Abstract
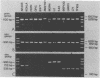
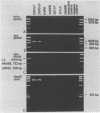
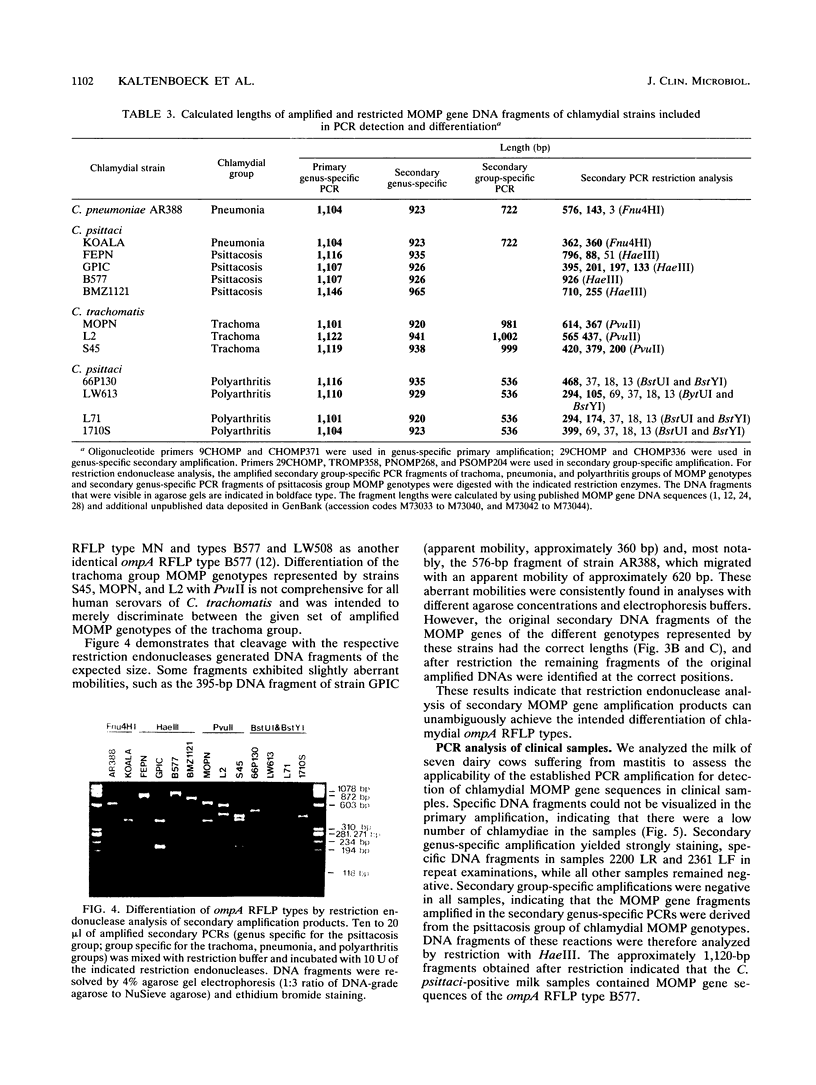
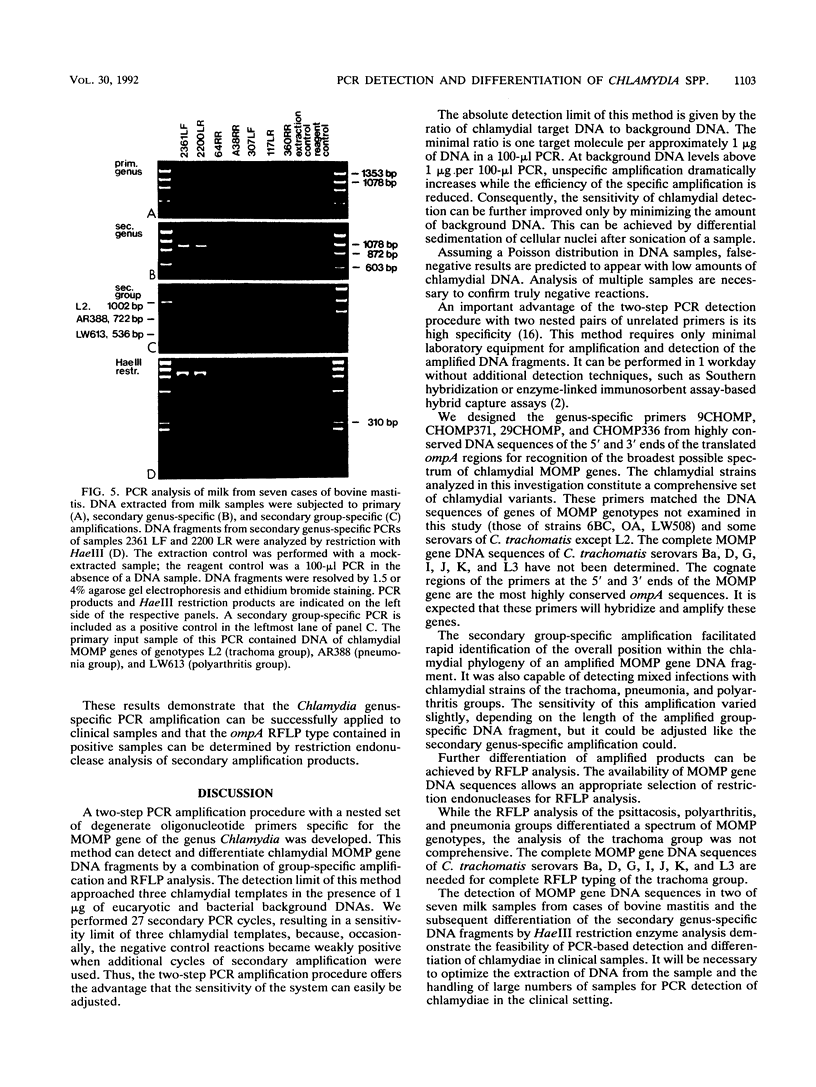
Specific and sensitive amplification of major outer membrane protein (MOMP) gene (ompA) DNA sequences of Chlamydia species with various MOMP genotypes was achieved by a two-step polymerase chain reaction (PCR). Degenerate, inosine-containing oligonucleotide primers homologous to the 5' and 3' ends of the translated regions of all chlamydial MOMP genes were used in a PCR to amplify a DNA fragment of approximately 1,120 bp. A portion of this DNA fragment was amplified in a second genus-specific reaction that yielded a DNA fragment of approximately 930 bp. A pair of degenerate oligonucleotide primers homologous to internal sequences of the primary DNA fragment was used in this PCR. This method detected three cognate chlamydial genomes in a background of 1 microgram of unrelated DNA. MOMP genes of 13 representative chlamydial MOMP genotypes of the species C. trachomatis, C. pneumoniae, and C. psittaci were amplified. In a secondary PCR, group-specific detection was achieved by the simultaneous use of one genus-specific primer and three primers derived from different fingerprint regions of three major groups of chlamydiae. This multiplex PCR differentiated the groups by the length of the amplified DNA fragments and detected the simultaneous presence of DNA sequences of the Chlamydia spp. with different MOMP genotypes. Further differentiation as ompA restriction fragment length polymorphism types among all chlamydial strains with the various MOMP genotypes analyzed here was achieved by restriction endonuclease analysis of the secondary PCR products. DNA sequences corresponding to the ompA restriction fragment length polymorphism type B577 of C. psittaci were detected in two of seven milk samples from cases of bovine mastitis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baehr W., Zhang Y. X., Joseph T., Su H., Nano F. E., Everett K. D., Caldwell H. D. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci U S A. 1988 Jun;85(11):4000–4004. doi: 10.1073/pnas.85.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobo L., Coutlee F., Yolken R. H., Quinn T., Viscidi R. P. Diagnosis of Chlamydia trachomatis cervical infection by detection of amplified DNA with an enzyme immunoassay. J Clin Microbiol. 