Abstract
Axin, a key modulator of the Wnt/β-catenin pathway, acts as a scaffold protein in phosphorylating and degrading cytoplasmic β-catenin. Canonical Wnt proteins appear to stabilize β-catenin by inducing the interaction of LRP5/6 with Axin. This interaction requires the phosphorylation of the Ser or Thr residues in the PPPP(S/T)PX(T/S) motifs at the intracellular domain of LRP5/6. In this work, we identified a novel Axin-interacting protein, zinc-finger BED domain-containing 3 (Zbed3), by yeast two-hybrid screening. The interaction was confirmed in co-immunoprecipitation experiment in mammalian cells and in vitro pulldown assays. Moreover, we found Zbed3 also contains a PPPPSPT motif, which is crucial to its binding to Axin. The Ser and Thr residues in the motif appear to be also phosphorylated by glycogen synthase kinase 3β (GSK3β) and the CKI family kinases, as GSK3β and CKIε could enhance the interaction of Zbed3 with Axin. Mutation of the Ser (SA) or Thr (TA) residue to Ala in the motif markedly impaired its ability to interact with Axin. Expressing Zbed3, but not these mutants, led to inhibition of GSK3β-mediated β-catenin phosphorylation, cytoplasmic β-catenin accumulation, and activation of lymphoid enhancer binding factor-1-dependent reporter gene transcription. Furthermore, knockdown of Zbed3 with RNA interference attenuated Wnt-induced β-catenin accumulation, lymphoid enhancer binding factor-1-dependent luciferase reporter activity, and the Wnt target gene expression. These results together indicate that Zbed3 is a novel Axin-binding protein that is involved in Wnt/β-catenin signaling modulation.
Wnt family proteins activate cellular signals that are involved in both embryogenesis and carcinogenesis in mammals (1–3). The Wnt/β-catenin pathway, which is also termed as the canonical Wnt pathway, has been most intensively studied, and the framework of its signaling transduction has been well elucidated (4, 5). In the presence of Wnt ligands, the Frizzled family of serpentine receptors and low density lipoprotein-receptor-related proteins 5 or 6 (LRP5/6)3 are hetero-oligomerized and activated through binding with Wnt proteins (6). Subsequently, signals transduced to the cytoplasmic effecter, Dishevelled, to inhibit Axin-adenomatous polyposis coli (APC)-GSK3β-CKIα involved protein complex-phosphorylating β-catenin (7–11). Meanwhile, activated LRP5/6 recruits Axin to the plasma membrane with its intracellular domain and leading to dysfunction of Axin (12, 13). Both of these incidents break down the β-catenin destruction complex and increase the stability of β-catenin in cytoplasm. Accumulated β-catenin translocates to the nucleus and interacts with lymphoid enhancer binding factor-1 (LEF-1)/T cell factor family transcription factors to activate the transcription of specific target genes, such as c-myc and axin2 (14–17).
Axin, originally identified as the product of the mouse Fused gene, is one of the core proteins in Wnt/β-catenin signaling transduction (18). One homolog of Axin, Axin2/Conductin, was found in mammals (19). Despite their expression pattern, they are functionally equivalent as an inhibitor in Wnt/β-catenin signals transduction in vivo (20). Mutation of mouse Axin causes β-catenin accumulation and axis duplication in mouse homozygous embryo (18). On the other hand, sequence variants of the Axin gene have been found in several cancers with overactivated Wnt/β-catenin signals, which suggested that Axin is a tumor suppressor (21, 22).
The endogenous protein level of Axin is relatively low in contrast to other components of the Wnt/β-catenin pathway, suggesting that regulations on Axin would be more effective in modulating Wnt/β-catenin signaling (23). The function of Axin is usually regulated by its interacting partners (22), several of which have been demonstrated to regulate Wnt/β-catenin signals by modulating the Axin-centered destruction complex of β-catenin (24–26). Among them, LRP5/6, the specific receptor for the canonical Wnt signal pathway, is the most intensively studied Axin-interacting protein.
Recently, the mechanism of LRP5/6-Axin interaction has been elaborately elucidated (12, 27–29). The interaction required Wnt-induced aggregation and phosphorylation of LRP5/6 (27–31). GSK3β and CKI family kinases are responsible for phosphorylating the conserved motif of “PPPP(S/T)PX(T/S)” on LRP5/6 to increase its binding affinity for Axin (27–29). Mutant of LRP5/6 at either the Ser or Thr in the motif abolished the ability to efficiently interact with Axin and activate Wnt/β-catenin signals (27–29). Although it is not clear how LRP5/6 regulates Axin, a recent report shows that LRP5/6 interacting with Axin may inhibit GSK3β's kinase activity to phosphorylate β-catenin (32). Until now, PPPP(S/T)PX(T/S) is the only confirmed Axin-binding motif.
In searching for more Axin-interacting partner that may be involved in the Wnt/β-catenin pathway, we found zinc-finger BED domain-containing protein 3 (Zbed3). Zbed3 is a member of zinc-finger domain protein superfamily, whereas no function analysis has been taken on this molecule before. In this study, we identified Zbed3 as an Axin-binding protein that can regulate Wnt/β-catenin signaling through its PPPPSPT motif by adopting a similar mechanism as LRP5/6.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies—Full-length mouse Zbed3 was identified from the pPC86-based mouse embryonic E10.5 cDNA library as a positive clone, which showed interaction with Axin, and subcloned into pCMV-HA, pCMV-EE, pCMV-FLAG, pCMV-EGFP, and pET28C plasmids. Other plasmids have been used previously (10). Zbed3 antibody was raised against recombinant full-length mouse Zbed3 expressed in Escherichia coli. by the Antibody Research Center at the Institute of Biochemistry and Cell Biology, and purified with antigen affinity chromatography. Anti-HA, -Myc, and -EE (Covance), FLAG (Sigma), anti-Axin1 (Cell Signaling), anti-Ser-33/37Thr-41 phospho-β-catenin (Cell Signaling), anti-β-catenin (BD Biosciences), and β-tubulin antibodies were used in this work.
