Abstract
In response to oxidative stress, the transcription factor NF-E2-related factor 2 (Nrf2) controls the fate of cells through transcriptional upregulation of antioxidant response element (ARE)-bearing genes, including those encoding endogenous antioxidants, phase II detoxifying enzymes, and transporters. Expression of the Nrf2-dependent proteins is critical for ameliorating or eliminating toxicants/carcinogens to maintain cellular redox homeostasis. As a result, activation of the Nrf2 pathway, by naturally-occurring compounds or synthetic chemicals at sub-toxic doses, confers protection against subsequent toxic/carcinogenic exposure. Thus, the use of dietary compounds or synthetic chemicals to boost the Nrf2-dependent adaptive response to counteract environmental insults has emerged to be a promising strategy for cancer prevention. Interestingly, recent emerging data has revealed the “dark” side of Nrf2. Nrf2 and its downstream genes are overexpressed in many cancer cell lines and human cancer tissues, giving cancer cells an advantage for survival and growth. Furthermore, Nrf2 is upregulated in resistant cancer cells and is thought to be responsible for acquired chemoresistance. Therefore, it may be necessary to inhibit the Nrf2 pathway during chemotherapy. This review is primarily focused on the role of Nrf2 in cancer, with emphasis on the recent findings indicating the cancer promoting function of Nrf2 and its role in acquired chemoresistance.
1. Introduction
In 1994, Moi et al. first cloned and characterized Nrf2 based on its ability to bind the NFE2/AP-1 repeat in the promoter of the beta-globin gene [1]. Like NF-E2, Nrf2 is also a member of the cap n’ collar (CNC) subfamily of transcription factors and contains a basic leucine zipper DNA binding domain (bZip) at the C-terminus. It was found to be ubiquitiously expressed in many organs and dispensable for the normal development of mice [2].
Human Nrf2 is homologous to chicken and mouse and has six highly conserved domains, Neh1–6. The Neh1 domain contains a CNC-type basic leucine zipper that is necessary for DNA binding and dimerization with other transcription factors. Additionally, a functional NLS has been identified in this domain [3]. The Neh2 domain binds the Kelch domain of Keap1, a negative regulator of Nrf2, and has seven lysine residues that are responsible for ubiquitin conjugation, which leads to proteasomal degradation of Nrf2 g[4, 5]. Neh3 is necessary for transcriptional activation by recruiting a coactivator, CHD6; however, not much is known about the specific role of CHD6 [6]. Neh4 and Neh5, rich in acidic residues, are two independent transactivation domains that act synergistically and interact with the CREB-binding protein (CBP) [7]. Lastly, the Neh6 domain is heavily concentrated with serine residues, but not much is known about the role and/or significance of the Neh6 domain.
Upon exposure of cells to oxidative stress or chemopreventive compounds, Nrf2 translocates to the nucleus, forms a heterodimer with its obligatory partner Maf, and binds to the ARE sequence to activate transcription of several different types of genes [8]. The Nrf2 downstream genes identified so far can be grouped into several categories, including (i) intracellular redox-balancing proteins: glutamate cysteine ligase (GCL), glutathione peroxidase (GPx), thioredoxin (Trx), thioredoxin reductase (TrxR), and peroxiredoxin (Prx), and heme oxygenase-1 (HMOX-1) (ii) phase II detoxifying enzymes: glutathione Stransferase (GST), NAD(P)H quinone oxidoreductase-1 (NQO1), and UDP-glucuronosyltransferase (UGT), and (iii) transporters: multidrug resistance-associated protein (MRP) [9–17]. The primary function of intracellular redox-balancing proteins is to maintain cellular glutathione and Trx levels and reduce levels of reactive oxygen species (ROS). Phase II enzymes function in two aspects: (i) metabolize xenobiotics into less toxic forms, or (ii) catalyze conjugation reactions to increase the solubility of xenobiotics, thereby, facilitating their elimination. Lastly, the main function of transporters is to control uptake and efflux of endogenous substances and xenobiotics. The majority of the downstream genes of Nrf2 contain an ARE sequence in the promoter, which was discovered and characterized by Rushmore et al. in 1991 [18]. The consensus sequence, 5`-GTGACNNNGC-3` was first identified in the 5`-flanking region of the rat GST-Ya subunit and the NQO1 gene by mutation and deletion analysis [18]. Later, Wasserman et al. further characterized the ARE sequence to a “core” sequence of 5`-RTGACnnnGCR-3` using murine GST-Ya ARE and identified 16 other genes that contained the sequence in their promoters [19]. However, in 2003 a comprehensive mutational study on the NQO1-ARE revealed that the ARE sequence did not conform to the general consensus sequence [20].
Based on the function of Nrf2 target genes, one can easily conclude that activation of Nrf2 may protect cells from various stresses imposed by toxic exposure. Indeed, the Nrf2-mediated antioxidant response is one of the major cellular defense mechanisms that facilitate cell survival under toxic insults. This notion is best demonstrated in animal models, showing that Nrf2-null mice are more sensitive than wild-type mice to the toxic and carcinogenic effects of a wide variety of xenobiotics, including benzo[a]pyrene, diesel exhaust, cigarette smoke, N-nitrosobutyl (4-hydroxybutyl) amine, pentachlorophenol, and acetaminophen [21–27].
2. Mechanism of Nrf2 activation
The activity of Nrf2 is negatively regulated by Kelch-like ECH-associated protein 1 (Keap1), which was cloned by Yamamoto and colleagues in 1999 using the N-terminal domain of Nrf2 (Neh2) as bait in a yeast two-hybrid system. Keap1 contains two major domains, a BTB domain (broad complex, tramtrack, and bric-a-brac) and a Kelch domain. The crystal structures of the Kelch domain alone or in complex with N-terminal peptides of Nrf2 have been resolved [28–31]. Despite the notion that the Kelch domain-containing proteins may bind actin filaments, mounting evidence has shown that Keap1 is a shuttling protein. For example, Keap1 interacts with the abundant nuclear protein prothymosin α (ProTα), which implies its ability to translocate from the cytoplasm to the nucleus [32]. Furthermore, leptomycin B, an inhibitor of Crm-1-dependent nuclear export, restrains Keap1 in the nucleus [21]. Furthermore, a very strong leucine-rich nuclear export signal (NES) was identified in Keap1 and the importance of the NES in regulating Nrf2 was demonstrated [32–35].
Since the cloning of Keap1, great progress has been made in understanding the mechanism of Keap1-mediated negative regulation of Nrf2. It has been proposed that Keap1 acts as a molecular switch that is able to turn the Nrf2 signaling pathway on or off according to intracellular redox conditions. Serving as a molecular switch, Keap1 possesses dual functions: it is able to (i) “sense” a disturbance in the redox homeostasis and (ii) turn the Nrf2-mediated response on or off. Recent studies have dissected how these two functions are accomplished by Keap1.
(i) Keap1 functions as a sensor
Keap1 is rich in cysteine residues (27 cysteine residues in human Keap1 and 25 cysteines in mouse Keap1), which encodes the sensor mechanism. Three key cysteine residues (C151, C273, and C288) were identified by both in vitro alkylation and in vivo site-directed mutagenesis assays [36–43]. Cysteine 151 is likely the major site that is directly alkylated by Nrf2 inducers [37–39]. The ability of the Keap1 mutant, where cysteine-151 is replaced with a serine (Keap1-C151S), to repress the activity of Nrf2, is comparable to wild-type Keap1 (Keap1-wt). However, mutation of C151 completely abolished induction of Nrf2 by many Nrf2-activators, such as sulforaphane (SF) and tert-butylhydroquinone (tBHQ) [40]. Interestingly, activation of the Nrf2 response by arsenic seems to be independent of C151 in Keap1 since the C151 mutation did not prevent Nrf2 induction [44]. A single cysteine to serine mutation was also made in C273 or C288, which rendered Keap1 unable to repress Nrf2, even though the Keap1-C273S or Keap1-C288S mutants were highly expressed and were capable of binding Nrf2 [40, 42, 43]. In summary, these data indicate that C151 in Keap1 is required for activation of the Nrf2 pathway while C273 or C288 is necessary for repressing Nrf2. More significantly, the functional importance of these three cysteine residues under physiological conditions has recently been confirmed using animal models [45]. Keap1-C151 is able to rescue the phenotype presented by Keap1-null mice, such as overexpression of Nrf2 and postnatal lethality in Keap1−/−∷TgKeap1-C151 mice. Moreover, mouse embryonic fibroblasts (MEF) derived from Keap1−/−∷TgKeap1-C151 mice showed both lower basal and inducible expression of Nrf2 [45]. It was concluded from these experiments that these three cysteine residues may be the center of the Keap1 redox-sensing mechanism. The mechanism(s) by which these cysteines function by first sensing the redox-imbalance then transducing these signals to Nrf2 remain(s) elusive.