1990 Sep;28(9):1968–1973. doi: 10.1128/jcm.28.9.1968-1973.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L. A., Kuo C. C., Grayston J. T. Characterization of the new Chlamydia agent, TWAR, as a unique organism by restriction endonuclease analysis and DNA-DNA hybridization. J Clin Microbiol. 1987 Oct;25(10):1911–1916. doi: 10.1128/jcm.25.10.1911-1916.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh B., Bébéar C., Rodriguez P., Vekris A., Bonnet J., Garret M. Specific amplification of a DNA sequence common to all Chlamydia trachomatis serovars using the polymerase chain reaction. Res Microbiol. 1989 Jan;140(1):7–16. doi: 10.1016/0923-2508(89)90053-3. [DOI] [PubMed] [Google Scholar]
- Frutos R., Pages M., Bellis M., Roizes G., Bergoin M. Pulsed-field gel electrophoresis determination of the genome size of obligate intracellular bacteria belonging to the genera Chlamydia, Rickettsiella, and Porochlamydia. J Bacteriol. 1989 Aug;171(8):4511–4513. doi: 10.1128/jb.171.8.4511-4513.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi H., Hirai K. Genetic diversity of avian and mammalian Chlamydia psittaci strains and relation to host origin. J Bacteriol. 1989 May;171(5):2850–2855. doi: 10.1128/jb.171.5.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girjes A. A., Hugall A. F., Timms P., Lavin M. F. Two distinct forms of Chlamydia psittaci associated with disease and infertility in Phascolarctos cinereus (koala). Infect Immun. 1988 Aug;56(8):1897–1900. doi: 10.1128/iai.56.8.1897-1900.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayston J. T., Kuo C. C., Wang S. P., Altman J. A new Chlamydia psittaci strain, TWAR, isolated in acute respiratory tract infections. N Engl J Med. 1986 Jul 17;315(3):161–168. doi: 10.1056/NEJM198607173150305. [DOI] [PubMed] [Google Scholar]
- Holland S. M., Gaydos C. A., Quinn T. C. Detection and differentiation of Chlamydia trachomatis, Chlamydia psittaci, and Chlamydia pneumoniae by DNA amplification. J Infect Dis. 1990 Oct;162(4):984–987. doi: 10.1093/infdis/162.4.984. [DOI] [PubMed] [Google Scholar]
- Kaltenboeck B., Kousoulas K. G., Storz J. Detection and strain differentiation of Chlamydia psittaci mediated by a two-step polymerase chain reaction. J Clin Microbiol. 1991 Sep;29(9):1969–1975. doi: 10.1128/jcm.29.9.1969-1975.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenboeck B., Spatafora J. W., Zhang X., Kousoulas K. G., Blackwell M., Storz J. Efficient production of single-stranded DNA as long as 2 kb for sequencing of PCR-amplified DNA. Biotechniques. 1992 Feb;12(2):164, 166, 168-71. [PubMed] [Google Scholar]
