Abstract
Poly(ADP-ribose) polymerase (PARP) transfers ADP ribose groups from NAD+ to nuclear proteins after activation by DNA strand breaks. PARP overactivation by massive DNA damage causes cell death via NAD+ and ATP depletion. Heretofore, PARP has been thought to be inactive under basal physiologic conditions. We now report high basal levels of PARP activity and DNA strand breaks in discrete neuronal populations of the brain, in ventricular ependymal and subependymal cells and in peripheral tissues. In some peripheral tissues, such as skeletal muscle, spleen, heart, and kidney, PARP activity is reduced only partially in mice with PARP-1 gene deletion (PARP-1−/−), implicating activity of alternative forms of PARP. Glutamate neurotransmission involving N-methyl-d-aspartate (NMDA) receptors and neuronal nitric oxide synthase (nNOS) activity in part mediates neuronal DNA strand breaks and PARP activity, which are diminished by NMDA antagonists and NOS inhibitors and also diminished in mice with targeted deletion of nNOS gene (nNOS−/−). An increase in NAD+ levels after treatment with NMDA antagonists or NOS inhibitors, as well as in nNOS−/− mice, indicates that basal glutamate-PARP activity regulates neuronal energy dynamics.
Poly(ADP-ribose) polymerase-1 (PARP-1; EC 2.4.4.30) participates in DNA repair as DNA damage activates PARP-1 to catalyze extensive polymerization of ADP-ribose from its substrate NAD+ to nuclear proteins, most notably PARP-1 itself (1). Through PARP activation, massive DNA damage substantially reduces NAD+ levels and kills cells by critically depleting ATP (2). This cell death mechanism is important in septic shock (3), retinal ischemia (4), and head trauma (5, 6). Streptozotocin damage to pancreatic β cells, a model of insulin-dependent diabetes, is abolished in PARP-1−/− mice (7–9), and ischemic myocardial damage is substantially reduced in PARP-1−/− hearts and by PARP inhibitors in wild-type (WT) hearts (10–13).
Poly(ADP-ribosyl)ation is implicated in glutamate neurotoxicity and vascular stroke. Glutamate toxicity involving N-methyl-d-aspartate receptors (NMDA-R) is mediated by activation of neuronal nitric oxide synthase (nNOS; ref. 1). NO stimulates neuronal PARP (14), and PARP inhibitors block cortical neuron NMDA toxicity (14) and cerebellar granule cell glutamate toxicity (15). NMDA toxicity is abolished in PARP-1−/− cortical cultures (16). Neural damage after middle cerebral artery occlusion elicits poly(ADP-ribose) (PAR) synthesis (16–18), which is attenuated in nNOS−/− mice after cerebral ischemia/reperfusion (I/R; ref. 19). The PARP inhibitor 3,4-dihydro-5-[4–1(1-piperidinyl) butoxy]-1 (2H)-isoquinolinone (DPQ) inhibits ischemia-elicited PAR synthesis and markedly reduces infarct volume in rodent middle cerebral artery occlusion (20–22), and PARP-1−/− mice show 65% (17) to 80% (16) reduction in infarct volume after middle cerebral artery occlusion.
DNA damage and poly(ADP-ribosyl)ation have classically been considered negligible under basal conditions. We now report substantial basal DNA damage and PARP activation in specific neuronal regions of the brain and in peripheral tissues. Basal in vivo neuronal PARP activity and DNA damage are diminished, whereas basal NAD+ levels are elevated after treatment with NMDA-R antagonists, free radical scavengers, and nNOS inhibitors, as well as in nNOS−/− mice.
Materials and Methods
We employed, at 2 weeks, primary rat cerebral cortical neurons (16), cerebellar granule cells (23), and cerebral cortical astrocytes (24). NAD+ assay (25), PARP assay (26), unilateral cortical ischemia (27), in situ end labeling (ISEL), and PAR in situ (PARIS; ref. 7) were performed in 21-day-old male Sprague–Dawley rats. PARIS controls used sections dissected from rodent heads decapitated directly into liquid nitrogen. PARIS and ISEL signals were visualized by autoradiography and quantified with scintillation counting as described (7) or by electronic autoradiography with a Packard Instant Imager with which sections were manually outlined to ensure comparability in area and anatomy between compared conditions. At least five sections from at least five different animals in each group were analyzed. PARP protein and PAR were immunoprecipitated from NP-40-lysed tissue sections with monoclonal anti-PARP (1:100; Biomol, Plymouth Meeting, PA) or polyclonal anti-PAR (1:50; Trevigen, Gaithersburg, MD), followed with protein G/Sepharose (1:40). Controls were with protein G/Sepharose alone. We conducted immunohistochemical staining for PAR (13) and immunohistochemical staining for PARP with polyclonal anti-PARP (1:2,000, Biomol). For propidium iodide staining of DNA, tissue was incubated with 5 mg/ml propidium iodide in PBS.