(ii) Keap1 functions as a molecular switch
Under basal conditions, in which redox homeostasis is maintained in cells, the molecular switch of Keap1 is in an “off” position. This is achieved through constant Keap1-mediated degradation of Nrf2 by the ubiquitin-mediated proteasomal degradation system [4, 46–48]. Ubiquitin-mediated protein degradation plays an important role in controlling many cellular processes, such as cell cycle, cell growth/differentiation, and cellular response to stress. The ubiquitin-mediated degradation machinery involves many proteins and protein complexes that execute degradation of a target protein by two successive processes: (i) ubiquitin conjugation to substrates, and (ii) 26S proteasomal-mediated degradation of the polyubiquitinated substrates [49–51].
Ubiquitin is an evolutionarily conserved small protein with 76 amino-acids. The post-translational process in which ubiquitin is covalently added to lysine residues of a substrate is called ubiquitination. Ubiquitin becomes covalently attached to substrates either as a single molecule (monoubiquitination) or as a poly-ubiquitin chain (polyubiquitination). The major function of ubiquitination is to label proteins for proteasomal degradation. However, ubiquitin modification has also been shown to play a role in other processes such as DNA repair, endocytosis, and ribosome biogenesis. Covalent attachment of ubiquitin to substrates is accomplished through the coordinated action of three classes of enzymes: (i) ubiquitin activating enzyme, E1, (ii) ubiquitin conjugating enzyme, E2, and (iii) ubiquitin ligase, E3. In the initial step, ubiquitin is attached to a cysteine residue on E1 through a high energy thioester bond. Next, ubiquitin is transferred from E1 to E2 by transthiolation, and lastly, E3 mediates ubiquitin transfer from E2 to lysine residues of a substrate. The E3 ligase is responsible for substrate specificity and can function as a single protein, but a large number of them appear to be protein complexes containing multiple proteins [49–51].
Keap1 was proved to be a substrate adaptor protein for the Keap1-Cul3-Rbx1 E3 ubiquitin ligase complex, responsible for Nrf2 degradation, by four independent groups in 2004 [4, 48, 52, 53]. Under basal conditions, Keap1 brings Nrf2 into the E3 ligase complex through its two major domains: the BTB domain interacts with Cul3 and the Kelch domain binds Nrf2 [4]. Docking of Nrf2 into the E3 complex facilitates ubiquitin transfer from E2 to lysine residues of Nrf2. Consequently, ubiquitinated Nrf2 is quickly degraded by the 26S proteasome to keep the Nrf2 pathway off. A low constitutive level of the Nrf2-dependent response is maintained by low levels of ROS that are generated by physiological processes, such as the respiratory chain reaction. In summary, Nrf2 is a very unstable protein under basal conditions because Keap1 is actively targeting Nrf2 for ubiquitination and degradation. Furthermore, we have shown that the ubiquitin accepting lysine residues are within the Neh2 domain of Nrf2 and mutating these seven lysine residues to arginines renders the Nrf2 mutant to be more resistant to Keap1-dependent degradation [4]. Interestingly, the seven lysine residues are within the high-affinity ETGE and the low-affinity DLG motifs that bind the Kelch domain of Keap1, which is the proposed hinge and latch two-site binding model [54, 55].
In response to Nrf2 inducers, Nrf2 becomes very stable and the Nrf2 pathway is turned on. We observed that the half-life of endogenous Nrf2 increased from 19 min in untreated cells to 51 min in stressed MDA-MB-231 cells [56]. Keap1-mediated ubiquitination of Nrf2 was reduced significantly upon treatment with tBHQ or SF [56]. Interestingly, we have shown that in cells cotransfected with Keap1-C151, tBHQ or SF could no longer repress ubiquitination of Nrf2 and had no effect on the half-life of Nrf2 [4, 40]. These data demonstrate the importance of C151 in sensing ROS and in turning on the Nrf2 pathway. However, the mechanism by which Nrf2 inducers are able to block Keap1-mediated ubiquitination and degradation of Nrf2 is still unclear. It is likely that direct modification of C151 in Keap1 by tBHQ may cause a conformational change of the Keap1-Cul3 E3 complex, resulting in a switch from Nrf2 ubiquitination to auto-ubiquitination of Keap1 [57]. Intriguingly, different Nrf2 inducers seem to activate Nrf2 by distinct mechanisms. In support of this notion, SF was unable to elicit a switch of ubiquitination from Nrf2 to Keap1[57]. More dramatically, arsenic-induction of Nrf2 was shown to be independent of C151 in Keap1 [44]. The details and distinct mechanisms of how various Nrf2 inducers activate the Nrf2 signaling pathway remain to be deciphered.
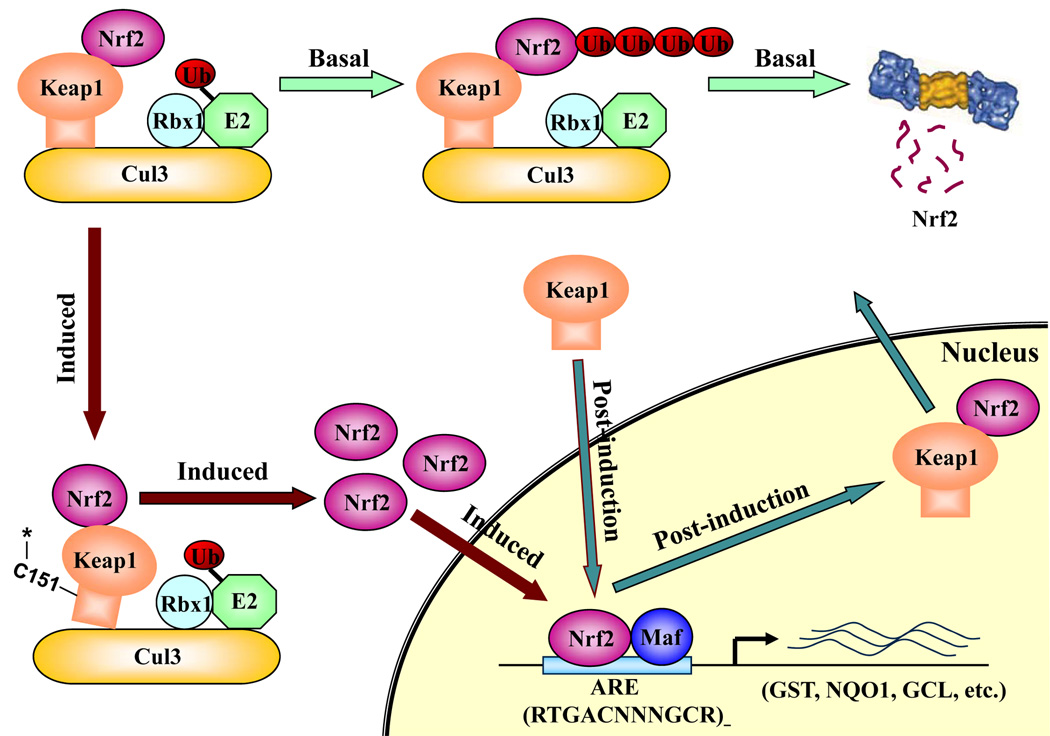
Recent results from our laboratory provide a better understanding of how the Nrf2-signaling pathway is turned off in the post-induction phase as intracellular redox levels approach homeostasis [35]. Our work was greatly facilitated by the findings that Keap1 is a shuttle protein [32–34]. Since Keap1 is a major key regulator of the Nrf2 signaling pathway, we performed our studies in cells coexpressing Keap1 and Nrf2. We concluded that the NES sequence in Keap1 is required for Nrf2 inactivation after redox homeostasis is re-obtained. In addition, we found that Keap1-mediated ubiquitination and degradation of Nrf2 occurred in the cytoplasm and that Keap1 was able to travel into the nucleus independent of Nrf2 [35]. Based on these findings, we proposed that post-induction repression of the Nrf2 activity is accomplished by Keap1 through two distinguished functions. (i) The NES in Keap1: Keap1 translocates into the nucleus, dissociates Nrf2 from ARE-binding, and then exports the Nrf2-Keap1 complex back into the cytoplasm. (ii) The substrate adaptor function of Keap1: once in the cytoplasm, Keap1 targets Nrf2 to the Cul3-containing E3 ubiquitin ligase for ubiquitination and degradation.
