Abstract
We previously demonstrated that salicylic acid-binding protein 2 (SABP2) of tobacco is an integral component of systemic acquired resistance (SAR). SABP2 is a methyl salicylate (MeSA) esterase that has high affinity for SA, which feedback inhibits its esterase activity. MeSA esterase activity is required in distal, healthy tissue of pathogen-infected plants to hydrolyze MeSA, which functions as a long-distance, phloem-mobile SAR signal; this hydrolysis releases the biologically active defense hormone SA. In this study, we examined the inhibitory interaction of SA with SABP2, and identified a synthetic SA analog, 2,2,2,2′-tetra-f luoroacetophenone (tetraFA) that, like SA, competitively inhibits the activity of SABP2 and targets esterases, which utilize MeSA as a substrate. However, in contrast to SA, tetraFA does not induce downstream defense responses and, therefore, is effective in planta at blocking SAR development in tobacco mosaic virus (TMV)-infected tobacco and Pseudomonas syringae-infected Arabidopsis. These results confirm the importance of SABP2 and MeSA for SAR development in tobacco and establish similar roles for MeSA and the orthologs of SABP2 in Arabidopsis. Moreover, they demonstrate that tetraFA can be used to determine whether MeSA and its corresponding esterase(s) play a role in SAR signaling in other plant species. In planta analyses using tetraFA, in conjunction with leaf detachment assays and MeSA quantification, were used to assess the kinetics with which MeSA is generated in pathogen-infected leaves, transmitted through the phloem, and processed in the distal healthy leaves. In TMV-infected tobacco, these studies revealed that critical amounts of MeSA are generated, transmitted, and processed between 48 and 72 h post primary infection.
Systemic acquired resistance (SAR)4 in plants is a state of heightened defense that provides long-lasting, broad spectrum resistance to microbial pathogens and is activated systemically following a primary (1°) infection (1). In many aspects, SAR resembles the immune response in animals, which is composed of both innate and adaptive components (2). The immediate, innate response is nonspecific and mediated by humoral, chemical, and cellular barriers, whereas the adaptive immune system involves the recognition of specific “non-self” antigens in the presence of “self”; this allows the development of immunological memory (3). However, plants lack mobile defender cells and instead rely on the innate immunity of each cell, which can be activated in uninfected tissues by systemic signal(s) originating from the site of infection (4).
A number of studies have provided important insights into the immune response occurring in infected plant cells (4-6). Plants have evolved several layers of immunity that recognize pathogen-associated molecular patterns or pathogen effector molecules (or their altered host targets) through receptors, such as receptor kinases containing a leucine-rich repeat domain or resistance proteins containing a nucleotide-binding site and leucine-rich repeats. This alarm system activates pathogen-associated molecular pattern-triggered immunity (non-host/basal resistance) or effector-triggered immunity (resistance gene-mediated resistance), respectively. Both forms of resistance are associated with physiological changes in the infected cells, such as a rapid increase in reactive oxygen species, ion fluxes, the accumulation of salicylic acid (SA), the synthesis of anti-microbial phytoalexins and the induction of defense-associated genes, including several families of pathogenesis-related genes. These immune responses also are often associated with programmed cell death at the sites of pathogen entry, which leads to the formation of necrotic lesions; this phenomenon is known as the hypersensitive response. In addition, the uninfected portions of the plant frequently develop SAR, which is accompanied by increases in SA levels and heightened pathogenesis-related gene expression.
A number of studies have demonstrated that SA plays a critical role in the resistance signaling pathway(s) (1, 7). Exogenously supplied SA enhances disease resistance and induces pathogenesis-related gene expression in a wide variety of plant species. Confirmation of SA as a critical resistance signal came from analyses of transgenic tobacco and Arabidopsis expressing the bacterial nahG gene, which encodes the SA-degrading enzyme salicylate hydroxylase. These plants failed to accumulate SA after pathogen infection, displayed reduced resistance to avirulent and virulent pathogens, and did not develop SAR or express pathogenesis-related genes in their distal leaves (8-10). Similar results were observed in tobacco deficient for phenylalanine ammonia-lyase, a key enzyme for SA biosynthesis (11), and in the sid1 and sid2/eds5 Arabidopsis mutants, which are impaired in SA biosynthesis (12). However, results from grafting experiments argued that SA was not the critical long-distance signal for SAR; tobacco leaves expressing the nahG transgene were able to transmit an SAR signal following infection by tobacco mosaic virus (TMV), despite suppressed SA levels (8). Rather, a recent study demonstrated that a key signal for SAR in tobacco is methyl salicylate (MeSA), a methyl ester of SA that moves from the infected tissue through the phloem to the distal, systemic tissue (13).
To elucidate the mechanism through which SA signals disease resistance, several potential effector proteins have been identified in tobacco. These SA-binding proteins (SABPs) include catalase, ascorbate peroxidase, the chloroplastic carbonic anhydrase (SABP3) and SABP2 (14-17). Silencing of SABP2 in tobacco resulted in the loss of SAR and suppression of local defense responses, indicating that SABP2 is integral for plant innate immunity (18). Grafting studies using SABP2-silenced rootstocks or scions further revealed that SABP2 is required for SAR signal perception in the distal tissue, but not for SAR signal production in the 1° infected leaves (13). Analysis of the crystal structure of SABP2, alone or in complex with SA, indicated that it belongs to the α/β-fold hydrolases superfamily and that it binds SA in its active-site pocket (19). Biochemical studies have demonstrated that its preferred substrate is MeSA, which it cleaves to release SA. SA competes with MeSA for binding in the active-site pocket of SABP2, which consists of catalytic triad residues Ser81, Asp210, and His238. During hydrolysis of MeSA, the hydroxyl group of Ser81 is perfectly positioned over the carboxyl carbon of MeSA to initiate nucleophilic attack, whereas the side chains of Ser81 and Asp210 form hydrogen bonds with His238 to complete the catalytic triad. In contrast, SA forms hydrogen bonds with Ala13 and His238, which disrupts hydrogen bond formation between the catalytic triad residues. Because the active-site pocket is too small to accommodate both the MeSA substrate and the SA product, SA is a potent inhibitor of the esterase activity of SABP2 (19).
To determine whether an SABP2-like esterase activity is required for SAR in general, we identified and characterized several synthetic SA analogs that inhibit the MeSA esterase activity of SABP2. This approach was chosen as it circumvented purifying the enzyme, cloning its gene, and/or silencing its expression. Note that although SA itself is an effective inhibitor for SABP2, it cannot be used for inhibitory studies because it triggers the defense signaling pathway downstream of SABP2 (7). Our results demonstrate that 2,2,2,2′-tetrafluoroacetophenone (tetraFA) rather specifically and competitively binds SABP2 and inhibits its esterase activity. In planta analyses demonstrated that tetraFA can be used on a variety of plant species to assess the involvement of MeSA in SAR. In addition, tetraFA was used to determine the time during which the MeSA signal moves to the distal tissue after the 1° infection.
