Abstract
Dimerization is fairly common in the G-protein-coupled receptor (GPCR) superfamily. First attempts to rationalize this phenomenon gave rise to an idea that two receptors in a dimer could be necessary to bind a single molecule of G protein or arrestin. Although GPCRs, G proteins and arrestins were crystallized only in their inactive conformations (in which they do not interact), the structures appeared temptingly compatible with this beautiful model. However, it did not survive the rigors of experimental testing: several recent studies unambiguously demonstrated that one receptor molecule is sufficient to activate a G protein and bind arrestin. Thus, to figure out the biological role of receptor self-association we must focus on other functions of GPCRs at different stages of their functional cycle.
G-protein-coupled receptors (GPCRs) are the largest and arguably the most diverse superfamily of proteins represented in every eukaryotic cell. GPCRs appeared before the separation of plants, animals and fungi ~1.2 billion years ago as either descendants or copycats of prokaryotic rhodopsins [1]. Their defining structural feature is the heptahelical domain (HD), a characteristically twisted bundle of seven transmembrane α-helices (TMHs). Evolution generously applied its usual tools: relatively minor tinkering that changed virtually every residue in the TMHs and produced intra- and extracellular loops of various lengths and domain swapping that yielded GPCRs with an incredible variety of additional domains shared with other protein families (Box 1). The great majority of mammalian GPCRs belong to class A (often called rhodopsin-like receptors), consist largely of a H D and bind small-molecule ligands via elements of this domain. Small classes B and C include GPCRs that carry large N-terminal extensions mediating ligand binding (Box 1). Because of the size of this family and its importance as a target for the majority of clinically used drugs [1], virtually every aspect of the GPCR function is extensively debated.
There is broad consensus that the HD is the key to the shared signaling function. As receptors, GPCRs have two interlinked jobs: they receive the external signal and ‘report’ it. The ability of the HD to exist in multiple conformations and to switch from one to another after a relatively mild ‘push’ underlies signal transduction across the plasma membrane (Box 2). The existing evidence suggests that in all GPCRs the HD is kept in the basal state by ionic ‘clasps’ that the agonist ‘unfastens’. The activation occurs within the HD and involves significant ‘shape-shifting’ [2–8]. Agonists initially engage different structural elements in various GPCR subtypes, but agonist binding promotes G protein activation only when it ultimately results in the transition of the HD into the active conformation. Simply put, no matter where natural agonists bind, the end point of GPCR activation involves conformational changes in the HD that couples to G proteins.
In contrast, the ability of class A GPCRs to dimerize and the role of GPCR dimers in signaling is currently a hot topic, with opinions ranging from ‘dimers of rhodopsin-like receptors don’t even exist’ [9] to ‘GPCRs always function as dimers’ [10]. Here we analyze existing structure–function information to gain insights into possible roles of receptor dimerization. Class C receptors that definitely exist as stable dimers offer crucial clues. The evidence indicates that whereas the HD is the key player in G protein coupling, self-association is mediated largely by other receptor elements. The structural separation is matched by functional independence: a single HD in active conformation is sufficient to activate G proteins. However, there is a wide range of other receptor functions (that are often overlooked) in which GPCR dimerization could play a role.
GPCR dimerization: the phenomenon and underlying mechanisms
Some GPCRs dimerize, but the functional role of receptor self-association in most cases is unclear. Despite dozens of publications on the subject, as far as the most numerous rhodopsin-like receptors are concerned, unambiguous evidence is sparse [24]. I n contrast, there is no doubt that class C receptors exist and function as stable dimers. The molecular mechanisms of their dimerization and signaling were studied extensively, providing valuable clues applicable to other GPCRs.
In addition to the signature HD that places them in GPCR family, class C receptors have an N-terminal Venus flytrap (VFT) module (Box 1) structurally and functionally homologous to bacterial periplasmic proteins that bind amino acids, sugars and ions [11]. Separately expressed VFTs from metabotropic glutamate (mGlu) receptors [12,13] and the unrelated single-transmembrane domain natriuretic peptide receptor [14] invariably crystallize as dimers with or without ligands. VFTs probably inherited their high propensity to dimerize from their bacterial ancestors that readily form dimers in crystal and in solution [15]. Cysteine-rich domains (CRDs), localized between VFT and HD, also carry an autonomous dimerization potential: VFT–CRD modules from the mGlu1 [16], mGlu4 [17] and calcium-sensing (CaS) receptors [18], when expressed without the rest of the receptor, exist as disulfide-linked dimers, readily forming the same S–S bonds between CRDs that participate in the dimerization of respective full-length receptors. In the CRD-less GABAB receptor, nature played a different trick to ensure its heterodimerization: the cytoplasmic tail of the GB1 subunit carries an RXR motif that ‘traps’ it in the endoplasmic reticulum (ER) [19]. The coiled-coil cytoplasmic domain of the GB2 subunit binds and masks this motif, allowing the release of the GB1–GB2 heterodimer from the ER [19].