Yeast Two-hybrid Screen and CPRG Assay—Yeast two-hybrid screens system and pPC86-based mouse embryonic E10.5 cDNA library were purchased from Invitrogen. Yeast-two hybrid screening was performed according to the manufacturer's manual. Quantitative assays for β-galactosidase activity in liquid cultures can be performed using chlorophenol red β-d-galactopyranoside (CPRG, Sigma) as a substrate. For each strain, three independent isolated colonies were used, and each sample extract was assayed in triplicate to reduce variability. Each isolated colony was cultured by shaking to A600 = 1.0–1.5. The A600 of each colony was determined and recorded. 1.5 ml of each colony was collected by centrifuging at 14,000 × g for 30 s, and then the yeast was washed once with Buffer 1 (2.38 g of HEPES, 0.9 g of NaCl, 0.065 g of l-aspartate (hemi-Mg salt), 1.0 g of bovine serum albumin, and 50μl of Tween 20 in 100 ml at pH 7.25–7.3, filter-sterilized). Each cell pellet was suspended in a final volume of 100 μl of Buffer 1. Autoclaved, acid-washed 0.5-mm glass beads were added to a final volume of 200 μl, and the mixture was vortexed for 2 min. 900 μl of Buffer 2 (27.1 mg of CPRG in 20 ml of Buffer 1) was added to each sample and mixed thoroughly. The elapsed time that the color changed from rusty yellow to red-brown was recorded. The reaction was stopped by addition of 250 μl of 6 mm ZnCl2. Samples were centrifuged 14,000 × g for 1 min to pellet cell debris, and the A574 of each sample was measured. The β-galactosidase units were calculated as previously done (33), and results were determined by folding other samples over that of pPC86.
Cell Culture, Transfection and Reporter Gene Assay—NIH3T3 or HEK293T cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum. Plasmids were transfected using Lipofectamine/Plus reagent (Invitrogen) according to the manufacturer's instructions. For reporter gene assay, NIH3T3 cells were used and seeded in 24-well plates. Each well was transfected with 500 ng of plasmids in total, including 75 ng of reporter plasmid, 25 ng of LEF-1 plasmid. 50 ng of GFP plasmid was co-transfected as the transfection control. After 24 h of transfection, cells were lysed and experiments were done as previously described (34).
Co-immunoprecipitation and Western Blotting—HEK293T cells were seeded in 35-mm dishes and cultured for 24 h before transfection. After 24 h post-transfection, cells were lysed in 500 μl of lysis buffer (1% Nonidet P-40, 10% glycerol, 138 mm NaCl, 20 mm Tris-HCl, pH 8.0), and 10 mm NaF, 2 mm NaVO4, 1 mm pyrophosphoric acid, and Complete™ protease inhibitors (Roche Applied Science) were added. The lysates were cleared by centrifugation for 15 min at 14,000 rpm. The supernatant were mixed with antibodies and Protein A/G plus-agarose (Santa Cruz Technology) for 3 h at 4 °C. Then immunoprecipitates were washed with lysis buffer for three times, denatured in SDS sample buffer for 10 min at 95 °C, and resolved by SDS-PAGE. For in vivo immunoprecipitation of Zbed3 and Axin, 120 million NIH3T3 cells were scraped from dishes and lysed in 2 ml of lysis buffer with inhibitors above and divided into two aliquots, which were added with rabbit IgG or Axin antibody, respectively. Lysates were mixed with antibodies overnight at 4 °C, and then 30 μl of Protein A/G plus-agarose was added and mixed for another 2 h. The immunoprecipitates were washed with lysis buffer, and the sample was prepared for Western blotting. For immunoblotting, proteins were transferred to nitrocellulose membranes (Schleicher and Schuell BioScience). After being blocked with 5% nonfat milk for 1 h, membrane was incubated with the primary antibodies for 1 h and then with IRDye800-conjugated secondary antibodies (Rockland) for 30 min at room temperature. Results were visualized by the Odessay Infrared Imaging System 9120 (LI-COR).
Immunofluorescence—NIH3T3 cells were transfected with Axin-HA and Zbed3-GFP expression constructs. 24 h after transfection, cells were washed once with phosphate-buffered saline and fixed with 4% paraformaldehyde for 15 min at room temperature. Then cells were washed three times with TBST (1.5 m NaCl, 0.5 m Tris-HCl, pH 7.5, 0.05% Tween 20) and incubated with mouse monoclonal anti-HA antibodies (1:100) for 1 h at room temperature. After washing by TBST for three times, multiple CY3-conjugated anti-mouse antibodies were incubated with for 1 h at room temperature, while avoiding light. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Cells were examined by fluorescence microscopy (Leica).