Results
Primary Cultures of Neurons but Not Astrocytes Display DNA Strand Breaks and Poly(ADP-Ribosyl)ation Reflecting NMDA and NO Neurotransmission.
We monitored PARP activity through conversion of [32P]NAD+ to PAR and DNA damage by DNA polymerase-I catalyzed incorporation of [32P]dCTP into DNA strand breaks. Substantial PARP activity and DNA ISEL are evident in primary cultures of cerebral cortical neurons (Table 1) and cerebellar granule neurons (data not shown). Cultured primary cortical astrocytes, however, display extremely low PARP activity and ISEL (Table 1). Western blots for PARP protein demonstrate only slightly more PARP-1 in neuronal cultures than in glial cultures (A.V., unpublished observation). Cerebral cortical astrocytes contain twice as much NAD+ as cortical neurons (Table 1) or cerebellar granule neurons (data not shown).
Table 1.
Primary cultured neurons have higher basal PARP activity and DNA damage and lower NAD+ levels than primary cultured astrocytes
| Cell | PARP activity, pmol NAD+ consumed/mg of protein | DNA damage, pmol [32P]dCTP incorporated/mg of protein | NAD+, μmol/mg of protein |
|---|---|---|---|
| Cortical neurons | 92.2 ± 5.2 | 3.25 ± 0.04 | 8.68 ± 0.22 |
| Cortical astrocytes | 1.33 ± 0.69 | 0.05 ± 0.0031 | 19.0 ± 0.74 |
Data are shown as mean ± SEM for six groups of 1 × 106 cells. Neuronal values exceed astrocyte levels for PARP activity and DNA damage (P < 0.001), whereas astrocyte NAD+ levels exceed neuronal levels (P < 0.001).
We wondered whether glutamate-NMDA neurotransmission causes basal DNA damage and poly(ADP-ribosyl)ation. With 1 h of exposure, DNA strand breaks decrease 20–30% with the NMDA-R antagonists MK801 and aminophosphonovalerate; PARP activity declines 40–45%; and NAD+ levels increase 20% (Table 2).
Table 2.
Inhibition of NMDA-R signaling events decreases basal PARP activity and DNA damage and elevates NAD+ in primary cultured neurons
| Treatment, 1 h | PARP activity, % control | ISEL signal, % control | NAD+, % control |
|---|---|---|---|
| MK801 (100 μM) | 55 ± 5 | 71 ± 1 | 144 ± 2 |
| Aminophosphonovalerate (100 μM) | 61 ± 9 | 74 ± 5 | 181 ± 3 |
| 7-NI (50 μM) | 67 ± 5 | 80 ± 3 | 128 ± 5 |
| l-nitroarginine (50 μM) | 58 ± 10 | 80 ± 6 | 180 ± 7 |
| MnTBAP (100 μM) | 68 ± 12 | 63 ± 3 | 157 ± 7 |
Differences between control and drug groups for PARP activity (P < 0.05). ISEL (P < 0.05) and NAD+ values (P < 0.001) are means ± SEM for five groups of 1 × 106 cells. Control values varied by 2–3%. MnTBAP, Mn(III)tetrakis (4-benzoic acid) porphyrin.
Glutamate-NMDA-R neurotoxicity is mediated by NO (1), and within 1 h, 7-nitroindazole (7-NI), a selective nNOS inhibitor, and l-nitroarginine, a more general NOS inhibitor, both reduce in vivo DNA strand breaks and poly(ADP-ribosyl)ation while elevating NAD+ levels (Table 2). NOS inhibitors are slightly less effective than NMDA-R antagonists. As observed with NMDA-R antagonists, NOS inhibitors lower poly(ADP-ribosyl)ation more than DNA strand breaks. PARP activity is reduced 30% and 40% by 7-NI and l-nitroarginine, respectively, and ISEL is reduced 20% with each drug. Downstream of NO, the superoxide and peroxynitrite scavenger MnTBAP (28) decreases ISEL by 35%, PARIS by 65%, and elevates NAD+ by 50% in cortical neurons treated for 1 h (Table 2).
Basal DNA Strand Breaks and PARP Activation Are Discretely Localized in the Brain.