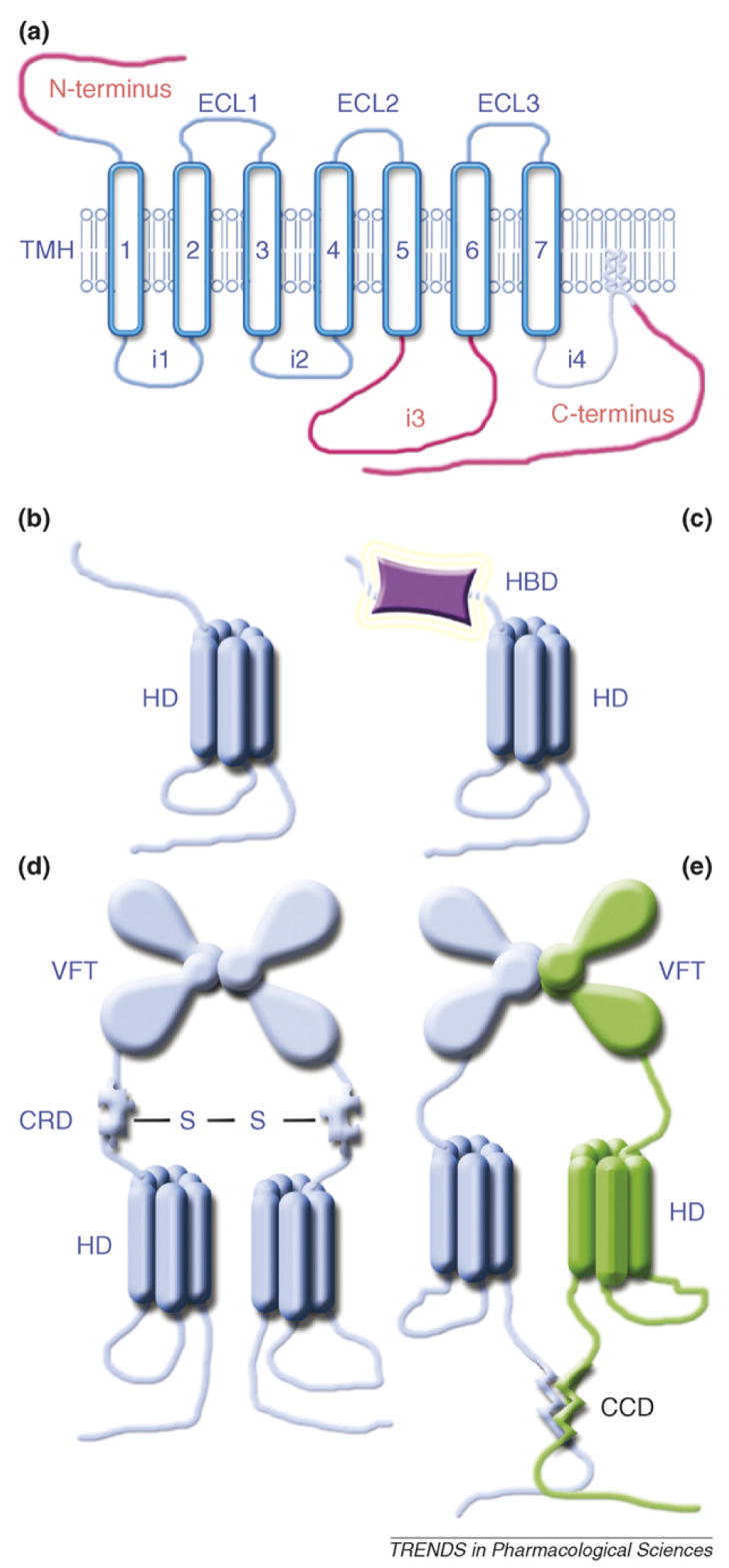
Box 1. Structural variability of G-protein-coupled receptors
The HD is the defining feature of the GPCR superfamily [65,66]. Because some GPCRs do not couple to G proteins, an alternative family name, ‘seven-transmembrane receptors (7TMRs)’, was proposed. Originally, proteins with seven transmembrane α-helices (TMHs) were divided into six groups, A through F, with >20% sequence identity of homologous TMHs within each group [66]. Groups (also known as ‘families’ or ‘classes’) A, B and C included all mammalian GPCRs known at the time. After an explosive increase in GPCR sequences driven by genome projects, more than 800 human GPCRs were reclassified (based solely on the HD sequence) into five families: glutamate, rhodopsin, adhesion, frizzled/taste2 and secretin (GRAFS) [65]. Each family has characteristic conserved motifs. No single motif is shared by all five families, but they are connected through motifs shared by two or more, suggesting the existence of a common ancestor [65]. Classes A, B and C roughly correspond to the rhodopsin, secretin and glutamate families in GRAFS. The HD consists of seven TMHs arranged in a characteristically twisted ‘barrel’ (Figure Ia). The helices are connected by three intracellular (i1–i3) and three extracellular loops (ECL1–ECL3). In many GPCRs one or two cysteines in the proximal C-terminus are palmitoylated, creating a membrane anchor. The sequence between TMH7 and the palmitoylation site is often called the fourth intracellular loop (i4). The N-terminus is the most variable element, ranging from seven residues in the adenosine A2 receptor to ~5 900 in the ‘very large G-protein-coupled receptor’ (VLGR), the largest known membrane protein [67]. In some GPCRs the N-terminus carries one or more additional domains, such as VFT, EGF, cadherin, immunoglobulin, lectin or laminin. The third intracellular loop varies widely even within class A, from 19 residues in the IL-8 receptor to 190 in the M3 muscarinic acetylcholine receptor. The length of the C-tail ranges from <20 residues in many receptors to 165 in the rat α1B-adrenoceptor. The three most variable elements are highlighted in red. A ‘minimal’ class A GPCR (the majority of rhodopsin-like receptors) largely consists of the HD (Figure Ib), although glycoprotein hormone receptors carry the hormone-binding domain (HBD) in the 300–500-residue-long N-terminus (Figure Ic). Class C receptors come in two flavors: metabotropic glutamate and calcium-sensing receptors are homodimers (Figure Id), whereas GABAB and sweet and umami taste receptors are obligatory heterodimers (Figure Ie). Several interactions hold the two monomers together: dimerization of the N-terminal ligand-binding VFT modules, disulfide bridges between CRDs and/or the interaction between C-terminal coiled-coil domains (CCDs) of the two GABAB receptor subunits, GB1 and GB2.