Based on the current knowledge, we propose the following model of how Keap1 functions in switching the Nrf2 signaling pathway on/off (Figure 1). Under basal conditions, Keap1 switches the Nrf2 signaling pathway off and maintains low basal levels of Nrf2 by constantly targeting Nrf2 for ubiquitin-mediated protein degradation. When Keap1 “senses” a disturbance in the redox balance, the cysteine residues in Keap1 are modified, resulting in a conformational change of the E3 ubiquitin ligase to a configuration not conducive for Nrf2 ubiquitination. Consequently, Nrf2 accumulates under oxidative conditions, which allows free Nrf2 to translocate to the nucleus and transcriptionally activate downstream genes by binding to the ARE sequences and switching the Nrf2 signaling pathway on. Upon recovery of the redox balance, Keap1 travels into the nucleus, where it causes dissociation of Nrf2 from the ARE sequence. Subsequently, Keap1 escorts Nrf2 out of the nucleus to the cytoplasmic Cul3-dependent E3 ubiquitin ligase machinery for degradation. Thus, a low level of Nrf2 is re-attained, turning the Nrf2 signaling pathway off.
Schematic model of Nrf2 regulation by Keap1.
Keap1 is a key regulator of the Nrf2 signaling pathway and serves as a molecular switch to turn on and off the Nrf2-mediated antioxidant response. (i) The switch is in off position: under basal conditions, Keap1, functioning as an E3 ubiquitin ligase, constantly targets Nrf2 for ubiquitination and degradation. As a consequence, there are minimal levels of Nrf2. (ii) The switch is turned on: oxidative stress or chemopreventive compounds inhibit activity of the Keap1-Cul3-Rbx1 E3 ubiquitin ligase, resulting in increased levels of Nrf2 and activation of its downstream target genes. (iii) The switch is turned off again: Upon recovery of cellular redox homeostasis, Keap1 travels into the nucleus to remove Nrf2 from the ARE. The Nrf2-Keap1 complex is then transported out of the nucleus by the NES in Keap1. In the cytosol, the Nrf2-Keap1 complex associates with the Cul3-Rbx1 core ubiquitin machinery, leading to degradation of Nrf2. For clarity, the constitutive cytoplasmic-nuclear shuttling of Nrf2, Keap1, and the complex is omitted.
Although Keap1 is the major regulator of Nrf2 activation, there is further evidence indicating multiple levels of Nrf2 regulation. For example, phosphorylation of Nrf2 by several different kinases, which include protein kinase C (PKC), extracellular-regulated kinase (ERK), Jun N-terminal kinase (JNK), and phosphatidylinositol3-kinase (PI3K), has been implicated in regulating Nrf2 activation [58]. However, the functional significance of the reported phosphorylation residues in Nrf2, using cell-based mutagenesis analysis, has not been defined yet. Additionally, coactivator CBP and P300 may provide another level of regulation. These coactivators have been shown to bind Nrf2 at the Neh4 and Neh5 domain to regulate transcription of ARE-containing genes [7, 59]. (For a more detailed review on the regulation of Nrf2, refer to [58].)
3. Nrf2 in cancer prevention
The concept of chemoprevention through the use of dietary compounds or synthetic chemicals has been rooted half a century ago when the first report demonstrated that systemic administration of small quantities of xenobiotics, such as 3-methylcholanthrene, decreased the incidence of cancer in rats that were subsequently fed large doses of carcinogenic azo dyes [60]. Work over the last 50 years has identified many compounds from plants, referred to as phytochemicals, possessing chemopreventive activities [61–69]. Interestingly, many well-studied chemopreventive compounds have been identified as Nrf2 inducers. Examples of potent Nrf2 inducers from plants include sulforaphane (cruciferous vegetables) [62], curcumin (a widely used spice) [70–72], epigallocatechin-3-gallate (EGCG) (green tea) [73, 74], resveratrol (grapes) [75, 76], caffeic acid phenethyl ester (conifer trees) [70], wasabi (Japanese horseradish) [77], cafestol and kahweol (coffee) [78, 79], cinnamonyl-based compounds (cinnamon) [80], zerumbone (ginger) [81], garlic organosulfur compounds (garlic) [82, 83], lycopene (tomato) [84], carnosol (rosemany) [85, 86], and avicins (Bentham plant) [87]. Besides phytochemicals, certain synthetic chemicals such as oltipraz (a substituted 1, 2-dithiole-3-thione) [21], 2-indol-3-yl-methylenequinuclidin-3-ols (an indole analogue) [88], and the synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid (CDDO) and its derivative 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl] imidazole (CDDO-Im) [89, 90], are also potent Nrf2 inducers. This list of Nrf2 inducing chemopreventive compounds and synthetic chemicals is continuously growing.
These chemopreventive compounds or synthetic chemicals exert their chemopreventive activity by inducing the Nrf2-dependent adaptive response, including phase II detoxifying enzymes, antioxidants, and transporters that defend cells from subsequent carcinogenic insults. Therefore, Nrf2 has been viewed as a “good” protein that protects humans from genotoxic damage caused by carcinogens. Several in vivo studies using Nrf2-null mice further verified the pivotal role of Nrf2 in cancer protection. Nrf2-null mice have reduced basal and induced levels of phase II genes such as GST, NQO1, and glutamate cysteine ligase (GCL). [91–95]. Nrf2 knockout mice display increased sensitivity to chemical toxicants and carcinogens and are refractory to the protective actions of chemopreventive compounds [21–23, 26, 96–99].
4. Nrf2 in cancer promotion
Surprisingly, new emerging data has revealed the “dark” side of Nrf2. Nrf2 protects not only normal cells from transforming into cancer cells, but also promotes the survival of cancer cells under a deleterious environment. The first evidence indicating the involvement of Nrf2 in cancer promotion came from the finding that Nrf2 and GSTP1 were upregulated during development of hepatocellular carcinoma [100]. GSTP1 is a mark for neoplastic lesions because it is absent in normal tissues but overexpressed in cancerous tissues. In the same paper, Nrf2 was shown to regulate expression of GSTP1 through an ARE in the promoter of GSTP1 [100]. Thereafter, more evidence has indicated a positive role of Nrf2 in cancer tumorigenesis and chemoresistance. For instance, many Keap1 mutations or loss of heterozygosity in the Keap1 locus have been identified in lung cancer cell lines or cancer tissues [30, 101]. Keap1 mutations or loss of heterozygosity resulted in inactivation of Keap1 or a reduced expression of Keap1, which upregulated the protein level of Nrf2 and transactivation of its downstream genes [30, 101]. In a similar study, the status of Keap1 was investigated in 65 Japanese patients with lung cancer, which identified and showed a high incidence of Keap1 somatic mutations in patients with lung adenocarcinoma [102]. In another study, a mutation of Keap1 (C23Y), that was found in breast cancer, was reported to have impaired ability in repressing Nrf2 [103]. Under a hypoxia/reoxygenation condition, which mimics a tumor microenvironment, Keap1 expression was decreased whereas Nrf2 and Prx1, were upregulated, resulting in removal of ROS and protection of cancer cells [104]. Interestingly, a recent report also indicates that Keap1 expression was reduced in lung cancer cell lines and tissues, compared to that in normal bronchial epithelial cell line [105]. However, the reduced expression was due to hypermethylation of the Keap1 promoter, an epigenetic mechanism [105]. In accordance with the previous findings, we found that Nrf2 was overexpressed at later stages of cancer in lung tissue [106]. Recently, upregulation of Nrf2 was detected in an arsenic-transformed human keratinocyte cell line, compared to its parental cell line [107]. Collectively, these results suggest that loss of function of Keap1 may result in prolonged activation of Nrf2 providing cancer cells with a growth advantage due to upregulation of Nrf2 downstream genes.