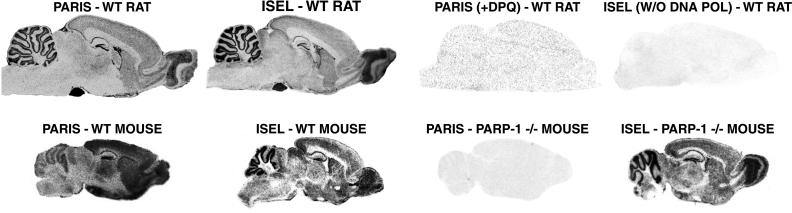
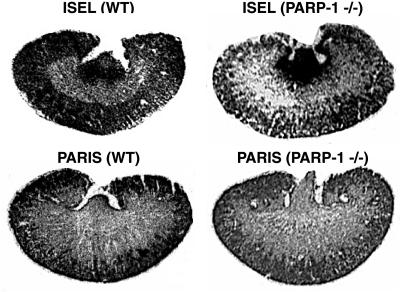
To investigate PARP activation in vivo, we developed the PARIS approach to monitor PARP activity microscopically (7, 13). Tissue sections are incubated with [33P]NAD+ and extensively washed in trichloroacetic acid to remove radiolabel not covalently bound to protein. Novobiocin, a specific inhibitor of mono(ADP-ribosyl)ation, eliminates background mono(ADP-ribosyl)ation. PARIS provides autoradiographic visualization of poly(ADP-ribosyl)ation in fresh-frozen tissue by monitoring conversion of [33P]NAD+ to labeled PAR. Highly discrete localizations of radiolabel are abolished by the PARP inhibitors DPQ (Fig. 1 and Table 3) and benzamide (Table 3). WT and PARP-1−/− mice exhibit similar levels of ISEL, but PARIS is abolished in PARP-1−/− brain. The PARP inhibitor nicotinamide also depletes PARIS, and NAD+ (1 mM) eliminates PARIS by exceeding the Km for PARP-1 (Table 3). Benzoic acid, structurally related to benzamide but not a PARP-1 inhibitor, does not affect PARIS (Table 3). PARP-1−/− brain PARIS is 7% of WT (Table 3) and diffusely distributed (Fig. 1).
Figure 1.
Basal PARP activity and DNA strand breaks are colocalized in rat brain. PARIS was autoradiographically visualized in rat brain sections. High labeling sensitive to DPQ (10 μM) occurs in cerebellar cortex, dentate gyrus of the hippocampus, olfactory bulb, ventricular ependymal cells, cerebral cortex, and striatum. Labeling is relatively absent from white matter regions, such as the corpus callosum. DNA strand breaks, assessed by ISEL with [33P]dCTP, show localizations in rat brain and in WT mouse brain identical to those of PARIS in WT rat brain. Although PARIS is absent from PARP-1−/− brain, ISEL is equally prominent in both WT and PARP-1−/− brain. There is substantially lower PARIS in mouse cerebellum relative to rat cerebellum and mouse forebrain. Results shown are representative of those obtained from at least five trials of at least 10 mice per group.
Table 3.
Specificity of PARIS assay
| PARIS assay modification | PARIS signal, % control |
|---|---|
| DPQ (10 μM) | 0 |
| Benzamide (1 mM) | 0 |
| Nicotinamide (1 mM) | 0 |
| Benzoic acid (1 mM) | 100 |
| NAD+ (1 mM) | 0 |
| Novobiocin (0.5 mM) | 98 |
| Immunoprecipitation with anti-PAR | 0.5 |
| Immunoprecipitation with anti-PARP | 0.5 |
| Protein G/Sepharose | 100 |
Values are means of replicated triplicate determinations.
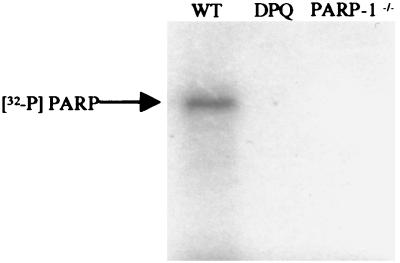
All radiolabel in PARIS is immunoprecipitated with polyclonal antibodies to PAR (Table 3), ensuring that the signal reflects PAR. Radiolabel is also immunoprecipitated with polyclonal antibodies to PARP-1 protein (Table 3), indicating that PARP-1 is predominantly labeled in rodent brain. All PARIS signal migrates with SDS/PAGE as a single intense band at about 110 kDa, identical to PARP-1 mobility. Labeling is abolished by DPQ and is absent in PARP-1−/− brains (Fig. 2).
Figure 2.
PARP-1 is the predominant protein labeled in the PARIS assay. Autoradiographic visualization of SDS/PAGE resolution of PARIS-labeled protein from WT mouse brain sections demonstrates that all signal migrates at approximately 110 kDa, identical to PARP-1 migration. Labeling is abolished by DPQ (10 μM). Specificity of the PARIS assay is further ensured through demonstration that protein labeling is absent in PARP-1−/− brain tissue.
The highest concentrations of PARIS radiolabel in rat brain are in pituitary and pineal glands. High levels also occur in cerebellar granule cells, hippocampal dentate gyrus, olfactory bulb, and ependymal and subependymal ventricular cells (Fig. 1). Under high magnification, cerebellar signal is mostly associated with Purkinje and granule cells, whereas label is in pinealocytes in the pineal gland and in anterior lobe in the pituitary gland. (data not shown). Rat and mouse brain display very little basal PARIS signal in white matter (Fig. 3), despite significant immunostaining for PARP-1 in white matter tracts and glial cells (data not shown).
Figure 3.