Figure I.
Structural elements of GPCRs and their arrangement in different receptors.
The presence of two dimerization-promoting elements in every class C GPCR suggests that evolution did not expect HDs to contribute appreciably to this process. Indeed, elimination of the ER-retention motif results in cell-surface expression of the GB1 subunit exceeding that of the wild type GB1–GB2 heterodimer [19]. The HD plays no role in this mechanism: unrelated membrane protein fused to the C-terminus of the GB1 subunit is retained in the ER in the absence and successfully reaches the cell surface in the presence of the coexpressed GB2 subunit [19]. The GB2 subunit expressed alone reaches the cell surface without any trouble as a fully functional GPCR: it is activated by the known positive allosteric modulator of the GABAB receptor CGP7930 and activates the same G proteins as the native GB1–GB2 heterodimer [20]. In this situation VFT becomes unnecessary: separately expressed HD of the GB2 subunit has the same affinity for CGP7930 and mediates the full cellular response, behaving like a class A receptor, which it closely resembles structurally [20]. This appears to be the case in every class C GPCR subjected to rigorous structure–function analysis: a separately expressed HD of the mGlu5 receptor is activated by positive allosteric modulators and couples to G proteins [21]; dimerization-deficient mutants of the CaS receptor are functional calcium-sensing receptors [22].
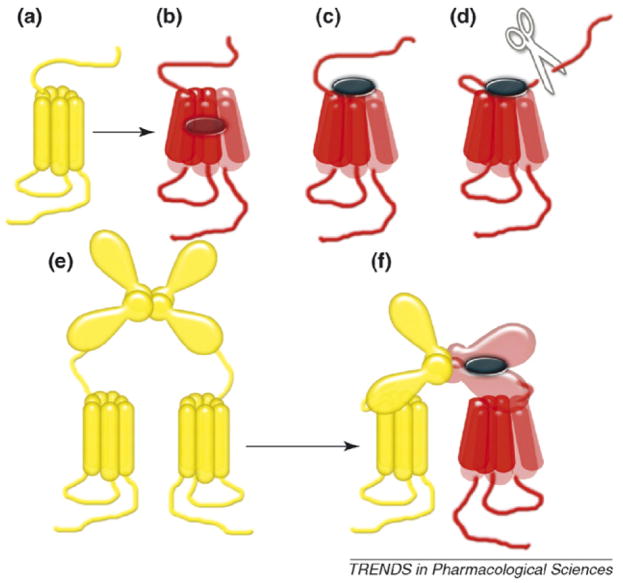
Box 2. Molecular mechanism of receptor activation: conformational change in the heptahelical domain
HDs of class A receptors are stabilized by salt bridges (fairly strong interactions in the hydrophobic environment) [2–4] that are rearranged in the process of receptor activation enabling large conformational changes [4–6]. Although individual charges are not conserved in other families, rearrangement of salt bridges during activation also occurs in the HD of the GB2 in class C [7] and in the yeast α-factor pheromone receptor Ste2 [8], which is so different from animal GPCRs that even their common ancestry is uncertain [1]. Inactive receptors (shown in yellow in Figure Ia) are activated by a wide variety of agonists (shown as black ovals in the diagram). Retinal is localized in the middle of the rhodopsin TMH ‘barrel’ [26,27] (Figure Ib), suggesting that its isomerization simply ‘pushes’ TMHs around, forcing the receptor into the active state (red). Small molecules bind within the same cavity of other class A receptors [28], ‘pushing’ TMHs directly. Some pep tides engage ECLs, probably using them as levers to provide a similar ‘push’ (Figure Ic). Proteinase-activated receptors contain their own ‘tethered agonist’ (unmasked by the proteolysis of the N-terminus) that binds ECLs and activates the receptor like an exogenous peptide (Figure Id). In glycoprotein hormone receptors (class A) and all class C receptors, agonists interact with the N-terminus so that the conformational change in the HD must be induced in a less direct way (Figure Ie). The N-terminus of class C receptors contains a VFT domain consisting o f two lobes and can assume ‘closed’ and ‘open’ conformations [12,13]. Agonist binding promotes VFT closure, which triggers receptor activation (Figure If). Mutations t hat eliminate steric or ionic clashes, thereby allowing the VFT to close with a bound antagonist, convert this antagonist into an agonist [68]. VFTs do not have a rigid connection with the HD. The deletion of VFTs eliminates regulation by native ligands, but does not preclude G-protein coupling, often increasing constitutive activity [20,21]. Thus, the extracellular ligand-binding domain interacts with the HD, serving as a tethered inverse agonist in inactive (yellow) and a tethered agonist in active (red) conformation. In the GABAB receptor only one VFT has to be in ‘closed’ conformation [19,32,34], whereas in mGlu5 receptors ‘open–closed’ and ‘closed–closed’ pairs work as partial and full agonists, respectively [69]. The ligand binding to the N-terminus leaves the ‘normal’ site in the HD ostensibly ‘unemployed’. Many small molecules bind the HDs of class C receptors and act as positive and negative allosteric modulators [20,21]. Although endogenous allosteric modulators of these GPCRs have not been described, it seems extremely unlikely that nature failed to create molecular tools targeting these perfectly functional sites.
Figure I.
To activate the receptor, ligand binding must change HD conformation.
