Abstract
Regulation of hepatic cholesterol biosynthesis, lipogenesis, and insulin signaling intersect at the transcriptional level by control of SREBP and Insig genes. We previously demonstrated that peroxisome-deficient PEX2-/- mice activate SREBP-2 pathways but are unable to maintain normal cholesterol homeostasis. In this study, we demonstrate that oral bile acid treatment normalized hepatic and plasma cholesterol levels and hepatic cholesterol synthesis in early postnatal PEX2 mutants, but SREBP-2 and its target gene expressions remained increased. SREBP-2 pathway induction was also observed in neonatal and longer surviving PEX2 mutants, where hepatic cholesterol levels were normal. Abnormal expression patterns for SREBP-1c and Insig-2a, and novel regulation of Insig-2b, further demonstrate that peroxisome deficiency widely affects the regulation of related metabolic pathways. We have provided the first demonstration that peroxisome deficiency activates hepatic endoplasmic reticulum (ER) stress pathways, especially the integrated stress response mediated by PERK and ATF4 signaling. Our studies suggest a mechanism whereby ER stress leads to dysregulation of the endogenous sterol response mechanism and concordantly activates oxidative stress pathways. Several metabolic derangements in peroxisome-deficient PEX2-/- liver are likely to trigger ER stress, including perturbed flux of mevalonate metabolites, altered bile acid homeostasis, changes in fatty acid levels and composition, and oxidative stress.
Cholesterol is an essential lipid in vertebrate cell membranes and an obligatory precursor for synthesis of steroid hormones, bile acids, and regulatory oxysterols (1, 2). Mammals adjust their sterol content by both transcriptional and posttranscriptional feedback systems that regulate cholesterol metabolism. Cholesterol homeostasis is achieved through the coordinated regulation of dietary cholesterol absorption, de novo biosynthesis, and disposal in the form of bile acids (3, 4, 5). The sterol regulatory element-binding protein (SREBP)3 family of transcription factors is central to this feedback system. Mammalian cells produce three SREBP isoforms, called SREBP-1a, SREBP-1c, and SREBP-2 (3). SREBP-1a and -1c are produced from the same gene by use of different promoters and alternative splicing. SREBP-1c activates predominantly genes involved in fatty acid and triglyceride biosynthesis, whereas SREBP-2 activates primarily genes required for cholesterol synthesis. SREBPs are present as transcriptionally inactive precursor proteins in the endoplasmic reticulum membrane by their interaction with SREBP cleavage-activating protein (SCAP), which functions as a sterol sensor. In sterol-depleted cells, SCAP escorts SREBPs from the ER to the Golgi, where the SREBPs are processed proteolytically to generate their nuclear forms that activate SREBP target genes (6).
Insigs (Insig-1 and -2; insulin-induced genes) are polytopic ER membrane proteins that reduce cholesterol synthesis by blocking the translocation of the SCAP·SREBP complex and by enhancing degradation of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase in response to sterols (3). Insig-1 is an obligatory SREBP target gene, whereas Insig-2 is constitutively expressed at a low level, at least in cultured cells (7). By use of different promoters, the Insig-2 gene produces two transcripts in the liver, Insig-2a, which is liver-specific and selectively down-regulated by insulin, and Insig-2b, which is ubiquitously expressed in various tissues (8).
Peroxisomes are ubiquitous organelles of eukaryotic cells that are predominantly involved in lipid metabolism (9, 10). The pre-squalene segment of the cholesterol biosynthetic pathway is localized to peroxisomes, and acetyl-CoA derived from peroxisomal β-oxidation of very long-chain fatty acids and medium-chain dicarboxylic acids is channeled preferentially to cholesterol synthesis inside the peroxisomes (11).
We recently examined cholesterol homeostasis in mice lacking functional peroxisomes (PEX2-/- mice), and demonstrated that: 1) PEX2-/- mice had significantly decreased total cholesterol and high density lipoprotein (HDL) cholesterol levels in plasma; 2) the steady-state cholesterol content in PEX2-/- liver was decreased by 40% relative to control mouse liver; 3) PEX2-/- mice did not lose cholesterol by catabolism to bile acids or excretion in the stool; and 4) PEX2-/- mice failed to maintain normal cholesterol balance, despite a dramatic increase in SREBP-2, its target genes, and cholesterol biosynthesis in most tissues (12).
Bile acids (BAs) are synthesized from cholesterol in the liver and are important for the digestion and absorption of lipid nutrients (5). To evaluate whether the lack of mature bile acids plays a role in the altered cholesterol homeostasis in peroxisome-deficient mice, control and PEX2-/- mice were fed a mixture of cholic and ursodeoxycholic acid daily by means of an orogastric tube feeding (13). The effects on cholesterol levels and its biosynthesis and transcriptional regulatory controls were evaluated. Our studies demonstrate that BA feeding normalized cholesterol levels in liver and plasma of early postnatal PEX2-/- mice, but the activities, expression and protein levels of SREBP-2 pathway genes were still significantly up-regulated, indicating a persistently abnormal cholesterol homeostasis. In untreated and BA-fed PEX2-/- mice at postnatal day 36 (P36), liver cholesterol levels were normal, but SREBP-2 target genes remained induced. Therefore, cholesterol biosynthesis pathway abnormalities persist in peroxisome-deficient mice even when cholesterol balance is maintained. We propose a mechanism whereby activation of the hepatic ER unfolded protein response (UPR) in peroxisome deficiency leads to dysregulation of the endogenous sterol response mechanism.
EXPERIMENTAL PROCEDURES
Animals—Homozygous PEX2-/- mice were obtained by breeding PEX2 heterozygotes (Swiss Webster (SW)×129SvEv mouse genetic background) (14-16). Mice had access to food and water ad libitum and were exposed to a 12-h light-dark cycle. For the purposes of this study, control mice consisted of either PEX2+/+ or PEX2+/- genotypes, as biochemical measures did not differ (12). Starting on P1, mice were fed a solution containing cholic acid and ursodeoxycholic acid (Sigma), each at 3 mg/ml, in sterile 1.5% sodium carbonate. A 15 mg/kg daily dose of each bile acid was administered by orogastric gavage as described (13). All protocols for animal use and experiments were reviewed and approved by the Institutional Animal Care and Use Committee of San Diego State University and Columbia University.
Plasma and Hepatic Lipid Analysis—Tissue sterols and plasma total cholesterol, HDL and LDL cholesterol, phospholipids, and triglycerides were measured as described previously (12).
Enzyme Assays—HMG-CoA reductase (HMGCR) (EC 1.1.1.34), farnesyl pyrophosphate synthase (EC 2.5.1.10), isopentenylpyrophosphate isomerase (IDI1) (EC 5.3.3.2), and squalene synthase (EC 2.5.1.21) activities were assayed as described previously (12). Protein concentration was determined by the BCA method (Pierce).
Western Blot Analysis—Proteins were separated on SDS-polyacrylamide gels. Immunoblot analysis was performed by enhanced chemiluminescence (PerkinElmer Life Sciences) with the following antibodies: anti-HMGCR, anti-mevalonate kinase, anti-IDI1, anti-squalene synthase (BD Biosciences), and anti-SREBP-1 (Santa Cruz Biotechnology, Santa Cruz, CA), with the appropriate horseradish peroxidase-linked secondary antibody (Bio-Rad). Blots were exposed to Kodak X-Omat LS film (Rochester, NY), scanned on a densitometer (GE Healthcare) and analyzed with ImageQuant® software (Amersham Biosciences).
Quantitative Real-time RT-PCR—Total RNA was prepared from frozen mouse tissues with TRIzol (Invitrogen) and treated with DNase I (DNA-free; Ambion, Austin, TX). First-strand cDNA was synthesized with random hexamer primers using Ready-To-Go You-Prime First-Strand Beads (Amersham Biosciences). The real-time RT-PCR reaction was set up in a final volume of 20 μl using 2× LightCycler 480 SYBR Green I Mastermix (Roche Diagnostics). PCR reactions were performed in triplicate using a Roche LightCycler 480. The relative mRNA amount was calculated using the comparative threshold cycle (CT) method. 18S rRNA was used as the invariant control. Primer sequences are available on request. Northern blot analysis was performed as described previously (12). RT-PCR analysis of XBP-1 splicing was performed as described previously (17, 18).
Immunohistochemistry—Mice were cardiac-perfused with 4% paraformaldehyde-phosphate-buffered saline. The liver was post-fixed overnight in paraformaldehyde-phosphate-buffered saline and either processed for paraffin embedding or cryoprotected in 30% sucrose for frozen section preparation. 2-4 μm of paraffin or 10 μm of frozen liver sections were mounted on Superfrost Plus slides. For antigen retrieval of paraffin-embedded tissue, deparaffinized and rehydrated sections were digested with 0.01% trypsin for 10 min at 37 °C followed by microwaving in 10 mm citrate buffer, pH 6.0, three times for 5 min at 800 watts. Nonspecific binding was blocked with 10% normal donkey serum, 1% bovine serum albumin, and 0.5% Triton X-100 in phosphate-buffered saline for 2 h, and sections were then incubated overnight at 4 °C with goat anti-Grp78 (SC-1051; Santa Cruz Biotechnology). Cy3-conjugated donkey anti-goat IgG was applied for 2 h (Jackson ImmunoResearch Laboratories). Negative control sections were incubated in parallel by omitting the primary antibody. Images were taken with a Zeiss LSM510 confocal laser-scanning microscope.