Very recently, several independent studies indicate that Nrf2 may be responsible for chemoresistance. Using genetic manipulation, we have demonstrated a strong positive correlation between Nrf2 levels and resistance of three cancer cell lines to chemotherapeutic drugs such as cisplatin, doxorubicin, and etoposide [106]. Chemical activation of Nrf2 by pretreatment with tBHQ also increased survival of neuroblastoma cells in response to the three drugs tested [106]. Consistent with our findings, the role of Nrf2 in determining efficacy of cisplatin was also demonstrated in ovarian cancer cells using siRNA knockdown of Nrf2 [108]. When the molecular mechanism of acquired resistance to tamoxifen was investigated, a MCF-7 derived tamoxifen resistance cell line was found to have an elevated expression of Nrf2 and its downstream genes, such as HMOX-1, Trx, Prx, and GCL [109]. Furthermore, knockdown of Nrf2, using Nrf2-siRNA, reversed resistance of the cells lines to tamoxifen [109]. Similarly, the Nrf2 pathway was activated in an imatinib-resistant cell line. Ascorbic acid, a reducing reagent capable of blocking Nrf2 activation, reversed imatinib sensitivity [110]. Using MEF cells from wild-type and Nrf2-null mice, Nrf2 was shown to be important in determining the sensitivity of cells in response to the GSH inhibitors L-buthionine-(S, R)-sulfoximine and doxorubicin [111].
Based on their ability to function as antioxidants and detoxifying enzymes, many Nrf2 downstream genes have been shown to contribute to the observed Nrf2-dependent chemoresistance and cancer promotion. The role of HMOX-1 in cancer promotion and drug resistance has been extensively investigated [112]. HMOX-1 is an enzyme that degrades pro-oxidant heme into ferrous iron, carbon monoxide, and biliverdin which is quickly converted into bilirubin. The end-products of HMOX-1 have antioxidant activities that are able to defend cells from oxidative stress. Therefore, activation of HMOX-1 plays a key role in the anti-inflammatory response and in cell survival. Similar to Nrf2, the protective effect of HMOX-1 in normal cells may protect us from oxidative stress-related diseases. However, it is undesirable in cancer since it provides a selective advantage for cancer cells to survive. Consistent with this notion, HMOX-1 has been found to be overexpressed in various tumor types. It is believed that overexpression of HMOX-1 facilitates cancer cell growth and survival in many aspects, such as stimulating rapid growth of cancer cells, enhancing cancer cell resistance to stress and apoptosis, promoting angiogenesis of tumors, and aiding in metastasis of tumors [112]. In support of this, overexpression of HMOX-1 in melanoma cells by viral infection or transient transfection resulted in an increase in cell proliferation, resistance to H2O2-induced oxidative stress, and increase in endothelial cell division leading to angiogenesis [113]. Similar results were seen in vivo when mice were injected with HMOX-1 overexpressing melanoma cells, compared to mice injected with melanoma cells [113]. Overexpression of HMOX-1 has also been implicated in resistance of cancer cells to conventional cancer therapies. For example, A549 is more resistant to cisplatin and EGCG induced cell death than any other lung cancer cell line. This was contributed to the high expression of Nrf2 and HMOX-1 because inhibition of HMOX-1 by siRNA or by zinc protoporphyrin, a pharmacological HMOX-1 inhibitor, rendered cancer cells to be more sensitive to cisplatin or EGCG-mediated cytotoxicity, associated with elevation of intracellular ROS [114–116]. Nitric oxide (NO) also induces the expression of HMOX-1 and has a potent anti-apoptotic function. However, when zinc protoporphyrin was utilized, apoptosis was induced and tumor growth was reduced in rat hepatoma cells in response to NO [117, 118]. In another study, auditory cells were protected by piperine, a major component of black pepper, from cisplatin-induced apoptosis. The protection was due to piperine-mediated induction of Nrf2 and HMOX-1, since blockage of induction of HMOX-1 by either antisense oligos or zinc protoporphyrin abrogated protection [119]. Additionally, in acute myeloid leukemia cells, TNFα was able to upregulate Nrf2 and HMOX-1, which makes acute myeloid leukemia cells refractory to TNFα-induced apoptosis [120]. Lastly, upregulation of HMOX-1 attenuates the cytotoxicity induced by cisplatin or photodynamic therapy [121, 122].
In addition to HMOX-1, other Nrf2-downstream genes such as Prx1, GPx, and TrxR, were also upregulated in many cancer cells or tissues and may contribute to chemoresistance [104, 123, 124]. Prxs are thiol-specific antioxidant proteins that detoxify peroxides. There have been several studies showing elevated expression of Prx1 in human cancers. For example, Prx1 was significantly higher in abnormal biopsies from thyroid lesions compared to normal tissues [125]. In another study involving 90 patients who had non-small cell lung cancer and underwent surgical resection, expression of Prx1 and Nrf2 was elevated in more than 60% of patients [124]. Additionally, a strong correlation was found between overexpression of Prx1 and recurrence/decreased survival [124]. Taken together, these data imply that Prx1 has an effect on cancer development and progression.
Another Nrf2 downstream gene is GPx, which is part of a large family of selenoproteins. There are five known isoforms that all have the same general function of detoxifying hydroperoxides to water or an alcohol using reduced glutathione [126]. The difference between the five isoforms is in substrate specificity and localization [127]. One particular GPx, GI-GPx or GPx2, is mainly expressed in the epithelium of the gastrointestinal system, but can also be found in other types of epithelial cells. It has been implicated in the control of inflammation and malignant growth [127]. An ARE sequence was identified in the promoter region of GI-Gpx and upregulation of GI-Gpx by overexpression of Nrf2 or by tBHQ, SF, and curcumin induction was demonstrated [11]. An increase in GI-GPx has been associated with carcinogenesis by supporting growth, facilitating proliferation, and inhibiting oxidant-mediated apoptosis. mRNA levels of GI-GPx are elevated in Barrett’s esophageal mucosa and in advance stages of colorectal adenomas [128]. In addition, other isoforms of GPxs have also been implicated in both cancer prevention and cancer progression, but whether or not they are regulated by Nrf2 is still uncertain.
Another selenoprotein, TrxR, is ubiquitously expressed in mammalian tissues and catalyzes the reduction of the active site disulfide of Trx [123]. Trx and TrxR are important in regulating the redox status of cells and are involved in many cellular functions including redox control of transcription factors, protection against oxidative stress, and cell growth [123]. TrxR has also been shown to be induced by SF and tBHQ both in vivo and in vitro [129–131]. Sakurai et al. showed that cadmium, along with other Nrf2 activators, including arsenite, DEM, and hydrogen peroxide, induced gene expression, and activated the activity of TrxR1, which suggests that TrxR has a protective role against cancer [12, 13]. However, it has been shown that TrxR1 is elevated in several cancer lines and in human gastrointestinal cancer tissues [132, 133]. Other studies in support of TrxR’s role in cancer progression showed that inhibition of TrxR prevents cancer cell growth in vivo and knockdown of TrxR in lung carcinoma cells reverses the tumorigenicity and invasion [134]. Several groups have shown that cisplatin induces intracellular expression of human Trx and that enhanced expression is closely associated with the development of cellular resistance to cisplatin [135]. It has also been shown that cells resistant to cisplatin have an elevated expression of TrxR and that inhibiting TrxR activity using a chemical inhibitor or siRNA increases the cellular sensitivity to cisplatin [135].
Other downstream genes of Nrf2 that are upregulated in response to oxidative stress include GSTs and GCL. There are three super-families of GSTs: microsomal (MAPEG), mitochondrial, and cytosolic [136]. GSTs are phase II conjugating enzymes that detoxify reactive electrophilic metabolites and their effectiveness relies on the level of glutathione (GSH), which is determined by GCL and GSH synthase (GSHS) [136]. Not only do GSTs detoxify electrophilic xenobiotics, but they also inactivate endogenous compounds, such as aldehydes, quinones, epoxides, and hydroperoxides [137]. The largest super-family, cytosolic GSTs, is comprised of seven classes: Alpha, Mu, Omega, Pi, Sigma, Theta, and Zeta, with a broad spectrum of substrate specificity and different expression patterns. (For a more detailed review on the activities of the various human cytosoloc, mitochondrial and microsomal GSTs, refer to [137].)
