcal mol− 1; t m = 61 °C),24 then we recover sufficient ΔH vH in the second (major) transition (70–86 kcal mol− 1) to account for unfolding of the entire molecule. From these fits, we calculate that the WT RNA is destabilized by 3.0 kcal mol− 1 relative to the ΔA35 mutant, while the three second-site revertants are destabilized by 2.1 kcal mol− 1 (ΔA35/U33C), 4.5 kcal mol− 1 (ΔA35/C34U), and 4.7 kcal mol− 1 (ΔA35/A36U) . These data reveal that each of the recovered revertant RNAs shares a common physical property with the WT RNA (i.e., all are thermodynamically destabilized relative to the ΔA35 mutant).
Table 2.
Thermodynamic parameters derived for the unfolding of WT*, SL1-ΔA35, SL1-ΔA35/U33C, SL1-ΔA35/C34U, and SL1-ΔA35/A36U RNAs
| RNA | Transition 1a |
Transition 2 |
||||
|---|---|---|---|---|---|---|
| ΔH1 | tmm1 | ΔH2 | tm2 | ΔGo37 | ΔΔGo37 | |
| WT* | 37.9 | 60.8 | 81.0 | 74.5 | − 11.5 | 3.0 |
| ΔA35 | 118.3 | 80.3 | – | – | − 14.5 | – |
| ΔA35/U33C | 37.9 | 60.8 | 85.7 | 76.7 | − 12.4 | 2.1 |
| ΔA35/C34U | 37.9 | 60.8 | 72.0 | 72.2 | − 10.0 | 4.5 |
| ΔA35/A36U | 37.9 | 60.8 | 69.8 | 72.2 | − 9.8 | 4.7 |
Enthalpy and free energy are reported in kilocalories per mole, and tm is reported in degrees Celsius. Thermodynamic parameters derived from the optical melting profiles determined using the two-state van't Hoff unfolding model, with ΔCp = 0.22
Parameters for transition 1 were fixed to the predicted values for the unfolding of the bottom four base pairs of SL1, with transition 2 parameters optimized during the fit (see the text for details). The ΔG37,i for the ith individual transition was obtained from ΔGo37 = ΔH − 310.15 × ΔS, where ΔS = ΔH/tm. ΔGo37 = ∑ΔG37i, and ΔΔGo37 is expressed relative to the ΔGo37 measured for the SL1-ΔA35 RNA.
Mutant SL1 RNAs experience enhanced dynamics and conformational heterogeneity
Since thermodynamic destabilization is a global property of a molecule, we used NMR spectroscopy in an effort to localize changes in the structure and/or dynamics that could account for this. One-dimensional (1D) imino proton spectra (10 °C, pH 6.0) for all five RNAs are shown in Fig. 7. What is immediately apparent for these RNAs is that the anticipated noncanonical base pairings [i.e., G10•U34 in ΔA35/C34U RNA (Fig. 6), and U9•U36 in ΔA35/A36U (Fig. 6)] are present, since the imino protons of U34 and U36 are strongly protected from exchange with solvent in each case.25, 26, 27 Although we did not directly determine whether the A33U substitution resulted in a new A•C base pair, this is expected, since this substitution would give rise to exactly the same three noncanonical pairings that characterize SL1 of a related group 2 CoV HCoV-OC43; in that RNA, a protonated A + •C base pair is formed.20 In addition, the finding that this RNA is the least destabilized relative to the ΔA35 mutant is not compatible with the presence of extrahelical nucleotides in each of these positions. Thus, all three revertant RNAs are characterized by multiple noncanonical pairings in an otherwise perfectly base-paired helical stem.
Inspection of these spectra (Fig. 7), as well as a compilation of the imino proton solvent exchange rates (Fig. 8), reveals that thermodynamic destabilization manifests itself in multiple complex ways. First, the WT RNA imino proton spectrum reveals a superposition of at least two conformations in slow exchange on the 1H NMR timescale, as evidenced by peak doubling of the U40, G6, and U38 imino resonances at the base of the stem. Exactly the same type of heterogeneity is observed in the ΔA35/U33C and ΔA35/C34U RNAs, but to a far greater degree—with the ΔA35/A36U RNA being more like the WT* RNA in this regard. All heterogeneity is lost in the ΔA35 parent RNA.