Statistical Analyses—Data are expressed as means ± S.D. Statistical significance was evaluated either by an unpaired Student's t test or by ANOVA followed by Fisher's least-squares difference post hoc test to compare four sample group means at each time.
RESULTS
Plasma Lipid Analysis of Bile Acid-fed Control and PEX2-/- Mice—We first investigated whether BA feeding changed the plasma lipid alterations seen in P10 untreated PEX2-/- mice, where total plasma cholesterol and HDL cholesterol were 0.57- and 0.27-fold the corresponding values of untreated controls, respectively (12). BA feeding normalized total plasma cholesterol level in PEX2-/- mice but did not alter the level in controls (Table 1). Plasma HDL cholesterol levels decreased with BA feeding in both control (65%) and PEX2-/- (55%) mice, resulting in a similar reduction of HDL cholesterol in mutants versus controls for both untreated (0.39-fold) and bile acid-fed (0.34-fold) PEX2-/- mice. BA feeding increased plasma LDL cholesterol concentrations 1.25- and 2.3-fold in control and PEX2-/- mice, respectively, as compared with untreated mice. Plasma triglyceride levels were similar in controls versus PEX2 mutants in either treatment group, although BA feeding increased triglyceride levels around 2.5-fold versus that in untreated mice for both genotypes (Table 1). Plasma phospholipids levels were reduced by 34% in untreated mutants and did not significantly change with BA feeding, whereas BAs reduced plasma phospholipids by 25% in control mice (Table 1).
TABLE 1.
Plasma analysis of untreated and BA-fed control and PEX2 knock-out mice
Each value represents the mean ± S.D. Numbers in parentheses denote the number of samples analyzed. Plasma LDL cholesterol was calculated according to the Friedewald formula applying the values of total cholesterol, HDL cholesterol, and triglycerides. One-way ANOVA was used to determine statistical significance of changes followed by Fisher's least-squares difference post hoc test to evaluate multiple group comparisons. Significance at the 0.01 level: *, significant difference vs. untreated control mice; #, significant difference vs. bile acid-fed control mice; †, significant difference vs. untreated PEX2−/− mice. Significance at the 0.05 level: ‡, significant difference vs. untreated PEX2−/− mice.
|
Concentration
|
|||||
|---|---|---|---|---|---|
| Age/Parameter |
Untreated mice
|
Bile acid-fed mice
|
|||
| Control | PEX2−/− | Control | PEX2−/− | ||
| mg/dl | |||||
| P9-P10 | |||||
| Total plasma cholesterol | 172.8 ± 31.9 (28) | 98.3 ± 30.0 (9)*# | 161.8 ± 34.3 (15)† | 168.7 ± 58.5 (12)† | |
| Plasma HDL cholesterol | 80.5 ± 28.8 (22) | 31.6 ± 18.8 (11)* | 33.8 ± 14.3 (14)* | 11.6 ± 6.7 (12)*#‡ | |
| Plasma triglycerides | 88.8 ± 45.1 (40) | 83.5 ± 26.4 (13)# | 179.5 ± 81.4 (14)*† | 201.7 ± 61.7 (9)*† | |
| Plasma phospholipids | 412.1 ± 52.1 (31) | 273.9 ± 93.7 (11)* | 311.6 ± 30.8 (11)* | 321.9 ± 95.5 (8)* | |
| Plasma LDL cholesterol | 74.5 | 50 | 92.1 | 116.8 | |
| P36 | |||||
| Total plasma cholesterol | 121.4 ± 22.4 (10) | 222.0 ± 20.4 (2)* | 147.0 ± 23.1 (8) | 178.1 ± 47.1 (4) | |
BA feeding enabled a subset of PEX2 mutants (∼10%) to survive past the early postnatal period (13). Surprisingly, the total plasma cholesterol level from two rare P36 untreated PEX2-/- mice was elevated 1.8-fold compared with that in controls (Table 1; p < 0.001), which contrasts markedly with the decreased level in early postnatal untreated mutants. BA feeding mildly increased total plasma cholesterol level in P36 control mice (p = 0.065 versus untreated controls) and again normalized it in P36 PEX2-/- mice.
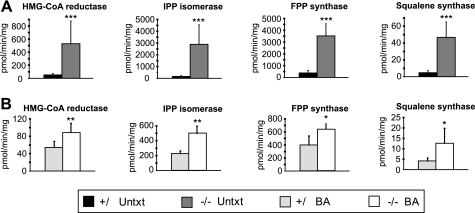
Hepatic Cholesterol Biosynthetic Enzyme Activities and Protein Levels Remain Increased after BA Feeding—We next examined the effect of BA feeding on the activities of cholesterol biosynthetic enzymes, including HMGCR, IDI1, farnesyl pyrophosphate synthase, and squalene synthase, in the liver and kidney of P9 control and PEX2-/- mice. Although the activities of these enzymes were increased ∼10-18-fold in the liver of untreated PEX2-/- mice compared with control mice (12), BA feeding significantly attenuated, but did not completely normalize, these enzyme activities in PEX2-/- mice (Fig. 1). In the PEX2 mutants the fold increase in activity of HMGCR and farnesyl pyrophosphate synthase changed from 10- to 1.6-fold, of IDI1 from 18- to 2.2-fold, and of squalene synthase from 10- to 3-fold. BA feeding did not alter the cholesterol biosynthetic enzyme activities in control mice.
FIGURE 1.
A-B, activities of selected cholesterol biosynthetic enzymes in liver homogenates of P7-P10 untreated (A) and P9 BA-fed (B) control and PEX2-/- mice. Enzyme activities from PEX2 wild-type and heterozygous mice were similar and were combined (control mice). Values are mean ± S.D. (Untreated mice: n = 10 for control mice, n = 7 for PEX2-/- mice; BA-fed mice: n = 5 for control and PEX2-/- mice; n = 7 for HMGCR activity measurements). *, p < 0.05; **, p < 0.005; ***, p < 0.001 (Student's t test). The data presented in A are reproduced from Kovacs et al. (12) with permission from the American Society for Microbiology. IPP, isopentenylpyrophosphate; FPP, farnesyl pyrophosphate; Untxt, untreated; BA, bile acid-fed.
We previously demonstrated a different regulatory pattern of cholesterol enzyme activities in PEX2-/- kidneys, with a 2-fold increase for farnesyl pyrophosphate synthase and squalene synthase and a 42% decrease for HMGCR (12). BA feeding did not affect the cholesterol biosynthetic enzyme activities in control and PEX2-/- kidneys (supplemental Table S1), leading to a similarly abnormal enzyme profile in BA-fed PEX2-/- mice.
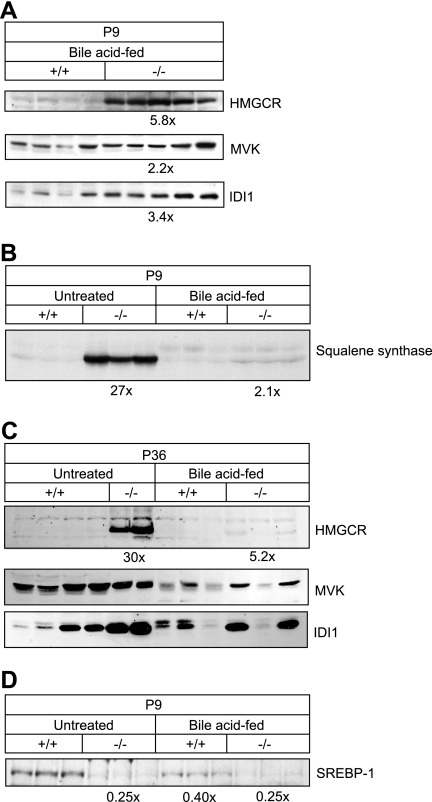
Western blot analysis of cholesterol biosynthetic enzymes was performed to determine whether the measured activities were a reflection of the protein levels. HMGCR protein was still elevated 5.8-fold in livers of P9 BA-fed PEX2-/- mice (Fig. 2A; p < 0.001), which is greater than the 1.6-fold increase in HMGCR activity (Fig. 1B). Mevalonate kinase, IDI1, and squalene synthase protein levels also remained increased in BA-fed PEX2-/- mice (Fig. 2, A and B; p < 0.005).
FIGURE 2.
A-C, immunoblot of cholesterol biosynthetic enzymes in extract of livers of individual control and PEX2-/- mice at the indicated ages and treatment using 200 μg of total liver protein (A and C) or 50 μg liver microsomal fraction (B). D, immunoblot of SREBP-1c precursor in microsomal fraction of P9 livers. The -fold change in the protein level in PEX2-/- mice was expressed relative to that in control mice, which in each case was arbitrarily set at 1.0. MVK, mevalonate kinase.