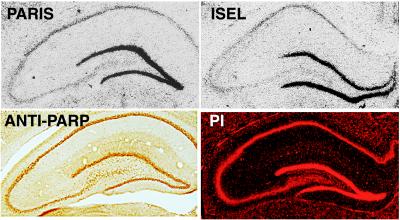
Prominent PARP activity in dentate gyrus of hippocampus reflects regionally high levels of DNA damage. Immunohistochemical staining for PARP demonstrates similar protein levels in hippocampal pyramidal cell layers and dentate gyrus, and propidium iodide (PI) DNA staining shows similar levels of DNA in these regions. However, PARIS and ISEL reveal much higher poly(ADP-ribosyl)ation and DNA damage in dentate gyrus relative to hippocampal pyramidal cell layers CA 1–3. Results shown are representative of those obtained from at least five trials of at least five mice per group.
Because PARP is activated by DNA damage, we directly monitored DNA strand breaks by ISEL with [33P]dCTP. ISEL localization is identical to that of PARIS in rat brain. Thus, basal PARP activity in rat brain seems to reflect basal DNA damage. In mouse brain, the regional distribution of ISEL is the same as in rat brain (Fig. 1). Though PARIS localization in WT mouse brain resembles that in rat brain, relative levels in forebrain and cerebellum differ somewhat from ratios in rat brain and for DNA strand breaks in rat and mouse brain (Fig. 1).
PARIS and ISEL are selectively localized in rat hippocampus, with the most intense signal in dentate gyrus granule cells and less signal in pyramidal cell layers CA 1–3 (Fig. 3). Immunohistochemical staining for PARP protein reveals similar densities in pyramidal cell layers and dentate gyrus (Fig. 3), thus PARIS signal reflects PARP activation associated with DNA damage rather than simply distribution of PARP protein. Although DNA staining with propidium iodide is more intense in dentate gyrus than CA 1–3, the relative enrichment in dentate is much less than it is with ISEL and PARIS (Fig. 3).
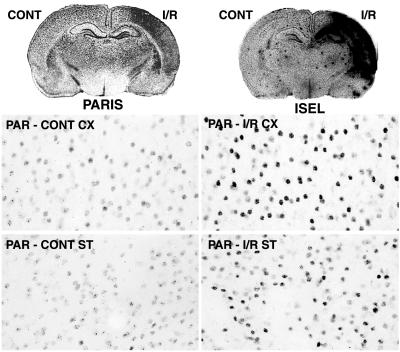
Unilateral cerebral ischemia experiments demonstrate that in vivo PARP activation parallels in vivo DNA damage. After I/R, DNA damage and PARIS are unilateral and similarly distributed in hippocampus, striatum, and cerebral cortex (Fig. 4). PARIS does not increase until 5 min after reperfusion, 65 min after initiation of ischemia (data not shown), fitting with other observations that PARP activation reflects reperfusion damage after cerebral ischemia (22). I/R also increases PAR staining in neurons of cerebral cortex and striatum (Fig. 4), as well as hippocampus (data not shown).
Figure 4.
Unilateral brain ischemia/reperfusion (I/R) increases ISEL (DNA damage) and PARIS signals. Unilateral brain ischemia for 1 h followed by 5 days of reperfusion (27) results in increased ISEL and PARIS in cerebral cortex, hippocampus, and striatum. Basal and I/R-induced accumulation of poly(ADP-ribose) (PAR) in cerebral cortex and striatum is seen in neuronal nuclei and is more abundant after I/R. Results shown are representative of those obtained from at least five trials of at least five rats per group.
Basal PARP Activity and DNA Damage in Vivo Reflect NMDA-R and NO Neurotransmission.
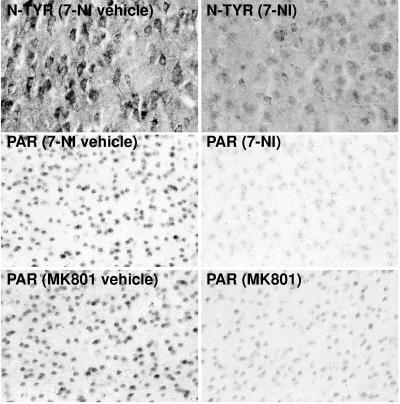
PARIS distribution in mouse forebrain is similar to that in rat forebrain, with high densities in pituitary gland, cerebellum, and dentate gyrus of hippocampus (Fig. 1). PARIS and ISEL are reduced by 15% in nNOS−/− forebrain, and NAD+ is elevated by 25% (Table 4). Thus, in intact animals as in brain cultures, nNOS influences basal NAD+ levels via PARP activity. 7-NI (50 mg/kg i.p.) reduces ISEL and PARIS in WT forebrain by 10% and 30%, respectively (Table 4). After 5 days of treatment with 7-NI (50 mg/kg s.c. every 12 h), immunohistochemically monitored nitrotyrosine and PAR are dramatically decreased in rat cortex (Fig. 5), as well as in striatum and hippocampus (data not shown). Treatment of WT rats with MK801 (1 mg/kg i.p.) for 1 h reduces ISEL by 25% and PARIS by 45% in forebrain (Table 4) and also reduces PAR staining (Fig. 5).