Thus, the results of ingenious structure–function studies of diverse homo- and heterodimeric class C receptors t ell a remarkably consistent story: the ability to couple to proteins resides in the HD, whereas molecular ‘equipment’ responsible for the formation of stable dimers and their preferential delivery t o the plasma membrane is localized in other parts of the receptor. Therefore, if we use class C receptors as a model for the GPCR family, it predicts that the HD is a key player in the G protein interaction, whereas dimerization is largely mediated by other elements.
How does this conclusion square with numerous cross-linking studies that implicated virtually every transmembrane α-helix in homo- or heterodimerization of different GPCRs (reviewed in Refs [10,23])? It is important to remember that crosslinking is a one-way street: it can ‘trap’ transient ‘kiss-and-run’ encounters as well as stable dimers(discussed in Ref. [24]). Thus, we need to decide which complex we call a dimer: one that exists for milliseconds, seconds, minutes or hours? We also shouldn’t forget that physical chemistry provides a useful reality check: there are simple relationships between the interaction energy and affinity (ΔG° = − RTlnKA, where ΔG° is free energy of association, R is a gas constant, T is the temperature in Kelvin and KA is the association constant, which is the inverse of the equilibrium dissociation constant, KA = 1/KD)and between affinity and the half-life of the complex (t1/2 = 6.93 × 107 KA). The interaction of monomers with KD = 1 μM translates into a half-life of less than 1 s; KD = 2–20 nM (as reported for the NTS1 receptor [25]) yields half-life of 35–350 s. To exist in a stable dimer with a half-life comparable to that of even short-lived GPCRs (2–20 hr), the monomers must interact with a KD from 10 to 100 pM. These affinities require a binding energy of ~60 kJ/mol (compared with 220 kJ/mol of a covalent S–S bond), which is unlikely to be achieved by interactions between hydrophobic residues that project from the HD into the membrane environment [26–28]. These simple calculations explain why two receptors that are meant to stay in a stable dimer (like class C GPCRs) are usually covalently linked. What do numerous studies of GPCR self-association that used crosslinking and energy transfer-based methods show? Their results often are presented as proof positive of obligatory dimerization of every GPCR known to man [10,23]. In reality these data probably reflect a dynamic equilibrium between monomers and dimers of varying stability. This equilibrium might be affected by receptor activation and its interactions with G proteins, kinases, arrestins and internalization machinery. The transient nature of GPCR dimers does not exclude their biological importance (many regulatory protein–protein interactions have nanomolar affinities and last for seconds or even less), but it should be kept firmly in mind when we discuss self-association of GPCRs that do not form covalent bonds between monomers.
Receptor coupling to G proteins
How many active HDs are necessary to activate a G protein? This question actually was asked in physiological studies testing the limits of two sensory systems: vertebrate vision [29] and invertebrate olfaction [30]. These experiments, performed in the 1970s when nobody even suspected that in both cases the response is mediated by structurally related receptors, gave identical answers: just one. Sensory cells of Bombyx mori, male silkworm moths, respond to a single molecule of female pheromone bombykol [30]. Single photons produce the response in individual toad rod photoreceptors [29].
However, rhodopsin and odorant receptors belong to class A, where the issue of dimerization is highly controversial [9,10,23,24]. Therefore, let us turn to class C receptors, where the activation mechanism of stable GPCR dimers that undoubtedly exist can be tested. Heterodimeric GABAB receptors yielded very interesting clues. Although both GB1 and GB2 subunits have VFT modules, only GB1 VFT binds GABA [31]. Separately expressed GB1 binds GABA, although the presence of two different VFTs increases the affinity [32]. Interestingly, it does not matter which VFT is attached to which HD: two chimeric subunits with swapped VFTs form a GABAB receptor functionally indistinguishable from the wild-type [32,33]. The length of the connector between VFT and HD does not matter either: the addition of an 11-residue, glycine-rich peptide predicted to form a random coil in the middle of each linker preserves receptor function [33]. The GB2 subunit is solely responsible for G-protein coupling [32,34]. The only structural manipulation that the cytoplasmic side of the GB2 subunit tolerates is the deletion of the coiled-coil C-terminal element that masks an ER-retention signal in the GB1 subunit [32,34]. In contrast, in GB1 the deletion of the C-terminus, replacement of the intracellular loops with their GB2 or mGlu1 receptor equivalents and replacement of a large portion of the i3 loop with a random coil peptide have no functional consequences [34]. Thus, one monomer within the GABAB receptor is responsible for all specific G protein contacts. In full agreement with this conclusion, both the N-terminal VFT and the C-terminal coiled-coil domain of the GB2 subunit can be completely eliminated: the GB2 HD expressed alone constitutes a fully functional receptor that normally couples to G proteins, retaining G protein specificity of the wild-type heterodimer [20]. Like class A receptors, this ‘minimal’ GB2 is regulated by ligands that bind to the HD: positive and negative allosteric modulators of the original GABAB receptor act as agonists and inverse agonists, respectively [20]. Separately expressed HDs of the naturally homodimeric mGlu5 receptor [21] or dimerization-deficient CaS receptor mutants [22] also behave like class A receptors. These data strongly suggest that only one HD in active conformation is necessary for the G-protein activation.