EXPERIMENTAL PROCEDURES
Reagents—Synthetic ketones, 2,2,2,2′-tetraFA (tetraFA, ≥99.0% pure [GC], Chemical Abstract Service Registry Number 124004-75-7) and 2′-methoxy-2,2,2-triFA (methoxytriFA, ≥99.0% [GC], CASRN. 26944-43-4) were obtained from Rieke Metals, Inc. (Lincoln, NE), and 2,2,2-triFA (triFA, ≥99.0% [GC], CASRN. 434-45-7) was obtained from Sigma. SA (≥99.0%, CASRN. 54-21-7) and MeSA (≥99.0% [GC], CASRN. 119-36-8) were obtained from Sigma.
Plant Material, Growth, and Pathogen Infection—Nicotiana tabacum (tobacco) cv. Xanthi-nc (NN) was grown and inoculated with TMV as described by Guo et al. (20). Arabidopsis thaliana ecotype Colombia-0 (Col-0) was grown in a 14-h photo period (140 μE m-2 s-1) at 22 °C in 60% relative humidity. The inoculation of bacteria was carried out by syringe infiltration as described by Maldonado et al. (21). See “In Planta TetraFA Assay” for more details.
Enzyme and Inhibition Assays—The MeSA esterase activity was determined as described by Forouhar et al. (19). Briefly, a standard reaction mixture of 50 μl consisted of 0.1 mm MeSA in 0.1 m potassium phosphate buffer (pH 7.5), and 5 mm β-mercaptoethanol. The reaction was initiated by adding esterase and incubated for 30 min at ambient temperature. After the reaction was stopped by boiling for 5 min, SA, the product, was coupled with radioactive [14C]S-adenosylmethionine by using a recombinant Clarkia breweri SA carboxylmethyltransferase (SAMT). Radiolabeled [14C]MeSA was then extracted with ethyl acetate, and radioactivity was determined in a scintillation counter. Steady-state kinetic parameters were determined by an initial velocity experiment. Measurements of the kcat and Km(app) values for MeSA (5-250 μm) were made at 2.5 nmol of [14C]MeSA and 10 μg of C. breweri SAMT. Kinetic parameters were calculated to fit untransformed data to v = kcat[S]/(Km + [S]) using SigmaPlot (Systat Software, Inc.).
For inhibitory activity of SA or FA to MeSA esterases, initial velocities were determined using the standard assay system. Enzyme activities were determined after addition of SA (1-50 μm) or FA (0.01-1 mm) to assay solution containing various MeSA concentrations (5-250 μm). Global fitting analysis was used to simultaneously fit all data to the equation for competitive inhibition, v = Vmax ([S]/Km (1 + [I]/Ki) + Vmax using SigmaPlot.
The esterase and lipase activities were determined colorimetrically by measuring the liberation of para-nitrophenol (pNP) from pNP-butyrate (C4) and pNP-myristate (C14) as described in Kumar and Klessig (18) with a slight modification. Briefly, the standard 1-ml reaction mixture consisted of 1 mm substrate in 50 mm Bicine (pH 8.0), 0.05% Triton X-100. The reaction was allowed to proceed for 15 min at 24 °C, and the amount of pNP was measured at 410 nm. Inhibition assays were performed by adding various concentrations of SA or FA to the standard reaction system. Values from control reactions without SABP2 were subtracted from each reaction.
Structural Modeling of SABP2—An atomic model with suitable conformation for tri- and tetraFA was obtained using The Dundee PRODRG2 Server (davapc1.bioch.dundee.ac.uk/prodrg). Using the XtalView program (22), each inhibitor was modeled into the active site of the SABP2 crystal structure in complex with SA (PDB accession code 1Y7I), using the position and orientation of SA as the reference. However, to obtain the most favored model for each inhibitor, several orientations of each were considered and subject to two cycles of rigorous rigid-body refinement using the Crystallography and NMR System (23).
In Planta TetraFA Assay—To address the utility of tetraFA in planta, we examined if it could inhibit SAR development by blocking SABP2, or its orthologous proteins, in TMV-infected tobacco and/or Pseudomonas-infected Arabidopsis. Before or after the 1° infection (-24, 0, 24, 48, 72, 96, and 120 h post 1° infection (hp1°i)), which was carried out by inoculating TMV in the three lower leaves of 6-week old plants, upper-uninfected leaves were treated with 10 mm HEPES (pH 7.0) in the presence or absence of 1 mm tetraFA. The treatment of tobacco with tetraFA was performed by syringe infiltration. Six days after the 1° TMV infection, those tetraFA- or buffer-treated (control) leaves were challenged by TMV inoculation (2° infection). The establishment of SAR was then determined by measuring and comparing the lesion sizes of TMV-infected leaves with a vernier caliper at 5 days after the infection.
Arabidopsis was treated with tetraFA by using a fine glass chromatography sprayer. SAR was induced in 2-3-week-old Arabidopsis plants by infection of 2-3 leaves per plant with coronatine-deficient Pseudomonas syringae pv. maculicola (Psm) carrying AvrRpt2 at a concentration of 1 × 106 colony forming units per ml in 10 mm MgCl2. Mock inoculation was performed using 10 mm MgCl2. Ten mm HEPES buffer (pH 7.0) with or without tetraFA (2, 10, or 20 mm) was applied to distal leaves by repeated spraying at 3, 24, and 48 hp1°i. The 2° challenge was then performed in tetraFA- or buffer-treated leaves with 1 × 105 colony forming units/ml of P. syringae pv. tomato (Pst) DC3000 at 3 days after a 1° infection. In planta bacterial titers were determined by shaking leaf dics, 4 mm in diameter, from 2° infected leaves in 10 mm MgCl2 supplemented with 0.1 m sucrose at 12 °C for 4 h. The resulting bacterial suspensions were serially diluted and spots of 10 μl per dilution were grown on KB medium and counted.
Leaf Detachment Assay—After induction of SAR in 6-week-old tobacco plants by the inoculation of the three lower leaves per plant with TMV, these infected leaves were excised from plants at various time points after a 1° infection (0-120 hp1°i). Mock inoculation was performed using 10 mm HEPES (pH 7.0). Six days after SAR induction, three distal leaves, located directly above the 1° infected ones, were challenged with TMV. To determine SAR development, lesion sizes formed in 2° challenged leaves were measured with a vernier caliper at 5 days after 2° infection.
Collection of Phloem Exudates and MeSA Quantification—Petiole/phloem exudates were collected in 1 mm EGTA (pH 7.0) for 24 h and MeSA levels were measured using gas chromatography-mass spectrometry (CP-3800/Quadrupode-1200L system, Varian) as described previously (13).