Free β-Catenin Assay and Cell Fractionation—For free β-catenin assay, 35-mm dishes of NIH3T3 cell were used. For cell fractionation in Fig. S1, about 50 million of NIH3T3 cells were applied. Cells were washed once with cold phosphate-buffered saline and scraped into 1 ml of phosphate-buffered saline. Then cells were collected by spinning at 3000 rpm for 10 min, and resuspended in buffer A (10 mm Hepes-KOH, pH 7.9, 10 mm KCl). After 10 min on ice, cells were homogenized and centrifuged at 3000 rpm at 4 °C for 5 min, and the deposited part was preserved for nuclear extraction. The supernatant was collected and centrifuged at 100,000 × g for 1 h at 4 °C. After centrifugation, the supernatant was collected as cytoplasmic fraction and subjected to Western blot to detect free β-catenin. For nuclear extraction, the pellet was suspended in Buffer C (20 mm Hepes-KOH, pH 7.9, 25% glycerol, 500 mm NaCl). After cooling on ice for 30 min, the nuclear extract was centrifuged at 14,000 rpm for 15 min, and the supernatant was obtained as nuclear fraction. For membrane fractionation, the pellet from ultracentrifugation was resuspended in Buffer A containing 1% Triton X-100, rotated back and forth at 4 °C for 30 min, and the spun at 14,000 rpm for 15 min at 4 °C, and the supernatant was collected as the fraction of membrane.
RNAi—We applied the pAS vector-based shRNA system in knocking down endogenous mouse Zbed3 in NIH3T3. pAS was modified based on pSuper (35) by incorporating a GFP-luciferase fusion protein expression unit. Zbed3 RNAi was designed according to the target sequence 5′-GAGCGTGAAGCACTGAGTA-3′. NIH3T3 cells transfected with pAS-Zbed3 were harvested after 48 h post-transfection for a later assay. Another synthesized siRNA designed according to the sequence 5′-TACTCCGAGGCCTGGGGCTACTTCC-3′ was used. The siRNA was transfected with Lipofectamine 2000 reagent (Invitrogen).
c-Myc and axin2 Expression—Semi-quantified PCR assay of c-myc and axin2 were done to evaluate their mRNA level. Total RNA was extracted with TRIzol reagent (Invitrogen), and reverse transcription-PCR was carried out by using the Superscript II™ RT system (Invitrogen). PCR reactions were carried out with specific primers as follows: mouse c-myc,5′-CAGCAGAGCGAGCTGCA-3′ (forward) and 5′-AGAGTCGCTGCTGGTGGT-3′ (reverse); mouse axin2,5′-CATACAGGAGGATGCT-3′ (forward) and 5′-AGCACTGTCTCGTCGT-3′ (reverse); mouse glyceraldehyde-3-phosphate dehydrogenase: 5′-GCCTGCTTCACCACCTTC-3 (forward) and 5′-CAAGGTCATCCATGACAACT-3′ (reverse). Glyceraldehyde-3-phosphate dehydrogenase was used as an inner control for normalization. PCR results were visualized and quantified by using the UVP image system (UltraViolet Products Ltd.).
RESULTS AND DISCUSSION
Zbed3 Interacts with Axin—To identify novel Axin-interacting proteins that may be involved in Wnt/β-catenin signaling, we performed yeast two-hybrid screening of a mouse embryonic cDNA library using a truncated form of Axin, ΔN1-Axin (Fig. 1A), as bait. One of 81 positive clones was characterized to encode the full-length mouse Zbed3, zinc-finger containing BED 3 (GenBank™ accession number BC085478).
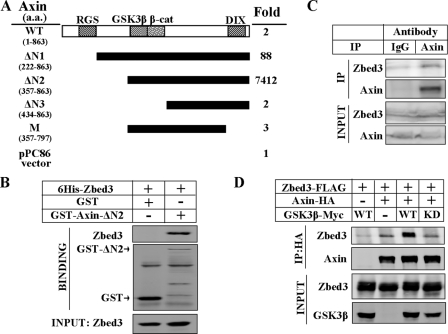
FIGURE 1.
Zbed3 interacts with Axin. A, schematic representation of full-length and truncated Axin used in CPRG assays is showed. Amino acid residues of Axins are presented on the left. The numbers on the right stand for the fold of each CPRG activity over negative control (pPC86). B, in vitro binding of GST-fused ΔN2-Axin and 6His-Zbed3 is shown. Recombinant His6-tagged Zbed3 and GST-fused ΔN2-Axin were expressed in E. coli. GST or GST-ΔN2-Axin was mixed with purified His6-Zbed3, respectively, and immunoprecipitated with GST antibody. The precipitates were subjected to Western blotting with His6 and GST antibodies. C, endogenous Zbed3 could be co-immunoprecipitated by Axin antibody. Endogenous co-immunoprecipitation was performed as described under “Experimental Procedures” and immunoblotted with relevant antibodies. D, exogenous Zbed3 and Axin with GSK3β (WT) or GSK3βK85A (KD) were co-immunoprecipitated in 293T cells. Immunoprecipitation was carried out with HA antibody and detected by Western blotting.
To determine which region of Axin is responsible for its binding to Zbed3, we carried out the CPRG-based interaction assay. The full-length Axin showed a lower apparent affinity for Zbed3 than ΔN1-Axin (Fig. 1A). Another N-terminal truncated Axin mutant (ΔN2-Axin, amino acids 357–863) showed the highest CPRG activity. We further shortened ΔN2-Axin from either its N terminus or C terminus to generate ΔN3-Axin (amino acids 434–863, deleting the GSK3β binding region) or M-Axin (amino acids 357–797, deleting the DIX domain). The ability of both mutants to interact with Zbed3 was markedly reduced, suggesting that ΔN2-Axin contains all of the critical sites required for binding to Zbed3 (Fig. 1A). This result is similar to our previous finding of the interaction between LRP5 and Axin, in which ΔN2-Axin showed the highest apparent affinity for LRP5 (12).