One might argue that the behavior of these artificial constructs does not necessarily reflect the situation in normal class C receptors with two HDs. However, elegant studies of mGlu1 [35] and mGlu5 receptor [36] homodimers also support the conclusion that a G protein needs just one active HD and go even further, demonstrating that two HDs in active conformation side-by-side impede signaling. The fact that engineered GABAB receptors with two GB2 HDs competent to couple to G proteins have significantly reduced ability to signal [32] tells the same story. This evidence per se does not exclude the possibility that the second HD in a dimer could play some kind of supporting role, although the magnitude of reengineering that the cytoplasmic surface of the GB1 subunit in the GABAB receptor can take without functional consequences [34] makes this rather unlikely.
This issue can be resolved definitively by comparing G-protein interaction with the same native receptor in monomeric and dimeric form, which can be accomplished only with class A GPCRs. The participation of class A receptor dimers in G-protein coupling is often inferred from changes in signaling observed upon coexpression of particular GPCR combinations (reviewed in Ref. [23]). However, these data do not exclude alternative interpretations, such as crosstalk between signaling pathways [24]. Similarly, neither energy transfer (FRET or BRET) between coexpressed receptors tagged with fluorescent moieties nor receptor crosslinking can distinguish between ‘crowding’ [37] or frequent kiss-and-run encounters and stable bona fide oligomers (discussed in Ref. [24]). Definitive experiments that address this issue under strictly controlled conditions excluding artifacts and alternative interpretations were performed recently with rhodopsin [38,39], the β2-adrenoceptor [40] and the neurotensin NTS1 receptor [25] (i.e. three receptors that couple to three different G proteins). In solubilized preparations, themonomeric NTS1 receptor activates Gq more efficiently than the receptor dimer [25]. Monomeric rhodopsin in detergent solution activates transducin as fast as the rate of protein diffusion allows [39]. Importantly, the two best studied GPCRs, rhodopsin and β2-adrenoceptor, were tested in preparations in which these receptors were reconstituted into lipid bilayer (mimicking their natural membrane milieu) in high-density lipoprotein particles [38,40]. The beauty of this system is that one or two receptor molecules can be inserted, and particles with one receptor can be separated from those containing two. Thus, the receptor is certainly monomeric in the former case, whereas in the latter two receptor molecules forced into close proximity may be in monomeric or dimeric state. Rhodopsin monomers were found to activate transducin with the same efficiency as two rhodopsins in the same particle [38], indicating that either dimerization reduces the efficiency o f G protein coupling by half or only one rhodopsin is accessible when two are close together. The monomeric β2-adrenoceptor effectively activates Gs and forms a biologically relevant receptor–G-protein complex with characteristic high affinity for agonists [40].
These data unambiguously show that a single active GPCR is sufficient for G-protein binding and activation and that the presence of a second receptor in a dimer [25] or in close proximity [38] impedes signaling. These results rule out the models with one G protein ‘docked’ to two receptors [10], lending strong support to the traditional one-to-one models [9] (see Figure 1). To summarize, carefully controlled studies of structurally diverse class A and C receptors yield exactly the same answer: one HD in active conformation couples to a G protein, whereas the neighbor simply gets in the way, more so if it is activated.
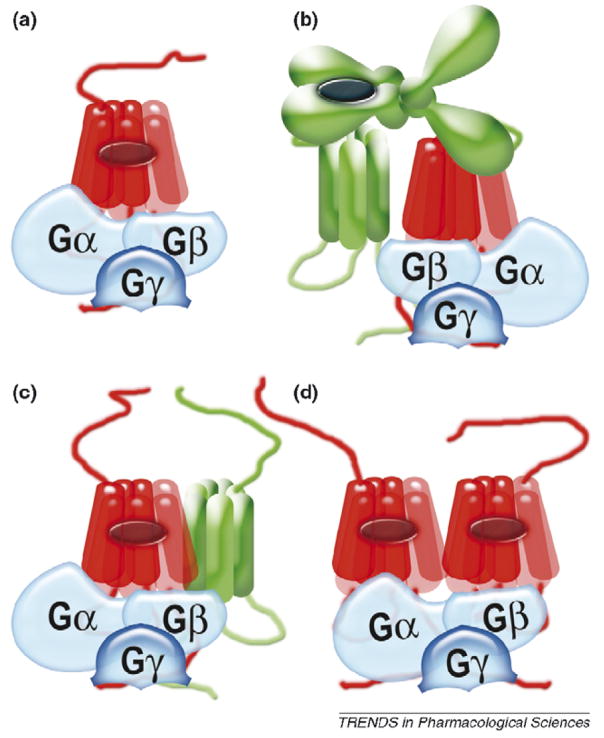
Figure 1.
Receptor coupling to G proteins. (a) The traditional model of one heterotrimeric G protein interacting with one active (red) receptor [9]. (b) A single HD in active conformation (red) in a dimeric receptor (a heterodimeric GABAB receptor is shown as an example; in this receptor VFT of the GB1 subunit binds GABA, whereas the HD of the GB2 subunit couples to G proteins) is strongly supported by the evidence obtained in strictly controlled experiments [32–34, 38–40]. (c) G-protein coupling to a dimeric receptor appears to be less efficient [25,38]; it works better when one of the receptors is inactive (green) [36], as shown here. Note that a pentameric complex of two receptors with one G protein described for the leukotriene receptor B4 [70] is equally consistent with this arrangement and with that shown in panel (d). (d) A model in which one G protein simultaneously docks to both receptors in a dimer [10] was proposed on the basis of crystal structures of an inactive receptor and an inactive (in a complex with GDP) heterotrimeric G protein, although the two proteins in these conformations do not interact. Several recent studies [38–40] convincingly ruled out the idea that the receptor dimer is required for G-protein coupling. A similar two-to-one model proposed for receptor–arrestin interaction [10] was also ruled out by the demonstration that each receptor molecule binds its own arrestin [49,52]. However, it is still unclear whether arrestin (like G proteins) binds monomeric receptors better, prefers a dimer or does not care one way or the other.