HMGCR protein level was dramatically increased in P36 untreated PEX2-/- liver (30-fold, p < 0.001) (Fig. 2C). As in early postnatal mutants, HMCGR protein level was still increased about 5-fold in P36 BA-fed PEX2-/- mice. A significant increase in mevalonate kinase and IDI1 proteins was also observed in the livers of P36 untreated and BA-fed PEX2-/- mice (Fig. 2C). These data show a persistent elevation of cholesterol synthesis enzymes in P36 untreated PEX2-/- mice and attenuation with BA feeding similar to that seen in early postnatal mutants.
Altered Catalytic Efficiency of Cholesterol Biosynthetic Enzymes in PEX2-/- Mice—Because enzyme activity measurements and immunoblots were done on the same liver samples, we normalized the specific activities to the enzyme protein content to obtain an estimate of “catalytic efficiency.” We demonstrated previously that the catalytic efficiency of HMGCR was decreased by 55% in P10 PEX2-/- mice (12). In P9 BA-fed PEX2-/- mice, the catalytic efficiency of HMGCR was reduced by about 70% (p < 0.001) (Table 2) and thus was more severely affected in BA-fed PEX2-/- mice compared with untreated mutants (p < 0.05). The catalytic efficiency of IDI1 in P9 BA-fed PEX2-/- mice was decreased by ∼60%, similar to the 70% decrease seen in untreated PEX2 mutants (data not shown). These data illustrate that peroxisome deficiency consistently alters the catalytic efficiency of hepatic cholesterol biosynthetic enzymes in PEX2-/- mice, a decrement that may be exacerbated by BA feeding.
TABLE 2.
Specific activity of HMGCR normalized to HMGCR protein content in the livers of P9 BA-fed control and PEX2−/− mice
Measurements of HMGCR activities and immunoblots were done on the same liver samples. For Western blot analysis, 200 μg of protein was separated on a SDS-7.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis using a polyclonal anti-HMGCR antibody. *, p < 0.001 (Student's t test). Sp. act., specific activity.
| Genotype | Sp. act. | Vol. density | Sp. act./vol. density | Normalized average sp. act./vol. density |
|---|---|---|---|---|
| pmol/min/mg | per mg of protein | fmol/min | ||
| +/+ | 47.8 | 217,437 | 220 | |
| +/+ | 76.2 | 31,6048 | 241 | |
| +/+ | 52.5 | 15,9508 | 329 | |
| +/+ | 70.3 | 14,5511 | 483 | 1.0 ± 0.4 |
| +/+ | 98.7 | 26,3780 | 374 | |
| +/+ | 41.0 | 22,7160 | 180 | |
| +/+ | 47.7 | 29,6570 | 161 | |
| +/+ | 43.6 | 26,8290 | 163 | |
| −/− | 105.5 | 88,1295 | 120 | |
| −/− | 99.7 | 91,6150 | 109 | |
| −/− | 118.4 | 161,7222 | 73 | |
| −/− | 80.2 | 155,4749 | 52 | 0.3 ± 0.1* |
| −/− | 86.1 | 112,2032 | 77 | |
| −/− | 116.0 | 137,4880 | 84 | |
| −/− | 70.5 | 103,4210 | 68 | |
| −/− | 59.0 | 126,0890 | 47 |
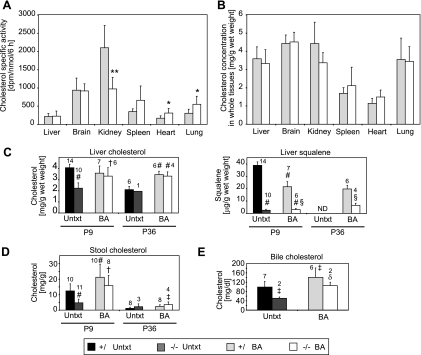
In Vivo Synthesis Rates of Sterols and Tissue Cholesterol Levels in BA-fed PEX2 Mice—To test the effect of BA feeding on cholesterol synthesis in vivo, we measured the rate of incorporation of subcutaneously injected [3H]acetate into nonsaponifiable lipids in various tissues of P9 control and PEX2-/- mice. Although the specific activity of cholesterol in livers of P7-P9 untreated PEX2-/- mice was 13-fold higher than in control mice (12), BA feeding reduced hepatic cholesterol synthesis in PEX2-/- mice to control levels (Fig. 3A). Cholesterol specific activity in the spleen, heart, and lungs of BA-fed PEX2-/- mice was attenuated compared with that in untreated PEX2-/- mice (12) but was still significantly increased 1.8-fold (p < 0.05; p = 0.06 for spleen) compared with BA-fed control mice. In kidney, BA feeding did not affect the cholesterol synthesis rate in controls, and there was a similar 2-fold decreased rate in both the untreated and BA-fed PEX2-/- mice. Surprisingly, whereas cholesterol specific activity in the brain was 2.4-fold lower in untreated PEX2-/- versus control mice (12), specific activity was normalized in PEX2-/- mice by BA feeding. These findings further emphasize that peroxisome deficiency and BA treatment differentially affect cholesterol synthesis in different organs.
FIGURE 3.
Rate of cholesterol synthesis and concentration of cholesterol in various tissues of control and PEX2-/- mice. A and B, [3H]acetate was administered by subcutaneous injection to BA-fed mice, and the animals were killed 6 h later. The amount of [3H]acetate incorporated into cholesterol (A) and cholesterol content (B) in the indicated tissues was determined (n = 7 for control mice; n = 6 for PEX2-/- mice). *, p < 0.05; **, p < 0.005 versus BA-fed control mice (Student's t test). C, levels of liver cholesterol and squalene in P9 and P36 untreated (Untxt) and BA-fed mice (BA). D, cholesterol levels in stool of P9 and P36 untreated and BA-fed mice. E, cholesterol levels in bile of P36 untreated and BA-fed mice. Significance at the 0.01 level was analyzed by ANOVA with Fisher's post test: #, significantly different from untreated control mice; †, significantly different from untreated PEX2-/- mice; §, significantly different from BA-fed control mice. Significance at the 0.05 level was analyzed by ANOVA with Fisher's post test: ‡, significantly different from untreated control mice; δ, significantly different from untreated PEX2-/- mice. Values represent the mean ± S.D. The number of animals/group indicated above the bars.
The total cholesterol level was similar in the brain, kidney, spleen, heart, and lung of untreated (12) and BA-fed PEX2-/- mice (Fig. 3B) when compared with control mice. In addition to normalizing total plasma cholesterol in early postnatal PEX2-/- mice (Table 1), BA feeding also corrected the 40% decrease in hepatic total cholesterol (Fig. 3, B and C) (12). Although fat malabsorption, present in untreated PEX2 mutants (13), may adversely affect intestinal cholesterol absorption (19, 20), fecal cholesterol content was reduced by 62% in P10 untreated PEX2-/- mice (Fig. 3D) (12). Fecal cholesterol content increased 1.7-fold in P9 BA-fed control mice (Fig. 3D), which may reflect feeding with unconjugated bile acids that are less efficient for cholesterol absorption. BA feeding of PEX2 mutants increased fecal cholesterol by 3.3-fold, leading to a level similar to that seen in BA-fed controls. In sum, these findings demonstrate that BA replenishment dramatically improved cholesterol homeostasis in early postnatal PEX2-/- mice.
Unexpectedly, the hepatic cholesterol level was similar in P36 untreated PEX2-/- and control mice, which may have been enabled by these older mutants obtaining small amounts of BAs through coprophagy. Despite the normal hepatic cholesterol content in the P36 untreated PEX2 mutant, bile cholesterol level was decreased 0.5-fold (Fig. 3E). Thus, the increased total plasma cholesterol level in P36 untreated mutants (Table 1) reflects this defect in hepatic cholesterol excretion into bile. In all of the P36 BA-fed mice, the hepatic and biliary cholesterol level was significantly increased compared with that in untreated mice (Fig. 3, C and E) and was similar between controls and PEX2 mutants. Interestingly, although P36 BA-fed PEX2 mutants can increase bile cholesterol content to control levels, biliary BA concentration is still markedly deficient (13). Stool cholesterol levels were much lower in P36 than in P9 mice (Fig. 3D), likely reflecting the decrement in dietary fat content between suckling pups and weaned animals. Fecal cholesterol content was similar in P36 untreated control and mutant mice and was mildly increased in BA-fed mice, paralleling the increases in their hepatic and bile cholesterol content.
We reported previously a markedly reduced amount of the cholesterol precursor squalene in livers of early postnatal untreated PEX2-/- mice compared with control mice (12). Hepatic squalene levels were still markedly decreased both in P9 and P36 BA-fed PEX2-/- mice (Fig. 3C), despite normalization of the cholesterol content in livers of BA-fed PEX2-/- mice.
Bile Acids Attenuate Expression of Cholesterogenic Genes—We reported previously that hepatic expression of SREBP-2-regulated cholesterol biosynthetic enzymes was markedly increased in P10 PEX2-/- compared with control mice (12). Furthermore, gene expression profiling studies using the Codelink Mouse 20K microarray (GE Healthcare) showed that all direct SREBP target genes were significantly increased in the liver of P9 untreated PEX2-/- mice (supplemental Table S2).