There have been several studies demonstrating increased GST expression in cancer cell lines and tumors that are multidrug resistant [136]. In an epidemiological study, patients with chronic lymphocytic leukemia had a higher expression of GST-Pi, Alpha, and Mu when compared to normal lymphocytes [138]. In addition, there was higher GST activity in chlorambucil-resistant patients [138]. It has been shown that transient transfection of GST-Pi into COS cells caused an increase in drug resistance to chlorambucil, and transfection of GST-Pi caused a human colon cancer cell line to be resistant to adriamycin, cisplatin, melphalan, and etoposide [139]. Also, inhibiting GST expression by antisense cDNA and specific inhibitors of GCL in several other cancer cell lines increased sensitivity to chemotherapeutics like doxorubicin and vincristine [140–143]. Levels of GSH and the expression of GCL, both mRNA and protein, were higher in human cancer cells that were resistant to cisplatin or doxorubicin compared to that in drug-sensitive cells [144]. In conjunction, depletion of GSH elevated the sensitivity of esophageal tumors to cisplatin, suggesting a role for GSH in drug resistance [145]. In a review discussing the role of glutathione-dependent enzymes, it listed a great deal of evidence supporting how these enzymes affect drug resistance in cancer cells: (i) GSH-dependent enzymes and enzymes involved in maintaining cellular GSH are increased in cancer cell lines and are resistant to alkylating agents or drugs that generate free radicals, (ii) drug resistant bone marrow or lung cancer cells and preneoplastic foci in rat liver have higher GSH levels, (iii) depletion of cellular GSH levels sensitizes cells to the toxic effects of a wide range of chemotherapeutics, and (iv) amplification of cellular GSH levels, both in vivo and in vitro, protects against toxic effects of cytotoxic drugs [146].
Overexpression of detoxifying enzymes is not the only culprit thought to be involved in the progression of cancer. It has also been hypothesized that transporters, like MRPs, may also play a role because they too are increased in cancer cell lines and tumor samples. MRPs transport exogenous and endogenous organic anions, including conjugated metabolites derived from detoxification by phase II conjugating enzymes, out of the cell [147]. This causes tumor cells to acquire a resistance to different drugs because increased expression of transporters leads to a decrease in drug accumulation in the cell [136]. Furthermore, Nrf2 was shown to be necessary for the constitutive and inducible expression of MRP1 in MEFs [148]. Nrf2 also enhances the expression of MRP2 by binding to its defined ARE sequence upon treatment with Nrf2 inducers [15]. It has been suggested that there is a shared coordinated regulatory mechanism between GST and MRP and that oxidative stress, some xenobiotics, and anticancer agents, like cisplatin and alkylating agents, co-induce GCL and MRP1 [136]. AREs have been identified in some isoforms of GSTs, UGTs, and MRPs, which provides a good indication that they are coordinately regulated by Nrf2 [10, 14–16] [149]. Coordinated regulation of GST-Pi and MRP1 by Nrf2 has also been confirmed by utilizing several known Nrf2-inducers, such as tBHQ, oltripaz, and SF [136].
NQO1 is another gene which protects against oxidative stress and can be induced by thiol-active agents in an Nrf2-dependent fashion. It has long been regarded as a chemoprotective enzyme that catalyzes the reduction and detoxification of highly reactive quinones [150, 151]. Recently, it has been discovered that NQO1 might be involved in the stabilization of the tumor suppressor gene p53, which supports the protective role of NQO1 against cancer [152]. However, there is mounting evidence supporting a positive role for NQO1 in cancer progression. NQO1 is overexpressed in various tumors including those of the adrenal gland, bladder, breast, colon, liver, lung, ovary, and thyroid [106, 150, 153–155]. In another study, protein expression and enzymatic activity of NQO1 was increased in colon and gastric carcinoma cell lines and in colorectal tumor samples compared to peripheral normal samples [156]. In addition, there was higher expression of NQO1 in metastatic tumors than tumors that were not. Together, these findings suggest a correlation between high levels of NQO1 and tumorigenesis as well as malignant progression of cancer [156]. Overexpression of NQO1 may also play a role in cancer drug resistance. In a study conducted in our laboratory, knockdown of Nrf2 using siRNA in A549 lung cancer cells, showed a decrease in NQO1 mRNA expression along with a dramatic decrease in NQO1 enzyme activity, which sensitized the cells to the toxic effects of three chemotherapeutic drugs, cisplatin, doxorubicin, and etoposide [106]. Collectively, these data confirm that Nrf2 regulates the expression of NQO1 and that NQO1 has potential dual roles in cancer. Lastly, it is thought that an increase in NQO1 is accompanied by an increase in other antioxidant enzymes, such as HMOX-1 and GST, providing tumors with increased protection against cytotoxic agents allowing for rapid cancer progression [157].
5. Conclusion
Since the discovery of Nrf2, there has been mounting evidence exhibiting the positive role of Nrf2 in cancer protection and how it is an essential transcription factor in protecting humans from oxidative stress-related diseases. The main focus of research thus far has been to find activators of Nrf2 for chemoprevention, but recent findings suggest that there is a “dark” side of Nrf2. In vitro studies show that overexpression of Nrf2 can lead to the increased expression of several intracellular redox-balancing proteins, phase II detoxifying enzymes, and transporters, which can provide cancer cells with a growth advantage and cause resistance to chemotherapies. It is important to verify the cancer promoting role of Nrf2 observed in cancer cell lines and tumor biopsies, using in vivo animal models.
Finding the “dark” side of Nrf2 provides an opportunity for therapeutic intervention against chemoresistance. An inhibitor of Nrf2 can be used in conjunction with chemotherapy to sensitize cancer cells to chemical treatment. Since Nrf2 is a transcription factor that regulates the expression of several downstream genes that protect cancer cells from apoptosis, it would be a more efficient drug target than the individual downstream gene, such as HMOX-1 or TrxR.
Acknowledgments
This study was supported by the NIH grant ES015010 and American Cancer Society grant RSG-07-154-01-CNE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 4.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nioi P, Nguyen T, Sherratt PJ, Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 9.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 10.Moinova HR, Mulcahy RT. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem Biophys Res Commun. 1999;261:661–668. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- 11.Banning A, Deubel S, Kluth D, Zhou Z, Brigelius-Flohe R. The GI-GPx gene is a target for Nrf2. Mol Cell Biol. 2005;25:4914–4923. doi: 10.1128/MCB.25.12.4914-4923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YC, Masutani H, Yamaguchi Y, Itoh K, Yamamoto M, Yodoi J. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J Biol Chem. 2001;276:18399–18406. doi: 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 13.Sakurai A, Nishimoto M, Himeno S, Imura N, Tsujimoto M, Kunimoto M, et al. Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: role of NF-E2-related factor-2. J Cell Physiol. 2005;203:529–537. doi: 10.1002/jcp.20246. [DOI] [PubMed] [Google Scholar]
- 14.Yueh MF, Tukey RH. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J Biol Chem. 2007;282:8749–8758. doi: 10.1074/jbc.M610790200. [DOI] [PubMed] [Google Scholar]
- 15.Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos. 2005;33:956–962. doi: 10.1124/dmd.105.003798. [DOI] [PubMed] [Google Scholar]
- 17.Ishii T, Yanagawa T. Stress-induced peroxiredoxins. Subcell Biochem. 2007;44:375–384. doi: 10.1007/978-1-4020-6051-9_18. [DOI] [PubMed] [Google Scholar]