The imino proton solvent exchange rates, k ex, reveal additional insight into SL1 dynamics. Although k ex is a complex function of the rate constants for the opening (k op) and closing (k cl) of the base pair (where the two-state equilibrium that is constant for base-pair opening, K op, is defined by k op/k cl), as well as of the intrinsic rate constant for proton exchange by base catalyst (k tr), the magnitude of k ex often tracks with the magnitude of k op and K op,28, 29, 30 since exchange will occur only very slowly from the base-paired state.31, 32 These data reveal that the WT* RNA is dynamically asymmetric, with imino protons above the U13–U31 base pair characterized by slow k ex (≤ 2 s− 1), while those below this base pair are ≈ 2- to ≈ 50-fold faster. For the ΔA35 RNA, imino proton solvent exchange rates are globally quenched, but most strongly near the site of the deletion. For example, the k ex values of U9, G10, and U31 are strongly attenuated in the ΔA35 RNA by ≈ 10-, ≈ 32-, and ≈ 5-fold, respectively. Inspection of the base-pair dynamics for the three mutant SL1 RNAs reveals that each reintroduces or accentuates one or more specific features of the kinetic lability that characterizes the WT* RNA. For example, in all three RNAs, the imino proton solvent exchange rates of the U13•U31 base pair are elevated relative to the ΔA35 RNA by 4- to 10-fold, with some more so than in the WT* RNA. This perturbation is local in the ΔA35/U33C RNA, but next nearest neighbor and long range in the ΔA35/C34U and ΔA35/A36U RNAs, respectively. The solvent exchange rate of G10 is also increased in all RNAs, most prominently in the ΔA35/C34U RNA in the context of the new G10•U34 base pairing, but also in the other two revertant RNAs (by 3- to 4-fold). These data argue that the base of SL1 through the pyrimidine base-pairing region in both the WT* and the second-site mutant SL1s recovered from the genetically unstable SL1-ΔA35 virus must be conformationally heterogeneous and dynamically unstable; this, in turn, allows this region to become transiently unfolded, so that a long-range interaction with the 3′ UTR in MHV can occur.
Discussion
The molecular mechanisms by which CoVs carry out sgRNA synthesis and ultimately coordinate this process with replication of the gRNA and translation of the genome remain poorly understood. An early event in these processes, however, may well be genome circularization, which places the 5′ and 3′ termini of the viral genome in close physical proximity to facilitate template switching during sgRNA minus-strand synthesis. Template switching is simply hybridization of the nascently synthesized minus strand corresponding to the intergenic or TRS-B sequences with the complementary leader TRS, a process expected to be strongly stimulated by the close physical proximity of the two complementary sequences. This process is almost certainly regulated by highly specific RNA structural motifs found in the 5′ and 3′ UTRs and associated interactions with virally encoded and/or host proteins. The 5′ leader RNA appended to all sgRNAs corresponds to the extreme 5′ end of the 5′ UTR and is composed of two stem–loop structures termed SL1 and SL2 just upstream of the TRS that defines the leader–body junction.19 Characterization of nonviable viruses that harbor mutations in SL1 (this work) or SL220 reveals that these genomes are generally WT with respect to the synthesis of full-length gRNA and anti-gRNAs; however, all are absolutely impaired in sgRNA synthesis.