- Kaneko S., Feinstone S. M., Miller R. H. Rapid and sensitive method for the detection of serum hepatitis B virus DNA using the polymerase chain reaction technique. J Clin Microbiol. 1989 Sep;27(9):1930–1933. doi: 10.1128/jcm.27.9.1930-1933.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth K., Roberds S., Poteet C., Tamkun M. Highly degenerate, inosine-containing primers specifically amplify rare cDNA using the polymerase chain reaction. Nucleic Acids Res. 1988 Nov 25;16(22):10932–10932. doi: 10.1093/nar/16.22.10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg C. AN UNIDENTIFIED VIRUS WHICH PRODUCES PNEUMONIA AND SYSTEMIC INFECTION IN MICE. Science. 1942 Jan 9;95(2454):49–50. doi: 10.1126/science.95.2454.49-a. [DOI] [PubMed] [Google Scholar]
- Ostergaard L., Birkelund S., Christiansen G. Use of polymerase chain reaction for detection of Chlamydia trachomatis. J Clin Microbiol. 1990 Jun;28(6):1254–1260. doi: 10.1128/jcm.28.6.1254-1260.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez J. A., Storz J. Antigenic diversity of Chlamydia psittaci of mammalian origin determined by microimmunofluorescence. Infect Immun. 1985 Dec;50(3):905–910. doi: 10.1128/iai.50.3.905-910.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez J. A., Storz J. Persistent infection of L cells with an ovine abortion strain of Chlamydia psittaci. Infect Immun. 1985 Nov;50(2):453–458. doi: 10.1128/iai.50.2.453-458.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R. S., Sanchez-Pescador R., Wagar E. A., Inouye C., Urdea M. S. Diversity of Chlamydia trachomatis major outer membrane protein genes. J Bacteriol. 1987 Sep;169(9):3879–3885. doi: 10.1128/jb.169.9.3879-3885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Zhang Y. X., Manning D. S., Caldwell H. D. Multiple tandem promoters of the major outer membrane protein gene (omp1) of Chlamydia psittaci. Infect Immun. 1990 Sep;58(9):2850–2855. doi: 10.1128/iai.58.9.2850-2855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. X., Morrison S. G., Caldwell H. D., Baehr W. Cloning and sequence analysis of the major outer membrane protein genes of two Chlamydia psittaci strains. Infect Immun. 1989 May;57(5):1621–1625. doi: 10.1128/iai.57.5.1621-1625.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]