- 18.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 19.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aoki Y, Sato H, Nishimura N, Takahashi S, Itoh K, Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 23.Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64:6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 24.Umemura T, Kuroiwa Y, Kitamura Y, Ishii Y, Kanki K, Kodama Y, et al. A crucial role of Nrf2 in in vivo defense against oxidative damage by an environmental pollutant, pentachlorophenol. Toxicol Sci. 2006;90:111–119. doi: 10.1093/toxsci/kfj076. [DOI] [PubMed] [Google Scholar]
- 25.Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O'Connor T, et al. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 26.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iizuka T, Ishii Y, Itoh K, Kiwamoto T, Kimura T, Matsuno Y, et al. Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells. 2005;10:1113–1125. doi: 10.1111/j.1365-2443.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Zhang DD, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 29.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. Embo J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Padmanabhan B, Scharlock M, Tong KI, Nakamura Y, Kang MI, Kobayashi A, et al. Purification, crystallization and preliminary X-ray diffraction analysis of the Kelch-like motif region of mouse Keap1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:153–155. doi: 10.1107/S1744309104032506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, et al. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol Cell Biol. 2005;25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol Cell Biol. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem. 2005;280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- 35.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 37.Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eggler AL, Luo Y, van Breemen RB, Mesecar AD. Identification of the Highly Reactive Cysteine 151 in the Chemopreventive Agent-Sensor Keap1 Protein is Method-Dependent. Chem Res Toxicol. 2007 doi: 10.1021/tx700217c. [DOI] [PubMed] [Google Scholar]
- 39.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang XJ, Sun Z, Chen W, Li Y, Villeneuve NF, Zhang DD. Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction. Toxicol Appl Pharmacol. 2008;230:383–389. doi: 10.1016/j.taap.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart D, Killeen E, Naquin R, Alam S, Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J Biol Chem. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 47.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 50.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 51.Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 52.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 56.Du Y, Villeneuve NF, Wang XJ, Sun Z, Chen W, Li J, et al. Oridonin confers protection against arsenic-induced toxicity through activation of the Nrf2-mediated defensive response. Environ Health Perspect. 2008 doi: 10.1289/ehp.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 58.Jeong WS, Jun M, Kong AN. Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal. 2006;8:99–106. doi: 10.1089/ars.2006.8.99. [DOI] [PubMed] [Google Scholar]
- 59.Zhu M, Fahl WE. Functional characterization of transcription regulators that interact with the electrophile response element. Biochem Biophys Res Commun. 2001;289:212–219. doi: 10.1006/bbrc.2001.5944. [DOI] [PubMed] [Google Scholar]
- 60.Richardson HL, Cunningham L. The inhibitory action of methylcholanthrene on rats fed the azo dye 3'-methyl-4-dimethylaminobenzene. Cancer Res. 1951;11:274. [Google Scholar]
- 61.Wolf CR. Chemoprevention: increased potential to bear fruit. Proc Natl Acad Sci U S A. 2001;98:2941–2943. doi: 10.1073/pnas.071042698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kensler TW, Curphey TJ, Maxiutenko Y, Roebuck BD. Chemoprotection by organosulfur inducers of phase 2 enzymes: dithiolethiones and dithiins. Drug Metabol Drug Interact. 2000;17:3–22. doi: 10.1515/dmdi.2000.17.1-4.3. [DOI] [PubMed] [Google Scholar]
- 63.Shapiro TA, Fahey JW, Wade KL, Stephenson KK, Talalay P. Chemoprotective glucosinolates and isothiocyanates of broccoli sprouts: metabolism and excretion in humans. Cancer Epidemiol Biomarkers Prev. 2001;10:501–508. [PubMed] [Google Scholar]
- 64.Talalay P, Fahey JW. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J Nutr. 2001;131:3027S–3033S. doi: 10.1093/jn/131.11.3027S. [DOI] [PubMed] [Google Scholar]
- 65.Kelloff GJ. Perspectives on cancer chemoprevention research and drug development. Adv Cancer Res. 2000;78:199–334. doi: 10.1016/s0065-230x(08)61026-x. [DOI] [PubMed] [Google Scholar]
- 66.Sporn MB, Suh N. Chemoprevention of cancer. Carcinogenesis. 2000;21:525–530. doi: 10.1093/carcin/21.3.525. [DOI] [PubMed] [Google Scholar]
- 67.Yang CS, Landau JM, Huang MT, Newmark HL. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu Rev Nutr. 2001;21:381–406. doi: 10.1146/annurev.nutr.21.1.381. [DOI] [PubMed] [Google Scholar]
- 68.Kelloff GJ, Crowell JA, Steele VE, Lubet RA, Malone WA, Boone CW, et al. Progress in cancer chemoprevention: development of diet-derived chemopreventive agents. J Nutr. 2000;130:467S–471S. doi: 10.1093/jn/130.2.467S. [DOI] [PubMed] [Google Scholar]
- 69.Kelly VP, Ellis EM, Manson MM, Chanas SA, Moffat GJ, McLeod R, et al. Chemoprevention of aflatoxin B1 hepatocarcinogenesis by coumarin, a natural benzopyrone that is a potent inducer of aflatoxin B1-aldehyde reductase, the glutathione S-transferase A5 and P1 subunits, and NAD(P)H:quinone oxidoreductase in rat liver. Cancer Res. 2000;60:957–969. [PubMed] [Google Scholar]
- 70.Balogun E, Hoque M, Gong P, Killeen E, Green CJ, Foresti R, et al. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem J. 2003;371:887–895. doi: 10.1042/BJ20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pae HO, Choi BM, Oh GS, Lee MS, Ryu DG, Rhew HY, et al. Roles of heme oxygenase-1 in the antiproliferative and antiapoptotic effects of nitric oxide on Jurkat T cells. Mol Pharmacol. 2004;66:122–128. doi: 10.1124/mol.66.1.122. [DOI] [PubMed] [Google Scholar]
- 72.Garg R, Gupta S, Maru GB. Dietary curcumin modulates transcriptional regulators of phase I and phase II enzymes in benzo[a]pyrene-treated mice: mechanism of its anti-initiating action. Carcinogenesis. 2008;29:1022–1032. doi: 10.1093/carcin/bgn064. [DOI] [PubMed] [Google Scholar]
- 73.Na HK, Surh YJ. Modulation of Nrf2-mediated antioxidant and detoxifying enzyme induction by the green tea polyphenol EGCG. Food Chem Toxicol. 2008;46:1271–1278. doi: 10.1016/j.fct.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 74.Shen G, Xu C, Hu R, Jain MR, Nair S, Lin W, et al. Comparison of (−)-Epigallocatechin-3-Gallate Elicited Liver and Small Intestine Gene Expression Profiles Between C57BL/6J Mice and C57BL/6J/Nrf2 (−/−) Mice. Pharm Res. 2005;22:1805–1820. doi: 10.1007/s11095-005-7546-8. [DOI] [PubMed] [Google Scholar]
- 75.Kode A, Rajendrasozhan S, Caito S, Yang SR, Megson IL, Rahman I. Resveratrol induces glutathione synthesis by activation of Nrf2 and protects against cigarette smoke-mediated oxidative stress in human lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2008;294:L478–L488. doi: 10.1152/ajplung.00361.2007. [DOI] [PubMed] [Google Scholar]
- 76.Chen CY, Jang JH, Li MH, Surh YJ. Resveratrol upregulates heme oxygenase-1 expression via activation of NF-E2-related factor 2 in PC12 cells. Biochem Biophys Res Commun. 2005;331:993–1000. doi: 10.1016/j.bbrc.2005.03.237. [DOI] [PubMed] [Google Scholar]
- 77.Morimitsu Y, Nakagawa Y, Hayashi K, Fujii H, Kumagai T, Nakamura Y, et al. A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. J Biol Chem. 2002;277:3456–3463. doi: 10.1074/jbc.M110244200. [DOI] [PubMed] [Google Scholar]
- 78.Higgins LG, Cavin C, Itoh K, Yamamoto M, Hayes JD. Induction of cancer chemopreventive enzymes by coffee is mediated by transcription factor Nrf2. Evidence that the coffee-specific diterpenes cafestol and kahweol confer protection against acrolein. Toxicol Appl Pharmacol. 2008;226:328–337. doi: 10.1016/j.taap.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 79.Cavin C, Holzhaeuser D, Scharf G, Constable A, Huber WW, Schilter B. Cafestol and kahweol, two coffee specific diterpenes with anticarcinogenic activity. Food Chem Toxicol. 2002;40:1155–1163. doi: 10.1016/s0278-6915(02)00029-7. [DOI] [PubMed] [Google Scholar]