A recent model for CoV replication13 postulated that circularization of the genome is a necessary early step in sgRNA synthesis. However, there is little direct evidence for or against a physical association of the 5′ and 3′ ends of the genome that might be required for sgRNA synthesis, as well as its nature (RNA mediated, protein mediated, or both), although it has been proposed that circularization mediated by a protein bridge composed of cap-binding protein, eIF4E, eIF4G, and poly(A) binding protein33 might be important for initiation of the replicase gene translation.34 During the course of testing the functional impact of mutations deposited in the MHV SL1, we have uncovered two critical aspects of SL1 that are required for sgRNA synthesis. Here, we present the first genetic evidence in support of a direct interaction between SL1 and the extreme 3′ end of the genome; furthermore, the physical analysis of RNAs harboring second-site revertants in SL1 recovered from the genetically unstable SL1-ΔA35 viruses allows us to pinpoint clear structural requirements for the lower region of SL1 (below the pyr–pyr base pairs mapped by NMR spectroscopy) that are essential for supporting viral replication.
Our functional data indicate that the base pairing of the upper portion of the stem is required for optimal viral replication, while the lower portion is weakly base paired or even unpaired. Genomes containing A5G/U40C or A7G/U38C mutations that stabilize the extreme lower portion of the stem could not be recovered as viable viruses. While deletion of A35 is still viable, this virus is genetically unstable and gives rise to second-site mutations in both the 5′ UTR and the 3′ UTR. Interestingly, all the second-site mutations in the 5′ UTR are located below the pyr–pyr base pairs and introduce noncanonical base pairs to maintain stem formation. Each of these mutants of SL1 stem structures are less thermodynamically stable compared with the genetically unstable SL1-ΔA35 mutant, which forms a fully base-paired helical stem. These mutations also introduce conformational heterogeneity in this region as well. Interestingly, the MHV SL1-ΔA35 revertant mutants bear some resemblance to the predicted secondary structure of a WT SL1 from a related group 2 CoV, HKU1.19, 35 HKU1 SL1 does not contain an extrahelical nucleotide, but instead is characterized by tandem-predicted A•C and G•A mismatches in precisely the same region where 5′ UTR SL1 second-site mutations map in MHV.
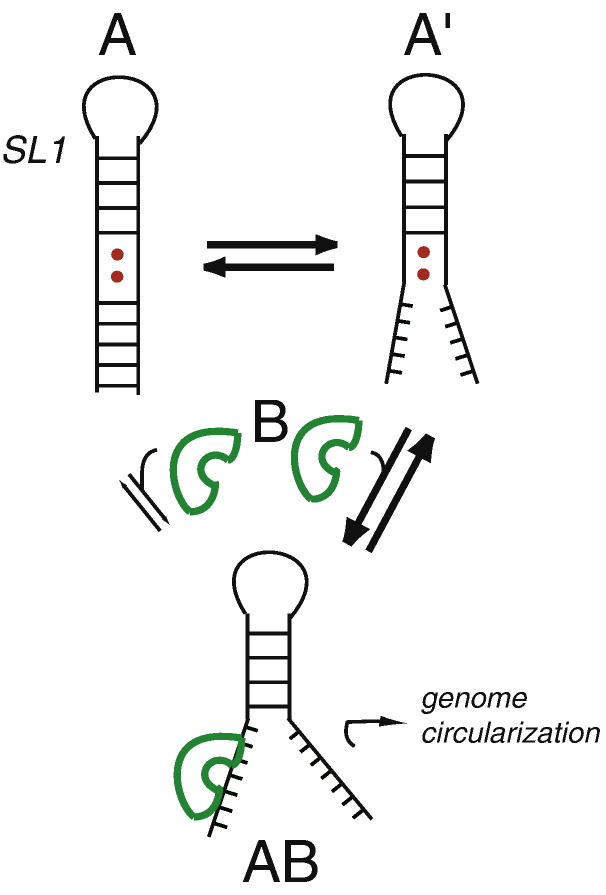
In addition to the global thermodynamic stability information, the measurement of imino proton exchange gives important insights into the differences in the dynamics or flexibility of individual base pairs in different ΔA35 RNAs. The k ex results show that the lower half of the SL1 is kinetically labile, and the three second-site revertant mutants recover one or more dynamic characteristics of the WT* RNA relative to the ΔA35 mutant. Indeed, the closer that the site of mutation localizes to A35, the more similar the dynamic properties become relative to the WT* RNA. Therefore, a less stable and more flexible SL1 might facilitate a specific interaction between the single-stranded RNA in this region and a host-encoded or virally encoded protein(s) that is crucial for genome circularization and replication. A cartoon model that graphically illustrates this idea is shown in Fig. 9 , where the structure, conformational ensemble, or longer-lived partially opened form(s) of SL1, schematized by A1′ relative to fully base-paired A structure, lowers the energy barrier for formation of a complex with hypothetical protein B, which in turn drives genome circularization and sgRNA synthesis.