RESULTS
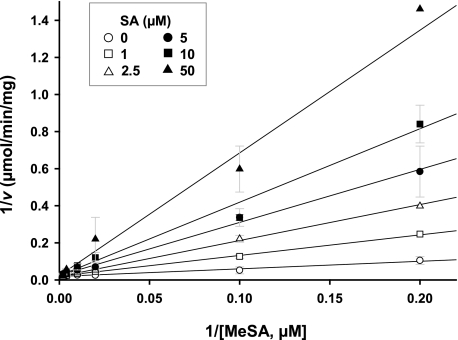
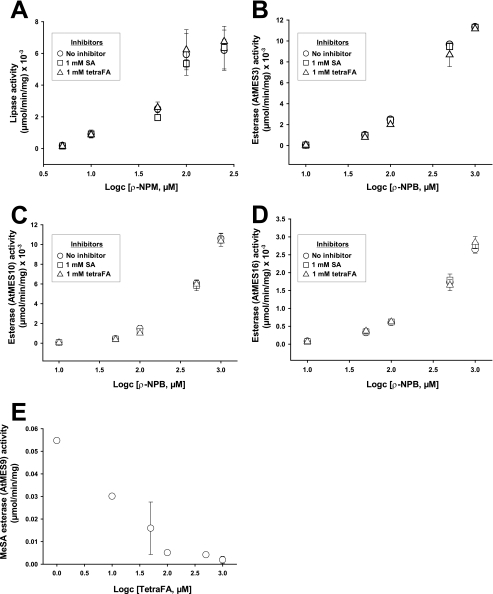
SA Competitively Inhibits the MeSA Esterase Activity of SABP2—Inhibition studies are often used to assess the physiological role and/or functional involvement of catalytic proteins. Previously, SA was shown to be an endogenous inhibitor of the MeSA esterase SABP2; SA binds in the active-site pocket of SABP2 (Kd = 90 nm), which results in the inhibition of the catalytic activity of SABP2 (15, 19). To further understand this interaction, we determined the effect of increasing SA levels on the MeSA esterase activity of SABP2 (Fig. 1). The steady-state kinetic parameters (kcat and Km(app)) of SABP2 for MeSA were 0.16 s-1 and 21.5 μm, respectively. SA competitively inhibited this activity with a Ki value of 16.4 μm. Because this Ki value falls within the range of reported SA levels in tobacco leaves after 1° infection by TMV (∼20-100 μm; 8, 10, and 24), it is likely that the MeSA esterase activity of SABP2 is increasingly inhibited as infection proceeds and SA levels rise. Indeed, the MeSA esterase activity of SABP2 in the infected leaves is predicted to be completely inhibited by 72 hp1°i, based on a calculation performed by integrating the kinetic parameters with the level kinetics of MeSA and SA, where vi = [S]·Vmax/{((Km(1 + [I]/Ki)) + [S]}, v0 = [S]·Vmax/(Km + [S]), i% = 100 (1 - (vi/v0)).
FIGURE 1.
Inhibition of the MeSA esterase activity of SABP2 by SA. Lines represent the global fit of all data (n = 3) to the equation for competitive inhibition. Initial velocities were determined as described under “Experimental Procedures” at 0 μm (open circles), 1 μm (open squares), 2.5 μm (open triangles), 5 μm (closed circles), 10 μm (closed squares), and 50 μm (closed triangles) SA.
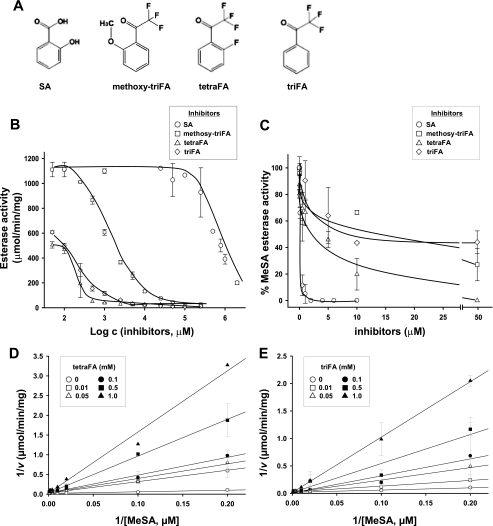
Identification of Synthetic SA Analogs and Their Inhibitory Activity on SABP2—Although SA is an effective inhibitor of SABP2, it cannot be used for in planta studies because it triggers defense responses downstream of SABP2 (for review see Ref. 7). Thus, we utilized the data base from PubChem. Substance to identify candidate compounds that could inhibit SABP2 esterase activity without activating defense responses. Candidate synthetic molecules were similar in structure to SA and benzoic acid, except that their hydroxyl groups were replaced with fluorine (fluoroacetophenone; FA). Due to the small atomic radius and extreme electronegativity of fluorine, it was anticipated that fluorine replacement for the hydroxyl groups might enhance binding in the active-site pocket of SABP2 (25). Among candidate compounds, three synthetic compounds were chosen for further analyses based on commercial availability. These were 2′-methoxy-2,2,2-triFA (methoxy-triFA), 2,2,2,2′-tetraFA (tetraFA), and 2,2,2-triFA (triFA) (Fig. 2A).
FIGURE 2.
Inhibition of the esterase activity of SABP2 by FAs. A, structure of SA and the FAs. B and C, inhibitory activities of SA (open circles), tetraFA (open triangles), triFA (open diamond), and methoxy-triFA (open squares) toward the general esterase activity of SABP2, using 1 mm pNP-butyrate (C4) as the substrate (B), or toward MeSA esterase activity, using 0.1 mm MeSA as the substrate (C). D and E, double-reciprocal plots of the effects of tetraFA (D) or triFA (E) on the MeSA esterase activity of SABP2. Lines represent the global fit of all data (n = 3) to the equation for competitive inhibition. Initial velocities were determined as described under “Experimental Procedures” at 0 mm (open circles), 0.01 mm (open squares), 0.05 mm (open triangles), 0.1 mm (closed circles), 0.5 mm (closed squares), and 1 mm (closed triangles) FA.