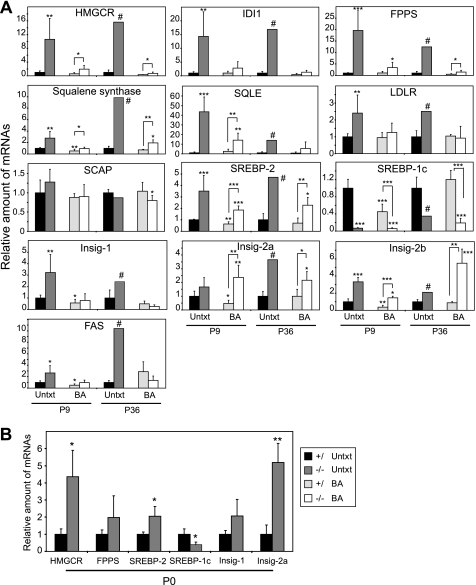
To further characterize the cholesterol-normalizing effect of BA feeding in PEX2-/- mice, we used quantitative real-time RT-PCR to study the expression of genes involved in hepatic lipid homeostasis (Fig. 4A, supplemental Fig. S1). In both P9 and P36 untreated PEX2 mutants, the mRNA levels of HMGCR, IDI1, farnesyl pyrophosphate synthase (FPPS), squalene synthase (SS), and squalene epoxidase (SQLE) were increased 3-44-fold (Fig. 4A). BA feeding dramatically reduced the expression of these cholesterol synthesis genes in PEX2-/- livers, but mRNA levels were still significantly increased 2-6-fold compared with BA-fed control mice at both ages (Fig. 4A, supplemental Fig. S1). The mRNA for the LDL receptor (LDLR) was increased 2.5-fold in the livers of P9 and P36 untreated PEX2-/- mice, and LDLR expression levels were similar in BA-fed control and PEX2-/- mice (Fig. 4A). SCAP expression was not affected in PEX2-/- mice.
FIGURE 4.
Quantitative RT-PCR analysis of various mRNAs in livers from untreated and BA-fed control and PEX2-/- mice. The values obtained were normalized to 18S rRNA values. Each value represents the amount of mRNA relative to that in untreated controls at that age, which was arbitrarily defined as 1. A, P9 and P36 liver. Values are mean ± S.D. from RNA samples of at least five individual mice, except for P36 untreated PEX2-/-, where n = 1. B, P0 liver. Values are mean ± S.D. from RNA samples of four individual mice. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus untreated control mice or versus BA-fed control mice (indicated by brackets; Student's t test); #, outside 95% confidence interval of P36 untreated control mice. FPPS, farnesyl pyrophosphate synthase; SQLE, squalene epoxidase; LDLR, LDL receptor; FAS, fatty acid synthase.
SREBP-2 expression changes at both P9 and P36 paralleled those for cholesterol biosynthesis genes, with the increased levels seen in untreated PEX2-/- mice being attenuated but not completely normalized by BA feeding (Fig. 4A). In contrast, the expression of SREBP-1c was reduced dramatically in both untreated (15-fold at P9; 3-fold at P36) and BA-fed (6.3-8-fold) PEX2-/- mice. Analysis of the SREBP-1c precursor protein level in P9 livers (Fig. 2D) further confirmed the gene expression profiles. The SREBP-1c mRNA level was also decreased 2-fold by BA feeding in P9 control mice, consistent with BA-mediated induction of the small heterodimer partner that inhibits the stimulatory effect of liver X-activated receptor α (LXRa) on SREBP-1c expression (21). However, this mechanism is not operative in PEX2 mutants, as the small heterodimer partner is not activated even in BA-fed mutants.4 BA feeding did not affect SREBP-1c mRNA expression in P36 control mice, most likely because of the relatively small amount of BA administered (13). The expression of LXRa and its target genes, ABCA1 and ABCG1, which are important regulators of cholesterol efflux, was unaffected in P10 PEX2-/- compared with control mice (12). LXRa expression was normal in P36 PEX2-/- mice and was unaffected by BA feeding in both control and PEX2 mutant mice at both ages (data not shown). These findings suggest that the LXR pathway is not altered in PEX2-/- mice and thus cannot explain the SREBP-1c gene expression changes in mutant mice.
Surprisingly, the expression of the SREBP-1c target gene fatty acid synthase (FAS) was increased 2.6- and 10-fold in the livers of P9 and P36 untreated PEX2-/- mice, respectively, compared with age-matched controls (Fig. 4A). This FAS expression pattern was also confirmed by Northern hybridization (data not shown). The increased FAS expression may be explained by the up-regulation of SREBP-2, which can partially substitute for SREBP-1c when overexpressed in liver (22). FAS expression levels were similar in BA-fed control and PEX2-/- mice.
Insig-1 and Insig-2 are essential elements in the feedback control of lipid synthesis in animal cells. Insig-1 is a direct target of SREBP action, and its mRNA rises and falls coordinately with nuclear SREBP (nSREBP) levels (23, 24), especially SREBP-1c. Despite the marked reduction in SREBP-1c transcripts in PEX2-/- livers, Insig-1 mRNA levels were significantly increased 3.2- and 2.4-fold in P9 and P36 untreated PEX2-/- mice, respectively, and declined below basal levels upon BA feeding in both control and PEX2 mutant mice (Fig. 4A). In early postnatal control mice, this BA-induced decrease in Insig-1 mRNA levels parallels the decline in SREBP-1c mRNA and protein levels. Surprisingly, both Insig-2a and Insig-2b mRNA levels were significantly increased in both untreated and BA-fed PEX2-/- mice at both ages.
As hepatic cholesterol levels were similar in newborn (P0) PEX2-/- and control mice (data not shown), we examined whether SREBP pathway genes were also activated in P0 mutants (Fig. 4B). The expression of SREBP-2 and its target genes, HMGCR, farnesyl pyrophosphate synthase (FPPS), and Insig-1, were all significantly increased in P0 PEX2-/- compared with control mouse liver. The activities and protein levels of cholesterol biosynthetic enzymes were similar or slightly elevated in the livers of P0 PEX2-/- versus control mice (12). SREBP-1c expression was reduced by 60% in P0 PEX2-/- mice. Similar to the expression pattern in P9 and P36 mutants, Insig-2a mRNA was increased 5.2-fold in P0 PEX2-/- mice. Collectively, the P0, P9, and P36 expression data suggest that SREBP pathways are abnormally regulated in peroxisome-deficient livers despite normal cholesterol levels.
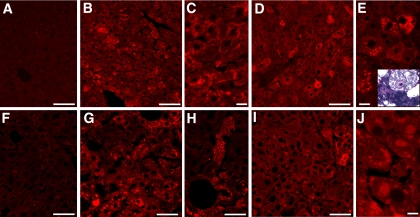
Activation of the UPR in the Liver of PEX2-/- Mice—Previous studies have demonstrated that ER stress induces SREBP activation independently of intracellular cholesterol concentration (25-27). To determine whether peroxisome deficiency causes ER stress and thereby induces the unfolded protein response, we first examined the expression of the ER chaperone proteins glucose-regulated protein 78 (Grp78) (Fig. 5) and Grp94 (data not shown) by immunohistochemistry in control and PEX2-/- livers. Whereas the levels of Grp78 and Grp94 were relatively low in control sections, the staining was significantly increased in the livers of all PEX2-/- mice. In early postnatal untreated mutants, Grp78 accumulation was most prominent in subplasmalemal or larger cytosolic aggregates (Fig. 5, B and C). Grp78 was in larger particles throughout the cytosol in early postnatal BA-fed mutants (Fig. 5, D and E), with the most strongly labeled cells representing heavily lipid-laden and vacuolated hepatocytes (Fig. 5E). Grp78 staining was dramatically increased in P36 untreated mutants and was present in variably sized cytosolic vesicular aggregates or larger vacuoles throughout the hepatic lobule (Fig. 5, G and H). Thus, the highly vacuolated degenerating hepatocytes, previously identified by ultrastructural analysis in P10 BA-fed and P36 untreated PEX2-/- mice (13), represent markedly dilated ER due to UPR activation. The large, often rounded Grp78-positive aggregates seen in P36 BA-fed PEX2-/- mice (Fig. 5, I and J) also correlate morphologically with the marked accumulations of ER seen by ultrastructure.
FIGURE 5.
Induction of ER chaperone expression in untreated and BA-fed PEX2-/- mouse livers. Liver sections from untreated control (A and F) and PEX2-/- (B, C, G, and H) mice and BA-fed PEX2-/- (D, E, I, and J) mice at P9-P13 (A-E) and P36 (F-J) were stained with an antibody to Grp78 and imaged by confocal microscopy. Grp78 was increased in an irregular mosaic pattern in both untreated and BA-fed PEX2 mutant livers. In early postnatal untreated mutants (B and C), staining was particularly prominent in clusters of vesicles beneath the plasma membrane and occasionally in larger cytosolic aggregates. In early postnatal BA-fed mutants (D and E), prominent vesicles were seen throughout the hepatocyte cytoplasm along with very heavily Grp78-labeled cells that correspond to highly vacuolated, degenerate hepaotcytes (inset in E, toluidine blue-stained semithin section). Large cytosolic aggregates (G) and multivacuolated cells (H) were prominently labeled in P36 untreated mutants. Labeling was more focal in P36 BA-fed PEX2 mutant livers (I), but large rounded clusters of ER were evident (J). Immunohistochemical staining patterns were similar for untreated and BA-fed control mice at either age (not shown). Scale bar:50 μm for panels A, B, D, and F-I;10 μm for panels C, E, and J.