Do GPCR dimers have special functions?
The requirement of a single HD for G-protein coupling does not mean that active receptors never self-associate. Dimerization could serve as a desensitization mechanism, rapidly suppressing G-protein-mediated signaling when there are too many active receptors around. The nanomolar self-association constant of the neurotensin NTS1 receptor and the significant reduction of its signaling efficiency upon dimerization [25], as well as more efficient coupling of monomeric rhodopsin to transducin [38], are consistent with this idea (Figure 1).
GPCR dimerization also was proposed to participate in arrestin-mediated desensitization: based on the size of the cytoplasmic tip of rhodopsin [26,27] and the presence of two cup-like domains in arrestins [41–43], both of which were implicated in receptor binding [44–46], a model in which one arrestin binds to two receptors in a dimer was suggested [10]. Twofold difference in the amount of arrestin that ‘saturates’ the receptor predicted by 1:1 [47] and 1:2 [10] models is experimentally tractable. Functional radiolabeled arrestins can be expressed in cell-free translation and used as radioligands to determine their affinity and the number of binding sites [48]. Scatchard analysis of the binding of radiolabeled arrestin to the purified M2 muscarinic acetylcholine receptor reconstituted into liposomes showed that the number of arrestin-binding sites approaches 90% of the number of receptors present in the assay [49], supporting the 1:1 model. Obviously, these experiments were performed in a highly artificial situation, so one might argue that the data cannot be unquestioningly extrapolated to physiological conditions. Luckily, the visual system provides a unique opportunity to perform this experiment in vivo. In photoreceptors rhodopsin is permanently localized to the outer segment, whereas the bulk of arrestin translocates to this compartment only in bright light and remains there as long as it is bound to rhodopsin [50]. Mouse rods express enormous amounts of arrestin and rhodopsin at ~0.8:1 molar ratio [51,52]. The availability of hemizygous knockout mice allows one to achieve a wide range of ratios of perfectly normal wild-type arrestin and rhodopsin in the native environment. The stoichiometry of the arrestin–rhodopsin interaction in living animals was recently tested by using arrestin translocation as the readout [52]. Virtually complete translocation was observed in every line expressing more rhodopsin than arrestin, with the maximum amount of translocated arrestin exceeding 80% of rhodopsin present [52]. T o ascertain that no other protein is ‘anchoring’ arrestin in the outer segment, the binding at similar arrestin:rhodopsin ratios was measured with two purified proteins in vitro, and the number of arrestin-binding sites turned out to be equal to the number of rhodopsin molecules [52]. Although rod arrestin readily self-associates at physiological concentrations, the results cannot be explained by one dimer binding to another because only the arrestin monomer binds rhodopsin [53]. Obviously, these experiments tell us nothing about the receptor self-association state: for all we know it could have been a monomer, dimer, higher order oligomer or any mix of these forms. However, they prove that in living animals one receptor molecule is sufficient to bind arrestin and each rhodopsin molecule does bind its own arrestin.
In the visual system arrestin binding to rhodopsin simply stops the signaling, whereas non-visual arrestins interact with dozens of diverse non-receptor partners with the arrestin–receptor complex serving as a scaffold that orchestrates the next round of signaling [54,55]. The relative sizes of the arrestin, receptor and proteins that interact with receptor-bound arrestin suggest that a unitary complex can simultaneously accommodate very few partners [56]. Therefore, it is tempting to speculate that multiple arrestin molecules bound to oligomeric GPCRs could provide larger and more versatile scaffolds for the assembly of signaling complexes.
Dimerization may also play a role in the progression of nascent GPCRs through the ‘assembly line’ from ER to Golgi to cell surface. For example, the delivery of two subtypes of α-adrenoceptors to the plasma membrane is facilitated by the coexpressed β2-adrenoceptor [57,58]; a mutant rhodopsin lacking the targeting sequence is transported to the outer segment only with wild-type rhodopsin [59]. Dimerization could underlie phosphorylation of two coexpressed GPCR subtypes when only one is activated [60,61]. However, phosphorylation of hundreds of rhodopsin molecules for every one activated by light [62] is more consistent with rapid sequential kiss-and-run encounters than with stable oligomers. Overexpressed GPCRs, which may concentrate in microdomains constituting as little as 1% of the plasma membrane [37], also can be crossphosphorylated due to frequent random encounters. Crossphosphorylation easily explains simultaneous internalization of coexpressed receptors: GPCRs are recruited to coated pits via bound arrestins, and even inactive phosphoreceptors bind non-visual arrestins fairly well [48,49]. However, postinternalization trafficking of both receptors via the same pathway that one of them does not use when expressed alone [61,63,64] is easier to explain by heterodimerization.
Conclusions
Many GPCRs form oligomers of varying stability, from permanent covalently linked dimers to kiss-and-run encounters. Recent rigorous studies showed that a single GPCR (or a single heptahelical domain of a naturally dimeric receptor) is necessary and sufficient to bind a G protein or arrestin, ruling out the hypothesis that two receptors in a dimer are required for these interactions. Therefore, to elucidate the biological significance of receptor dimerization, we need to focus on other GPCR functions at various stages of their complicated life cycle.