As a first step to examine the potential inhibitory activities of the FAs, various concentrations of the FAs and SA (0.05-2.0 mm) were added to an SABP2 enzymatic assay using the synthetic substrate pNP-butyrate (C4). As shown in Fig. 2B, the three FAs inhibited the esterase activity of SABP2 in a concentration-dependent manner, and this inhibition was more effective than that mediated by SA. Unexpectedly, the FAs were less efficient than SA at inhibiting the esterase activity of SABP2 when MeSA was the substrate (Fig. 2C). The inhibitory kinetics of tetraFA and triFA, which were stronger inhibitors than methoxy-triFA, were further analyzed in the presence of MeSA (Fig. 2, D and E). TetraFA and triFA displayed the same inhibitory mechanism as SA, with all three competitively inhibiting the esterase activity of SABP2. The Ki values for tetraFA and triFA were 131.2 and 303.1 μm, respectively (Fig. 2, D and E). Although these values are >8-fold higher than that of SA (16.4 μm), they raised the possibility that the FAs, particularly tetraFA, might block the esterase activity in planta of SABP2. Given that SABP2 is only required in uninfected distal tissues for SAR development and MeSA levels in these tissues peak at ∼1.5 μm between 48 and 72 hp1oi (13), 0.5 mm tetraFA should inhibit ∼75% of the esterase activity of SABP2 in uninfected distal tissues, whereas 1.0 mm tetraFA should inhibit ∼90%. The application of 1.0 mm tetraFA to tobacco leaves did not have any detectable toxic effect (Figs. 5 and 7C), but higher concentrations (>5.0 mm) caused cell death at the infiltrated site within 72 h (data not shown).
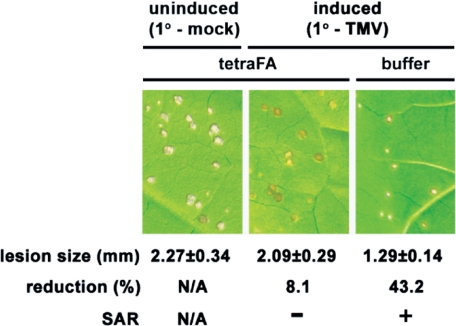
FIGURE 5.
TetraFA blocks SAR development in TMV-infected tobacco. Induced plants received a 1° inoculation of TMV on the three lower leaves per plant to activate SAR, whereas uninduced plants received a mock 1° inoculation with 10 mm HEPES (pH 7.0) buffer. At 48 and 72 hp1°i, distal leaves were treated with 1 mm tetraFA or buffer. Six days post 1° TMV infection (dp1°i), these distal leaves were challenged by a 2° TMV infection. The size of lesions was measured (in mm ± S.D.) and photographed at 5 days post 2° infection. N/A, not applicable. Reduction (%), percent reduction in the size of 2° TMV lesions formed on induced versus uninduced plants.
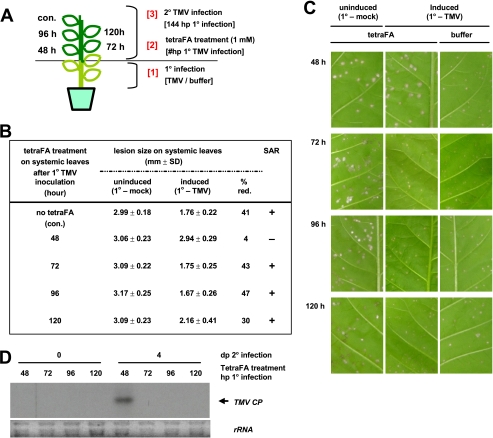
FIGURE 7.
The MeSA esterase activity of SABP2 is required in distal leaves between 48 and 72 hp1°i for SAR development. A, schematic design for the time course of tetraFA (1 mm) application. Control leaf (con.) received a buffer (10 mm HEPES, pH 7.0) treatment without tetraFA at 48 hp1°i. B, determination of lesion sizes in millimeters on the distal leaves at 5 days post 2° infection. Uninduced plants were mock inoculated on the three lower leaves, whereas induced plants received TMV inoculation on three lower leaves 6 days prior to 2° TMV infection. % red., percent reduction in the size of 2° TMV lesions formed on induced versus uninduced plants. C, photographs of TMV-induced lesions on distal leaves described in B at 5 days post 2° infection. Note induced plants without tetraFA treatment were used as a positive control for SAR (right column). D, RNA-blot analysis of TMV coat protein (CP) transcripts in the distal leaves, at 0 and 4 days post 2° infection, of plants received tetraFA treatment at various times after 1° infection. Five μg of total RNA were loaded per lane and ethidium bromide-stained ribosomal RNA (rRNA) was used as a loading control.
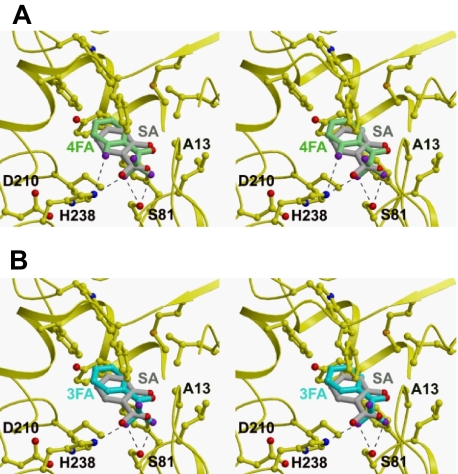
Structural Modeling of SABP2 with TetraFA and TriFA—To investigate how tetraFA and triFA inhibit the activity of SABP2, we used the three-dimensional structure of SABP2 complexed with SA to model these inhibitors in the active-site pocket of SABP2 (Fig. 3). Both tetraFA and triFA can be accommodated readily in the active site and require few conformational changes in the enzyme. However, one difference in their predicted interaction with SABP2 is that they are flipped 180° in comparison to SA. The modeling suggests that the trifluoro group of the FAs interacts with both Ser81 and His238 of the catalytic triad, whereas SA hydrogen bonds with Ala13 and His238. His238 makes two hydrogen bonds with tetraFA, but only one hydrogen bond with triFA; this difference may account for the different Ki values for these inhibitors. Despite the possible interactions of the tri-fluoro group with both Ser81 and His238, its bulkiness likely compromises binding, and thus explains why these inhibitors exhibit higher Ki values than SA.
FIGURE 3.
Model of the active-site pocket of SABP2 and the binding modes of FAs compared with SA. The binding modes of SA (gray), tetraFA (green, A), and triFA (blue, B) in the active site of SABP2 are depicted; the catalytic triad residues (Ser81, Asp210 and His238) and Ala13 are noted, and the hydrogen bonds are indicated as dashed lines.