Fig. 9.
Model of a dynamic SL1 that is consistent with the functional and structural data presented here. The fully based-paired SL1 (A; modeled by the ΔA35 RNA) exists in equilibrium with one or more higher-energy conformers (A′; WT* and ΔA35 second-site revertants) that are partially unfolded or that experience dynamic destabilization as a result of noncanonical pairing. A hypothetical protein (B) binds to both A and A′ to form the same partially unwound AB complex, but the affinity of B for A′ will be higher, since the full energetic cost of unfolding the lower stem will not have to be paid; this interaction then mediates a long-distance RNA–RNA, RNA–protein, or protein–protein interaction, which is crucial for the viral replication.
Genetic interaction between the 5′ UTR and the 3′ UTR
We note that four of five SL1 second-site mutations recovered from SL1-ΔA35 viruses (U33C, C34U, A36U, and C37U) map specifically to the 3′ strand of SL1, with the lone exception being G10A; all recovered viruses therefore maintain a base-paired 5′-6GAGYR10 sequence in the 5′ portion of SL1 in MHV. This result suggests that the nucleotide sequence of the 5′ region of SL1 may also be important for viability. While this motif is not absolutely essential when the base of SL1 is predicted to be unpaired (6GAG8 to 6CUC8 in the SL1-C virus; Table 1), a lethal mutation results when the same mutation is introduced while maintaining base pairing here (in the SL1-CD virus; Table 1) (i.e., in a structural context most similar to that of WT MHV). This short sequence motif is conserved and predicted to be base paired in all group 2 CoV genomes, with the exception of SARS-CoV, which has an 5′-AGGU sequence in what is predicted to be a region of SL1 weakly paired with the A extrahelical.19 Strikingly, this SL1 5′-6GAG8 sequence is identical with the 5′-GAG sequence recovered from second-site mutations codeposited in 3′ UTR in the same viruses; this suggests the possibility that an oligomer of the same protein(s) that is capable of recognizing and/or “melting out” this short purine-rich motif may well play an important role in mediating a physical interaction between the 5′ UTR and the 3′ UTR. An excellent candidate for this role is hnRNAP A1, since both 3′ UTR mutations occur in exactly the same sequence context (5′-GAAG, a near-consensus binding site for A1), and each replaces the second A with a G (a consensus hnRNAP A1 binding site),36, 37 with the 5′-AG as the key specificity determinant. hnRNAP A1 possesses RNA chaperone or helix destabilization activity and has been shown to bind the 3′ UTR in MHV, while hnRNP A1 and PTB both bind to the complementary strands at the 5′ end of MHV RNA. There is some evidence to suggest that these proteins together mediate the formation of an ribonucleoprotein complex involving the 5′- and 3′-end fragments of MHV RNA in vitro.38
Our data are compatible with the three-step working model of CoV transcription recently refined by Zuniga et al., in which the first step is the formation of a 5′–3′ UTR complex through protein–RNA and protein–protein interactions, by which the TRS-L would be located in close proximity to sequences located at the 3′ end of gRNA and, in turn, make template jumping from individual TRS-B sequences favored.13 Overall, we hypothesize that SL1 must possess an optimum stability or kinetic lability required to mediate a key long-range physical interaction between the 5′ UTR and 3′ the UTR that is critical specifically for sgRNA synthesis, but not required for genomic minus-strand synthesis. This finding suggests that the structural requirements and/or the nature of the assembled replication complexes that are capable of synthesizing full-length gRNA is distinct from those that are capable of synthesizing sgRNAs. Such a functionally bipartite structure, where a subregion of a helical stem below tandem wobble pairs is tuned to an optimum stability, is reminiscent of stem I of the U2 snRNA; here, these features have been hypothesized to allow U2 to adopt a number of mutually exclusive folded conformations during spliceosome assembly and catalysis.39
Materials and Methods
Virus and cells
DBT cells were maintained at 37 °C and 5% CO2 in DMEM supplemented with 10% calf serum (HyClone, Logan, UT). L2 cells were maintained at 37 °C and 3% CO2 in DMEM supplemented with 10% calf serum. Baby hamster kidney-21 cells expressing the MHV receptor (BHK-R cells) were grown in DMEM supplemented with 10% calf serum, 3% tryptose phosphate broth, and G418 (800 μg ml− 1) to select for cells expressing the MHV receptor. MHV-A59 1000 was used as a WT control virus for comparison with chimeric viruses.