Table 4.
In vivo inhibition of NMDA-R neurotransmission or nNOS decreases basal PARP activity and DNA damage in rat forebrain
| In vivo treatment | PARIS signal, % control | ISEL signal, % control |
|---|---|---|
| MK801 (1 mg/kg, 1 h) | 50 ± 2 | 74 ± 4 |
| 7-NI (50 mg/kg, 1 h) | 72 ± 3 | 89 ± 1 |
| PARIS signal, % WT | ISEL signal, % WT | NAD+, % WT | |
|---|---|---|---|
| nNOS−/− mice | 82 ± 1 | 88 ± 4 | 123 ± 8 |
MK801, 7-NI, and nNOS−/− values for PARIS, ISEL, and NAD+ differ from control (P < 0.05–0.01). Data are means ± SEM of five animals.
Figure 5.
In vivo accumulation of PAR and nitrotyrosine is markedly decreased in rat cortex after 5 days of 7-NI treatment (50 mg/kg s.c. every 12 h). In vivo accumulation of PAR is also decreased in rat cortex 1 h after MK801 treatment (1 mg/kg i.p.). Results are representative of at least five trials of at least six rats per group.
Basal DNA Damage and Poly(ADP-Ribosyl)ation in Peripheral Tissues.
ISEL assays for DNA damage reveal a wide range in peripheral tissues, with levels in the bladder 50 times higher than those in the pancreas. Tissues with substantial cellular turnover, such as bladder, thymus, colon, small intestine, and kidney display highest ISEL, and ISEL signal is unaltered in PARP-1−/− animals. PARIS levels reflecting basal poly(ADP-ribosyl)ation also vary widely, with highest values being found in testis that are about 30 times the lowest values that are found in liver (data not shown). Like the brain, PARIS and ISEL are colocalized in the eye, with highest levels in retina and relatively lower levels in cornea. PARIS and ISEL are colocalized in thymus, skeletal muscle, bladder, testis, liver, and lung (data not shown). PARIS and ISEL are also colocalized in heart tissue, with highest levels in the subendocardium and decreased signal in adjacent myocardium. PARIS and ISEL values do not correlate well among all organs, however, suggesting that in some tissues poly(ADP-ribosyl)ation is not determined solely by DNA damage. The discrepancy between ISEL and PARIS is highlighted by their patterns in the kidney (Fig. 6). ISEL is pronounced most in the pelvis and calyces, least in the medulla, and intermediately in the cortex. By contrast, PARIS is low in the pelvis and highest in cortex. Substantial PARIS remains in PARP-1−/− kidney, and ISEL does not differ at all. Although in the stomach both PARIS and ISEL are most intense in the mucosa and relatively absent in submucosa and muscular wall, PARIS is relatively absent from the mucosa of small intestine and colon, whereas ISEL remains high in these regions (data not shown). In the pancreas, PARIS is diffusely distributed with focal increases in islets, whereas ISEL does not show these same focal increases. In the spleen, both PARIS and ISEL are prominent in the subcapsular compartment. Although PARIS is enriched in white pulp more than in red pulp, ISEL is more homogeneously distributed throughout splenic parenchyma (data not shown).
Figure 6.
Poly(ADP-ribosyl)ation in mouse kidney localizes differently than DNA damage and is substantial in PARP-1−/− kidney. ISEL signal in WT and PARP-1−/− kidney is highest in renal pelvis and calyces, light in the medulla, and moderate in cortex. PARIS signal is much less intense than ISEL in renal pelvis and calyces of WT and PARP-1−/− kidney. Like ISEL, PARIS is very light in medulla and moderate in cortex. PARIS in PARP-1−/− kidney is localized similarly to PARIS in WT but much less intense overall.
Most strikingly, although PARIS is fully depleted in some organs of PARP-1−/− mice, in other organs, gene deletion leads to little or no decrease (Table 5). Skeletal muscle, whose PARIS activity is second highest, eye, and spleen show no loss of PARIS signal with PARP-1 gene deletion. PARIS signal declines only 35–70% in kidney, thymus, heart, and lung of PARP-1−/− animals. By contrast, PARIS activity is largely depleted in liver, colon, small intestine, brain, testis, pancreas, bladder, and stomach of PARP-1−/− mice. We also measured NAD+ in peripheral tissues, whose relative levels in various organs do not correlate with variations in ISEL or PARIS and which are not altered in PARP-1−/− mice (data not shown).
Table 5.