References
- 1.Rompler H, et al. G protein-coupled time travel: evolutionary aspects of GPCR research. Mol Interv. 2007;7:17–25. doi: 10.1124/mi.7.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Cohen GB, et al. Mechanism of activation and inactivation of opsin: role of Glu113 and Lys296. Biochemistry. 1992;31:12592–12601. doi: 10.1021/bi00165a008. [DOI] [PubMed] [Google Scholar]
- 3.Porter JE, et al. Activation of the alpha1b-adrenergic receptor is initiated by disruption of an interhelical salt bridge constraint. J Biol Chem. 1996;271:28318–28323. doi: 10.1074/jbc.271.45.28318. [DOI] [PubMed] [Google Scholar]
- 4.Ballesteros JA, et al. Activation o f t he beta 2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J Biol Chem. 2001;276:29171–29177. doi: 10.1074/jbc.M103747200. [DOI] [PubMed] [Google Scholar]
- 5.Farrens DL, et al. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 6.Kim JM, et al. Structural origins of constitutive activation in rhodopsin: role of the K296/E113 salt bridge. Proc Natl Acad Sci U S A. 2004;101:12508–12513. doi: 10.1073/pnas.0404519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binet V, et al. Common structural requirements for heptahelical domain function in class A and class C G protein-coupled receptors. J Biol Chem. 2007;282:12154–12163. doi: 10.1074/jbc.M611071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eilers M, et al. Comparison of class A and D G protein-coupled receptors: common features in structure and activation. Biochemistry. 2005;44:8959–8975. doi: 10.1021/bi047316u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chabre M, le Maire M. Monomeric G-protein-coupled receptor as a functional unit. Biochemistry. 2005;44:9395–9403. doi: 10.1021/bi050720o. [DOI] [PubMed] [Google Scholar]
- 10.Fotiadis D, et al. Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr Opin Struct Biol. 2006;16:252–259. doi: 10.1016/j.sbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 11.O’Hara PJ, et al. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993;11:41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- 12.Kunishima N, et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 13.Muto T, et al. Structures of the extracellular regions of the groupII/III metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2007;104:3759–3764. doi: 10.1073/pnas.0611577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Akker F, et al. Structure of the dimerized hormone-binding domain of a guanylyl-cyclase-coupled receptor. Nature. 2000;406:101–104. doi: 10.1038/35017602. [DOI] [PubMed] [Google Scholar]
- 15.Mowbray SL, Petsko GA. The x-ray structure of the periplasmic galactose binding protein from Salmonella typhimurium at 3.0-A resolution. J Biol Chem. 1983;258:7991–7997. doi: 10.2210/pdb1gbp/pdb. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto T, et al. Expression and purification o f the extracellular ligand binding region of metabotropic glutamate receptor subtype 1. J Biol Chem. 1998;273:13089–13096. doi: 10.1074/jbc.273.21.13089. [DOI] [PubMed] [Google Scholar]
- 17.Han G, Hampson DR. Ligand binding to the amino-terminal domain of the mGluR4 subtype of metabotropic glutamate receptor. J Biol Chem. 1999;274:10008–10013. doi: 10.1074/jbc.274.15.10008. [DOI] [PubMed] [Google Scholar]
- 18.Goldsmith PK, et al. Expression, purification, and biochemical characterization of the amino-terminal extracellular domain of the human calcium receptor. J Biol Chem. 1999;274:11303–11309. doi: 10.1074/jbc.274.16.11303. [DOI] [PubMed] [Google Scholar]
- 19.Margeta-Mitrovic M, et al. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 20.Binet V, et al. The heptahelical domain of GABA(B2) is activated directly by CGP7930, a positive allosteric modulator of the GABA(B) receptor. J Biol Chem. 2004;279:29085–29091. doi: 10.1074/jbc.M400930200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goudet C, et al. Heptahelical domain of metabotropic glutamate receptor 5 behaves like rhodopsin-like receptors. Proc Natl Acad Sci U S A. 2004;101:378–383. doi: 10.1073/pnas.0304699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray K, et al. Identification of the cysteine residues in the amino-terminal extracellular domain of the human Ca(2+) receptor critical for dimerization. Implications for function of monomeric Ca(2+) receptor. J Biol Chem. 1999;274:27642–27650. doi: 10.1074/jbc.274.39.27642. [DOI] [PubMed] [Google Scholar]
- 23.Milligan G. G protein-coupled receptor dimerisation: molecular basis and relevance to function. Biochim Biophys Acta. 2007;1768:825–835. doi: 10.1016/j.bbamem.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 24.Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends Neurosci. 2008;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White JF, et al. Dimerization of the class A G protein-couple d neurotensin receptor NTS1 alters G protein interaction. Proc Natl Acad Sci U S A. 2007;104:12199–12204. doi: 10.1073/pnas.0705312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 27.Li J, et al. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen SG, et al. Crystal structure of the human beta(2) adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 29.Baylor DA, et al. Responses of retinal rods to single photons. J Physiol. 1979;288:613–634. [PMC free article] [PubMed] [Google Scholar]
- 30.Kaissling KE, Priesner E. Smell threshold of the silkworm. Naturwissenschaften. 1970;57:23–28. doi: 10.1007/BF00593550. [DOI] [PubMed] [Google Scholar]
- 31.Galvez T, et al. Mapping the agonist-binding site of GABAB type 1 subunit sheds light on the activation process of GABAB receptors. J Biol Chem. 2000;275:41166–41174. doi: 10.1074/jbc.M007848200. [DOI] [PubMed] [Google Scholar]