Assembly of a full-length MHV-A59 infectious construct
The reverse genetic system for MHV-A59 has been described.40 cDNAs representing the entire MHV-A59 genome with either the WT sequence or the MHV SL1 mutant sequences were constructed by ligation of the A fragments to fragments B–G, transcribed with T7 RNA polymerase, and electroporated into cells as described previously.19, 20 Cultures were observed for up to 72 h for the development of cytopathic effect and harvested by freezing at − 70 °C. Cultures that did not develop cytopathic effect were blind passed three times through DBT cells in a further attempt to recover infectious virus. At least three independent experiments, including at least one experiment in which electroporated cells were incubated at 34 °C and 40 °C, were performed before a mutant genome was considered nonviable.
Plasmid constructions
The plasmid carrying the MHV-A59 A clone (plasmid A)40 was used to introduce mutations into the MHV 5′ UTR. Briefly, a 0.55-kb BamHI–MluI fragment of A plasmid was amplified and cloned into pGEM-T vector (Promega) to produce pWt5′ UTR-PG, which harbors a T7 promoter, the 209-nt MHV-A59 5′ UTR, and 257 nt of the 5′ Orf1a coding sequence. Most mutations were introduced into pWt5′ UTR-PG with the Quick Change site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer's instructions and confirmed by sequencing. An oligonucleotide assembly strategy20 was used to introduce the A5G, U40C, and Δ(C16,C19,C20,A35) mutations into pWt5′ UTR-PG. The MluI–BamHI fragments containing the desired mutations were introduced into plasmid A by restriction fragment exchange. The region between the MluI and the BamHI sites in the resulting cloned plasmids was sequenced to verify that the desired mutation was recovered. The sequences of the mutagenic oligonucleotides are shown in Supplementary Table 1.
5′ rapid amplification of cDNA ends
Mutant viruses were subjected to one round of plaque purification and were expanded once in DBT cells. Total RNA (2 μg) was reverse transcribed using oligo 8 (Supplementary Table 1). Resultant RT products were purified using the QIAquick PCR Purification Kit (Qiagen), then 3′ tailing was conducted using Terminal Transferase (Roche) to add a poly(A) tail to the 3′ ends of the purified RT products. The poly(A)-tailed RT products were repurified using the QIAquick PCR Purification Kit. A first round of PCR was performed using 10 μl of the poly(A)-tailed RT products with the adapter primer (AP-dT17) and the MHV-1RV5 primer. A second round of PCR was performed with 1 μl of a 1:50 dilution of the previous reaction using the adapter primer and the SL1 check (−) primer (Supplementary Table 1). The PCR products were gel purified and sequenced using the SL1 check (−) primer.