Residual basal poly(ADP-ribosyl)ation in PARP-1−/− tissues
| PARP-1−/− tissue | Percentage of WT basal PARIS signal |
|---|---|
| Brain | 6.9 ± 2.1 |
| Pancreas | 2.9 ± 3.0 |
| Testis | 4.8 ± 0.9 |
| Liver | 9.9 ± 3.3 |
| Colon | 14.0 ± 10.5 |
| Small intestine | 12.5 ± 6.1 |
| Stomach | 19.8 ± 0.4 |
| Bladder | 19.2 ± 9.3 |
| Thymus | 31.5 ± 11.7 |
| Lung | 40.9 ± 15.0 |
| Heart | 64.0 ± 2.6 |
| Kidney | 63.3 ± 11.0 |
| Spleen | 82.1 ± 20.7 |
| Skeletal muscle | 94.2 ± 7.3 |
| Eye | 105.8 ± 12.9 |
Percentages of WT PARIS signal are means ± SEM for the average of five assays from five different animals. WT values varied from 4% to 7%.
Discussion
PARP has long been thought to be virtually inactive under basal physiologic conditions. We now show in cell culture and intact animals that basal NMDA-R mediated neurotransmission is responsible in part for a previously undescribed high degree of basal neuronal DNA damage and PARP activity, which influences neuronal NAD+.
To visualize in vivo PARP activation, we developed the PARIS method. After unilateral I/R in rat brain, increased PARIS signal in hippocampus, striatum, and cerebral cortex parallels increased immunohistochemically visualized PAR and increased DNA damage as measured with ISEL. Thus, PARIS visualizes in situ PARP activity induced by DNA damage after neurotoxic insult. For reasons that are not clear, PARIS signal in WT mouse cerebellum is less than that in forebrain, despite higher ISEL in mouse cerebellum relative to that in forebrain.
PARIS signal, which in PARP-1−/− brain is 7% of that WT, is abolished by PARP inhibitors. Although new enzymes with poly(ADP-ribosyl)ation capability have been identified, including VPARP (29), tankyrase (30) and PARP-2 (31–37), our findings indicate that basal poly(ADP-ribosyl)ation in the brain almost exclusively reflects PARP-1.
NAD+ levels increase after reduction of NMDA-R–NO neurotransmission, suggesting that this signaling pathway acts through poly(ADP-ribosyl)ation to regulate neuronal NAD+ and energy dynamics physiologically. Lower NAD+ levels after augmented NMDA-R–NO neurotransmission may limit neuronal capacity to initiate other activities that would threaten energy reserves. The notion that synaptic transmission in neurons depletes NAD+ fits with the levels of NAD+ in primary cultured cerebral cortical astrocytes that are 2-fold higher than in primary cultured cerebral cortical neurons.
Regional enrichments in basal DNA damage and PARP activity suggest a basis by which some regions of the brain may be more vulnerable than others to metabolic disturbance. Besides NMDA-R mediated neurotransmission and NO signaling, what other events determine basal DNA damage and PARP activation? Candidates include perturbations of calcium signaling systems, which could activate DNase, or other DNA events involving poly(ADP-ribosyl)ation, such as transcription (38–44), telomere length regulation (30, 45), or DNA replication (46, 47). Another likely source of endogenous DNA damage is basal free radical production other than NO, because 1 h of exposure to MnTBAP decreases basal DNA damage and PARP activity and elevates NAD+ in neuronal cultures. In addition to scavenging peroxynitrite, MnTBAP also scavenges superoxide anion, which is generated via pathways independent of NMDA-R signaling and NO generation.
PARIS is virtually abolished in PARP-1−/− brain, pancreas, liver, small intestine, colon, and testis, is not reduced in PARP-1−/− skeletal muscle, spleen, or eye, and is depleted only partially in PARP-1−/− heart, lung, and kidney. Low levels of residual PARIS exist in PARP-1−/− stomach, bladder, and thymus. In those tissues, other forms of PARP presumably mediate the PARIS signal. PARP-2 lacks some of the nuclear localization sequences of PARP-1 (31). Incongruities in localization of basal in vivo PARIS and ISEL in various tissues implicate an in vivo role for poly(ADP-ribosyl)ation other than DNA repair. Conceivably in skeletal muscle, the function of poly(ADP-ribosyl)ation may relate more to the preeminent energy dynamics of this tissue than DNA repair.
Acknowledgments
We thank NEN Life Science Products for synthesis and donation of [33P]NAD+, Pushpa Sharma for assistance with cell cultures and NAD+ determinations, Rajiv Ratan, Jay Baraban, David Ginty, Mark Molliver, and Elizabeth O'Hearn for helpful discussions, and Z.-Q. Wang for breeding pairs of PARP−/− mice. Under an agreement between the Johns Hopkins University and Guilford, S.H.S. is entitled to a share of sales royalties related to PARP received by the University from Guilford. The University owns stock in Guilford with S.H.S. having an interest in the University Share under University policy. S.H.S. serves on the Board of Directors and the Scientific Advisory Board of Guilford; he is a consultant to the company and owns additional equity in Guilford. This arrangement is being managed by the University in accordance with its conflict-of-interest policies. This work was supported by U.S. Public Health Service Grants MH18501 and DA00266 (to S.H.S.), Research Scientist Award DA00074 (to S.H.S.), Uniformed Services University of the Health Sciences Grant R09271 (to A.V.), a grant from the Defense and Veteran's Head Injury Program (to A.V.), and National Institute of Mental Health Training Grant MH418 (to A.A.P.).