- 80.Liao BC, Hsieh CW, Liu YC, Tzeng TT, Sun YW, Wung BS. Cinnamaldehyde inhibits the tumor necrosis factor-alpha-induced expression of cell adhesion molecules in endothelial cells by suppressing NF-kappaB activation: effects upon IkappaB and Nrf2. Toxicol Appl Pharmacol. 2008;229:161–171. doi: 10.1016/j.taap.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 81.Nakamura Y, Yoshida C, Murakami A, Ohigashi H, Osawa T, Uchida K. Zerumbone, a tropical ginger sesquiterpene, activates phase II drug metabolizing enzymes. FEBS Lett. 2004;572:245–250. doi: 10.1016/j.febslet.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 82.Gong P, Hu B, Cederbaum AI. Diallyl sulfide induces heme oxygenase-1 through MAPK pathway. Arch Biochem Biophys. 2004;432:252–260. doi: 10.1016/j.abb.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 83.Chen C, Pung D, Leong V, Hebbar V, Shen G, Nair S, et al. Induction of detoxifying enzymes by garlic organosulfur compounds through transcription factor Nrf2: effect of chemical structure and stress signals. Free Radic Biol Med. 2004;37:1578–1590. doi: 10.1016/j.freeradbiomed.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 84.Ben-Dor A, Steiner M, Gheber L, Danilenko M, Dubi N, Linnewiel K, et al. Carotenoids activate the antioxidant response element transcription system. Mol Cancer Ther. 2005;4:177–186. [PubMed] [Google Scholar]
- 85.Satoh T, Kosaka K, Itoh K, Kobayashi A, Yamamoto M, Shimojo Y, et al. Carnosic acid, a catechol-type electrophilic compound, protects neurons both in vitro and in vivo through activation of the Keap1/Nrf2 pathway via Salkylation of targeted cysteines on Keap1. J Neurochem. 2008;104:1116–1131. doi: 10.1111/j.1471-4159.2007.05039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin D, Rojo AI, Salinas M, Diaz R, Gallardo G, Alam J, et al. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J Biol Chem. 2004;279:8919–8929. doi: 10.1074/jbc.M309660200. [DOI] [PubMed] [Google Scholar]
- 87.Haridas V, Kim SO, Nishimura G, Hausladen A, Stamler JS, Gutterman JU. Avicinylation (thioesterification): a protein modification that can regulate the response to oxidative and nitrosative stress. Proc Natl Acad Sci U S A. 2005;102:10088–10093. doi: 10.1073/pnas.0504430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sekhar KR, Crooks PA, Sonar VN, Friedman DB, Chan JY, Meredith MJ, et al. NADPH oxidase activity is essential for Keap1/Nrf2-mediated induction of GCLC in response to 2-indol-3-yl-methylenequinuclidin-3-ols. Cancer Res. 2003;63:5636–5645. [PubMed] [Google Scholar]
- 89.Liby K, Hock T, Yore MM, Suh N, Place AE, Risingsong R, et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 90.Dinkova-Kostova AT, Liby KT, Stephenson KK, Holtzclaw WD, Gao X, Suh N, et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci U S A. 2005;102:4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, et al. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 92.Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, et al. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem Soc Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 93.Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, Elcombe CR, et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan JY, Kwong M. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim Biophys Acta. 2000;1517:19–26. doi: 10.1016/s0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 95.Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol Med. 2001;7:135–145. [PMC free article] [PubMed] [Google Scholar]
- 96.Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, et al. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 97.Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci U S A. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor NRF2 protects against pulmonary fibrosis. Faseb J. 2004;18:1258–1260. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- 100.Ikeda H, Nishi S, Sakai M. Transcription factor Nrf2/MafK regulates rat placental glutathione S-transferase gene during hepatocarcinogenesis. Biochem J. 2004;380:515–521. doi: 10.1042/BJ20031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 103.Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun. 2007;362:816–821. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 104.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 105.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 106.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pi J, Diwan BA, Sun Y, Liu J, Qu W, He Y, et al. Arsenic-induced malignant transformation of human keratinocytes: Involvement of Nrf2. Free Radic Biol Med. 2008 doi: 10.1016/j.freeradbiomed.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cho JM, Manandhar S, Lee HR, Park HM, Kwak MK. Role of the Nrf2-antioxidant system in cytotoxicity mediated by anticancer cisplatin: Implication to cancer cell resistance. Cancer Lett. 2008;260:96–108. doi: 10.1016/j.canlet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 109.Kim SK, Yang JW, Kim MR, Roh SH, Kim HG, Lee KY, et al. Increased expression of Nrf2/ARE-dependent anti-oxidant proteins in tamoxifen-resistant breast cancer cells. Free Radic Biol Med. 2008;45:537–546. doi: 10.1016/j.freeradbiomed.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 110.Tarumoto T, Nagai T, Ohmine K, Miyoshi T, Nakamura M, Kondo T, et al. Ascorbic acid restores sensitivity to imatinib via suppression of Nrf2-dependent gene expression in the imatinib-resistant cell line. Exp Hematol. 2004;32:375–381. doi: 10.1016/j.exphem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 111.Lee HR, Cho JM, Shin DH, Yong CS, Choi HG, Wakabayashi N, et al. Adaptive response to GSH depletion and resistance to L: -buthionine-(S,R)-sulfoximine: involvement of Nrf2 activation. Mol Cell Biochem. 2008 doi: 10.1007/s11010-008-9853-y. [DOI] [PubMed] [Google Scholar]
- 112.Jozkowicz A, Was H, Dulak J. Heme oxygenase-1 in tumors: is it a false friend? Antioxid Redox Signal. 2007;9:2099–2117. doi: 10.1089/ars.2007.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Was H, Cichon T, Smolarczyk R, Rudnicka D, Stopa M, Chevalier C, et al. Overexpression of heme oxygenase-1 in murine melanoma: increased proliferation and viability of tumor cells, decreased survival of mice. Am J Pathol. 2006;169:2181–2198. doi: 10.2353/ajpath.2006.051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim HR, Kim S, Kim EJ, Park JH, Yang SH, Jeong ET, et al. Suppression of Nrf2-driven heme oxygenase-1 enhances the chemosensitivity of lung cancer A549 cells toward cisplatin. Lung Cancer. 2008;60:47–56. doi: 10.1016/j.lungcan.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 115.Loboda A, Was H, Jozkowicz A, Dulak J. Janus face of Nrf2-HO-1 axis in cancer-Friend in chemoprevention, foe in anticancer therapy. Lung Cancer. 2008;60:1–3. doi: 10.1016/j.lungcan.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 116.Kweon MH, Adhami VM, Lee JS, Mukhtar H. Constitutive overexpression of Nrf2-dependent heme oxygenase-1 in A549 cells contributes to resistance to apoptosis induced by epigallocatechin 3-gallate. J Biol Chem. 2006;281:33761–33772. doi: 10.1074/jbc.M604748200. [DOI] [PubMed] [Google Scholar]
- 117.Tanaka S, Akaike T, Fang J, Beppu T, Ogawa M, Tamura F, et al. Antiapoptotic effect of haem oxygenase-1 induced by nitric oxide in experimental solid tumour. Br J Cancer. 2003;88:902–909. doi: 10.1038/sj.bjc.6600830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Doi K, Akaike T, Fujii S, Tanaka S, Ikebe N, Beppu T, et al. Induction of haem oxygenase-1 nitric oxide and ischaemia in experimental solid tumours and implications for tumour growth. Br J Cancer. 1999;80:1945–1954. doi: 10.1038/sj.bjc.6690624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choi BM, Kim SM, Park TK, Li G, Hong SJ, Park R, et al. Piperine protects cisplatin-induced apoptosis via heme oxygenase-1 induction in auditory cells. J Nutr Biochem. 2007;18:615–622. doi: 10.1016/j.jnutbio.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 120.Rushworth SA, MacEwan DJ. HO-1 underlies resistance of AML cells to TNF-induced apoptosis. Blood. 2008;111:3793–3801. doi: 10.1182/blood-2007-07-104042. [DOI] [PubMed] [Google Scholar]
- 121.Nowis D, Legat M, Grzela T, Niderla J, Wilczek E, Wilczynski GM, et al. Heme oxygenase-1 protects tumor cells against photodynamic therapy-mediated cytotoxicity. Oncogene. 2006;25:3365–3374. doi: 10.1038/sj.onc.1209378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim HJ, So HS, Lee JH, Lee JH, Park C, Park SY, et al. Heme oxygenase-1 attenuates the cisplatin-induced apoptosis of auditory cells via down-regulation of reactive oxygen species generation. Free Radic Biol Med. 2006;40:1810–1819. doi: 10.1016/j.freeradbiomed.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 123.Brigelius-Flohe R. Selenium compounds and selenoproteins in cancer. Chem Biodivers. 2008;5:389–395. doi: 10.1002/cbdv.200890039. [DOI] [PubMed] [Google Scholar]