RNA analysis by RT-PCR
Replicate cultures of BHK-R cells were electroporated in parallel with either MHV SL1 mutant genomes, WT genomes, or the nsp12-FS mutant (a construct containing a frameshift mutation in nsp12 rendering it incapable of directing virus-specific RNA synthesis20) genome, and total RNAs were extracted at various times (hours) postelectroporation (hpe). To determine whether the input RNAs, plus any replicated genome RNA, were present in the electroporated cells, the extracted RNAs were primed for RT by oligo A59(−) 16596–16577 and amplified by PCR using oligo A59(+) 14639–14658 and oligo A59(−) 16596–16577. Resultant PCR products were further amplified by nested PCR using oligo A59(+) 16038–16059 and oligo A59(−) 16596–16577. The RNA species present in cells were further characterized by RT-PCR as described previously.19, 20 Parallel reactions in which reverse transcriptase was omitted from the cDNA step were always performed to ensure that the PCR assay did not detect residual DNA transcription templates that entered the cells during electroporation. Quantitative RT-PCR was conducted to determine whether nonviable MHV SL1 mutant genomes synthesize positive-stranded gRNA (+ gRNA). Total RNAs were extracted at 4 hpe, 8 hpe, and 12 hpe, and poly(A) + mRNAs were purified using the Oligotex mRNA Midi Kit (Qiagen). Poly(A) + mRNAs (180 ng) were used as template for the synthesis of genomic cDNA, glyderaldehyde-3-phosphate dehydrogenase (G3PDH) cDNA with primer A59(−) 16577–16596, and GAPDH-R2, respectively, using the SuperScript II RNase H-Reverse Transcription Kit (Invitrogen). Then, quantitative PCR was followed to amplify + gRNA with primer A59(+) 16038–16059 using iQ™ SYBR Green Supermix (Bio-Rad). In parallel, GAPDH was also amplified with primers GAPDH-F2 and GAPDH-R2. After 40 cycles, the PCR products were displayed by gel electrophoresis. Quantitative PCRs with poly(A) + selected RNA demonstrated that samples were free of detectable DNA.
Preparation of RNA samples for physical studies
RNAs were obtained by in vitro runoff transcription using SP6 RNA polymerase and purified by denaturing PAGE essentially as previously described.41 The NMR samples were subjected to exhaustive dialysis into a final buffer of 10 mM potassium phosphate (pH 6.0) and 100 μM 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), with RNA concentrations ranging from ≈ 1 mM to ≈ 2.5 mM in 300 μl (10% D2O). The RNA samples for thermal denaturation experiments and calorimetry experiments were prepared by dilution into a final dialysis buffer of 10 mM potassium phosphate (pH 6.0), 100 mM KCl, and 5 mM MgCl2. Before each experiment, the RNA samples were annealed by heating at 65 °C for 10 min, followed by slow cooling at room temperature. All samples were ≥90% monomeric, as judged by nondenaturing PAGE.
Thermal denaturation experiments
RNA melts were collected on a Cary 1 scanning spectrophotometer operating in double-beam mode. The RNA concentrations were between 1 μM and 20 μM, and all melting profiles were shown to be independent of RNA concentration over this range. The first derivative data of absorbance at 260 nm and 280 nm with respect to temperature (dA/dT) were subjected to a simultaneous nonlinear least-squares fit of ΔH i, t m,i, and A i, for each ith unfolding transition via sequential interacting two-state unfolding transition model using the t-melt program running on a Silicon Graphics O2 workstation as previously described.22 Melting profiles were subjected to single- or two-transition unfolding models, as described in the text. Parallel thermal melts (50–100 μM RNA strand) were carried out on a Microcal VP-DSC scanning calorimeter under the same solution conditions as previously described.42 Analysis of these baseline-corrected data with single-transition two-state unfolding model gave fitted parameters in qualitative agreement with those obtained from analysis of the optically monitored thermal unfolding experiments (data not shown).