Abbreviations
- WT
wild type
- PARP
poly(ADP-ribose) polymerase
- PAR
poly(ADP-ribose)
- nNOS
neuronal nitric oxide synthase
- NMDA
N-methyl-d-aspartate
- NMDA-R
NMDA receptor
- DPQ
3,4-dihydro-5-[4–1(1-piperidinyl) butoxy]-1 (2H)-isoquinolinone
- PARIS
PAR in situ
- ISEL
in situ end labeling
- I/R
ischemia/reperfusion
- 7-NI
7-nitroindazole
- MnTBAP
Mn(III)tetrakis (4-benzoic acid) porphyrin
References
- 1.Pieper A A, Verma A, Zhang J, Snyder S H. Trends Pharmacol Sci. 1999;20:171–181. doi: 10.1016/s0165-6147(99)01292-4. [DOI] [PubMed] [Google Scholar]
- 2.Berger N A. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 3.Oliver F J, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet J C, de Murcia G. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam T T. Res Commun Mol Pathol Pharmacol. 1997;95:241–252. [PubMed] [Google Scholar]
- 5.LaPlaca M C, Raghupathi R, Verma A, Pieper A A, Saatman K E, Snyder S H, McIntosh T K. J Neurochem. 1999;73:205–213. doi: 10.1046/j.1471-4159.1999.0730205.x. [DOI] [PubMed] [Google Scholar]
- 6.Whalen M J, Clark R S, Dixon C E, Robichaud P, Marion D W, Vagni V, Graham S H, Virag L, Hasko G, Stachlewitz R, Szabo C, Kochanek P M. J Cereb Blood Flow Metab. 1999;19:835–842. doi: 10.1097/00004647-199908000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Pieper A A, Brat D J, Krug D K, Watkins C C, Gupta A, Blackshaw S, Verma A, Wang Z-Q, Snyder S H. Proc Natl Acad Sci USA. 1999;96:3059–3064. doi: 10.1073/pnas.96.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkart V, Wang Z-Q, Radons J, Heller B, Herceg Z, Stingl L, Wagner E F, Kolb H. Nat Med. 1999;5:314–319. doi: 10.1038/6535. [DOI] [PubMed] [Google Scholar]
- 9.Masutani M, Suzuki H, Kamada N, Watanabe M, Ueda O, Nozaki T, Jishage K, Watanabe T, Sugimoto T, Nakagama H, et al. Proc Natl Acad Sci USA. 1999;96:2301–2304. doi: 10.1073/pnas.96.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zingarelli B, Salzman A L, Szabo C. Circ Res. 1998;83:85–94. doi: 10.1161/01.res.83.1.85. [DOI] [PubMed] [Google Scholar]
- 11.Bowes J, Ruetten H, Martorana P A, Stockhausen H, Thiemermann C. Eur J Pharmacol. 1998;359:143–150. doi: 10.1016/s0014-2999(98)00638-4. [DOI] [PubMed] [Google Scholar]
- 12.Thiemermann C, Bowes J, Myint F P, Vane J R. Proc Natl Acad Sci USA. 1997;94:679–683. doi: 10.1073/pnas.94.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pieper, A. A., Walles, T., Wei, G., Clements, E. E., Verma, A., Snyder, S. H. & Zweier, J. (2000) Mol. Med., in press. [PMC free article] [PubMed]
- 14.Zhang J, Dawson V L, Dawson T M, Snyder S H. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- 15.Cosi C, Suzuki H, Milani D, Facci L, Menegazzi M, Vantini G, Kanai Y, Skaper S D. J Neurosci Res. 1994;39:38–46. doi: 10.1002/jnr.490390106. [DOI] [PubMed] [Google Scholar]
- 16.Eliasson M J, Sampei K, Mandir A S, Hurn P D, Traystman R J, Bao J, Pieper A A, Wang Z-Q, Dawson T M, Snyder S H, Dawson V L. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 17.Endres M, Wang Z-Q, Namura S, Waeber C, Moskowitz M A. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Love S, Barber R, Wilcox G K. Neuropathol Appl Neurobiol. 1999;25:98–103. doi: 10.1046/j.1365-2990.1999.00179.x. [DOI] [PubMed] [Google Scholar]
- 19.Endres M, Scott G, Namura S, Salzman A L, Huang P L, Moskowitz M A, Szabo C. Neurosci Lett. 1998;248:41–44. doi: 10.1016/s0304-3940(98)00224-9. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Greenberg J H, Jackson P, Maclin K, Zhang J. J Cereb Blood Flow Metab. 1997;17:1137–1142. doi: 10.1097/00004647-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K, Pieper A A, Croul S E, Zhang J, Snyder S H, Greenberg J H. Brain Res. 1999;829:46–54. doi: 10.1016/s0006-8993(99)01335-9. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K, Greenberg J H. NeuroReport. 1999;10:2017–2022. doi: 10.1097/00001756-199907130-00005. [DOI] [PubMed] [Google Scholar]