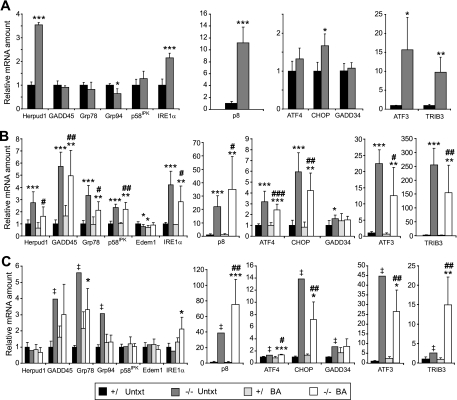
Recent studies have demonstrated considerable overlap between the subsets of genes regulated by the three principal stress-sensing ER-transmembrane proteins: protein kinase RNA-like ER kinase (PERK), inositol-requiring protein (IRE)-1α, and activating transcription factor (ATF)-6α (28-31). The response of these various targets to chronic stress in vivo has not been well characterized. mRNA expression analysis of several UPR target genes revealed an evolving age- and treatment-dependent pattern of the hepatic stress response in peroxisome deficiency (Fig. 6). For instance, Herpud1 (homocysteine-inducible ER stress-inducible ubiquitin-like domain member 1) mRNA was most prominently increased in P0 PEX2-/- mice (Fig. 6A; 3.5-fold), somewhat less so in P9 mutants (Fig. 6B; 1.8-2.8-fold), and not at all in P36 mutants (Fig. 6C), independently of BA feeding. Both Herpud1 and Edem1 (ER degradation enhancer mannosidase α-like 1) are components of the ER-associated protein degradation (ERAD) pathway (32). Thus, the progressive postnatal loss of Herpud1 overexpression and the absence of an increase in Edem1 in P9 and P36 PEX2 mutants (Fig. 6, B and C) suggest a declining function and/or role of ERAD in peroxisome-deficient liver.
FIGURE 6.
Expression of ER stress markers in untreated (Untxt) and BA-fed control and PEX2-/- mouse livers. RNA was analyzed by quantitative real-time RT-PCR as described in the legend for Fig. 4. A, P0 liver. B, P9 liver. Relative mRNA levels of Herpud1, CHOP, GADD45, Grp78, and p8 were also quantified from Northern blots (supplemental Fig. 2) and yielded highly similar results. C, P36 liver. Values represent the amount of mRNA relative to that in untreated control mice, which was arbitrarily defined as 1. Values are mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control mice. #, p < 0.05; ##, p < 0.01; p < 0.001 versus BA-fed control mice (Student's t test). ‡, outside 95% confidence interval of P36 untreated control mice.
Although ER chaperone (Grp78 and Grp94) and GADD45 (growth arrest and DNA damage-inducible 45) mRNA levels were not increased in P0 PEX2-/- mice (Fig. 6A), their expression was strongly induced in livers from P9 (Fig. 6B) and P36 (Fig. 6C) untreated and BA-fed PEX2 mutants. The expression of ER chaperones and the cochaperone protein p58IPK are regulated to some degree by all three arms of the UPR (29, 33), but p58IPK expression was only transiently increased in P9 PEX2 mutants (Fig. 6B).
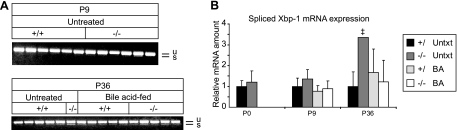
IRE1α is a dual function serine-threonine protein kinase and endoribonuclease. Activation of its RNase domain results in the unconventional splicing of XBP-1 (X-box-binding protein 1) mRNA (sXBP-1) and feedback down-regulation of IRE1α mRNA (34). RT-PCR analysis for XBP-1 mRNAs (Fig. 7A) and quantitative RT-PCR analysis for sXBP-1 (Fig. 7B) showed normal sXBP-1 and precursor mRNA levels in P0 and P9 PEX2-/- mice. sXBP-1 expression was significantly elevated only in the P36 untreated PEX2 mutant (Fig. 7) but still represented a minor component of total XBP-1 mRNAs. Although the IRE1α mRNA level was increased in P0 and P9 PEX2-/- mice, expression was variable in P36 mutants, being somewhat reduced in the untreated mutant (consistent with some XBP-1 splicing) but still increased in BA-fed mutants. These findings show that IRE1α signaling and it's RNase activity are not prominently induced in PEX2-/- livers.
FIGURE 7.
Analysis of spliced XBP-1 mRNA. XBP-1 mRNA splicing was determined either by RT-PCR and subsequent gel electrophoresis (A) or by quantitative real-time RT-PCR (B). Unspliced (u) and spliced (s) Xbp-1 mRNA products are indicated. Values represent the amount of mRNA relative to that in untreated control mice, which was arbitrarily defined as 1. Values are mean ± S.D. ‡, outside 95% confidence interval of P36 untreated control mice (Untxt).
PERK activation and subsequent eIF2α phosphorylation lead to production of the ATF4 transcription factor, which plays a crucial role for the adaptation to stress (28). ATF4 mRNA level was most prominently increased in P9 PEX2-/- mice and less so in P36 mutants, independently of BA treatment (Fig. 6). Next, we determined the expression of transcriptional targets of ATF4, namely ATF3, CHOP (C/EBP-homologous protein), and TRIB3 (tribbles homolog 3). CHOP expression (predominantly reflects PERK activation but is also modulated by ATF6α) was mildly increased in P0 PEX2-/- liver but was strongly elevated in both untreated and BA-fed PEX2 mutants at P9 and P36 (Fig. 6). The expression of ATF3 was highly up-regulated in the livers of both untreated and BA-fed PEX2-/- mice at all ages (Fig. 6). p8 (stress-associated protein p8), also a downstream target of the PERK pathway (35), showed a different pattern, with strong up-regulation at P0 and an ever increasing mRNA induction exacerbated by BA treatment in postnatal PEX2 mutants (Fig. 6). TRIB3, a direct transcriptional target of ATF4 and CHOP, is induced after prolonged ER stress and acts as a transcriptional repressor to attenuate CHOP- and ATF4-mediated transcription (36, 37). TRIB3 expression was already increased 10-fold in P0 PEX2-/- mice (Fig. 6A). TRIB3 mRNA levels were dramatically elevated 250- and 150-fold in untreated and BA-fed P9 PEX2-/- mice, respectively, correlating with the highest ATF4 expression (Fig. 6B). At P36, the TRIB3 mRNA level in BA-fed PEX2 mutants was again similar to that in P0 mutants but was much less induced in the untreated mutant (Fig. 6C). Thus, the persistent elevation of Grp78 and CHOP mRNA in P36 PEX2 mutants likely reflects modulatory effects of other UPR pathways. There was also a minimal expression change for the CHOP target GADD34 in P9 PEX2-/- liver, a finding best explained by ATF6α-mediated suppression of GADD34 production (33).
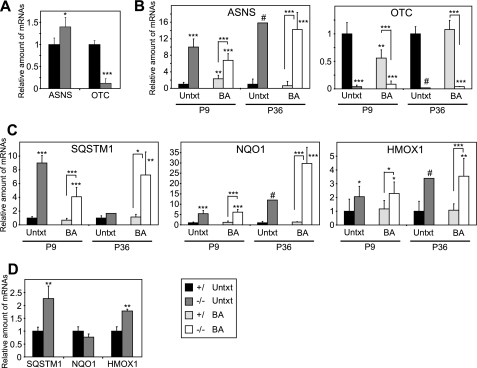
As ER stress results in the accumulation of reactive oxygen species, the PERK and ATF4-mediated integrated stress response (ISR) transcriptionally activates genes involved in amino acid metabolism and antioxidant-detoxifying enzymes that may protect against oxidative stress (38). As part of the amino acid response, ATF4 regulates expression of the amino acid biosynthetic enzyme asparagine synthetase, which was highly increased in the livers of both untreated and BA-fed PEX2-/- mice at all ages (Fig. 8, A and B, ASNS). Amino acid catabolism genes, including ornithine transcarbamylase, were highly decreased in the livers of both untreated and BA-fed PEX2-/- mice at all ages (Fig. 8, A and B, OTC). Furthermore, PERK-mediated phosphorylation of the transcription factor NF-E2-related factor 2 (Nrf2) activates several oxidative stress genes to promote cellular redox homeostasis (39) and thereby connects ER stress and oxidative stress signaling pathways. Expression of several Nrf2 target genes was highly increased in untreated and BA-fed PEX2-/- mice at all ages, including Sqstm1 (sequestosome 1), Nqo1 (NAD(P)H:quinine oxidoreductase 1), and Hmox1 (heme oxygenase 1) (Fig. 8, C and D). These data suggest that excessive reactive oxygen species in livers of PEX2-/- mice lead to the activation of the ISR and transcriptional activation of PERK and ATF4 oxidative stress pathways.
FIGURE 8.