Saturation transfer solvent exchange experiments
NMR spectra were acquired on a Varian Inova 500-MHz spectrometer at the Biomolecular NMR Laboratory, Texas A&M University. The data were processed using NMRPipe43 and analyzed using SPARKY (SPARKY 3; T.D. Goddard and D.G. Kneller, University of California, San Francisco)§. The imino proton resonances were assigned by the jump–return echo 1D and Watergate homonuclear 1H–1H nuclear Overhauser enhancement spectroscopy spectra (τmix = 300 ms), with reference to an internal standard DSS at 10 °C. The imino proton exchange rates were obtained by transfer of magnetization from water.44 The exchange was initiated by selectively inverting the water proton resonance using a Gaussian 180° pulse (5.3 ms duration), with exchange delay times ranging from 2 ms to 850 ms, followed by a weak gradient (0.1 G cm− 1) applied during the exchange delay to minimize effects due to radiation damping. At the end of the exchange delay, a second Gaussian pulse (2.8 ms) was applied to bring the water magnetization back to the z-axis. Imino proton resonances were detected using a gradient-enhanced spin-echo sequence. The acquired spectral array was processed using NUTS (Acorn NMR, Inc.), and the intensities of the imino proton resonances of interest were fitted to obtain the imino proton solvent exchange rate k ex using Kaleidagraph (Synergy Software) as previously described.28, 29
Acknowledgements
This work was supported by National Institutes of Health grants AI040187, AI067416, and AI051493. We thank Drs. A. Arunkumar and Susan Weiss for helpful comments on the manuscript.
Edited by M. F. Summers
Footnotes
Although A35 or A36 (or both, one at a time) could theoretically be extruded from the SL1 helix, extrusion of either one gives rise to the same structure of paired bases in the SL1 stem. In this work, we characterized SL1-ΔA35 MHV mutants, but SL1-ΔA36 viruses would give rise to exactly the same sequence in SL1 and thus would not be functionally distinguishable from the SL1-ΔA35 virus.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2008.01.068
Contributor Information
Julian L. Leibowitz, Email: jleibowitz@tamu.edu.
David P. Giedroc, Email: giedroc@indiana.edu.
Appendix A. Supplementary Data
References
- 1.Gonzalez J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Weiss S.R., Leibowitz J.L. Pathogenesis of murine coronavirus infections. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; Washington, DC: 2007. pp. 259–278. [Google Scholar]
- 5.Breedenbeek P.J., Pachuk C.J., Noten A.F.H., Charité J., Luyjtes W., Weiss S.R., Spaan W.J.M. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leibowitz J.L., Weiss S.R., Paavola E., Bond C.W. Cell-free translation of murine coronavirus RNA. J. Virol. 1982;43:905–913. doi: 10.1128/jvi.43.3.905-913.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leibowitz J.L., Wilhelmsen K.C., Bond C.W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-A59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1344–1839. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai M.M.C., Baric R.S., Brayton P.R., Stohlman S.A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc. Natl Acad. Sci. USA. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawicki S.G., Sawicki D.L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sethna P.B., Hung S.-L., Brian D.A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl Acad. Sci. USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baric R.S., Yount B. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 2000;74:4039–4046. doi: 10.1128/jvi.74.9.4039-4046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuniga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raman S., Bouma P., Williams G.D., Brian D.A. Stem–loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003;77:6720–6730. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raman S., Brian D.A. Stem–loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005;79:12434–12446. doi: 10.1128/JVI.79.19.12434-12446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang R.Y., Hofmann M.A., Sethna P.B., Brian D.A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y.-N., Jeong Y.S., Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalton K., Casais R., Shaw K., Stirrups K., Evans S., Britton P. cis-Acting sequences required for coronavirus infectious bronchitis virus defective—RNA replication and packaging. J. Virol. 2001;75:125–133. doi: 10.1128/JVI.75.1.125-133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang H., Feng M., Schroeder M.E., Giedroc D.P., Leibowitz J.L. Putative cis-acting stem–loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J. Virol. 