We next investigated whether Zbed3 could directly interact with Axin. In vitro binding experiments were performed using recombinant Axin and Zbed3 proteins expressed in Escherichia coli. As shown in Fig. 1B, GST-ΔN2-Axin, but not GST, could pull down Zbed3, demonstrating that Axin could directly bind to Zbed3.
Although many of zinc-finger domain-containing proteins localize in nucleus as transcriptional factors, Zbed3 is not one of them. Overexpressed Zbed3 mainly resided in cytosol and co-localized with Axin (supplemental Fig. S1A). Moreover, endogenous Zbed3 also appears mostly in the cytoplasmic fraction, and a little in the membrane fraction as Axin does (supplemental Fig. S1B), which indicates endogenous Zbed3 and Axin could form a complex in cytoplasm. Indeed, as shown in Fig. 1C, Zbed3 could be pulled down by Axin-specific antibody, which suggests that the interaction of Zbed3 and Axin happens in vivo.
The interaction between Axin and Zbed3 was further confirmed by immunoprecipitation of these two proteins expressed in 293T cells (Fig. 1D). Interestingly, as in the case of Axin-LRP5 interaction (12), co-expression of GSK3β could enhance the interaction, but GSK3β-K85R, a kinase-dead mutant of GSK3β, could not potentiate the interaction (Fig. 1D). This result suggests that the kinase activity of GSK3β is critically involved in the increased interaction of Zbed3 and Axin. Moreover, GSK3β did not interact with Zbed3 (data not shown), which also excluded the possibility that GSK3β acts as a scaffold between the two proteins. Because GSK3β enhances the interaction between LRP5/6 and Axin through phosphorylation, Zbed3 might also be modified by GSK3β in binding to Axin.
The PPPPSPT Motif Is Crucial to Zbed3 Interaction with Axin—It was recently reported that the PPPP(S/T)PX(T/S) motif in the intracellular domain of LRP5/6 is essential for the interaction with Axin, and that GSK3β and CKI family protein kinases phosphorylate the Ser and Thr in the motif to enhance the interaction (28, 29). In searching for the Axin binding site on Zbed3, we found a “PPPPSPT” motif in its amino acid sequence from 107 to 113 (Fig. 2A).
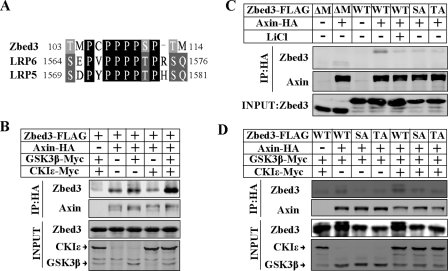
FIGURE 2.
The PPPPSPT motif is crucial for Zbed3 interacting with Axin. A, alignment of homologous motif in mouse Zbed3 and LRP5/6. Amino acid numbers of these motifs are shown. B, GSK3β and CKIε synergistically enhanced Zbed3 and Axin interaction. 24 h after transfection, 293T cells were subjected to co-immunoprecipitation and Western blotting as shown. C, Zbed3 (wild-type, WT), with LiCl (25 mm, 1-h treatment before lysing the cell) or its mutants (ΔM, SA,or TA) exhibited different interacting affinities in binding to Axin and (D) in the presence of co-transfected GSK3β and CKIε. Experiments were done as described in B.
We next tested whether GSK3β and CKI can positively regulate the interaction between Zbed3 and Axin as they did for LRP5/6-Axin interaction (28, 29). GSK3β has been shown to enhance Zbed3 binding to Axin (Fig. 1C), whereas CKIε alone had little effect on this interaction. However, CKIε could markedly potentiated the interaction between Zbed3 and Axin only when GSK3β was present (Fig. 2B), suggesting that GSK3β and CKI play the same role in regulating Axin-Zbed3 interaction as they do in regulating LRP5/6-Axin interaction.
With these similarities, we can expect that the PPPPSPT motif would also be important to the interaction of Zbed3 and Axin. We deleted the motif from Zbed3 and named the mutant as Zbed3-ΔM. As we expected, Zbed3-ΔM has an extremely weak interaction with Axin compared with wild-type Zbed3 (Fig. 2C, lanes 2 and 4), suggesting that PPPPSPT motif is not the only but is the most crucial site for the interaction of Zbed3 and Axin. In addition, phosphorylation of the PPPP-SPXT motif in LRP5/6 creates a stronger binding site for Axin. Mutation of the S or T residue in the motif markedly attenuates the interaction of LRP5/6 with Axin (28, 29). Therefore, we also generated the corresponding S to A and T to A mutations in the PPPPSPT motif of Zbed3. These mutants were named as Zbed3-SA and Zbed3-TA, respectively. Zbed3-SA and -TA also showed much lower affinity as Zbed3-ΔM to bind Axin (Fig. 2C, lanes 6 and 7). Furthermore, wild-type Zbed3 could not efficiently interact with Axin in the existence of LiCl (25 mm), which is a specific inhibitor of GSK3β to reduce its kinase activity (Fig. 2C, lane 5). Thus, we conclude that phosphorylation really happens to potentiate Zbed3 binding with Axin. On the other hand, the interaction of the Zbed3 mutants with Axin was much weaker than that of wild-type Zbed3 even in the presence of GSK3β or GSK3β and CKIε together (Fig. 2D). Taken together, like what happens in LRP5/6, phosphorylation of the Ser and Thr residues in the PPPPSPT motif of Zbed3 are crucial to the interaction with Axin. Zbed3 and LRP5/6 may share the same mechanism in binding Axin, and Zbed3 is the first identified PPPPSPXT-like motif containing protein besides LRP5/6.