- 124.Kim JH, Bogner PN, Ramnath N, Park Y, Yu J, Park YM. Elevated peroxiredoxin 1, but not NF-E2-related factor 2, is an independent prognostic factor for disease recurrence and reduced survival in stage I non-small cell lung cancer. Clin Cancer Res. 2007;13:3875–3882. doi: 10.1158/1078-0432.CCR-06-2893. [DOI] [PubMed] [Google Scholar]
- 125.Yanagawa T, Ishikawa T, Ishii T, Tabuchi K, Iwasa S, Bannai S, et al. Peroxiredoxin I expression in human thyroid tumors. Cancer Lett. 1999;145:127–132. doi: 10.1016/s0304-3835(99)00243-8. [DOI] [PubMed] [Google Scholar]
- 126.Margis R, Dunand C, Teixeira FK, Margis-Pinheiro M. Glutathione peroxidase family - an evolutionary overview. Febs J. 2008;275:3959–3970. doi: 10.1111/j.1742-4658.2008.06542.x. [DOI] [PubMed] [Google Scholar]
- 127.Brigelius-Flohe R. Glutathione peroxidases and redox-regulated transcription factors. Biol Chem. 2006;387:1329–1335. doi: 10.1515/BC.2006.166. [DOI] [PubMed] [Google Scholar]
- 128.Chu FF, Esworthy RS, Doroshow JH. Role of Se-dependent glutathione peroxidases in gastrointestinal inflammation and cancer. Free Radic Biol Med. 2004;36:1481–1495. doi: 10.1016/j.freeradbiomed.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 129.Li J, Johnson JA. Time-dependent changes in ARE-driven gene expression by use of a noise-filtering process for microarray data. Physiol Genomics. 2002;9:137–144. doi: 10.1152/physiolgenomics.00003.2002. [DOI] [PubMed] [Google Scholar]
- 130.Hintze KJ, Wald KA, Zeng H, Jeffery EH, Finley JW. Thioredoxin reductase in human hepatoma cells is transcriptionally regulated by sulforaphane and other electrophiles via an antioxidant response element. J Nutr. 2003;133:2721–2727. doi: 10.1093/jn/133.9.2721. [DOI] [PubMed] [Google Scholar]
- 131.Campbell L, Howie F, Arthur JR, Nicol F, Beckett G. Selenium and sulforaphane modify the expression of selenoenzymes in the human endothelial cell line EAhy926 and protect cells from oxidative damage. Nutrition. 2007;23:138–144. doi: 10.1016/j.nut.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 132.Arner ES, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol. 2006;16:420–426. doi: 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 133.Mork H, Lex B, Scheurlen M, Dreher I, Schutze N, Kohrle J, et al. Expression pattern of gastrointestinal selenoproteins--targets for selenium supplementation. Nutr Cancer. 1998;32:64–70. doi: 10.1080/01635589809514720. [DOI] [PubMed] [Google Scholar]
- 134.Yoo MH, Xu XM, Carlson BA, Gladyshev VN, Hatfield DL. Thioredoxin reductase 1 deficiency reverses tumor phenotype and tumorigenicity of lung carcinoma cells. J Biol Chem. 2006;281:13005–13008. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- 135.Sasada T, Nakamura H, Ueda S, Sato N, Kitaoka Y, Gon Y, et al. Possible involvement of thioredoxin reductase as well as thioredoxin in cellular sensitivity to cis-diamminedichloroplatinum (II) Free Radic Biol Med. 1999;27:504–514. doi: 10.1016/s0891-5849(99)00101-x. [DOI] [PubMed] [Google Scholar]
- 136.Meijerman I, Beijnen JH, Schellens JH. Combined action and regulation of phase II enzymes and multidrug resistance proteins in multidrug resistance in cancer. Cancer Treat Rev. 2008 doi: 10.1016/j.ctrv.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 137.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 138.Schisselbauer JC, Silber R, Papadopoulos E, Abrams K, LaCreta FP, Tew KD. Characterization of glutathione S-transferase expression in lymphocytes from chronic lymphocytic leukemia patients. Cancer Res. 1990;50:3562–3568. [PubMed] [Google Scholar]
- 139.Ban N, Takahashi Y, Takayama T, Kura T, Katahira T, Sakamaki S, et al. Transfection of glutathione S-transferase (GST)-pi antisense complementary DNA increases the sensitivity of a colon cancer cell line to adriamycin, cisplatin, melphalan, and etoposide. Cancer Res. 1996;56:3577–3582. [PubMed] [Google Scholar]
- 140.Kisara S, Furusawa S, Murata R, Ogata M, Hikichi N, Takayanagi Y, et al. Combined effects of buthionine sulfoximine and cepharanthine on cytotoxic activity of doxorubicin to multidrug-resistant cells. Oncol Res. 1995;7:191–200. [PubMed] [Google Scholar]
- 141.Vanhoefer U, Cao S, Minderman H, Toth K, Skenderis BS, 2nd, Slovak ML, et al. d,l-buthionine-(S,R)-sulfoximine potentiates in vivo the therapeutic efficacy of doxorubicin against multidrug resistance protein-expressing tumors. Clin Cancer Res. 1996;2:1961–1968. [PubMed] [Google Scholar]
- 142.Chuman Y, Chen ZS, Seto K, Sumizawa T, Furukawa T, Tani A, et al. Reversal of MRP-mediated vincristine resistance in KB cells by buthionine sulfoximine in combination with PAK-104P. Cancer Lett. 1998;129:69–76. doi: 10.1016/s0304-3835(98)00083-4. [DOI] [PubMed] [Google Scholar]
- 143.Akan I, Akan S, Akca H, Savas B, Ozben T. Multidrug resistance-associated protein 1 (MRP1) mediated vincristine resistance: effects of N-acetylcysteine and Buthionine sulfoximine. Cancer Cell Int. 2005;5:22. doi: 10.1186/1475-2867-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Iida T, Mori E, Mori K, Goto S, Urata Y, Oka M, et al. Co-expression of gamma-glutamylcysteine synthetase sub-units in response to cisplatin and doxorubicin in human cancer cells. Int J Cancer. 1999;82:405–411. doi: 10.1002/(sici)1097-0215(19990730)82:3<405::aid-ijc14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 145.Kaur T, Khanduja KL, Gupta R, Gupta NM, Vaiphei K. Changes in antioxidant defense status in response to cisplatin and 5-FU in esophageal carcinoma. Dis Esophagus. 2008;21:103–107. doi: 10.1111/j.1442-2050.2007.00742.x. [DOI] [PubMed] [Google Scholar]
- 146.Black SM, Wolf CR. The role of glutathione-dependent enzymes in drug resistance. Pharmacol Ther. 1991;51:139–154. doi: 10.1016/0163-7258(91)90044-m. [DOI] [PubMed] [Google Scholar]
- 147.Zaman GJ, Lankelma J, van Tellingen O, Beijnen J, Dekker H, Paulusma C, et al. Role of glutathione in the export of compounds from cells by the multidrug-resistance-associated protein. Proc Natl Acad Sci U S A. 1995;92:7690–7694. doi: 10.1073/pnas.92.17.7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem Biophys Res Commun. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 149.Aleksunes LM, Slitt AL, Maher JM, Augustine LM, Goedken MJ, Chan JY, et al. Induction of Mrp3 and Mrp4 transporters during acetaminophen hepatotoxicity is dependent on Nrf2. Toxicol Appl Pharmacol. 2008;226:74–83. doi: 10.1016/j.taap.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schlager JJ, Powis G. Cytosolic NAD(P)H:(quinone-acceptor)oxidoreductase in human normal and tumor tissue: effects of cigarette smoking and alcohol. Int J Cancer. 1990;45:403–409. doi: 10.1002/ijc.2910450304. [DOI] [PubMed] [Google Scholar]
- 151.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–14975. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc Natl Acad Sci U S A. 2001;98:1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Malkinson AM, Siegel D, Forrest GL, Gazdar AF, Oie HK, Chan DC, et al. Elevated DT-diaphorase activity and messenger RNA content in human non-small cell lung carcinoma: relationship to the response of lung tumor xenografts to mitomycin Cl. Cancer Res. 1992;52:4752–4757. [PubMed] [Google Scholar]
- 154.Siegel D, Ross D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic Biol Med. 2000;29:246–253. doi: 10.1016/s0891-5849(00)00310-5. [DOI] [PubMed] [Google Scholar]
- 155.Basu S, Brown JE, Flannigan GM, Gill JH, Loadman PM, Martin SW, et al. Immunohistochemical analysis of NAD(P)H:quinone oxidoreductase and NADPH cytochrome P450 reductase in human superficial bladder tumours: relationship between tumour enzymology and clinical outcome following intravesical mitomycin C therapy. Int J Cancer. 2004;109:703–709. doi: 10.1002/ijc.20005. [DOI] [PubMed] [Google Scholar]
- 156.Mikami K, Naito M, Ishiguro T, Yano H, Tomida A, Yamada T, et al. Immunological quantitation of DT-diaphorase in carcinoma cell lines and clinical colon cancers: advanced tumors express greater levels of DT-diaphorase. Jpn J Cancer Res. 1998;89:910–915. doi: 10.1111/j.1349-7006.1998.tb00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nioi P, Hayes JD. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat Res. 2004;555:149–171. doi: 10.1016/j.mrfmmm.2004.05.023. [DOI] [PubMed] [Google Scholar]