Expression of genes involved in amino acid metabolism (A and B) and oxidative stress-activated Nrf2-regulated genes (C and D) in untreated (Untxt) and BA-fed (BA) control and PEX2-/- mouse livers. RNA was analyzed by quantitative real-time RT-PCR as described in the legend for Fig. 4. A and D, P0 liver. B and C, P9 and P36 liver. Values represent the amount of mRNA relative to that in untreated control mice, which was arbitrarily defined as 1. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus untreated control mice or versus BA-fed control mice (indicated by brackets; Student's t test); #, outside 95% confidence interval of P36 untreated control mice. ASNS, asparagine synthetase; OTC, ornithine transcarbamylase; SQSTM1, sequestosome 1; NQO1, NAD(P)H: quinine oxidoreductase 1; HMOX1, heme oxygenase 1.
DISCUSSION
Our previous studies demonstrated that peroxisome-deficient PEX2-/- mice have a complex cholesterol phenotype in which activation of hepatic SREBP-2 and its target genes occurs, as expected, in association with reductions in hepatic and plasma cholesterol levels but is unable to maintain cholesterol homeostasis. As cholesterol and BA synthetic pathways are normally tightly and coordinately regulated in the liver, the question remained whether the concurrent disruption to BA synthesis and resulting marked paucity of mature C24 bile acids in PEX2-/- mice (13) was central to the pathogenesis of the observed cholesterol abnormalities. In the present study we evaluated the effects of BA replenishment on cholesterol metabolism in both neonatal and longer surviving PEX2-/- mice. A striking finding of our data is that the cholesterol biosynthetic pathway remains up-regulated in peroxisome-deficient PEX2-/- liver even when plasma and hepatic cholesterol levels are normalized by BA feeding. Induction of several stress-related pathways in the livers of both untreated and BA-fed PEX2-/- mice suggest a mechanism whereby ER stress leads to dysregulation of the endogenous sterol response mechanism.
ER Stress and Activation of SREBP-2 Genes Independent of Cholesterol Content in PEX2-/- Liver—PEX2-/- mice displayed persistent up-regulation of cholesterogenic pathways in liver even when hepatic cholesterol levels were normal, as seen in newborn and P36 mutants and in BA-fed early postnatal mutants. Our studies revealed several abnormalities in gene expression pathways regulated by cholesterol levels through SREBPs and Insigs in PEX2-/- mice. Although SCAP overexpression leads to noninhibitable SREBP processing by saturating endogenous Insigs (40, 41), SCAP expression was normal in PEX2-/- mice at all ages and thus cannot be used to explain the observed cholesterol-independent SREBP-2 activation. Recent studies report that cells can bypass the cholesterol inhibition of SREBP processing in response to ER stress and activate SREBP-2 (25-27). Various biochemical and physiologic stimuli may impose stress to the ER and lead to the accumulation of unfolded or misfolded proteins in the ER lumen (28, 30). These disturbances trigger the UPR to cope with the changing environment and reestablish normal ER function. Our data clearly demonstrate that the UPR was activated in the liver of both untreated and BA-fed PEX2-/- mice.
Insigs inhibit SREBP processing and enhance the degradation of HMGCR in response to sterols (3). We provide novel data that link ER dysfunction in peroxisome-deficient cells to defects in Insig-SCAP-SREBP signaling. Cholesterol-independent SREBP-2 activation in response to ER stress has been attributed to the rapid turnover of Insig-1 protein, even if Insig-1 mRNA is increased (26). Decreased Insig-1 expression in BA-fed PEX2-/- mice could create a relative resistance to the effects of sterols in blocking SREBP processing (42) and accelerating HMGCR degradation (43-45). ER stress does not alter Insig-2 expression, and Insig-2 cannot substitute for Insig-1 in blocking SREBP cleavage under conditions of hypotonic stress (26). Unfortunately we could not determine whether Insig-1, a protein with a very rapid rate of turnover, is depleted in the liver of PEX2-/- mice. However, depletion of rapidly degraded proteins such as Insig-1 in PEX2 mutant liver is likely, given the inhibition of protein translation that occurs with ER stress due to PERK activation. Our HMGCR data are also consistent with an altered ER environment, as the activity of HMGCR in the liver of both untreated and BA-fed PEX2-/- mice was significantly lower than anticipated from the highly increased mRNA and protein levels (Table 2) (12). HMGCR is the rate-limiting enzyme in cholesterol synthesis, and its transcription, translation, and degradation are governed by a complex multivalent regulatory system (3). Microsomal abnormalities have been observed in peroxisome-deficient hepatocytes in both PEX2-/- (13) and PEX5-/- mice (46). In this study, we have demonstrated that vesicular aggregates, large vacuoles, and proliferated ER seen in untreated and BA-fed PEX2-/- hepatocytes (Fig. 5) label strongly with antibodies to Grp78, associating marked activation of the UPR with hepatocyte degeneration. In addition, as HMGCR degradation is regulated by ERAD (3), the declining postnatal expression of Herpud1 and lack of Edem1 up-regulation in PEX2-/- livers (Fig. 6) may well contribute to the persistently elevated hepatic HMGCR protein levels. In contrast to liver, HMGCR activity was reduced in PEX2-/- kidneys (supplemental Table S1) (12) even in BA-fed mutants. This appears to be a posttranscriptional regulation, as the mRNA expression of HMGCR, Insig-1, and SREBP-2 was increased 2-fold in untreated P10 PEX2-/- kidney (p < 0.05; data not shown). However, there was no ER stress response in PEX2-/- kidney, as Grp78 immunostain and quantitative real-time RT-PCR expression analysis of Grp78, Herpud1, CHOP, GADD45, and p8 failed to reveal any abnormalities (data not shown).
Analysis of ER stress-associated genes in PEX2-/- liver revealed an evolving, age- and treatment-dependent pattern of the hepatic stress response (Fig. 6). The mRNA expression of ER chaperones and the CHOP gene increased in early postnatal PEX2-/- liver and persisted in longer surviving P36 mutants, consistent with activation of the ATF6α and/or PERK pathway (30). Interplay between these pathways was observed in P9 mutants by paucity of GADD34 up-regulation, despite a prominent increase in its upstream regulator, CHOP, suggesting that suppression by ATF6α activity may enable persistent PERK/eIF2α-mediated translation attenuation. At all ages, there was a paucity of XBP1 mRNA splicing in the PEX2-/- liver, as has been observed in cell lines and in vivo with chronic stress exposure (47, 48). In chronic retinal degeneration, down-regulation of the IRE1α/XBP1 pathway was also seen in vivo, but CHOP induction was maintained in the absence of Grp78 activation, suggesting persistent activation of PERK but attenuated ATF6α activity (17). Thus, the regulatory pattern differs in chronically stressed PEX2-/- livers, as both CHOP and Grp78 induction are maintained, likely reflecting different disease mechanisms and/or tissue-specific responses to stress.
CHOP expression serves as a sentinel for apoptosis and the failure of cells to adapt to chronic stress (48). However, despite a strong increase in caspase-3 activation by BA feeding in P9 and P36 PEX2-/- liver (13), CHOP mRNA levels did not differ significantly between these mutants. Thus, BA feeding did not generally increase UPR activation in PEX2-/- mice, and the increased hepatocyte cell death is more likely due to BA-mediated mitochondrial toxicity (49). Interestingly, p8 gene expression was prominently increased even in P0 PEX2-/- livers and was further induced in postnatal mutant livers and with BA treatment. p8 is up-regulated in response to several stresses, including lipopolysaccharide and proapoptotic agents (50); our data demonstrate that bile acids may also regulate this stress protein.
Peroxisome Deficiency Induces the Integrated Stress Response—The integrated stress response is one of the three known components of the UPR whereby PERK activation and ATF4 induction links ER stress and other cellular adapative pathways. The ATF4 targets ATF3 and TRIB3 were highly increased in both untreated and BA-fed PEX2-/- mice at all ages. Hepatic expression of ATF3 has been shown to result in several symptoms of liver dysfunction, including hypoglycemia and increased serum levels of bilirubin, bile acids, and liver transaminases (51, 52). It has been established that the ISR is subject to negative regulation, mediated in part by GADD34 and TRIB3, which are induced after prolonged ER stress. However, whereas GADD34 expression was not prominently affected in PEX2-/- mice, TRIB3 was highly induced at all ages but did not attenuate CHOP- and ATF4-mediated transcription. Further studies are needed to characterize the negative feedback regulation of the ISR under chronic stress.
ER stress, nutrient deprivation, oxidative stress, double-stranded RNA accumulation, and heme depletion converge on eIF2α phosphorylation and elicit an ISR pathway designed to resolve stress (38, 53). Four different kinases that respond to distinct upstream stress signals are known to phosphorylate eIF2α (i.e. PERK, GCN2, PKR, and HRI). Stress from accumulation of unfolded or misfolded proteins in the ER activates the ISR through ER-localized PERK, leading to ATF4 production and Nrf2 phosphorylation (31). Amino acid starvation and diverse metabolic perturbations activate the ISR through GCN2 (54). The exact mechanisms triggering ER stress in peroxisome-deficient PEX2-/- livers remain to be fully defined but likely involve multiple signals, including perturbed flux of mevalonate metabolites, altered bile acid homeostasis, changes in fatty acid levels and composition, and oxidative stress (Fig. 9).