Zbed3 Expression Activates Wnt/β-Catenin Signaling by Stabilizing Cytoplasmic β-Catenin—Wnt proteins and its downstream components, such as Dishevelled, can activate LEF-1/T cell factor-dependent transcription by stabilizing cytosolic β-catenin. To determine whether Zbed3 is involved in the Wnt/β-catenin pathway, we examined the effect of Zbed3 overexpression on the activity of a Wnt/β-catenin reporter gene in NIH3T3 cells. Expression of Zbed3 alone activated the LEF-1 binding site-containing reporter gene just as the Wnt-3a-conditioned medium did, whereas neither of them had any effect on the activity of the LEF-1 binding site-mutated reporter gene (Fig. 3A). In addition, neither Zbed3-SA nor Zbed3-TA could activate LEF-1-dependent transcriptional activity compared with wild-type Zbed3 (Fig. 3B), indicating that Zbed3 activates Wnt signaling through binding to Axin.
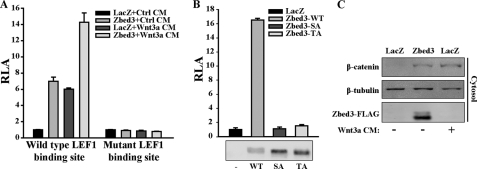
FIGURE 3.
Overexpressing Zbed3 activates Wnt/β-catenin signaling by stabilizing cytoplasmic β-catenin. A, overexpression of Zbed3 activates LEF-1-dependent reporter gene. NIH3T3 cells were transfected with wild-type or mutant LEF-1 binding site reporter gene, respectively. The plasmid of LacZ was transfected as the negative control. One day later, transfected cells were treated with control or Wnt-3a conditioned medium (CM) for 6 h, and then luciferase activity was determined as described under “Experimental Procedures.” B, Zbed3 wild-type (WT) and mutants (SA or TA) exhibited different abilities in activating LEF-1-dependent reporter activities. LEF-1-dependent reporter plasmid was co-transfected with WT, SA, or TA forms of Zbed3 into NIH3T3 cells. 24 h later, cells were lysed for reporter assay. The expression of Zbed3 Wild-typed (WT), SA, and TA mutants are shown in the lower panel. C, Zbed3 could stabilize cytoplasmic β-catenin. Transfected NIH3T3 cells were stimulated with Wnt-3a CM for 3 h, and cytoplasmic fraction was collected and detected by anti-β-catenin antibody, and β-tubulin was used as cytoplasmic marker.
Zbed3 is a cytoplasmic protein and interacts with Axin, so it is predictable that Zbed3 might activate Wnt signals by stabilizing cytoplasmic β-catenin. Indeed, in Zbed3-overexpressing cells, cytosolic β-catenin accumulated as in those treated with Wnt-3a-conditional medium (Fig. 3C). These data demonstrate that gain-of-function of Zbed3 in NIH3T3 cells leads to activation of Wnt signaling by accumulating cytoplasm-free β-catenin.
Knocking Down Endogenous Zbed3 Decreases Wnt/β-Catenin Signaling—To determine if Zbed3 regulates Wnt signaling in vivo, we performed loss-of-function experiments using the method of RNAi. We had applied a vector-based shRNA. An antibody against mouse Zbed3 was generated to evaluate the effectiveness of the RNAi by Western blotting. After 48-h transfection with either pAS-Zbed3 (Zbed3-shRNA) or pAS vector control plasmid (Ctrl-shRNA), cells were harvested. Zbed3 expression was markedly knocked down in pAS-Zbed3-transfected cells (Fig. 4A, lower panel). Then we used the pAS-Zbed3 plasmids to analyze the function of Zbed3 in the Wnt/β-catenin signaling pathway in NIH3T3 cells. Knockdown of Zbed3 significantly decreased the Wnt-3a-stimulated reporter gene activity (Fig. 4A, upper panel) and accumulation of cytoplasmic β-catenin (Fig. 4B). Meanwhile, pAS-Zbed3 had no effect on the transcriptional activity induced by ΔN-β-catenin, an active form of catenin (data not shown).
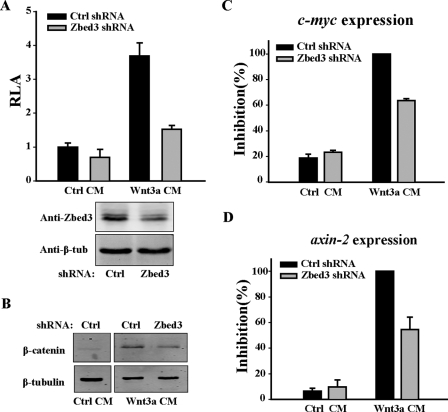
FIGURE 4.