TetraFA Inhibits Esterases with Strong Preference for MeSA as a Substrate—SABP2 belongs to a superfamily of α/β-fold hydrolases that share both a similar mode of catalysis and overall three-dimensional structure (26). Thus, before the SA analogs could be used for in planta studies, their specificity for SABP2 inhibition needed to be established. To assess specificity, the effect of tetraFA on the activities of other α/β-fold hydrolases within the same subgroup of lipases/esterases, which differ only in their substrate preferences, was determined (Fig. 4, A-D). These hydrolases included a lipase from Phixomucor miehei and three Arabidopsis methyl esterases (AtMES3 (At2g23610), AtMES10 (At3g50440), and AtMES16 (At4g16690)). The AtMESs (numbers 1-18) were initially identified based on their high sequence homology to SABP2; several members were characterized biochemically and genetically as potential orthologs of SABP2 (27). AtMES3, AtMES10, and AtMES16, however, showed no catalytic activity with MeSA; rather they preferentially de-methylated methyl indole acetic acid or methyl jasmonate (28). pNP-myristate (C14) and pNP-butyrate (C4) were utilized as substrates for the P. meihei lipase and the three methyl esterases, respectively. Neither tetraFA nor SA affected their enzymatic activities. In contrast, AtMES9 (At4g37150), an SABP2 ortholog that displays SA-inhibitable MeSA esterase activity (27), was inhibited by tetraFA (IC50 = 19 μm; Fig. 4E). Arabidopsis contains four additional MeSA esterases, which are inhibitable by SA (27). Because AtMES9 has equal or higher specific activity for MeSA than the other four MeSA esterases and is inhibited by tetraFA, it is likely that the activity of all five Arabidopsis esterases is suppressed by tetraFA. These results suggest that tetraFA specifically inactivates SABP2, as well as its orthologs in Arabidopsis.
FIGURE 4.
Specific inactivation of MeSA esterases by tetraFA. The effects of tetraFA and SA on the activities of P. miehei lipase (A), AtMES3 (B), AtMES10 (C), AtMES16 (D), and AtMES9 (E). A-D, the assays for lipase and esterase activity were performed utilizing pNP-myristate (C14; A) or pNP-butyrate (C4; B-D), respectively, as a substrate in the absence of an inhibitor (open circles) or in the presence of 1 mm inhibitor, SA (open squares) or tetraFA (open triangles). E, inhibition of the esterase activity of AtMES9 by tetraFA was determined using 0.25 mm MeSA as the substrate.
TetraFA Blocks SAR in TMV-infected Tobacco—To address the utility of tetraFA in planta, we first determined whether it could inhibit SAR development in TMV-infected tobacco, because both MeSA and the MeSA esterase of SABP2 activity are essential for this process. In tobacco, SAR is manifested as a reduction in the size of lesions formed after 2° TMV infection of the distal leaves on plants that previously received a 1° TMV infection, as compared with the lesions developed by plants that were mock-inoculated for the 1° infection and therefore were encountering the virus for the first time. The reduction in lesion size occurs because SAR elicited by the 1° infection enables the plant to restrict viral replication and spread more efficiently the second time it encounters the virus. Following 1° TMV infection of three lower leaves, the efficacy of tetraFA (1 mm) was assessed by its application to healthy distal leaves at 48 and 72 hp1°i; these times were chosen because overexpression of a synthetic SABP2, which is not susceptible to RNA interference-mediated silencing, in the distal leaves of SABP2-silenced plants at 24 hp1°i was sufficient to restore SAR (13, 18, 27). At 144 hp1°i, the tetraFA-treated leaves were then challenged with TMV (2° infection). SAR was observed in plants that received a 1° and 2° infection with TMV (SAR-induced) but no tetraFA treatment, as their 2° lesions were 43% smaller than those formed on plants that were mock inoculated for the 1° infection (uninduced; Fig. 5). By contrast, SAR-induced plants that were treated with tetraFA failed to develop SAR; their lesions were nearly as large as those on uninduced plants. Note that tetraFA treatment did not affect lesion size in uninduced plants, suggesting that tetraFA does not directly affect either the multiplication and spread of TMV or the innate immune response.
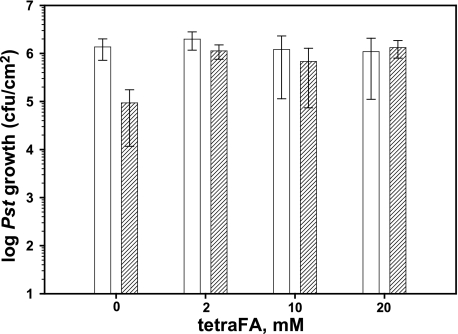
MeSA Esterase-mediated SAR Development Is Conserved in Arabidopsis—Whether tetraFA could block SAR development in other plant species, such as Arabidopsis, was then tested (Fig. 6). Following 1° inoculation with the avirulent bacterial pathogen Psm ES4326 cfa6::Kmr (avrRpt2) (29) to induce SAR, the distal leaves of Col-0 plants were treated with various concentrations of tetraFA and then challenged with virulent Pst DC3000 at 72 hp1oi. SAR-induced plants that were not treated with tetraFA displayed ∼25-fold less Pst growth than untreated, uninduced control plants, which received neither the 1° infection nor tetraFA treatment. Bacterial growth in SAR-induced plants treated with tetraFA was comparable with that in the untreated, uninduced control plants, indicating that SAR was effectively blocked by all concentrations of tetraFA tested. Note that tetraFA did not show any direct effect on the multiplication of Pst, because similar levels of bacterial growth were observed in uninduced plants regardless of tetraFA treatment. Moreover, application of tetraFA from 2 to 100 mm to Arabidopsis leaves did not have detectable toxic effects (supplemental Fig. S1). The ability of tetraFA to inhibit SAR in Arabidopsis, which contains four additional SABP2 orthologs that are at least partially functionally redundant with AtMES9 for SAR development (27), confirms the importance of MeSA and MeSA esterases for SAR in Arabidopsis. It also demonstrates the utility of tetraFA for assessing the involvement of MeSA and its esterases during SAR activation in various plant species.
FIGURE 6.
TetraFA blocks SAR in Arabidopsis. SAR was analyzed in mock-inoculated (white bars) and avirulent Psm AvrRpt2-infected (striped bars) wild type Col-0 plants. After 1° infection, 10 mm HEPES buffer (pH 7.0) with 0, 2, 10, or 20 mm tetraFA was applied to distal leaves by repeated spraying at 3, 24, and 48 hp1°i. Distal leaves were infected at 72 hp1oi with virulent Pst DC3000 and titers of bacteria in the distal challenged tissue were determined at 3 days post 2° infection.
SAR Development Requires MeSA Esterase Activity of SABP2 in Distal Tissue between 48 and 72 hp1°i with TMV—To determine the kinetics of MeSA accumulation and processing/perception in the distal tissue of tobacco plants, distal leaves were treated with tetraFA at various times after a 1° inoculation with TMV or buffer, followed by a 2° TMV infection at 144 hp1°i (Fig. 7A). TetraFA treatment blocked SAR development when applied at 48 hp1°i, as the lesions on these plants were comparable in size to those on uninduced plants, regardless of tetraFA treatment (Fig. 7, B and C). In contrast, tetraFA treatment at 72, 96, or 120 hp1°i was not effective at blocking SAR development, suggesting that sufficient amounts of MeSA have been converted to SA in the distal tissues by 72 hp1°i, but not by 48 hp1°i. As previously shown (13), suppression of viral replication in SAR was confirmed at the molecular level through analysis of the levels of viral coat protein transcript after challenge infection of the distal leaves (Fig. 7D).