2006;80:10600–10614. doi: 10.1128/JVI.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P., Li L., Millership J.J., Kang H., Leibowitz J.L., Giedroc D.P. A U-turn motif-containing stem–loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA. 2007;13:763–780. doi: 10.1261/rna.261807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson R.F., Feng M., Liu P., Millership J.J., Yount B., Baric R.S., Leibowitz J.L. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 2005;79:14570–14585. doi: 10.1128/JVI.79.23.14570-14585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theimer C.A., Giedroc D.P. Equilibrium unfolding pathway of an H-type RNA pseudoknot which promotes programmed-1 ribosomal frameshifting. J. Mol. Biol. 1999;289:1283–1299. doi: 10.1006/jmbi.1999.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia T., SantaLucia J., Jr, Burkard M.E., Kierzek R., Schroeder S.J., Jiao X. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 24.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theimer C.A., Finger L.D., Feigon J. YNMG tetraloop formation by a dyskeratosis congenita mutation in human telomerase RNA. RNA. 2003;9:1446–1455. doi: 10.1261/rna.5152303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du Z., Yu J., Ulyanov N.B., Andino R., James T.L. Solution structure of a consensus stem–loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry. 2004;43:11959–11972. doi: 10.1021/bi048973p. [DOI] [PubMed] [Google Scholar]
- 27.Ohlenschlager O., Wohnert J., Bucci E., Seitz S., Hafner S., Ramachandran R. The structure of the stem–loop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure. 2004;12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Cornish P.V., Stammler S.N., Giedroc D.P. The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical. RNA. 2006;12:1959–1969. doi: 10.1261/rna.199006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhavan G.M., Lapham J., Yang S., Crothers D.M. Decreased imino proton exchange and base-pair opening in the IHF–DNA complex measured by NMR. J. Mol. Biol. 1999;288:659–671. doi: 10.1006/jmbi.1999.2690. [DOI] [PubMed] [Google Scholar]
- 30.Lee J.H., Pardi A. Thermodynamics and kinetics for base-pair opening in the P1 duplex of the Tetrahymena group I ribozyme. Nucleic Acids Res. 2007;35:2965–2974. doi: 10.1093/nar/gkm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snoussi K., Leroy J.L. Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry. 2001;40:8898–8904. doi: 10.1021/bi010385d. [DOI] [PubMed] [Google Scholar]
- 32.Gueron M., Leroy J.L. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 33.Tarun S.Z., Jr, Wells S.E., Deardorff J.A., Sachs A.B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl Acad. Sci. USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spagnolo J.F., Hogue B.G. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding J., Hayashi M.K., Zhang Y., Manche L., Krainer A.R., Xu R.M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers J.C., Shamoo Y. Human UP1 as a model for understanding purine recognition in the family of proteins containing the RNA recognition motif (RRM) J. Mol. Biol. 2004;342:743–756. doi: 10.1016/j.jmb.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 38.Huang P., Lai M.M.C. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′–3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 2001;75:5009–5017. doi: 10.1128/JVI.75.11.5009-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sashital D.G., Venditti V., Angers C.G., Cornilescu G., Butcher S.E. Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human. RNA. 2007;13:328–338. doi: 10.1261/rna.418407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yount B., Denison M.R., Weiss S.R., Baric R.S. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 2002;76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nixon P.L., Cornish P.V., Suram S.V., Giedroc D.P. Thermodynamic analysis of conserved loop–stem interactions in P1–P2 frameshifting RNA pseudoknots from plant Luteoviridae. Biochemistry. 2002;41:10665–10674. doi: 10.1021/bi025843c. [DOI] [PubMed] [Google Scholar]
- 42.Theimer C.A., Giedroc D.P. Contribution of the intercalated adenosine at the helical junction to the stability of the gag-pro frameshifting pseudoknot from mouse mammary tumor virus. RNA. 2000;6:409–421. doi: 10.1017/s1355838200992057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Coman D., Russu I.M. Site-resolved stabilization of a DNA triple helix by magnesium ions. Nucleic Acids Res. 2004;32:878–883. doi: 10.1093/nar/gkh228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.