Knocking down of Zbed3 with shRNA decreases Wnt/β-catenin signaling. A, knockdown of Zbed3 attenuated Wnt3a CM-induced transcription of LEF-1-dependent reporter genes. Control or Zbed3 shRNA plasmids were co-transfected with LEF-1-dependent reporter plasmid into NIH3T3 cells. After 48 h, cells were treated with control or Wnt-3a CM for another 6 h, and luciferase activity was determined. RNAi efficiency of Zbed3 is shown in Fig. 4A, lower panel. B, knockdown of Zbed3 decreased the Wnt3a CM triggered accumulation of β-catenin. NIH3T3 were transfected with plasmids of control or Zbed3 shRNA. 48 h later, transfected cells were treated with Wnt-3a CM for 3 h, and free β-catenin was fractionated and examined with Western blot. The control CM-treated sample from the same gel was presented as a negative control in Fig. 4B (left part). Zbed3 functions in Wnt induced expressions of c-myc (C) and axin2 (D) in NIH3T3 cells. Cells transfected with control or Zbed3 shRNA plasmids were treated with control or Wnt3a-CM before RNA extraction. Results in C and D were normalized by glyceraldehyde-3-phosphate dehydrogenase RNA levels. Shown are the average ± S.D. values of two individual experiments.
c-myc and axin2 are two target genes of the canonical Wnt signaling pathway (14–17). To further understand the function of endogenous Zbed3, we investigated whether the expression of c-myc and axin2 is affected by knocking down the endogenous Zbed3. c-myc and axin2 expression was increased by adding Wnt-3a CM to NIH3T3 cells. However, Wnt-3a-induced elevation of c-myc and axin2 transcript levels was suppressed in the pAS-Zbed3-transfected cells (Fig. 4, C and D). These data further support the idea that Zbed3 is involved in Wnt signal transduction in vivo. We also tested a synthesized siRNA oligonucleotide that targets a different sequence as the shRNA, which showed the same results as the shRNA in inhibiting Wnt-induced reporter gene activities (supplemental Fig. S2A) and the accumulation of β-catenin (supplemental Fig. S2B).
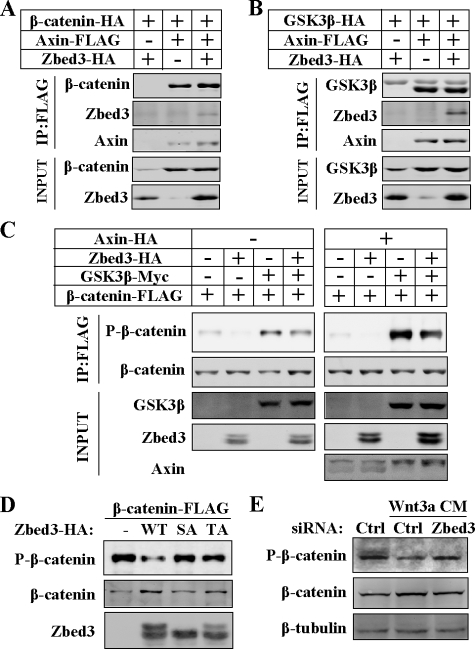
Zbed3 Interacting with Axin to Inhibit GSK3β Phosphorylating β-Catenin—To explore how Zbed3 modulates Axin-centered destruction complex of β-catenin, we first questioned whether Zbed3 could disrupt the interactions of Axin with its binding partners to activate Wnt signals. Although the Zbed3 binding region in Axin covers both GSK3β- and β-catenin-interacting motifs, Zbed3 did not interfere with the interaction of Axin with either β-catenin or GSK3β (Fig. 5, A and B). Next, we wonder whether Zbed3 could block the process of GSK3β-mediated phosphorylation of β-catenin by binding with Axin. Overexpressing Zbed3 indeed decreased the phosphorylation of β-catenin at S33/37Thr41 (Fig. 5C, lanes 2 and 6), which is known to depend on GSK3β (36). Overexpressed GSK3β increased the phosphorylation of β-catenin (Fig. 5C, lane 3) especially in the presence of Axin (Fig. 5C, lane 7), whereas Zbed3 expression inhibited the phosphorylation (Fig. 5C, lanes 4 and 8). The fact that Zbed3-SA or -TA showed little effect on the phosphorylation of β-catenin (Fig. 5D) suggests that the interaction between Zbed3 and Axin is important for Zbed3 to inhibit β-catenin phosphorylation. This result also suggests that Zbed3 is an Axin-dependent GSK3β inhibitor. This conclusion is further corroborated by the finding that knocking down of Zbed3 antagonized the dephosphorylation of β-catenin in response to Wnt-3a CM (Fig. 5E). This also indicates that endogenous Zbed3 is involved in the regulation of β-catenin phosphorylation by GSK3β. All of these results together demonstrate that Zbed3 is a positive regulator involved in Wnt/β-catenin signaling transductions.
FIGURE 5.
Zbed3 could inhibit GSK3β phosphorylating β-catenin through binding Axin. Axin binding with β-catenin (A) or GSK3β (B) was not affected by Zbed3. Co-immunoprecipitation and Western blotting were done with transfected 293T cells as described above. C, Zbed3 inhibited GSK3 phosphorylating β-catenin. 293T cells were transfected with plasmids as indicated. 24 h later, the cells were lysed, and FLAG-tagged β-catenin was immunoprecipitated and checked with FLAG or phosphor-β-catenin antibody, respectively. D, Zbed3-SA or -TA could not affect the phosphorylation of β-catenin by GSK3β. Experiments were manipulated as in C with 293T cells. E, Wnt-3a CM-stimulated dephosphorylation of β-catenin was blocked by Zbed3 siRNA in NIH3T3 cells. 48 h after transfection of the indicated siRNA, Wnt-3a CM was added for additional 3 h with MG132 (25 μm), and total cell lysates were used for Western blotting assay with the indicated antibodies.
In this work, we identified and characterized a novel Axin-interacting protein, Zbed3, which interacts with Axin via a motif that is very similar to those in LRP5/6. In addition, we have demonstrated that Zbed3 is a positive modulator of the Wnt/β-catenin signaling pathway.