Because SAR requires systemic movement of a signal from infected to distal tissue through the phloem (30), the level of signal delivered to distal tissues may differ depending on their position and/or distance from 1° infected leaves. Indeed, previous analyses showed that the leaf located directly above the inoculated leaf displayed the greatest increase in SA levels (31). To circumvent any possible effect due to leaf positioning, the experiments described above were performed by infecting three lower leaves. In addition, the possibility that leaf position affected these analyses was tested by applying tetraFA at different times to distal leaves in different positions relative to the three lower 1° inoculated leaves (Fig. 8). Regardless of leaf position, loss of SAR was consistently observed only when tetraFA was applied at 48 hp1°i. Application of tetraFA at earlier time points (24 h before 1° infection, 0 hp1°i or 24 hp1°i) also failed to inhibit SAR. Together these results suggest that cleavage of MeSA by SABP2 is critical in the distal leaves between 48 and 72 hp1°i, and that tetraFA cannot indefinitely block SABP2 activity, possibly because it is unstable in planta.
FIGURE 8.
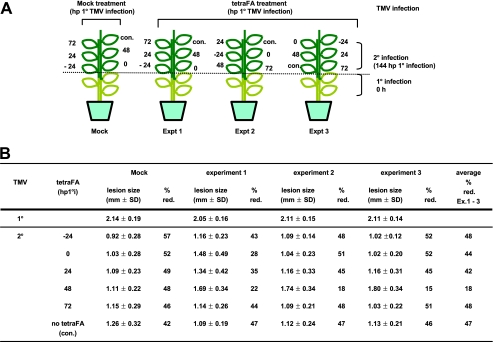
SABP2 activity in distal leaves is essential between 48 and 72 hp1°i, regardless of leaf position. A, schematic design for in planta tetraFA treatment (1 mm) of various leaves at different time points. Control leaf (con.) received a buffer treatment without tetraFA at 48 hp1°i. B, determination of lesion sizes in millimeters on the distal leaves at 5 days post 2° infection. All plants received a 1° TMV inoculation on three lower leaves. At the specified times post 1° infection, leaves at different positions were treated with buffer or tetraFA. Six days post 1° infection, the buffer- or tetraFA-treated distal leaves received a 2° TMV infection. % red., percent reduction in size of 2° versus 1° lesions on plants that received either buffer or tetraFA treatment.
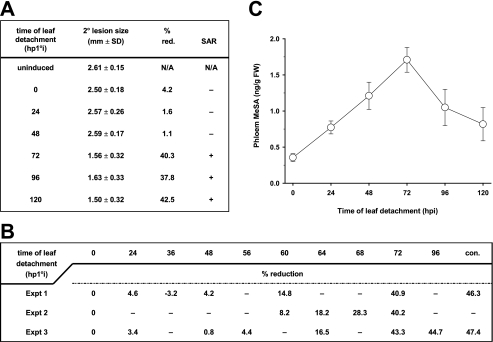
The MeSA SAR Signal Is Transmitted to Distal Tissues between 48 and 72 hp1°i with TMV—To assess whether the movement of an SAR signal, particularly MeSA, corresponds to the time period when the esterase activity of SABP2 is required, the kinetics of SAR signaling were monitored by a leaf detachment assay (Fig. 9, A and B). At various times after the 1° inoculation, the three TMV-infected leaves were excised; the upper distal leaves were then challenged with TMV at 144 hp1°i. Plants whose 1° TMV-inoculated leaves were detached at 0, 24, or 48 hp1°i failed to develop SAR; the 2° lesions on these plants were as large as those on uninduced plants, which had received a mock 1° inoculation (Fig. 9A). In contrast, SAR was clearly observed in plants whose 1° infected leaves were detached at or after 72 hp1°i, as indicated by a ∼40% reduction in lesion size. This result indicates that a sufficient amount of SAR signal was generated and transmitted from 1°-infected leaves before 72 hp1°i. Subsequent analysis using additional time points revealed that enough signal to induce partial SAR was transmitted from the inoculated leaf by 60 hp1°i (Fig. 9B). As the time before detachment increased, a parallel increase in SAR strength was observed up to 72 hp1°i, at which point SAR was maximal. The timing for transmission of the SAR signal correlated with the kinetics of MeSA accumulation in the phloem (petiole) from the 1° infected leaves, as MeSA levels increased gradually before peaking at 72 hp1°i (Fig. 9C). Together, these results indicate that a critical amount of the MeSA signal for SAR has been exported from the infected leaves between 48 and 72 hp1°i.
FIGURE 9.
Kinetics of SAR signal movement from 1° infected leaves and MeSA levels in phloem/petiole exudates of these leaves. A and B, SAR development was affected by the time at which TMV-infected leaves were detached. Primary inoculated leaves were detached from plants at the times indicated and the distal leaves were challenged with TMV at 6 days post 1° infection. Uninduced plants were mock inoculated on the lower leaves, which were not excised from plants. Control plants (con.) in B, which did not have their 1° TMV infected leaves detached prior to 2° TMV infection, exhibited the usual level of % reduction (red.) of size of 2° lesions versus size of 2° lesions on plants receiving a mock 1° inoculation. N/A, not applicable. C, levels of MeSA in petiole exduates of detached 1° TMV infected leaves from A.
DISCUSSION
In this study, we examined the inhibitory interaction of SA with SABP2, identified a synthetic analog of SA, tetraFA, and used it to further study SAR development. TetraFA inactivates SABP2 with a similar mode of action to that of SA, but, unlike SA, does not induce downstream defense responses. Although the inhibitory efficiency of tetraFA (Ki = 131.2 μm) is lower than that of SA (Ki = 16.4 μm) toward the MeSA esterase activity of SABP2, tetraFA specifically inhibited SABP2 and at least one of its functional Arabidopsis homologs. Among the tested α/β-fold hydrolases to which SABP2 catalytically and structurally belongs, tetraFA selectively inhibited only esterases that principally utilize MeSA as a substrate. Exogenous application of tetraFA was effective in planta in blocking SAR development in TMV-infected tobacco, where MeSA esterase activity and MeSA are essential for SAR.