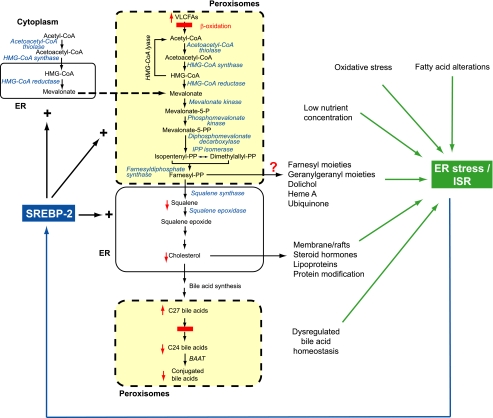
FIGURE 9.
Model illustrating the relationships between peroxisome deficiency, cholesterol biosynthesis, and ER stress/integrated stress response. The missing peroxisomal compartment is illustrated in yellow with dashed lines. Defective metabolic pathways due to peroxisome deficiency are indicated by solid red bars; red arrows indicate increased or decreased metabolite levels due to peroxisome deficiency. Blue fonts in italic indicate SREBP-2-regulated cholesterol biosynthetic enzymes. Peroxisome deficiency activates hepatic ER stress pathways, especially the ISR mediated by PERK and ATF4 signaling. Several metabolic derangements in peroxisome-deficient PEX2-/- liver are likely to trigger ER stress, including perturbed flux of mevalonate metabolites, altered bile acid homeostasis, changes in fatty acid levels and composition, and oxidative stress. BAAT, bile acid-coenzyme A:amino acid N-acyltransferase.
The mevalonate pathway produces isoprenoids that are critical for diverse cellular functions (55). We have demonstrated here that hepatic levels of squalene, an intermediate in the cholesterol biosynthetic pathway, are markedly decreased both in P9 and P36 untreated and BA-fed PEX2-/- mice despite normalization of the cholesterol content in BA-fed PEX2-/- mice. Therefore, levels of other mevalonate metabolites may also be perturbed in PEX2-/- mice leading to an activation of PERK and/or GCN2. For instance, inhibition of cholesterol biosynthesis can trigger the ISR, probably because of alterations of membrane cholesterol composition (56, 57). Perturbations of non-sterol isoprenoids downstream of the farnesyl pyrophosphate branch point also contribute to the development of ER stress. Dolichol is involved in the N-linked glycosylation of ER proteins (55), and inhibition of N-linked glycosylation by tunicamycin induces ER stress in a wide variety of cell types. Perturbations in farnesylated and geranylgeranylated Rho and Rab proteins, which play critical roles in ER, have been demonstrated to result in ER stress (58, 59). Ubiquinone is a powerful antioxidant and an important component of the mitochondrial respiratory chain, and its depletion may lead to oxidative cellular stress (55). Thus, a number of mevalonate metabolites trigger the ISR; future studies are needed to define whether these metabolite levels are altered in the PEX2-/- liver.
Bochkis et al. (60) showed that dysregulation of BA homeostasis in hepatocyte-specific Foxa2-/- mice leads to ER stress due to cumulative defects in expression of bile acid-CoA ligase, involved in BA conjugation, and BA transporters. Indeed, our previous study demonstrated that BA homeostasis is widely disturbed in PEX2-/- mice associated with alterations in BA transporters, leading to hypercholanemia and defects in BA conjugation (13), which strongly suggests that BA alterations contribute to the activation of the ISR.
Fatty acid alterations induced by peroxisome deficiency may also induce the ISR. Recently, it has been shown that changes in fatty acid composition in Scd1-/- mice fed a very low fat diet induce ER stress (61). Alterations in cellular fatty acid composition may induce ER stress due to a disturbed physical state of cellular membranes and altered function and/or localization of membrane transport proteins. Peroxisome deficiency leads to an accumulation of fatty acids that are degraded via peroxisomal β-oxidation (e.g. very long-chain and branched-chain fatty acids, dicarboxylic acids). Plasma and hepatic very long-chain fatty acids were significantly increased in PEX2-/- mice (14) and other peroxisome-deficient mouse models (62). Furthermore, levels of n-6 polyunsaturated fatty acids (i.e. 18:2n-6, 18:3n-6, 20:2n-6, 20:3n-6, 22:4n-6) were increased in the livers of P9 PEX2-/- mice compared with controls (data not shown).
Peroxisomes play an important role in the homeostasis of reactive oxygen species (63). Peroxisome-deficient mice may activate the ISR through oxidative stress caused by defective peroxisomal antioxidant mechanisms. Marked alterations of the mitochondria were observed in both PEX2-/- (13) and PEX5-/- (64) mice, which resemble closely those in disorders associated with oxidative stress. Although no evidence of oxidative damage to proteins or lipids or of increased peroxide production has been found in hepatocyte-specific PEX5-/- mice (46), the highly increased hepatic expression of several targets of the oxidative stress-activated transcription factor Nrf2 strongly suggests that increased oxidative stress is present in livers of PEX2-/- mice.
In conclusion, our study has revealed a dysregulation of SREBP-2, SREBP-1c, and Insig-2a expression and a novel regulation of Insig-2b. Our data support a mechanism by which peroxisome-deficient cells experience ER stress that leads to the activation and dysregulation of the endogenous sterol response pathway. The discovery that the regulation of SREBP/Insig genes is disturbed in peroxisome-deficient cells paves the way for new studies on the coordinated control of these closely regulated metabolic pathways. The overall pathology in the PEX2-/- mouse model has potential implications for future disease treatment interventions that target lipid metabolism.
Supplementary Material
Acknowledgments
We thank Herbert Stangl for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK58238 and DK58040 (to S. K. K.), HD36807 and NS050602 (to P. L. F.), and EY007361 (to S. J. F.). This work was also supported by an unrestricted departmental grant from Research to Prevent Blindness (to S. J. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
Footnotes
The abbreviations used are: SREBP, sterol regulatory element-binding protein; SCAP, SREBP cleavage-activating protein; ER, endoplasmic reticulum; ERAD, ER-associated protein degradation; PERK, protein kinase RNA-like ER kinase; IRE, inositol-requiring protein; ATF, activating transcription factor; Insig, insulin-induced gene; HMG, 3-hydroxy-3-methylglutaryl; HMGCR, HMG-CoA reductase; HDL, high density lipoprotein; LDL, low density lipoprotein; BA, bile acid; UPR, unfolded protein response; IDI, isopentenylpyrophosphate isomerase; P, postnatal day; LXR, liver X-activated receptor; CHOP, C/EBP-homologous protein; ISR, integrated stress response; RT, reverse transcription; ANOVA, analysis of variance.