Although Zbed3 was identified as an Axin-interacting protein, results from the yeast CPRG assay showed that the N-terminal-truncated Axin mutant has a higher affinity for Zbed3 than the full-length form (Fig. 1A). This is similar to what was shown previously for the Axin-LRP5 interaction (12). In both cases, the N-terminal region of Axin may exhibit a structurally inhibitory effect on the interaction of Axin with LRP5/6 or Zbed3. It was reported that Axin in the β-catenin-destruction complex was highly phosphorylated (37, 38). Wnt induces dephosphorylation of Axin, which has a lower affinity for β-catenin (37). These findings suggest that phosphorylation on Axin may cause some conformational changes that may alter its affinity for its binding partners. Therefore, N terminus-deleted Axin may be free from the N-terminal inhibitory effect and have a more exposed binding site to Zbed3 and LRP5/6 (Fig. 1A) (12).
Previous studies have demonstrated that the Ser and Thr residues in the PPPP(S/T)PX(T/S) motif of LRP5/6 are phosphorylated by GSK3β and CKI, respectively (28, 29). Our results shown in Fig. 2 suggest that the PPPPSPT motif on Zbed3 also plays a crucial role for its binding to Axin. Although the mutants of Zbed3 in this motif still have very weak interaction with Axin, they cannot activate Wnt signals as does wild-type Zbed3 (Fig. 3B), which indicates that the function of Zbed3 requires efficient binding with Axin. The regulation of GSK3β and CK1ε to the interaction of Zbed3 and Axin also suggests the Ser and Thr residues in the Zbed3 PPPPSPT motif may also be phosphorylated by these two kinases, and the phosphorylation is important for the interaction with Axin. It is still not clear how Axin binding to phosphorylated LRP5/6 PPP(S/T)PX(T/S) motifs would lead to suppression of the β-catenin destruction complex. However, recent study suggests the LRP5/6 intracellular domain containing the motif may inhibit the kinase activity of GSK3β (32), in the same way that we found Zbed3 did in this work. In respecting the similarity between Zbed3 and LRP5/6, we believe that Zbed3 regulates β-catenin stabilization most likely via the same mechanism as LRP5/6.
A recent micro-array study has found that the Zbed3 mRNA is extremely enriched in mice oocytes and fertilized eggs, where axin2 is also highly expressed (39). This may suggest that Zbed3 and Axin2 may both have important roles in regulating Wnt activity in these cells. It would be of great interest to characterize the in vivo significance of Zbed3 in model organisms.
Supplementary Material
Acknowledgments
We greatly appreciated the gift of CKIε plasmid from Dr. David Virshup (Huntsman Cancer Institute, University of Utah, Salt Lake City, UT). We are grateful to D. Wu for critical reading and commenting on this report.
This work was supported by Ministry of Science and Technology of China Grants 2007CB914500 and 2007CB947100, by Natural Science Foundation of China Grants 30821065 (to L. L.) and 30600305 (to J. W.), and by Science and Technology Commission of Shanghai Municipality and Chinese Academy of Sciences grants. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: LRP5/6, low density lipoprotein-receptor-related proteins 5 or 6; GSK3, glycogen synthase kinase 3; LEF-1, lymphoid enhancer binding factor-1; Zbed3, zinc-finger BED domain-containing 3; HA, hemagglutinin; CMV, cytomegalovirus; GFP, green fluorescent protein; EGFP, enhanced GFP; CPRG, chlorophenol red β-d-galactopyranoside; RNAi, RNA interference; siRNA, small interference RNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; shRNA, short hairpin RNA; CM, conditioned medium; GST, glutathione S-transferase; CKI, casein kinase I.
References
- 1.Wodarz, A., and Nusse, R. (1998) Annu. Rev. Cell Dev. Biol. 14 59–88 [DOI] [PubMed] [Google Scholar]
- 2.Polakis, P. (2000) Genes Dev. 14 1837–1851 [PubMed] [Google Scholar]
- 3.Peifer, M., and Polakis, P. (2000) Science 287 1606–1609 [DOI] [PubMed] [Google Scholar]
- 4.Clevers, H. (2006) Cell 127 469–480 [DOI] [PubMed] [Google Scholar]
- 5.Kimelman, D., and Xu, W. (2006) Oncogene 25 7482–7491 [DOI] [PubMed] [Google Scholar]
- 6.Cong, F., Schweizer, L., and Varmus, H. (2004) Development 131 5103–5115 [DOI] [PubMed] [Google Scholar]
- 7.Ikeda, S., Kishida, S., Yamamoto, H., Murai, H., Koyama, S., and Kikuchi, A. (1998) EMBO J. 17 1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kishida, S., Yamamoto, H., Ikeda, S., Kishida, M., Sakamoto, I., Koyama, S., and Kikuchi, A. (1998) J. Biol. Chem. 273 10823–10826 [DOI] [PubMed] [Google Scholar]
- 9.Hart, M. J., de los Santos, R., Albert, I. N., Rubinfeld, B., and Polakis, P. (1998) Curr. Biol. 8 573–581 [DOI] [PubMed] [Google Scholar]