TetraFA treatment also suppressed SAR development in P. syringae-infected Arabidopsis; this result argues that an SABP2-like esterase activity plays an essential role for SAR in this plant species (Fig. 6). Consistent with this conclusion, several members of the AtMES family were recently identified as functional homologs of SABP2 (27). These members share functional redundancy for MeSA hydrolysis and, most likely, SAR development. Three of these members, AtMES1, -7, and -9, were capable of complementing SAR deficiency in SABP2-silenced tobacco. Conversely, underexpression of AtMES genes, including those encoding functional MeSA esterases, compromised SAR. A data base search (32) revealed that genes sharing sequence homology with SABP2 are common in many other plant species, such as tomato (Solanum lycopersicum, TC165610), potato (S. tuberosum, CK270870), sweet potato (Ipomoea batatas, TA2555_4120), cotton (Gossypium hirsutum, TA41446_3635), grape (Vitis vinifera, TA51764_29760), barrel medic (Medicago truncatula, TA31451_3880), rice (Oryza sativa, Os01g0787600, AK061058, and CT83232), Zea mays (EU972429), and Nicotiana benthamiana (EH386450). These genes have a high degree of sequence similarity with SABP2 (>70%), suggesting that their encoded proteins share similar biochemical activities, and that MeSA esterase and its substrate, MeSA, are general components of plant innate immunity. Supporting this possibility, SAR in Phytophthora infestans-infected potato was effectively suppressed by treatment with tetraFA.5
We previously demonstrated that SABP2 is an integral component for SAR development in tobacco, and that it mediates SA signaling via demethylation of MeSA (13, 18, 19). The reverse reaction, the methylation of SA to MeSA, also is necessary for SAR (13); this reaction is catalyzed by SAMT (33). Successful establishment of SAR requires the methyltransferase activity of SAMT in the pathogen-infected, SAR signal-generating tissue and esterase activity of SABP2 in the distal, SAR signal-receiving tissue (13). To facilitate accumulation of sufficient levels of MeSA in the 1° infected tissue, we previously proposed that the activity of SABP2 is feedback inhibited by SA, its catalytic product (13, 19). Consistent with this model, overexpression of a mutant SABP2, which fails to bind SA and thus is not feedback inhibited, suppressed MeSA accumulation in SABP2-silenced plants and failed to restore SAR proficiency (13). The kinetic analyses performed in this article further support this model; we calculate that the increased level of SA synthesized in pathogen-infected leaves is sufficient to completely inhibit the activity of SABP2 by 72 hp1°i (Fig. 1). This finding correlates with the kinetics of MeSA accumulation in the phloem exudates of infected leaves, because MeSA levels peaked at 72 hp1°i (Fig. 9C).
In contrast to the inoculated leaf, SA levels in distal leaves developing SAR range from ∼0.5 to 9 μm (8, 10, 24). Because this concentration is too low to effectively inhibit the MeSA activity of SABP2, SABP2 appears to function exclusively or predominantly in perceiving/processing an SAR signal. Consistent with this model, MeSA is biologically inactive and must be converted to active SA to activate or prime downstream defense responses in the distal tissue (13, 34). Furthermore, the combined results from in planta inhibition of SABP2 and the leaf detachment assay indicate that the majority of the SAR signal is transported out of the inoculated leaf and perceived/processed in the distal tissue between 48 and 72 hp1°i. This time frame correlates with the kinetics of MeSA accumulation in the phloem exudate of 1° infected leaves (Fig. 9C). Interestingly, a similar timing for SAR signal transmission was observed in pathogen-infected cucumber, as SAR only developed when the infected leaves remained attached to the plant for 72-96 hp1°i (35). Our results also suggest that at or around 72 hp1oi, either little to no additional SAR signal is being transmitted to the systemic tissue and/or a threshold of SAR signal has been reached in this tissue, above which the additional signal does not enhance the strength of SAR development. The decreasing level of MeSA in phloem exudates after 72 hp1oi is consistent with a decrease in signal transmission, but does not exclude the possibility that a threshold of SAR signal has already been reached.
In comparison to the results presented in this paper and in previous studies, which argue that MeSA is a phloem-mobile signal for SAR development (13, 27), other studies have suggested that MeSA is an airborne signal that travels from a pathogen-infected plant to neighboring plants, where it activates defense responses (36, 37). However, no such plant-to-plant signaling was observed in our experiments. Our experiments were carried out in 30-square feet growth chambers, where control and test plants were intermingled and confined within a small space. If volatile MeSA, rather than its liquid phloem-mobile counterpart, was the predominant form of the SAR signal, we would have obtained very different results. This observation suggests that in nature, where conditions are not highly optimized for transmission of a volatile signal and plants are generally less confined than in growth chambers, the MeSA SAR signal is in a liquid form.
In addition to MeSA, several studies have suggested that other mobile, long-distance signals play a role in activating SAR. For example, analyses of Arabidopsis dir1-1, sfd1, and fad7 mutants implicate a lipid or lipid derivative in systemic signaling (21, 38, 39). The relationship between MeSA and a lipid-based signal(s) is unclear. The phytohormone jasmonic acid, which plays an important role(s) in defense against necrotrophic pathogens and insects, also has been implicated in SAR signaling (40). Jasmonic acid-mediated induction of the benzoic acid/SA methyltransferase gene in Arabidopsis and tomato (37, 41, 42) suggests one possible mode of interaction between these two signaling systems.
In summary, we have characterized tetraFA, a synthetic SA analog that specifically inhibits the MeSA esterase activities of SABP2 and its orthologs. Its use in planta confirmed both the importance of MeSA esterase and MeSA for SAR development in tobacco, and helped establish similar roles for these factors in Arabidopsis. In addition, the combined results from in planta tetraFA analyses, leaf detachment assays, and MeSA quantification allowed us to explore the dynamics of SAR signal transmission and perception/processing in tobacco. After pathogen attack, SA is synthesized at the site of infection through an isochorismate synthase and/or phenylalanine ammonia-lyase pathway (43, 44) and is a key mediator of immunity. In parallel, rising SA levels suppress and eventually fully inhibit the esterase activity of SABP2 by 72 hp1°i. This facilitates the build up of MeSA, which is synthesized from SA by SAMT. MeSA then moves to distal tissues through phloem and is processed to SA by active SABP2 in distal tissues; transmission and processing of this signal occurs mainly between 48 and 72 hp1°i. Moreover, the ability of tetraFA to block SAR in a variety of plant species argues that it will be a highly useful tool for assessing the extent to which different plant species use an SABP2-like esterase activity(s) and MeSA for SAR activation, regardless of the nature of the inducing pathogen and/or the local defense response.
Supplementary Material
Acknowledgments
We thank Dr. D'Maris Dempsey for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant Protein Structure Initiative P50 GM62413 (to L. T.). This work was also supported by National Science Foundation Grant IOS-0525360 (to D. F. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: SAR, systemic acquired resistance; SA, salicylic acid; MeSA, methyl salicylate; SABP2, salicylic acid-binding protein 2; TMV, tobacco mosaic virus; FA, fluoroacetophenone; tetraFA, 2,2,2,2′-tetrafluoroacetophenone; AtMES, Arabidopsis methyl esterases; SAMT, SA carboxylmethyltransferase; pNP, para-nitrophenol; Bicine, N,N-bis(2-hydroxyethyl)glycine; hp1°i, hours post primary infection.