T. Wikander and P. Faust, manuscript in preparation.
References
- 1.Yeagle, P. L. (ed) (1988) The Biology of Cholesterol, CRC Press, Boca Raton, FL
- 2.Fliesler, S. J. (ed) (2002) Sterols and Oxysterols: Chemistry, Biology and Pathobiology, Research Signpost, Kerala, India
- 3.Goldstein, J. L., DeBose-Boyd, R. A., and Brown, M. S. (2006) Cell 124 35-46 [DOI] [PubMed] [Google Scholar]
- 4.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) J. Clin. Investig. 109 1125-1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell, D. W. (2003) Annu. Rev. Biochem. 72 137-174 [DOI] [PubMed] [Google Scholar]
- 6.Horton, J. D., Shah, N. A., Warrington, J. A., Anderson, N. N., Park, S. W., Brown, M. S., and Goldstein, J. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12027-12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yabe, D., Brown, M. S., and Goldstein, J. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12753-12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yabe, D., Komuro, R., Liang, G., Goldstein, J. L., and Brown, M. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3155-3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacs, W. J., Olivier, L. M., and Krisans, S. K. (2002) Prog. Lipid Res. 41 369-391 [DOI] [PubMed] [Google Scholar]
- 10.Wanders, R. J. A., and Tager, J. M. (1998) Mol. Aspects Med. 19 71-154 [DOI] [PubMed] [Google Scholar]
- 11.Kovacs, W. J., Tape, K. N., Shackelford, J. E., Duan, X., Kasumov, T., Kelleher, J. K., Brunengraber, H., and Krisans, S. K. (2007) Histochem. Cell Biol. 127 273-290 [DOI] [PubMed] [Google Scholar]
- 12.Kovacs, W. J., Shackelford, J. E., Tape, K. N., Richards, M. J., Faust, P. L., Fliesler, S. J., and Krisans, S. K. (2004) Mol. Cell Biol. 24 1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keane, M. H., Overmars, H., Wikander, T. M., Ferdinandusse, S., Duran, M., Wanders, R. J. A., and Faust, P. L. (2007) Hepatology 45 982-997 [DOI] [PubMed] [Google Scholar]
- 14.Faust, P. L., and Hatten, M. E. (1997) J. Cell Biol. 139 1293-1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faust, P. L., Su, H.-M., Moser, A., and Moser, H. W. (2001) J. Mol. Neurosci. 16 289-297 [DOI] [PubMed] [Google Scholar]
- 16.Faust, P. L. (2003) J. Comp. Neurol. 461 394-413 [DOI] [PubMed] [Google Scholar]
- 17.Lin, J. H., Li, H., Yasumura, D., Cohen, H. R., Zhang, C., Panning, B., Shokat, K. M., LaVail, M. M., and Walter, P. (2007) Science 318 944-949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipson, K. L., Fonseca, S. G., Ishigaki, S., Nguyen, L. X., Foss, E., Bortell, R., Rossini, A. A., and Urano, F. (2006) Cell Metab. 4 245-254 [DOI] [PubMed] [Google Scholar]
- 19.Repa, J. J., Lund, E. G., Horton, J. D., Leitersdorf, E., Russell, D. W., Dietschy, J. M., and Turley, S. D. (2000) J. Biol. Chem. 275 39685-39692 [DOI] [PubMed] [Google Scholar]
- 20.Schwarz, M., Russell, D. W., Dietschy, J. M., and Turley, S. D. (2001) J. Lipid Res. 42 1594-1603 [PubMed] [Google Scholar]
- 21.Watanabe, M., Houten, S. M., Wang, L., Moschetta, A., Mangelsdorf, D. J., Heyman, R. A., Moore, D. D., and Auwerx, J. (2004) J. Clin. Investig. 113 1408-1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton, J. D., Shimomura, I., Brown, M. S., Hammer, R. E., Goldstein, J. L., and Shimano, H. (1998) J. Clin. Investig. 101 2331-2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, T., Espenshade, P. J., Wright, M. E., Yabe, D., Gong, Y., Aebersold, R., Goldstein, J. L., and Brown, M. S. (2002) Cell 110 489-500 [DOI] [PubMed] [Google Scholar]
- 24.Janowski, B. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12675-12680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werstuck, G. H., Lentz, S. R., Dayal, S., Hossain, G. S., Sood, S. K., Shi, Y. Y., Zhou, J., Maeda, N., Krisans, S. K., Malinow, M. R., and Austin, R. C. (2001) J. Clin. Investig. 107 1263-1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. N., and Ye, J. (2004) J. Biol. Chem. 279 45257-45265 [DOI] [PubMed] [Google Scholar]
- 27.Colgan, S. M., Tang, D., Werstuck, G. H., and Austin, R. C. (2007) Int. J. Biochem. Cell Biol. 39 1843-1851 [DOI] [PubMed] [Google Scholar]
- 28.Xu, C., Bailly-Maitre, B., and Reed, J. C. (2005) J. Clin. Investig. 115 2656-2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, J., Rutkowski, D. T., Dubois, M., Swathirajan, J., Saunders, T., Wang, J., Song, B., Yau, G. D., and Kaufman, R. J. (2007) Dev. Cell 13 351-364 [DOI] [PubMed] [Google Scholar]
- 30.Rutkowski, D. T., and Kaufman R. J. (2007) Trends Biochem. Sci. 32 469-476 [DOI] [PubMed] [Google Scholar]
- 31.Ron, D., and Walter, P. (2007) Nat. Rev. Mol. Cell Biol. 8 519-529 [DOI] [PubMed] [Google Scholar]
- 32.Ni, M., and Lee, A. S. (2007) FEBS Lett. 581 3641-3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rutkowski, D. T., Kang, S. W., Goodman, A. G., Garrison, J. L., Taunton, J., Katze, M. G., Kaufman, R. J., and Hegde, R. S. (2007) Mol. Biol. Cell 18 3681-3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tirasophon, W., Lee, K., Callaghan, B., Welihinda, A., and Kaufman, R. J. (2000) Genes Dev. 14 2725-2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Passe, C. M., Cooper, G., and Quirk, C. C. (2006) Endocrine 30 81-91 [DOI] [PubMed] [Google Scholar]
- 36.Ohoka, N., Yoshii, S., Hattori, T., Onozaki, K., and Hayashi, H. (2005) EMBO J. 24 1243-1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jousse, C., Deval, C., Maurin, A. C., Parry, L., Cherasse, Y., Chaveroux, C., Lefloch, R., Lenormand, P., Bruhat, A., and Fafournoux, P. (2007) J. Biol. Chem. 282 15851-15861 [DOI] [PubMed] [Google Scholar]
- 38.Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., Sadri, N., Yun, C., Popko, B., Paules, R., Stojdl, D. F., Bell, J. C., Hettmann, T., Leiden, J. M., and Ron, D. (2003) Mol. Cell 11 619-633 [DOI] [PubMed] [Google Scholar]
- 39.Ishii, T., Itoh, K., Takahashi, S., Sato, H., Yanagawa, T., Katoh, Y., Bannai, S., and Yamamoto, M. (2000) J. Biol. Chem. 275 16023-16029 [DOI] [PubMed] [Google Scholar]
- 40.Hua, X., Nohturfft, A., Goldstein, J. L., and Brown, M. S. (1996) Cell 87 415-426 [DOI] [PubMed] [Google Scholar]
- 41.Lee, P. C. W., Liu, P., Li, W. P., and DeBose-Boyd, R. A. (2007) J. Lipid Res. 48 1944-1954 [DOI] [PubMed] [Google Scholar]
- 42.Adams, C. M., Reitz, J., De Brabander, J. K., Feramisco, J. D., Li, L., Brown, M. S., and Goldstein, J. L. (2004) J. Biol. Chem. 279 52772-52780 [DOI] [PubMed] [Google Scholar]
- 43.Sever, N., Yang, T., Brown, M. S., Goldstein, J. L., and DeBose-Boyd, R. A. (2003) Mol. Cell 11 25-33 [DOI] [PubMed] [Google Scholar]
- 44.Lee, P. C. W., Sever, N., and DeBose-Boyd, R. A. (2005) J. Biol. Chem. 280 25242-25249 [DOI] [PubMed] [Google Scholar]
- 45.Song, B.-L., Javitt, N. B., and DeBose-Boyd, R. A. (2005) Cell Metab. 1 179-189 [DOI] [PubMed] [Google Scholar]
- 46.Dirkx, R., Vanhorebeek, I., Martens, K., Schad, A., Grabenbauer, M., Fahimi, D., Declerq, P., Van Veldhoven, P. P., and Baes, M. (2005) Hepatology 41 868-878 [DOI] [PubMed] [Google Scholar]
- 47.Marciniak, S. J., Yun, C. Y., Oyadomari, S., Novoa, I., Zhang, Y., Jungreis, R., Nagata, K., Harding, H. P., and Ron, D. (2004) Genes Dev. 18 3066-3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rutkowski, D. T., Arnold, S. M., Miller, C. N., Wu, J., Li, J., Gunnison, K. M., Mori, K., Sadighi Akha, A. A., Raden, D., and Kaufman, R. J. (2006) PLoS Biol. 4 e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmeira, C. M., and Rolo, A. P. (2004) Toxicology 203 1-15 [DOI] [PubMed] [Google Scholar]
- 50.Iovanna, J. L. (2002) Int. J. Gastrointest. Cancer 31 89-98 [DOI] [PubMed] [Google Scholar]
- 51.Allen-Jennings, A. E., Hartman, M. G., Kociba, G. J., and Hai, T. (2001) J. Biol. Chem. 276 29507-29514 [DOI] [PubMed] [Google Scholar]
- 52.Allen-Jennings, A. E., Hartman, M. G., Kociba, G. J., and Hai, T. (2002) J. Biol. Chem. 277 20020-20025 [DOI] [PubMed] [Google Scholar]
- 53.Dever, T. E. (2002) Cell 108 545-556 [DOI] [PubMed] [Google Scholar]
- 54.Hinnebusch, A. G., and Natarajan, K. (2002) Eukaryot. Cell 1 22-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldstein, J. L., and Brown, M. S. (1990) Nature 343 425-430 [DOI] [PubMed] [Google Scholar]
- 56.Harding, H. P., Zhang, Y., Khersonsky, S., Marciniak, S., Scheuner, D., Kaufman, R. J., Javitt, N., Chang, Y. T., and Ron, D. (2005) Cell Metab. 2 361-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niknejad, N., Morley, M., and Dimitroulakos, J. (2007) J. Biol. Chem. 282 29748-29756 [DOI] [PubMed] [Google Scholar]
- 58.Alvarez, C., and Sztul, E. S. (1999) Eur. J. Cell Biol. 78 1-14 [DOI] [PubMed] [Google Scholar]
- 59.Seabra, M. C., Mules, E. H., and Hume, A. N. (2002) Trends Mol. Med. 8 23-30 [DOI] [PubMed] [Google Scholar]
- 60.Bochkis, I. M., Rubins, N. E., White, P., Furth, E. E., Friedman, J. R., and Kaestner, K. H. (2008) Nat. Med. 14 828-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flowers, M. T., Keller, M. P., Choi, Y., Lan, H., Kendziorski, C., Ntambi, J. M., and Attie, A. D. (2008) Physiol. Genomics 33 361-372 [DOI] [PubMed] [Google Scholar]
- 62.Baes, M., and Van Veldhoven, P. P. (2006) Biochim. Biophys. Acta 1763 1785-1793 [DOI] [PubMed] [Google Scholar]
- 63.Schrader, M., and Fahimi, H. D. (2006) Biochim. Biophys. Acta 1763 1755-1766 [DOI] [PubMed] [Google Scholar]
- 64.Baumgart, E., Vanhorebeek, I., Grabenbauer, M., Borgers, M., Declercq, P. E., Fahimi, H. D., and Baes, M. (2001) Am. J. Pathol. 159 1477-1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.