- 32.Galvez T, et al. Allosteric interactions between GB1 and GB2 subunits are required for optimal GABA(B) receptor function. EMBO J. 2001;20:2152–2159. doi: 10.1093/emboj/20.9.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margeta-Mitrovic M, et al. Ligand-induced signal transduction within heterodimeric GABA(B) receptor. Proc Natl Acad Sci U S A. 2001;98:14643–14648. doi: 10.1073/pnas.251554798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margeta-Mitrovic M, et al. Function of GB1 and GB2 subunits in G protein coupling of GABA(B) receptors. Proc Natl Acad Sci U S A. 2001;98:14649–14654. doi: 10.1073/pnas.251554498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hlavackova V, et al. Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J. 2005;24:499–509. doi: 10.1038/sj.emboj.7600557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goudet C, et al. Asymmetric functioning of dimeric metabotropic glutamate receptors disclosed by positive allosteric modulators. J Biol Chem. 2005;280:24380–24385. doi: 10.1074/jbc.M502642200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer BH, et al. FRET imaging reveals that functional neurokinin-1 receptors are monomeric and reside in membrane microdomains of live cells. Proc Natl Acad Sci U S A. 2006;103:2138–2143. doi: 10.1073/pnas.0507686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayburt TH, et al. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 39.Ernst OP, et al. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci U S A. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirsch JA, et al. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 42.Han M, et al. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 43.Sutton RB, et al. Crystal structure of cone arrestin at 2.3Å: evolution of receptor specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 44.Vishnivetskiy SA, et al. Mapping the arrestin-receptor interface: structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 45.Hanson SM, et al. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci U S A. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J Biol Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:59–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Gurevich VV, et al. Binding of wild type and chimeric arrestins to the m2 muscarinic cholinergic receptor. J Biol Chem. 1993;268:16879–16882. [PubMed] [Google Scholar]
- 49.Gurevich VV, et al. Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 50.Nair KS, et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005:46. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strissel KJ, et al. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanson SM, et al. Each rhodopsin molecule binds its own arrestin. Proc Nat Acad Sci U S A. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanson SM, et al. Structure and function of the visual arrestin oligomer. EMBO J. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lefkowitz RJ, et al. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 55.Gurevich VV, Gurevich EV. The new face of active receptor bound arrestin attracts new partners. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 56.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G protein-coupled receptors. Pharmacol Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prinster SC, et al. Alpha2C-adrenergic receptors exhibit enhanced surface expression and signaling upon association with beta2-adrenergic receptors. J Pharmacol Exp Ther. 2006;318:974–981. doi: 10.1124/jpet.106.106526. [DOI] [PubMed] [Google Scholar]
- 58.Uberti MA, et al. Heterodimerization with beta2-adrenergic receptors promotes surface expression and functional activity of alpha1D-adrenergic receptors. J Pharmacol Exp Ther. 2005;313:16–23. doi: 10.1124/jpet.104.079541. [DOI] [PubMed] [Google Scholar]
- 59.Concepcion F, et al. The carboxyl-terminal domain is essential for rhodopsin transport in rod photoreceptors. Vision Res. 2002;42:417–426. doi: 10.1016/s0042-6989(01)00195-x. [DOI] [PubMed] [Google Scholar]
- 60.Huttenrauch F, et al. G protein-coupled receptor kinases promote phosphorylation and beta-arrestin-mediated internalization of CCR5 homo- and hetero-oligomers. J Biol Chem. 2005;280:37503–37515. doi: 10.1074/jbc.M500535200. [DOI] [PubMed] [Google Scholar]
- 61.Pfeiffer M, et al. Heterodimerization of substance P and mu-opioid receptors regulates receptor trafficking and resensitization. J Biol Chem. 2003;278:51630–51637. doi: 10.1074/jbc.M307095200. [DOI] [PubMed] [Google Scholar]
- 62.Binder BM, et al. Light activation of one rhodopsin molecule causes the phosphorylation of hundreds of others. A reaction observed in electropermeabilized frog rod outer segments exposed to dim illumination. J Biol Chem. 1990;265:15333–15340. [PubMed] [Google Scholar]
- 63.Jordan BA, et al. Oligomerization of opioid receptors with beta 2-adrenergic receptors: a role in trafficking and mitogen-activated protein kinase activation. Proc Natl Acad Sci U S A. 2001;98:343–348. doi: 10.1073/pnas.011384898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Terrillon S, et al. Heterodimerization of V1a and V2 vasopressin receptors determines the interaction with beta-arrestin and their trafficking patterns. Proc Natl Acad Sci U S A. 2004;101:1548–1553. doi: 10.1073/pnas.0305322101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fredriksson R, et al. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 66.Kolakowski LF., Jr GCRDb: a G-protein-coupled receptor database. Recept Channels. 1994;2:1–7. [PubMed] [Google Scholar]
- 67.McMillan DR, et al. Very large G protein-coupled receptor-1, the largest known cell surface protein, is highly expressed in the developing central nervous system. J Biol Chem. 2002;277:785–792. doi: 10.1074/jbc.M108929200. [DOI] [PubMed] [Google Scholar]
- 68.Bessis AS, et al. Closure of the Venus flytrap module of mGlu8 receptor and the activation process: insights from mutations converting antagonists into agonists. Proc Natl Acad Sci U S A. 2002;99:11097–11102. doi: 10.1073/pnas.162138699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kniazeff J, et al. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat Struct Mol Biol. 2004;11:706–713. doi: 10.1038/nsmb794. [DOI] [PubMed] [Google Scholar]
- 70.Baneres JL, Parello J. Structure-based analysis of GPCR function: evidence for a novel pentameric assembly between the dimeric leukotriene B4 receptor BLT1 and the G-protein. J Mol Biol. 2003;329:815–829. doi: 10.1016/s0022-2836(03)00439-x. [DOI] [PubMed] [Google Scholar]