Patricia Manosalva, S.-W. Park, and D. F. Klessig, unpublished results.
References
- 1.Durrant, W. E., and Dong, X. (2004) Annu. Rev. Phytopathol. 42 185-209 [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann, J. A., Kafatos, F. C., Janeway, C. A., and Ezekowitz, R. A. (1999) Science 284 1313-1318 [DOI] [PubMed] [Google Scholar]
- 3.Aderem, A., and Underhill, D. M. (1999) Annu. Rev. Immunol. 17 593-623 [DOI] [PubMed] [Google Scholar]
- 4.Jones, J. D., and Dangl, J. L. (2006) Nature 444 323-329 [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M. (2005) Nat. Immunol. 6 973-979 [DOI] [PubMed] [Google Scholar]
- 6.DeYoung, B. J., and Innes, R. W. (2006) Nat. Immunol. 7 1243-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dempsey, D. A., Shah, J., and Klessig, D. F. (1999) Crit. Rev. Plant Sci. 18 547-575 [Google Scholar]
- 8.Vernooij, B., Friedrich, L., Morse, A., Reist, R., Kolditz-Jawhar, R., Ward, E., Uknes, S., Kessmann, H., and Ryals, J. (1994) Plant Cell 6 959-965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L. B., Wdymann, K. B., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, H., and Ryals, J. (1994) Science 266 1247-1250 [DOI] [PubMed] [Google Scholar]
- 10.Gaffney, T., Friedrich, L. B., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, L. (1993) Science 261 754-756 [DOI] [PubMed] [Google Scholar]
- 11.Pallas, J. A., Paiva, N. L., Lamb, C., and Dixon, R. A. (1996) Plant J. 10 281-293 [Google Scholar]
- 12.Nawrath, C., and Métraux, J. P. (1999) Plant Cell 11 1393-1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, S.-W., Kaimoyo, E., Kumar, D., Mosher, S., and Klessig, D. F. (2007) Science 318 113-116 [DOI] [PubMed] [Google Scholar]
- 14.Chen, Z., Silva, H., and Klessig, D. F. (1993) Science 262 1883-1886 [DOI] [PubMed] [Google Scholar]
- 15.Du, H., and Klessig, D. F. (1997) Plant Physiol. 113 1319-1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durner, J., and Klessig, D. F. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11312-11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slaymaker, D. H., Navarre, D. A., Clark, D., del Poza, O., Martin, G. B., and Klessig, D. F. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11640-11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, D., and Klessig, D. F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 16101-16106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forouhar, F., Yang, Y., Kumar, D., Chen, Y., Fridman, E., Park, S.-W., Chiang, Y., Acton, T. B., Montelione, G. T., Pichersky, E., Klessig, D. F., and Tong, L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 1773-1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, A., Salih, G., and Klessig, D. F. (2000) Plant J. 21 409-418 [DOI] [PubMed] [Google Scholar]
- 21.Maldonado, A. M., Doerner, P., Dixon, R. A., Lamb, C. J., and Cameron, R. K. (2002) Nature 419 399-403 [DOI] [PubMed] [Google Scholar]
- 22.McRee, D. E. (1999) J. Struct. Biol. 125 156-165 [DOI] [PubMed] [Google Scholar]
- 23.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 24.Enyedi, A. J., Yalpani, N., Silverman, P., and Raskin, I. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 2480-2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillespie, R. J., and Robinson, E. A. (1992) Inor. Chem. 31 1960-1963 [Google Scholar]
- 26.Holmquist, M. (2000) Curr. Protein Pept. Sci. 1 209-235 [DOI] [PubMed] [Google Scholar]
- 27.Vlot, A. C., Liu, P.-P., Cameron, R. A., Park, S.-W., Yang, Y., Kumar, D., Zhou, F., Padukkavidana, T., Gustagsson, C., Pichersky, E., and Klessig, D. F. (2008) Plant J. 56 445-456 [DOI] [PubMed] [Google Scholar]
- 28.Yang, Y., Xu, R., Ma, C. J., Vlot, A. A., Klessig, D., F., and Pichersky, E. (2008) Plant Physiol. 147 1034-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui, J., Bahrami, A. K., Pringle, E. G., Hernandez-Guzman, G., Bender, C. L., Pierce, N. E., and Ausubel, F. M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 1791-1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenns, A. E., and Kuć, J. (1979) Phytopathology 69 753-756 [Google Scholar]
- 31.Shulaev, V., Leon, J., and Raskin, I. (1995) Plant Cell 7 1691-1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dudareva, N., Raguso, R. A., Wang, J., Ross, J. R., and Pichersky, E. (1998) Plant Physiol. 116 599-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seskar, M., Shulaev, V., and Raskin, I. (1998) Plant Physiol. 116 387-392 [Google Scholar]
- 35.Dean, R. A., and Kuć, J. (1986) Phytopathology 76 966-970 [Google Scholar]
- 36.Shulaev, V., Silverman, P., and Raskin, I. (1997) Nature 385 718-721 [Google Scholar]
- 37.Koo, Y. J., Kim, M. A., Kim, E. H., Song, J. T., Jung, C., Moon, J.-K., Kim, J.-H., Seo, H. S., Song, S. I., Kim, J.-K., Lee, J. S., Cheong, J.-J., and Choi, Y. D. (2007) Plant Mol. Biol. 64 1-15 [DOI] [PubMed] [Google Scholar]
- 38.Chaturvedi, R., Krothapalli, K., Makandar, R., Nandi, A., Sparks, A. A., Roth, M. R., Welti, R., and Shah, J. (2008) Plant J. 54 106-117 [DOI] [PubMed] [Google Scholar]
- 39.Nandi, A., Welti, R., and Shah, J. (2004) Plant Cell 16 465-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truman, W., Bennett, M. H., Kubigsteltig, I., Turnbull, C., and Grant, M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1075-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ament, K., Kant, M. R., Sabelis, M. W., Haring, M. A., and Schuurink, R. C. (2004) Plant Physiol. 135 2025-2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Torres-Zabala, M., Truman, W., Bennett, M. H., Lafforgue, G., Mansfield, J. W., Rodriguez Egea, P., Bögre, L., and Grant, M. (2007) EMBO J. 26 1434-1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yalpani, N., Silverman, P., Wilson, T. M., Kleier, D. A., and Raskin, I. (1991) Plant Cell 3 809-818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001) Nature 414 562-565 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.