- 10.Li, L., Yuan, H., Weaver, C. D., Mao, J., Farr, G. H., 3rd, Sussman, D. J., Jonkers, J., Kimelman, D., and Wu, D. (1999) EMBO J. 18 4233–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, C., Li, Y., Semenov, M., Han, C., Baeg, G. H., Tan, Y., Zhang, Z., Lin, X., and He, X. (2002) Cell. 108 837–847 [DOI] [PubMed] [Google Scholar]
- 12.Mao, J., Wang, J., Liu, B., Pan, W., Farr, G. H., 3rd, Flynn, C., Yuan, H., Takada, S., Kimelman, D., Li, L., and Wu, D. (2001) Mol Cell. 7 801–809 [DOI] [PubMed] [Google Scholar]
- 13.Tolwinski, N. S., Wehrli, M., Rives, A., Erdeniz, N., DiNardo, S., and Wieschaus, E. (2003) Dev. Cell 4 407–418 [DOI] [PubMed] [Google Scholar]
- 14.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B., and Kinzler, K. W. (1998) Science 281 1509–1512 [DOI] [PubMed] [Google Scholar]
- 15.Yan, D., Wiesmann, M., Rohan, M., Chan, V., Jefferson, A. B., Guo, L., Sakamoto, D., Caothien, R. H., Fuller, J. H., Reinhard, C., Garcia, P. D., Randazzo, F. M., Escobedo, J., Fantl, W. J., and Williams, L. T. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14973–14978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lustig, B., Jerchow, B., Sachs, M., Weiler, S., Pietsch, T., Karsten, U., Van de Wetering, M., Clevers, H., Schlag, P. M., Birchmeier, W., and Behrens, J. (2002) Mol. Cell. Biol. 22 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jho, E. H., Zhang, T., Domon, C., Joo, C. K., Freund, J. N., and Costantini, F. (2002) Mol. Cell. Biol. 22 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng, L., Fagotto, F., Zhang, T., Hsu, W., Vasicek, T. J., Perry, W. L., 3rd, Lee, J. J., Tilghman, S. M., Gumbiner, B. M., and Costantini, F. (1997) Cell 90 181–192 [DOI] [PubMed] [Google Scholar]
- 19.Behrens, J., Jerchow, B. A., Wurtele, M., Grimm, J., Asbrand, C., Wirtz, R., Kuhl, M., Wedlich, D., and Birchmeier, W. (1998) Science 280 596–599 [DOI] [PubMed] [Google Scholar]
- 20.Chia, I. V., and Costantini, F. (2005) Mol. Cell. Biol. 25 4371–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahmen, R. P., Koch, A., Denkhaus, D., Tonn, J. C., Sorensen, N., Berthold, F., Behrens, J., Birchmeier, W., Wiestler, O. D., and Pietsch, T. (2001). Cancer Res. 61 7039–7043 [PubMed] [Google Scholar]
- 22.Salahshor, S., and Woodgett, J. R. (2005) J. Clin. Pathol. 58 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, E., Salic, A., Kruger, R., Heinrich, R., and Kirschner, M. W. (2003) PLoS Biol. 1 E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusano, S., and Raab-Traub, N. (2002) Mol. Cell. Biol. 22 6393–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiomi, K., Uchida, H., Keino-Masu, K., and Masu, M. (2003) Curr. Biol. 13 73–77 [DOI] [PubMed] [Google Scholar]
- 26.Chen, H. J., Lin, C. M., Lin, C. S., Perez-Olle, R., Leung, C. L., and Liem, R. K. (2006) Genes Dev. 20 1933–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamai, K., Zeng, X., Liu, C., Zhang, X., Harada, Y., Chang, Z., and He, X. (2004) Mol. Cell. 13 149–156 [DOI] [PubMed] [Google Scholar]
- 28.Davidson, G., Wu, W., Shen, J., Bilic, J., Fenger, U., Stannek, P., Glinka, A., and Niehrs, C. (2005) Nature 438 867–872 [DOI] [PubMed] [Google Scholar]
- 29.Zeng, X., Tamai, K., Doble, B., Li, S., Huang, H., Habas, R., Okamura, H., Woodgett, J., and He, X. (2005) Nature 438 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilic, J., Huang, Y. L., Davidson, G., Zimmermann, T., Cruciat, C. M., Bienz, M., and Niehrs, C. (2007) Science 316 1619–1622 [DOI] [PubMed] [Google Scholar]
- 31.Baig-Lewis, S., Peterson-Nedry, W., and Wehrli, M. (2007) Dev. Biol. 306 94–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cselenyi, C. S., Jernigan, K. K., Tahinci, E., Thorne, C. A., Lee, L. A., and Lee, E. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 8032–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Platt, T., Muller-Hill, B., and Miller, J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, p. 352
- 34.Pan, W. J., Pang, S. Z., Huang, T., Guo, H. Y., Wu, D., and Li, L. (2004) Cell Res. 14 324–330 [DOI] [PubMed] [Google Scholar]
- 35.Brummelkamp, T. R., Bernards, R., and Agami, R. (2002) Science 296 550–553 [DOI] [PubMed] [Google Scholar]
- 36.Yost, C., Torres, M., Miller, J. R., Huang, E., Kimelman, D., and Moon, R. T. (1996) Genes Dev. 10 1443–1454 [DOI] [PubMed] [Google Scholar]
- 37.Willert, K., Shibamoto, S., and Nusse, R. (1999) Genes Dev. 13 1768–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto, H., Kishida, S., Kishida, M., Ikeda, S., Takada, S., and Kikuchi, A. (1999) J. Biol. Chem. 274 10681–10684 [DOI] [PubMed] [Google Scholar]
- 39.Su, A. I., Cooke, M. P., Ching, K. A., Hakak, Y., Walker, J. R., Wiltshire, T., Orth, A. P., Vega, R. G., Sapinoso, L. M., Moqrich, A., Patapoutian, A., Hampton, G. M., Schultz, P. G., and Hogenesch, J. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.