- 23.Nicoletti F, Wroblewski J T, Novelli A, Alho H, Guidotti A, Costa E. J Neurosci. 1986;7:1905–1911. doi: 10.1523/JNEUROSCI.06-07-01905.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimaldi M, Navarra P, Pozzoli G, Preziosi P, Schettini G. Biochem Biophys Res Commun. 1998;250:789–804. doi: 10.1006/bbrc.1998.9378. [DOI] [PubMed] [Google Scholar]
- 25.Nisselbaum J S, Green S. Anal Biochem. 1969;27:212–217. doi: 10.1016/0003-2697(69)90025-6. [DOI] [PubMed] [Google Scholar]
- 26.Schraufstatter I U, Hinshaw D B, Hyslop P A, Spragg R G, Cochrane C G. J Clin Invest. 1986;77:1312–1320. doi: 10.1172/JCI112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vannucci R C, Vannucci S J. Ann NY Acad Sci. 1997;835:234–249. doi: 10.1111/j.1749-6632.1997.tb48634.x. [DOI] [PubMed] [Google Scholar]
- 28.Cuzzocrea S, Zingarelli B, Costantino G, Caputi A P. Free Radical Biol Med. 1999;26:25–33. doi: 10.1016/s0891-5849(98)00142-7. [DOI] [PubMed] [Google Scholar]
- 29.Kickhoefer V A, Siva A C, Kedersha N L, Inman E M, Ruland C, Streuli M, Rome L H. J Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith S, Giriat I, Schmitt A, de Lange T. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 31.Ame J C, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 32.Shieh W M, Ame J C, Wilson M V, Wang Z-Q, Koh D W, Jacobson M K. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 33.Johansson M. Genomics. 1999;57:442–445. doi: 10.1006/geno.1999.5799. [DOI] [PubMed] [Google Scholar]
- 34.Kawamura T, Hanai S, Yokota T, Hayashi T, Poltronieri P, Miwa M, Uchida K. Biochem Biophys Res Commun. 1998;251:35–40. doi: 10.1006/bbrc.1998.9403. [DOI] [PubMed] [Google Scholar]
- 35.Lepiniec L, Babiychuk E, Kushnir S, Van Mantagu M, Inze D. FEBS Lett. 1995;364:103–108. doi: 10.1016/0014-5793(95)00335-7. [DOI] [PubMed] [Google Scholar]
- 36.Babiychuk E, Cottrill P B, Storozhenko S, Fuangthong M, Chen Y, O'Farrell M K, Van Montagu M, Inze D, Kushnir S. Plant J. 1998;15:635–645. doi: 10.1046/j.1365-313x.1998.00240.x. [DOI] [PubMed] [Google Scholar]
- 37.Berghammer H, Ebner M, Marksteiner R, Auer B. FEBS Lett. 1999;449:259–263. doi: 10.1016/s0014-5793(99)00448-2. [DOI] [PubMed] [Google Scholar]
- 38.Kameoka M, Tanaka Y, Ota K, Itaya A, Yoshihara K. Biochem Biophys Res Commun. 1999;262:285–289. doi: 10.1006/bbrc.1999.1146. [DOI] [PubMed] [Google Scholar]
- 39.Simbulan-Rosenthal C M, Rosenthal D S, Luo R, Smulson M E. Oncogene. 1999;18:5015–5023. doi: 10.1038/sj.onc.1202900. [DOI] [PubMed] [Google Scholar]
- 40.Oei S L, Griesenbeck J, Ziegler M, Schweiger M. Biochemistry. 1998;37:67–76. doi: 10.1021/bi9727390. [DOI] [PubMed] [Google Scholar]
- 41.Kannan P, Yu Y, Wankhade S, Tainsky M A. Nucleic Acids Res. 1999;27:866–874. doi: 10.1093/nar/27.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butler A J, Ordahl C P. Mol Cell Biol. 1999;19:296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Amours D, Desnoyers S, D'Silva I, Poirier G G. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 44.Meisterernst M, Stelzer G, Roeder R G. Proc Natl Acad Sci USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.d'Adda di Fagagna F, Hande M P, Tong W M, Lansdorp P M, Wang Z-Q, Jackson S P. Nat Genet. 1999;23:76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- 46.Simbulan-Rosenthal C M, Rosenthal D S, Boulares A H, Hickey R J, Malkas L H, Coll J M, Smulson M E. Biochemistry. 1998;37:9363–9370. doi: 10.1021/bi9731089. [DOI] [PubMed] [Google Scholar]
- 47.Simbulan-Rosenthal C M, Rosenthal D S, Iyer S, Boulares H, Smulson M E. Mol Cell Biochem. 1999;193:137–148. [PubMed] [Google Scholar]